Abstract
Plants are constantly beset by pathogenic organisms. To successfully infect their hosts, plant pathogens secrete effector proteins, many of which are translocated to the inside of the host cell where they manipulate normal physiological processes and undermine host defense. The way by which effectors cross the frontier to reach the inside of the host cell varies among different classes of pathogens. For oomycete plant pathogens – like the potato late blight pathogen Phytophthora infestans – it has been shown that effector translocation to the host cell cytoplasm is dependent on conserved amino acid motifs that are present in the N-terminal part of effector proteins. One of these motifs, known as the RXLR motif, has a strong resemblance with a host translocation motif found in effectors secreted by Plasmodium species. These malaria parasites, that reside inside specialized vacuoles in red blood cells, make use of a specific protein translocation complex to export effectors from the vacuole into the red blood cell. Whether or not also oomycete RXLR effectors require a translocation complex to cross the frontier is still under investigation. For one P. infestans RXLR effector named IPI-O we have found a potential host target that could play a role in establishing the first contact between this effector and the host cell. This membrane spanning lectin receptor kinase, LecRK-I.9, interacts with IPI-O via the tripeptide RGD that overlaps with the RXLR motif. In animals, RGD is a well-known cell adhesion motif; it binds to integrins, which are membrane receptors that regulate many cellular processes and which can be hijacked by pathogens for either effector translocation or pathogen entry into host cells.
Keywords: RXLR–dEER, Phytophthora, effector, oomycetes, RGD, lectin receptor kinase, LecRK-I.9
Introduction
Although plants have an effective immune system, plant pathogens have evolved mechanisms to overcome plant resistance, and are thus able to invade and cause disease. Vital to the success of plant pathogens is their ability to secrete so-called effectors that manipulate host plant defense (reviewed by Stassen and van den Ackerveken, 2010; De Jonge et al., 2011). Ergo, the pathogen can colonize plant cells with less hindrance. In the “arms race” plants acquired specific resistance (R) proteins that directly or indirectly recognize effectors. This recognition leads to effector-triggered immunity (ETI) that blocks further growth of the pathogen and that is often associated with a hypersensitive response (HR; Jones and Dangl, 2006).
Oomycete plant pathogens are notorious for causing devastating diseases in a large variety of plants. The well-known and best studied species are the potato late blight pathogen Phytophthora infestans, the soybean stem and root rot pathogen Phytophthora sojae, and the downy mildew Hyaloperonospora arabidopsidis that infects Arabidopsis. Plant resistance to oomycetes is mostly governed by R proteins comprising a N-terminal nucleotide-binding site (NBS) domain and a C-terminal leucine-rich-repeat (LRR) domain. Since these NBS–LRR proteins are located intracellular, it was hypothesized that oomycete avirulence (Avr) proteins should be translocated to the cytoplasm of the plant cell. Indeed, it has been shown that oomycetes secrete effectors that they deliver inside host cells. These intracellular effectors include Avr proteins that are recognized by their cognate R proteins (Rehmany et al., 2005; Whisson et al., 2007). They all share a signal peptide and a conserved N-terminal RXLR motif with often an dEER motif in its vicinity. These RXLR and dEER motifs were detected in 2004 during a Phytophthora genome annotation jamboree (Govers and Gijzen, 2006), a discovery that marked the beginning of a new episode in plant–pathogen interaction research. At that time, after many years of labor-intensive map based cloning procedures, only a few oomycete avirulence (Avr) genes were identified. As no conserved motifs were known for fungal Avr proteins the presence of a conserved motif in oomycete Avr proteins was unexpected. The number of RXLR–dEER effector genes in each of the three sequenced Phytophthora genomes is remarkably high, with 563 predicted for P. infestans (Haas et al., 2009) and 385 in P. sojae (Jiang et al., 2008). Significantly fewer RXLR–dEER effector genes were found in the genomes of two obligate oomycete pathogens sequenced so far, 115 in H. arabidopsidis and 25 in the white rust Albugo laibachii (Baxter et al., 2010; Kemen et al., 2011), whereas very few or none (≤2) were found in two species with a necrotrophic life style, Pythium ultimum and Aphanomyces euteiches (Gaulin et al., 2008; Lévesque et al., 2010). Even more surprisingly than its conservation and its abundance in Phytophthora spp., is the clear resemblance of the RXLR motif with the RXLXE/Q host cell targeting motif in effector proteins of the malaria parasite Plasmodium falciparum known as the Plasmodium export element (PEXEL). Transfer into the erythrocyte cytoplasm enables the PEXEL effectors to remodel and reprogram the host cell to establish successful asexual replication of the malaria parasite (Templeton and Deitsch, 2005; Tilley et al., 2011). The RXLXE/Q motif was shown to be crucial for effector translocation across the parasitophorous vacuole membrane, which surrounds the malaria parasite in red blood cells (Hiller et al., 2004; Marti et al., 2004). This led to the hypothesis that oomycete RXLR effectors are also targeted into host cells, and stimulated many researchers to address the question how oomycete effectors are delivered into plant cells (Haldar et al., 2006).
Crossing the Frontier; The Role of the RXLR Motif
Whisson et al. (2007) were the first to show that the RXLR–dEER domain is indeed required for effector translocation into host plant cells. P. infestans transformants expressing Avr3a in which either the RXLR or dEER motifs were mutated, were shown to be no longer detected by the resistance protein R3a, and as a result did not trigger an HR. Transient co-expression of the R3a gene with variants of Avr3a lacking the signal peptide sequence and with a mutated or deleted RXLR–dEER domain showed, however, that R3a-mediated cell death was not affected, and therefore it was concluded that the RXLR–dEER domain in Avr3a is needed for effector translocation to the host cytoplasm (Whisson et al., 2007). A similar approach was used for P. sojae Avr1b-1, the avirulence counterpart of the soybean R gene Rps1b and the first oomycete Avr gene that was cloned (Shan et al., 2004). The phenotype of a P. sojae strain changed from virulent to avirulent on Rps1b plants when complemented with the full length Avr1b-1 gene, but remained virulent when transformed with Avr1b-1 with mutations in RXLR and dEER. On the other hand, delivery of Avr1b-1 by particle bombardment for transient expression of the Avr gene in Rps1b-leaves triggered cell death even when the RXLR or dEER motifs were mutated or absent, and hence, a role for these motifs in delivery of Avr1b was apparent (Dou et al., 2008).
Although the two studies described above provided convincing evidence that the RXLR–dEER domains of Avr3 and Avr1b are crucial for uptake, the mechanisms underlying effector uptake by host cells remained unclear. To gain further insight in this process Dou et al. (2008) investigated what happens when Avr1b, produced in planta upon bombardment with an Avr1b-1 construct, has a signal peptide. Whereas secreted Avr1b triggered Rps1b-mediated cell death with the same efficiency as Avr1b lacking its signal peptide, secreted Avr1b with mutated RXLR and dEER motifs triggered no response. This suggested that the motifs mediate re-entry of Avr1b into the cell and, more strikingly, that the entry does not require the presence of the pathogen (Dou et al., 2008). Using similar approaches and supported by immunolocalization, Rafiqi et al. (2010) showed that also internalization in host cells of two Avr proteins produced by the flax rust fungus can occur in the absence of the pathogen. As with Avr1b, uptake is dependent on features in the N-terminal region of the Avr proteins; but unlike Avr1b, the rust Avr proteins have no obvious conserved motifs. For P. sojae Avr1b autonomous uptake in plant cells was also demonstrated with Avr1b–GFP fusion proteins synthesized in Escherichia coli (Dou et al., 2008). Soaking root tips of soybean seedlings for 12 h in a solution containing partly purified fusion protein resulted in uptake of the Avr1b–GFP protein into the root tip cells up to 10 cell layers. Also in this assay mutations in the RXLR or dEER motifs abolished uptake. These experiments suggested that the RXLR–dEER domain is all that is needed from the pathogen to enable translocation of oomycete effectors into host cells (Dou et al., 2008). This is quite different from the situation in certain Gram-negative bacteria that make use of the Type III secretion system (T3SS). These bacteria provide a suite of building blocks for a complex translocation machinery consisting of a multi-protein needle-like structure through which effectors are injected into the host cell cytoplasm (Figures 1A,B).
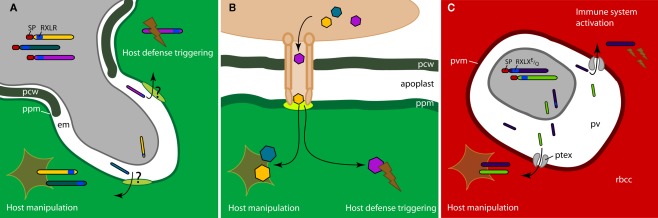
Figure 1.
Effectors crossing the host–pathogen interface. (A) Oomycete plant pathogens secrete RXLR–dEER effector proteins from haustoria into the extrahaustorial matrix (em). How these effectors cross the plant plasma membrane (ppm) to reach the host cell cytoplasm is unknown. (B) Bacterial pathogens exploit the Type III secretion system (T3SS) to inject effectors into the host cell cytoplasm. T3SS is a pathogen-derived multi-protein needle-like structure that penetrates the plant cell wall (pcw) and the plasma membrane. (C) Malaria parasites (Plasmodium spp.) invade red blood cells and secrete effector proteins into the parasitophorous vacuole (pv). PEXEL effectors are translocated to the red blood cell cytoplasm (rbcc) via a pathogen-derived translocon complex (PTEX), which resides in the parasitophorous vacuole membrane (pvm). Effectors are shown in different colored bars [in (A,C)] and hexagons [in (B)]. In the colored bars the red box is the signal peptide (SP) and the blue box the RXLR or RXLXE/Q motif.
The next question to be answered is how RXLR–dEER effectors orchestrate their own translocation. If it is true that no other pathogen components are required, an issue that is still under debate, are the pathogens then actively recruiting host-derived components to support effector entry into host cells? Or do they possibly exploit an existing uptake machinery? According to Kale et al. (2010) the RXLR motif is capable to interact with phosphoinositides (PPIs) at the host plasma membrane (PM), in particular phosphatidylinositol-3-phosphate (PI3P). In filter-binding and liposome-binding assays they showed interaction of the Avr protein Avr1b and two other members of the P. sojae RXLR family, Avh5 and Avh331, with PI3P. Since mutations in RXLR resulted in loss of binding it was concluded that the interaction with PPIs is specific. A puzzling issue, however, was the dogma that PPIs always reside on the cytoplasmic site of eukaryotic cell membranes whereas the RXLR effectors approach the PM from the outside. To address this Kale et al. (2010) exploited highly specific PPI biosensors, proteins composed of lipid-binding domains that target either PI3P or PI4P fused to a fluorescent tag. When exposed to soybean root suspension cultures they observed fluorescence with the PI3P biosensor, but not with the PI4P biosensor, on the extracellular leaflet of the PM, suggesting that PI3P is externally exposed. In a recent study Yaeno et al. (2011) also reported binding of an RXLR effector to PPI. However, in contrast to Kale et al. (2010), they found that mutations in the RXLR motif of Avr1b do not interfere with PI3P binding. Instead they observed that a positively charged lysine-rich patch in the C-terminus of Avr1b, that is conserved in the P. infestans homolog Avr3a, is responsible for the binding to PPI. The PPI binding to the C-terminus of Avr3a, which is thought to take place inside host cells, seems to be required for accumulation of Avr3a and for suppressing host immunity mediated by Avr3a via interaction with the E3 ubiquitin ligase CMPG1 (Yaeno et al., 2011). To what extent PPI binding plays a role in the activity of other RXLR effectors remains to be investigated. Taken together the data that are currently available on the role of PPI binding are conflicting and fragmented, and the frontier-crossing journey of RXLR effectors is still a black box.
Facing the Frontier; Phytophthora RXLR–dEER Versus Plasmodium PEXEL
For PEXEL effectors the frontier-crossing journey is much better pictured. The fact that the RXLR motif is highly similar to the RXLXE/Q translocation motif present in Plasmodium effectors and the observations that the RXLR and RXLXE/Q motifs are functionally interchangeable (Bhattacharjee et al., 2006; Grouffaud et al., 2008), raise questions about potential similarities in uptake machineries. Although PEXEL motif-containing proteins are shown to be taken up in the absence of the pathogen (Kale et al., 2010), a Plasmodium translocon complex (PTEX) is described to work as an active export machine to deliver PEXEL effectors into the cytosol of the erythrocyte host cell (Figure 1C; De Koning-Ward et al., 2009). Components comprising the PTEX complex are an AAA+-ATPase, the exported protein EXP2, thioredoxin 2, and two novel Plasmodium proteins, PTEX88 and PTEX150. In the model it is assumed that also host-derived proteins assist this translocon complex. So far, BLAST searches did not reveal obvious homologs of PTEX components in Phytophthora genomes (K. Bouwmeester and H. J. G. Meijer, unpublished results). This makes it less likely that Plasmodium and Phytophthora share highly conserved translocon machineries, although the existence of an analogous translocon complex for shuttling Phytophthora effectors is still conceivable.
Intriguingly, it was reported that the PEXEL motif is not involved in a direct interaction with the PTEX, but is functioning as a recognition site for cleavage (Figure 2; Chang et al., 2008; Boddey et al., 2009). Cleavage of the RXLXE/Q motif was shown to occur after the third residue (RXL↓), and when the conserved L residue was mutated, export was prevented (Boddey et al., 2009). Plasmepsin V, an endoplasmic reticulum (ER) aspartic protease (AP), was found responsible for PEXEL cleavage (Boddey et al., 2010; Russo et al., 2010). APs are well represented in oomycetes. A recent study showed that each of the three sequenced Phytophthora species has 11–12 AP genes distributed over five clades (Kay et al., 2011). One clade with Phytophthora AP10, AP11, and AP12 clusters with Plasmepsin V, but whether or not these are functional RXLR or PEXEL proteases remains to be tested.
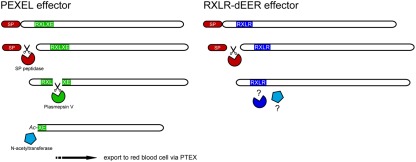
Figure 2.
Plasmodium export element effectors of Plasmodium spp. are proteolytically processed by a signal peptidase and the ER-localized aspartic protease plasmepsin V that cleaves the RXLXE/Q motif after the leucine (L). This results in a new N-terminus which is then N-acetylated. Phytophthora effectors have an RXLR motif that resembles the Plasmodium RXLXE/Q motif and the motifs are functionally interchangeable. What happens to RXLR effectors after removal of the signal peptide is not known. As indicated by the question marks, there is as yet no evidence for further processing, neither for cleavage within the RXLR motif nor for N-acetylation.
An additional step in PEXEL processing is N-acetylation of the new N-terminus, which could well be a crucial step to facilitate translocation to the red blood cell (Chang et al., 2008; Boddey et al., 2009; Figure 2). Both cleavage and N-acetylation are executed early in the trafficking pathway within the ER of Plasmodium, but not in the parasitophorous vacuole or during export to the erythrocyte, as was concluded based on inhibition of ER-to-Golgi protein transport with brefeldin A. As yet, the N-acetyltransferase in Plasmodium remains to be identified. So far, there are no indications for cleavage of oomycete RXLR effectors or N-acetylation and it is therefore questionable whether RXLR–dEER effectors from oomycetes are processed in a similar way.
Opening the Frontier; The Role of the RGD Motif
One of the 563 predicted P. infestans effectors that carries an RXLR motif is IPI-O. The encoding gene ipiO (with ipi referring to in planta induced) was one of the first oomycete genes that was isolated and studied in detail (Pieterse et al., 1993). At that time, the assignment of a putative function of ipiO in pathogenesis was solely based on its expression pattern, i.e., high expression in planta versus low or no expression in in vitro culture. Attempts to determine a function of IPI-O by gene silencing or overexpression failed (Van West, 2000). The finding that IPI-O is a potential Avr factor came many years later when ipiO was tested in a high-throughput effector genomics screening aimed at identifying novel pairs of P. infestans Avr genes and host plant R genes (Vleeshouwers et al., 2008). Subsequently, we confirmed that ipiO is Avr-blb1, the counterpart of the late blight R gene Rpi-blb1 which originates from the nightshade Solanum bulbocastanum (Champouret et al., 2009).
In the pre-genomic era comparative analysis that could have led to the discovery of the RXLR–dEER domain in IPI-O was not yet feasible; at that time the only potentially interesting feature that could be distinguished in IPI-O was the presence of a cell attachment motif (Pieterse et al., 1994). This motif consisting of the tripeptide sequence Arg-Gly-Asp (RGD) partly overlaps with the RXLR motif in IPI-O, i.e., RXLRGD. The RGD motif is found in a large number of extracellular proteins, such as the mammalian glycoproteins fibronectin and vitronectin that reside in the extracellular matrix (ECM). A wide variety of these proteins binds to integrins, a family of transmembrane receptors that function in “inside-out” and “outside-in” signaling and regulate many cellular processes. The fact that membrane-to-matrix interactions can be easily disrupted by adding synthetic RGD peptides shows that these interactions are important for the ECM–PM continuum (Gronowicz and Derome, 1994; Canut et al., 1998). Integrins are also chosen by pathogens as a target for binding and their role in animal innate immune responses has been emphasized in several reports (Isberg and Van Nhieu, 1994; Pribila et al., 2004; Stewart and Nemerow, 2007). Integrins, for instance, interact with the RGD-containing penton base proteins of adenoviruses (reviewed in Mercer et al., 2010). Following this interaction integrins initiate clathrin-mediated endocytosis and promote virus internalization. It was demonstrated, however, that the RGD motif is not necessary for initial virus attachment per se; adenoviruses with deleted RGD motifs could still attach but were impaired in endosomal internalization and in escape to the host cytoplasm (Shayakhmetov et al., 2005). Another example can be found in Helicobacter pylori, a bacterium causal to stomach inflammation, that exploits integrins for effector translocation into the host cell (Kwok et al., 2007). This process is dependent on CagL, a RGD-containing protein that is situated on the surface of the pilus and interacts in an RGD-dependent manner with the integrin α5ß1. Upon interaction, various downstream components are activated that were shown to be essential for effector translocation. Other potent effectors of integrin function are the disintegrins from snake venoms of various viper species (Calvete et al., 2007; Matsui et al., 2010). Disintegrins are usually short cysteine-rich peptides that contain a RGD motif on an exposed hairpin loop. Disintegrins selectively bind and activate integrin receptors, and hence compete with their natural RGD-containing ligands, such as fibrinogen and Von Willebrand factor, subsequently disturbing platelet aggregation and cell–ECM adhesion. Several RGD-disintegrins – such as contortrostatin and jarastatin – have been shown to activate the focal adhesion kinase FAK that regulates actin dynamics, and to trigger phosphoinositide 3-kinase and MAPK pathways upon integrin-binding (Coelho et al., 2001; Oliva et al., 2007).
It is conceivable that also plant pathogens take advantage of the RGD motif for manipulating their hosts. One example comes from the RGD-containing ToxA protein produced by Pyrenophora trititci-repentis and Stagonospora nodorum, two fungi that cause leaf spot disease on wheat. These pathogens require the host-selective toxin ToxA to induce necrosis in host cells (Sarma et al., 2005; Friesen et al., 2008). The ToxA gene encodes a pre-pro-protein of which the signal peptide (pre-region) and the N-terminal pro-region are both cleaved by the fungus prior to secretion of the mature 13.2 kDa protein. The RGD motif in ToxA was found to be required for internalization into wheat mesophyll cells (Meinhardt et al., 2002; Manning et al., 2008) and it was proposed that internalization occurs by means of receptor-mediated endocytosis. This is in line with the finding that a number of genes encoding endocytosis-related proteins and lectin receptor kinases were significantly upregulated in wheat treated with ToxA (Pandelova et al., 2009). Other studies reported that exogenously applied RGD peptides can change the structure and behavior of plant cells. When added, adhesions between the cell wall (CW) and plasma membrane (PM) were easily disrupted, and resulted in reduced levels of phytoalexins and phenolic compounds (Schindler et al., 1989; Canut et al., 1998; Kiba et al., 1998; Gao et al., 2007). Moreover, Mellersh and Heath (2001) showed that RGD-treated cowpea plants were attenuated in their defense response, which correlated with enhanced fungal penetration efficiency.
In our studies on the P. infestans effector IPI-O we also obtained strong evidence that the RGD motif is a functional determinant. Similar to RGD peptides, exogenously added IPI-O has the capacity to disrupt CW–PM adhesions through its RGD motif. Plasmolysed Arabidopsis cells from a suspension culture showed convex pockets when exposed to recombinant IPI-O protein but concave pockets when exposed to recombinant IPI-O protein with RGD mutated into RGE or RGA (Senchou et al., 2004; Figure 3A). Transgenic Arabidopsis plants with constitutive expression of the ipiO gene develop normally but at the microscopic level we again observed concave forms of plasmolysis in hypocotyl cells reminiscent of a disrupted adhesion between the CW and PM, this in contrast to wild-type plants and transgenic plants expressing a mutant form of ipiO (RGD > RGE; Bouwmeester et al., 2011; Figure 3B). Intriguingly, the ipiO expressing plants responded in a different manner to infection and elicitor treatment. They displayed gain of susceptibility to Phytophthora brassicae, and showed less callose deposition, thus suggesting that IPI-O functions as a suppressor of defense (Bouwmeester et al., 2011). Since also these phenotypes are RGD dependent, we conclude that RGD is essential for IPI-O to function at the host–pathogen interface. We imagine that plants perceive IPI-O either directly via the RGD motif or via a structural feature imposed on IPI-O by the RGD motif.
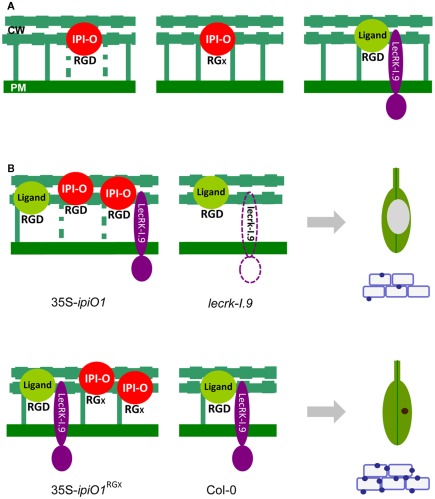
Figure 3.
The Phytophthora infestans RXLR effector IPI-O and its putative host cell receptor Arabidopsis LecRK-I.9. (A) Observed changes in Arabidopsis suspension cells upon exposure to IPI-O. The CW–PM continuum (shown as green vertical bars) is disturbed (dotted vertical bars) when IPI-O has an intact RGD motif (left panel) but when RGD is mutated into RGA or RGE the continuum is not disturbed (middle panel; Senchou et al., 2004). Model depicting the membrane-associated LecRK-I.9 in interaction with a putative RGD-containing extracellular ligand (right panel). Under normal circumstances (wild-type) the CW–PM continuum is in a balanced state. (B) Observed changes in the Arabidopsis phenocopy lines 35S-ipiO1 and lecrk-I.9 (top left and right panel, respectively). Both show gain of susceptibility to Phytophthora brassicae (the gray area on the leaf depicts lesion growth) and reduced callose deposition (shown as blue dots at the cell boundaries). Arabidopsis lines expressing ipiO with a mutated RGD motif (35S-ipiO1RGA or 35S-ipiO1RGE) behave as the wild-type Col-0 accession (lower left and right panel, respectively); no lesion formation upon infection with P. brassicae (the brown dot represents a hypersensitive response) and efficient callose deposition (Bouwmeester et al., 2011). The model predicts that IPI-O competes with the endogenous ligand of LecRK-I.9 leading to less adhesions between CW and PM, and gain of susceptibility. A mutation in the RGD motif abolishes the competition resulting in a wild-type phenotype. In the absence of LecRK-I.9 the proper balance between endogenous ligand and receptor is disturbed and results in gain of susceptibility.
Opening the Frontier; The Role of Host Cell Receptor Proteins
The fact that several animal and human pathogens exploit integrin-mediated-uptake for host cell entry or effector delivery raises the question whether plant pathogens use similar strategies. Plants however, lack integrins. In the 1980s and 1990s several researchers attempted to identify plant integrins using immunological and biochemical approaches but failed. Also the first plant genome sequence did not reveal the occurrence of integrin homologs (The Arabidopsis Genome Initiative, 2000). It is conceivable though that plant pathogens use other types of host receptors for cell entry. In a search for functional homologs of integrins in plants Canut et al. (1998) showed that Arabidopsis contains high affinity RGD-binding sites in the PM. Consistently, one of these binding sites had a very high affinity for IPI-O. Since this affinity was lost when RGD in IPI-O was mutated into RGA or RGE, the RGD motif seemed to be crucial for binding (Senchou et al., 2004). Subsequently, Gouget et al. (2006) performed a phage display screen to identify proteins that potentially interact with IPI-O via the RGD motif. This resulted in two heptamers that could inhibit the RGD-binding activity to the Arabidopsis PM and disrupt CW–PM membrane adhesions, possibly by competing with naturally occurring RGD-based ligand-receptor contacts. Screening the Arabidopsis proteome for the occurrence of these two peptide sequences led to the identification of the Arabidopsis legume-like lectin receptor kinase LecRK-I.9, which has two amino acid motifs in the N-terminal region (ASYY and PHPR) that are implicated in binding IPI-O via its RGD motif (Gouget et al., 2006). LecRK-I.9 belongs to a family of 45 members divided over nine clades and a few singletons (Bouwmeester and Govers, 2009). They are typical transmembrane receptors with an extracellular lectin domain and an intracellular kinase domain. As yet, little is known about the functions of LecRK-I.9 and other legume-like lectin receptor kinases, neither about their natural ligands. They are likely involved in monitoring CW integrity and it is conceivable that LecRK-I.9 interacts with endogenous RGD-containing proteins to balance CW–PM adhesions (Figure 3A). To elucidate the role of LecRK-I.9 at the Phytophthora–host interface we analyzed expression during compatible and incompatible interactions with P. brassicae isolates and determined the phenotypes of Arabidopsis LecRK-I.9 knock-out lines (lecrk-I.9) in disease assays. These studies convincingly demonstrated that LecRK-I.9 plays a role in Phytophthora disease resistance. LecRK-I.9 expression was induced in incompatible interactions (Bouwmeester et al., 2008) and two independent knock-out lines showed gain of susceptibility to P. brassicae and the bacterium Pseudomonas syringae. Strikingly, the lecrk-I.9 lines behaved as perfect phenocopies of the Arabidopsis lines that constitutively express the RXLR effector gene ipiO suggesting that an overload of IPI-O disables LecRK-I.9 to properly guard the frontier (Figure 3B). P. brassicae does not have an ipiO homolog and hence cannot effectively disable LecRK-I.9. In contrast, ipiO is highly expressed in P. infestans hyphae invading potato leaves (Van West et al., 1998). Competition of IPI-O with potential endogenous RGD-containing ligands of LecRK-I.9 may destabilize the CW–PM continuum helping P. infestans to colonize host tissue. Loosening CW–PM adhesions might facilitate the formation of haustoria, feeding structures that are not only important for nutrient uptake but also for effector translocation to host cells. We hypothesize that Phytophthora uses LecRK-I.9 as a gateway to establish infection and that one of the functions of IPI-O is to mediate this early step in the infection process. Other anticipated functions of IPI-O are more downstream in the process when IPI-O has reached the cytoplasm.
Strengthening the Frontier
In recent years comparative genomics and high-throughput functional analyses of Phytophthora effectors have boosted the discovery of translocation motifs but the molecular machinery that is used by oomycetes to deliver their armory into the host cell is still a black box. The recent identification of host components at the cell surface that bind translocation and adhesion motifs opened new avenues and the search for additional components is on-going. Insight into similarities and dissimilarities in transport mechanisms exploited by different groups of pathogens will help to shed light in the black box. The ultimate challenge is to find ways to block the uptake machinery thereby strengthening the frontier at the Phytophthora–host interface. Hopefully, this will provide novel strategies to control these notorious pathogens.
Conflict of Interest Statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Acknowledgments
Our research is financially supported by the Centre for BioSystems Genomics (Klaas Bouwmeester and Francine Govers) and the Technology Foundation STW (VIDI grant Harold J. G. Meijer).
References
- Baxter L., Tripathy S., Ishaque N., Boot N., Cabral A., Kemen E., Thines M., Ah-Fong A., Anderson R., Badejoko W., Bittner-Eddy P., Boore J. L., Chibucos M. C., Coates M., Dehal P., Delehaunty K., Dong S., Downton P., Dumas B., Fabro G., Fronick C., Fuerstenberg S. I., Fulton L., Gaulin E., Govers F., Hughes L., Humphray S., Jiang R. H. Y., Judelson H., Kamoun S., Kyung K., Meijer H. J. G., Minx P., Morris P., Nelson J., Phuntumart V., Qutob D., Rehmany A., Rougon-Cardoso A., Ryden P., Torto-Alalibo T., Studholme D., Wang Y., Win J., Wood J., Clifton S. W., Rogers J., van den Ackerveken G., Jones J. D. G., McDowell J. M., Beynon J., Tyler B. M. (2010). Signatures of adaptation to obligate biotrophy in the Hyaloperonospora arabidopsidis genome. Science 330, 1549–1551 10.1126/science.1195203 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bhattacharjee S., Hiller N. L., Liolios K., Win J., Kanneganti T.-D., Young C., Kamoun S., Haldar K. (2006). The malarial host-targeting signal is conserved in the Irish potato famine pathogen. PLoS Pathog. 2, e50. 10.1371/journal.ppat.0020050 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boddey J. A., Hodder A. N., Günther S., Gilson P. R., Patsiouras H., Kapp E. A., Pearce J. A., De Koning-Ward T. F., Simpson R. J., Crabb B. S., Cowman A. F. (2010). An aspartyl protease directs malaria effector proteins to the host cell. Nature 463, 627–631 10.1038/nature08728 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boddey J. A., Moritz R. L., Simpson R. J., Cowman A. F. (2009). Role of the Plasmodium export element in trafficking parasite proteins to the infected erythrocyte. Traffic 10, 285–299 10.1111/j.1600-0854.2008.00864.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bouwmeester K., de Sain M., Weide R., Gouget A., Klamer S., Canut H., Govers F. (2011). The lectin receptor kinase LecRK-I.9 is a novel Phytophthora resistance component and a potential host target for a RXLR effector. PloS Pathog. 7, e1001327. 10.1371/journal.ppat.1001327 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bouwmeester K., Govers F. (2009). Arabidopsis L-type lectin receptor kinases: phylogeny, classification, and expression profiles. J. Exp. Bot. 60, 4383–4396 10.1093/jxb/erp277 [DOI] [PubMed] [Google Scholar]
- Bouwmeester K., Klamer S., Gouget A., Haget N., Canut H., Govers F. (2008). “Lectin receptor kinase 79, a putative target of the Phytophthora infestans effector IPI-O,” in Biology of Plant-Microbe Interactions, eds Lorito M., Woo S. L., Scala F. (St. Paul, MN: International Society for Molecular Plant-Microbe Interactions; ), 1–6 [Google Scholar]
- Calvete J. J., Marcinkiewicz C., Sanz L. (2007). KTS and RTS-disintegrins: anti-angiogenic viper venom peptides specifically targeting the α1β1 integrin. Curr. Pharm. Des. 13, 2853–2859 10.2174/138161207782023766 [DOI] [PubMed] [Google Scholar]
- Canut H., Carrasco A., Galaud J. P., Cassan C., Bouyssou H., Vita N., Ferrara P., Pont-Lezica R. (1998). High affinity RGD-binding sites at the plasma membrane of Arabidopsis thaliana links the cell wall. Plant J. 16, 63–71 10.1046/j.1365-313x.1998.00276.x [DOI] [PubMed] [Google Scholar]
- Champouret N., Bouwmeester K., Rietman H., van der Lee T., Maliepaard C., Heupink A., van de Vondervoort P. J. I., Jacobsen E., Visser R. G. F., van der Vossen E. A. G., Govers F., Vleeshouwers V. G. A. A. (2009). Phytophthora infestans isolates lacking class I ipiO variants are virulent on Rpi-blb1 potato. Mol. Plant Microbe Interact. 22, 1535–1545 10.1094/MPMI-22-12-1535 [DOI] [PubMed] [Google Scholar]
- Chang H. H., Falick A. M., Carlton P. M., Sedat J. W., Derisi J. L., Marletta M. A. (2008). N-terminal processing of proteins exported by malaria parasites. Mol. Biochem. Parasitol. 160, 107–115 10.1016/j.molbiopara.2008.04.011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Coelho A. L., De Freitas M. S., Mariano-Oliveira A., Oliveira-Carvalho A. L., Zingali R. B., Barja-Fidalgo C. (2001). Interaction of disintegrins with human neutrophils induces cytoskeleton reorganization, focal adhesion kinase activation, and extracellular-regulated kinase-2 nuclear translocation, interfering with the chemotactic function. FASEB J. 15, 1643–1645 [DOI] [PubMed] [Google Scholar]
- De Jonge R., Bolton M. D., Thomma B. P. (2011). How filamentous pathogens co-opt plants: the ins and outs of fungal effectors. Curr. Opin. Plant Biol. 14, 400–406 10.1016/j.pbi.2011.03.005 [DOI] [PubMed] [Google Scholar]
- De Koning-Ward T. F., Gilson P. R., Boddey J. A., Rug M., Smith B. J., Papenfuss A. T., Sanders P. R., Lundie R. J., Maier A. G., Cowman A. F., Crabb B. S. (2009). A newly discovered protein export machine in malaria parasites. Nature 459, 945–949 10.1038/nature08104 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dou D., Kale S. D., Wang X., Jiang R. H. Y., Bruce N. A., Arredondo F. D., Zhang X., Tyler B. M. (2008). RXLR-mediated entry of Phytophthora sojae effector Avr1b into soybean cells does not require pathogen-encoded machinery. Plant Cell 20, 1930–1947 10.1105/tpc.107.056093 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Friesen T. L., Zhang Z., Solomon P. S., Oliver R. P., Faris J. D. (2008). Characterization of the interaction of a novel Stagonospora nodorum host-selective toxin with a wheat susceptibility gene. Plant Physiol. 146, 682–693 10.1104/pp.107.108761 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gao H., Gong Y. W., Yuan Y. J. (2007). RGD-dependent mechanotransduction of suspension cultured Taxus cell in response to shear stress. Biotechnol. Prog. 23, 673–679 10.1021/bp060329+ [DOI] [PubMed] [Google Scholar]
- Gaulin E., Madoui M. A., Bottin A., Jacquet C., Mathé C., Couloux A., Wincker P., Dumas B. (2008). Transcriptome of Aphanomyces euteiches: new oomycete putative pathogenicity factors and metabolic pathways. PLoS ONE 3, e1723. 10.1371/journal.pone.0001723 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gouget A., Senchou V., Govers F., Sanson A., Barre A., Rougé P., Pont-Lezica R., Canut H. (2006). Lectin receptor kinases participate in protein-protein interactions to mediate plasma membrane-cell wall adhesions in Arabidopsis. Plant Physiol. 140, 81–90 10.1104/pp.105.066464 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Govers F., Gijzen M. (2006). Phytophthora genomics: the plant destroyers’ genome decoded. Mol. Plant Microbe Interact. 19, 1295–1301 10.1094/MPMI-19-1295 [DOI] [PubMed] [Google Scholar]
- Gronowicz G. A., Derome M. E. (1994). Synthetic peptide containing Arg-Gly-Asp inhibits bone formation and resorption in a mineralizing organ culture system of fetal rat parietal bones. J. Bone Miner. Res. 9, 193–201 10.1002/jbmr.5650090208 [DOI] [PubMed] [Google Scholar]
- Grouffaud S., van West P., Avrova A. O., Birch P. R. J., Whisson S. C. (2008). Plasmodium falciparum and Hyaloperonospora parasitica effector translocation motifs are functional in Phytophthora infestans. Microbiology 154, 3743–3751 10.1099/mic.0.2008/021964-0 [DOI] [PubMed] [Google Scholar]
- Haas B. J., Kamoun S., Zody M. C., Jiang R. H. Y., Handsaker R. E., Cano L. M., Grabherr M., Kodira C. D., Raffaele S., Torto-Alalibo T., Bozkurt T. O., Ah-Fong A. M. V., Alvarado L., Anderson V. L., Armstrong M. R., Avrova A., Baxter L., Beynon J., Boevink P. C., Bollmann S. R., Bos J. I. B., Bulone V., Cai G., Cakir C., Carrington J. C., Chawner M., Conti L., Costanzo S., Ewan R., Fahlgren N., Fischbach M. A., Fugelstad J., Gilroy E. M., Gnerre S., Green P. J., Grenville-Briggs L. J., Griffith J., Grünwald N. J., Horn K., Horner N. R., Hu C. H., Huitema E., Jeong D. H., Jones A. M. E., Jones J. D. G., Jones R. W., Karlsson E. K., Kunjeti S. G., Lamour K., Liu Z., Ma L., MacLean D., Chibucos M. C., McDonald H., McWalters J., Meijer H. J. G., Morgan W., Morris P. F., Munro C. A., O’Neill K., Ospina-Giraldo M., Pinzón A., Pritchard L., Ramsahoye B., Ren Q., Restrepo S., Roy S., Sadanandom A., Savidor A., Schornack S., Schwartz D. C., Schumann U. D., Schwessinger B., Seyer L., Sharpe T., Silvar C., Song J., Studholme D. J., Sykes S., Thines M., van de Vondervoort P. J. I., Phuntumart V., Wawra S., Weide R., Win J., Young C., Zhou S., Fry W., Meyers B. C., van West P., Ristaino J., Govers F., Birch P. R. J., Whisson S. C., Judelson H. S., Nusbaum C. (2009). Genome sequence and analysis of the Irish potato famine pathogen Phytophthora infestans. Nature 461, 393–398 10.1038/nature08358 [DOI] [PubMed] [Google Scholar]
- Haldar K., Kamoun S., Hiller N. L., Bhattacharje S., van Ooij C. (2006). Common infection strategies of pathogenic eukaryotes. Nat. Rev. Microbiol. 4, 922–931 10.1038/nrmicro1549 [DOI] [PubMed] [Google Scholar]
- Hiller N. L., Bhattacharjee S., van Ooij C., Liolios K., Harrison T., Lopez-Estraño C., Haldar K. (2004). A host-targeting signal in virulence proteins reveals a secretome in malarial infection. Science 306, 1934–1937 10.1126/science.1102737 [DOI] [PubMed] [Google Scholar]
- Isberg R. R., Van Nhieu G. T. (1994). Binding and internalization of microorganisms by integrin receptors. Trends Microbiol. 2, 10–14 10.1016/0966-842X(94)90338-7 [DOI] [PubMed] [Google Scholar]
- Jiang R. H. Y., Tripathy S., Govers F., Tyler B. M. (2008). RXLR effector reservoir in two Phytophthora species is dominated by a single rapidly evolving superfamily with more than 700 members. Proc. Natl. Acad. Sci. U.S.A. 105, 4874–4879 10.1073/pnas.0810962106 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jones J. D. G., Dangl J. L. (2006). The plant immune system. Nature 444, 323–329 10.1038/nature05286 [DOI] [PubMed] [Google Scholar]
- Kale S. D., Gu B., Capelluto D. G. S., Dou D., Feldman E., Rumore A., Arredondo F. D., Hanlon R., Fudal I., Rouxel T., Lawrence C. B., Shan W., Tyler B. M. (2010). External lipid PI3P mediates entry of eukaryotic pathogen effectors into plant and animal host cells. Cell 142, 284–295 10.1016/j.cell.2010.08.035 [DOI] [PubMed] [Google Scholar]
- Kay J., Meijer H. J. G., ten Have A., van Kan J. A. L. (2011). The aspartic proteinase family of three Phytophthora species. BMC Genomics 12, 254. 10.1186/1471-2164-12-254 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kemen E., Gardiner A., Schultz-Larsen T., Kemen A. C., Balmuth A. L., Robert-Seilaniantz A., Bailey K., Holub E., Studholme D. J., Maclean D., Jones J. D. G. (2011). Gene gain and loss during evolution of obligate parasitism in the white rust pathogen of Arabidopsis thaliana. PLoS Biol. 9, e1001094. 10.1371/journal.pbio.1001094 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kiba A., Sugimoto M., Toyoda K., Ichinose Y., Yamada T., Shiraishi T. (1998). Interaction between cell wall and plasma membrane via RGD motif is implicated in plant defense responses. Plant Cell Physiol. 39, 1245–1249 [Google Scholar]
- Kwok T., Zabler D., Urman S., Rohde M., Hartig R., Wessler S., Misselwitz R., Berger J., Sewald N., Konig W., Backert S. (2007). Helicobacter exploits integrin for type IV secretion and kinase activation. Nature 449, 862–866 10.1038/nature06187 [DOI] [PubMed] [Google Scholar]
- Lévesque C. A., Brouwer H., Cano L., Hamilton J. P., Holt C., Huitema E., Raffaele S., Robideau G. P., Thines M., Win J., Zerillo M. M., Beakes G. W., Boore J. L., Busam D., Dumas B., Ferriera S., Fuerstenberg S. I., Gachon C. M. M., Gaulin E., Govers F., Grenville-Briggs L., Horner N., Hostetler J., Jiang R. H. Y., Johnson J., Krajaejun T., Lin H., Meijer H. J. G., Moore B., Morris P., Phuntmart V., Puiu D., Shetty J., Stajich J. E., Tripathy S., Wawra S., van West P., Whitty B. R., Coutinho P. M., Henrissat B., Martin F., Thomas P. D., Tyler B. M., De Vries R. P., Kamoun S., Yandell M., Tisserat N., Buell C. R. (2010). Genome sequence of the necrotrophic plant pathogen, Pythium ultimum, reveals original pathogenicity mechanisms and effector repertoire. Genome Biol. 11, R73. 10.1186/gb-2010-11-7-r73 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Manning V. A., Hamilton S. M., Karplus P. A., Ciuffetti L. M. (2008). The Arg-Gly-Asp-containing, solvent-exposed loop of PtrToxA is required for internalization. Mol. Plant Microbe Interact. 21, 315–325 10.1094/MPMI-21-3-0315 [DOI] [PubMed] [Google Scholar]
- Marti M., Good R. T., Rug M., Knuepfer E., Cowman A. F. (2004). Targeting malaria virulence and remodeling proteins to the host erythrocyte. Science 306, 1930–1933 10.1126/science.1102452 [DOI] [PubMed] [Google Scholar]
- Matsui T., Hamako J., Titani K. (2010). Structure and function of snake venom proteins affecting platelet plug formation. Toxins (Basel) 2, 10–23 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meinhardt S. W., Cheng W., Kwon C. Y., Donohue C. M., Rasmussen J. B. (2002). Role of the arginyl-glycyl-aspartic motif in the action of PtrToxA produced by Pyrenophora tritici-repentis. Plant Physiol. 130, 1545–1551 10.1104/pp.006684 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mellersh D. G., Heath M. C. (2001). Plasma membrane-cell wall adhesion is required for expression of plant defense responses during fungal penetration. Plant Cell 13, 413–424 10.1105/tpc.13.2.413 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mercer J., Schelhaas M., Helenius A. (2010). Virus entry by endocytosis. Annu. Rev. Biochem. 79, 803–833 10.1146/annurev-biochem-060208-104626 [DOI] [PubMed] [Google Scholar]
- Oliva I. B., Coelho R. M., Barcellos G. G., Saldanha-Gama R., Wermelinger L. S., Marcinkiewicz C., BenedetaZingali R., Barja-Fidalgo C. (2007). Effect of RGD-disintegrins on melanoma cell growth and metastasis: involvement of the actin cytoskeleton, FAK and c-Fos. Toxicon 50, 1053–1063 10.1016/j.toxicon.2007.07.016 [DOI] [PubMed] [Google Scholar]
- Pandelova I., Betts M. F., Manning V. A., Wilhelm L. J., Mockler T. C., Ciuffetti L. M. (2009). Analysis of transcriptome changes induced by PtrToxA in wheat provides insights into the mechanisms of plant susceptibility. Mol. Plant 2, 1067–1083 10.1093/mp/ssp045 [DOI] [PubMed] [Google Scholar]
- Pieterse C. M. J., Riach M. B. R., Bleker T., van den Berg-Velthuis G. C. M., Govers F. (1993). Isolation of putative pathogenicity genes of the potato late blight fungus Phytophthora infestans by differential hybridization of a genomic library. Physiol. Mol. Plant Pathol. 43, 69–79 10.1006/pmpp.1993.1041 [DOI] [Google Scholar]
- Pieterse C. M. J., van West P., Verbakel H. M., Brassé P. W. H. M., van den Berg-Velthuis G. C. M., Govers F. (1994). Structure and genomic organization of the ipiB and ipiO gene clusters of Phytophthora infestans. Gene 138, 67–77 10.1016/0378-1119(94)90784-6 [DOI] [PubMed] [Google Scholar]
- Pribila J. T., Quale A. C., Mueller K. L., Shimizu Y. (2004). Integrins and T cell-mediated immunity. Annu. Rev. Immunol. 22, 157–180 10.1146/annurev.immunol.22.012703.104649 [DOI] [PubMed] [Google Scholar]
- Rafiqi M., Gan P. H. P., Ravensdale M., Lawrence G. J., Ellis J. G., Jones D. A., Hardham A. R., Dodds P. N. (2010). Internalization of flax rust avirulence proteins into flax and tobacco cells can occur in the absence of the pathogen. Plant Cell 22, 2017–2032 10.1105/tpc.109.072983 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rehmany A. P., Gordon A., Rose L. E., Allen R. L., Armstrong M. R., Whisson S. C., Kamoun S., Tyler B. M., Birch P. R. J., Beynon J. L. (2005). Differential recognition of highly divergent downy mildew avirulence gene alleles by RPP1 resistance genes from two Arabidopsis lines. Plant Cell 17, 1839–1850 10.1105/tpc.105.031807 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Russo I., Babbitt S., Muralidharan V., Butler T., Oksman A., Goldberg D. E. (2010). Plasmepsin V licenses Plasmodium proteins for export into the host erythrocyte. Nature 463, 632–636 10.1038/nature08726 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sarma G. N., Manning V. A., Ciuffetti L. M., Karplus P. A. (2005). Structure of PtrToxA: an RGD-containing host-selective toxin from Pyrenophora tritici-repentis. Plant Cell 17, 3190–3202 10.1105/tpc.105.034918 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schindler M., Meiners S., Cheresh D. A. (1989). RGD-dependent linkage between plant cell wall and plasma membrane: consequences for growth. J. Cell Biol. 108, 1955–1965 10.1083/jcb.108.5.1955 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Senchou V., Weide R., Carrasco A., Bouyssou H., Pont-Lezica R., Govers F., Canut H. (2004). High affinity recognition of a Phytophthora protein by Arabidopsis via an RGD motif. Cell. Mol. Life Sci. 61, 502–509 10.1007/s00018-003-3394-z [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shan W., Cao M., Leung D., Tyler B. M. (2004). The Avr1b locus of Phytophthora sojae encodes an elicitor and a regulator required for avirulence on soybean plants carrying resistance gene Rps1b. Mol. Plant Microbe Interact. 17, 394–403 10.1094/MPMI.2004.17.4.394 [DOI] [PubMed] [Google Scholar]
- Shayakhmetov D. M., Eberly A. M., Li Z. Y., Lieber A. (2005). Deletion of penton RGD motifs affects the efficiency of both the internalization and the endosome escape of viral particles containing adenovirus serotype 5 or 35 fiber knobs. J. Virol. 79, 1053–1061 10.1128/JVI.79.2.1053-1061.2005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stassen J. H., van den Ackerveken G. (2010). How do oomycete effectors interfere with plant life? Curr. Opin. Plant Biol. 14, 1–8 [DOI] [PubMed] [Google Scholar]
- Stewart P. L., Nemerow G. R. (2007). Cell integrins: commonly used receptors for diverse viral pathogens. Trends Microbiol. 15, 500–507 10.1016/j.tim.2007.10.001 [DOI] [PubMed] [Google Scholar]
- Templeton T. J., Deitsch K. W. (2005). Targeting malaria parasite proteins to the erythrocyte. Trends Parasitol. 21, 399–402 10.1016/j.pt.2005.07.006 [DOI] [PubMed] [Google Scholar]
- The Arabidopsis Genome Initiative. (2000). Analysis of the genome sequence of the flowering plant Arabidopsis thaliana. Nature 408, 796–815 10.1038/35048692 [DOI] [PubMed] [Google Scholar]
- Tilley L., Dixon M. W. A., Kirk K. (2011). The Plasmodium falciparum-infected red blood cell. Int. J. Biochem. Cell Biol. 43, 839–842 10.1016/j.biocel.2011.03.012 [DOI] [PubMed] [Google Scholar]
- Van West P. (2000). Molecular tools to unravel the role of genes from Phytophthora infestans Ph.D. thesis, Wageningen University, Wageningen. [Google Scholar]
- Van West P., de Jong A. J., Judelson H. S., Emons A. M. C., Govers F. (1998). The ipiO gene of Phytophthora infestans is highly expressed in invading hyphae during infection. Fungal Genet. Biol. 23, 126–138 10.1006/fgbi.1998.1036 [DOI] [PubMed] [Google Scholar]
- Vleeshouwers V., Rietman H., Krenek P., Champouret N., Young C., Oh S. K., Wang M. Q., Bouwmeester K., Vosman B., Visser R. G. F., Jacobsen E., Govers F., Kamoun S., van der Vossen E. A. G. (2008). Effector genomics accelerates discovery and functional profiling of potato disease resistance and Phytophthora infestans avirulence genes. PLoS ONE 3, e2875. 10.1371/journal.pone.0002875 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Whisson S. C., Boevink P. C., Moleleki L., Avrova A. O., Morales J. G., Gilroy E. M., Armstrong M. R., Grouffaud S., van West P., Chapman S., Hein I., Toth I. K., Pritchard L., Birch P. R. J. (2007). A translocation signal for delivery of oomycete effector proteins into host plant cells. Nature 450, 115–118 10.1038/nature06203 [DOI] [PubMed] [Google Scholar]
- Yaeno T., Li H., Chaparro-Garcia A., Schornack S., Koshiba S., Watanabe S., Kigawa T., Kamoun S., Shirasu K. (2011). Phosphatidylinositol monophosphate-binding interface in the oomycete RXLR effector AVR3a is required for its stability in host cells to modulate plant immunity. Proc. Natl. Acad. Sci. U.S.A. 108, 14682–14687 10.1073/pnas.1106002108 [DOI] [PMC free article] [PubMed] [Google Scholar]