Abstract
Global transcript analyses based on publicly available microarray dataset have revealed that genes with similar function tend to be transcriptionally coordinated. Indeed, many genes involved in the formation of cellulose, hemicelluloses, and lignin have been identified using co-expression approaches in Arabidopsis. To facilitate these transcript analyses, several web-based tools have been developed that allow researchers to investigate co-expression relationships of their gene(s) of interest. In addition, several tools now also provide the possibility of comparative transcriptional analyses across species, which potentially increases the predictive power. In this short review, we describe recent developments and updates of plant-related co-expression tools, and summarize studies that have successfully used expression profiling in cell wall research. Finally, we illustrate the value of comparative co-expression relationships across species using genes involved in lignin biosynthesis.
Keywords: plant, co-expression, cell wall, cellulose, xylan, lignin, comparison across species
INTRODUCTION
In the last decade, the increased use of microarrays for global expression analyses made large-scale transcript analyses possible. Using publicly available microarray datasets, several studies showed that genes with similar function tended to be transcriptionally coordinated (Stuart et al., 2003; Ihmels et al., 2004). Based on this observation, co-expression approaches have been used to assign functions for genes involved in cell wall formation, isoprenoid and glucosinolate biosynthesis, and different metabolic pathways in Arabidopsis (Wille et al., 2004; Brown et al., 2005; Persson et al., 2005; Wei et al., 2006; Hirai et al., 2007). Interestingly, certain co-expression relationships appear to also be conserved across different species across the kingdoms of life (Stuart et al., 2003; Bergmann et al., 2004). For example, orthologous genes involved in protein synthesis, cell cycle, and protein degradation formed comparable co-expressed clusters in different species (Stuart et al., 2003). In the above studies, the microarray analyses still had to be quality-controlled and evaluated by the investigators themselves to obtain co-expressed relationships. However, web-based co-expression tools now enable users to easily mine publicly available microarray dataset to investigate their gene(s) of interest. In this review we present several co-expression tools and describe recent developments and updates for them. We also give an overview of how these tools have been used to identify novel cell wall-related genes, and exemplify the use of comparative co-expression analyses across species to infer lignin-related genes.
CO-EXPRESSION TOOLS
Several co-expression tools have been developed for plant biology, including ACT (Manfield et al., 2006), ASIDB (Rawat et al., 2008), ATTED-II (Obayashi et al., 2009), CressExpress (Srinivasasainagendra et al., 2008), CSB.DB (Steinhauser et al., 2004), and GeneCAT (Mutwil et al., 2008), which have been comprehensively described and compared elsewhere (Usadel et al., 2009). In addition, several recent platforms have emerged, such as AraNet, CORNET, GeneMANIA, PlaNet, and RiceArrayNet. AraNet1 aims at annotation of Arabidopsis genes by integrating available large-scale experimental data, e.g., co-expression and protein–protein interaction data, as well as gene associations inferred from other species and literature queried data (Lee et al., 2010). Similarly, GeneMANIA2 is predicting gene function based on available genomics and proteomics data sets for Arabidopsis, yeast, and several animal model species (Warde-Farley et al., 2010). CORNET3 allows for user-defined selection of microarray experiments and cut-offs for assessing co-regulated genes in Arabidopsis. These data may be supplemented with protein–protein interaction, functional annotation, and localization data (De Bodt et al., 2010). PlaNet provides a platform for gene co-expression network analysis for seven plant species and includes information about significant enrichment for functional annotation using MapMan ontology terms4 (Mutwil et al., 2011). Another interesting co-expression tool is RiceArrayNet, which calculates positive as well as negative correlation of gene expression profiles in rice (Lee et al., 2009). This tool has later been extended to Brassica and Arabidopsis, referred to as PlantArrayNet5.
In principle, the platforms above calculate co-expression relationships between two genes of interest by comparing their respective expression profiles. The Pearson’s correlation coefficient (PCC) is a commonly used measure to estimate transcriptional co-ordination. Using this measure as a basis, co-expression relationships between many genes can be determined, and can be visualized as networks in which nodes represent genes and the connection between nodes indicates the transcriptional co-ordination of the genes. Such co-expression networks can be divided into rank-based and value-based networks (Aoki et al., 2007). Previously, the most widely used approach was value-based networks, i.e., edges were established between genes that were co-expressed above a certain PCC-value threshold (Lee et al., 2004; Oldham et al., 2006). One major drawback of this method arises from the fact that some biological processes are tightly transcriptionally co-regulated, while other processes are not. Therefore, when a stringent global PCC-value cut-off is applied many genes involved in weakly transcriptionally coordinated processes that may be biologically relevant become disconnected. In contrast, a lower PCC-threshold will in many instances result in excessively large gene clusters, containing thousands of genes (Mao et al., 2009). To avoid such problems, some tools have introduced the rank-based method, which is based on the ranks of two given genes in their mutual co-expression lists (ATTED-II: Obayashi and Kinoshita, 2009; PlaNet: Mutwil et al., 2010,2011). Although both the value- and rank-based networks are derived from PCC, rank-based networks appear to lead to a network topology that closer resembles biological networks (Ruan et al., 2010). However, it is important to note that both approaches have their advantages and drawbacks (Usadel et al., 2009).
Most of the co-expression tools have focused on the model plant Arabidopsis, and have incorporated the main bulk of publicly available microarray datasets, thus representing condition-independent co-expression relationships (Usadel et al., 2009). However, some biologically relevant transcriptional relationships may be revealed only under specific experimental conditions, or in certain tissues. To find such relationships, tools like CORNET allow for user-defined selection of microarray experiments to calculate co-expression relationships. Another example of condition-dependent databases is the updated version of ATTED-II6, which enables the user to analyze co-expression relationships of genes under five predefined conditions: tissue and development, abiotic stress, biotic stress, hormone treatment, and different light regimes (Obayashi et al., 2011). Furthermore, SeedNet7 is a relatively new tissue-specific database, which returns co-expression relationships of genes during seed development (Bassel et al., 2011).
One possible caveat with co-expression analyses is the rate of “false positives,” i.e., co-expressed genes that might be co-expressed by chance rather than being functionally related. A useful approach to minimize the rate of such “false positives” may be to investigate whether orthologous genes are also co-expressed in related species. As mentioned above, co-expression relationships are often conserved across species (Stuart et al., 2003). Hence, two co-expressed genes from one species often have orthologs in another species that in turn are also co-expressed. In theory it should therefore be possible to enrich for co-occurring co-expression relationships across species, and hence minimize “false positives.” Therefore, across species co-expression analyses might improve the reliability of co-expression-based functional annotation. Consequently, several tools, such as StarNet8, CoP9, and ATTED-II, allow pairwise comparison between species (Jupiter et al., 2009; Ogata et al., 2009; Obayashi et al., 2011). Moreover, comparison of several species at the same time was introduced by the NetworkComparer-tool of PlaNet (Mutwil et al., 2011). This tool bins genes into gene families according to their Pfam annotation (Finn et al., 2010), and then finds recurring Pfams in co-expression networks across species (Mutwil et al., 2011).
TRANSCRIPTIONAL CO-ORDINATION OF GENES INVOLVED IN CELLWALL BIOSYNTHESIS
Cellulose is produced by the cellulose synthase (CESA) complex (CSC), which is comprised of CESA1, 3, and 6-related proteins in the primary wall and CESA4, 7, and 8 in the secondary wall (Gardiner et al., 2003; Desprez et al., 2007; Persson et al., 2007). Interestingly, these primary and the secondary CESAs display similar expression patterns, respectively (Brown et al., 2005; Persson et al., 2005). In addition, these studies were able to show that many genes involved in xylan and lignin synthesis were co-expressed with the secondary wall CESAs (Brown et al., 2005; Persson et al., 2005). Hence, co-expression analyses may be useful to identify new genes involved in secondary cell wall-related synthesis.
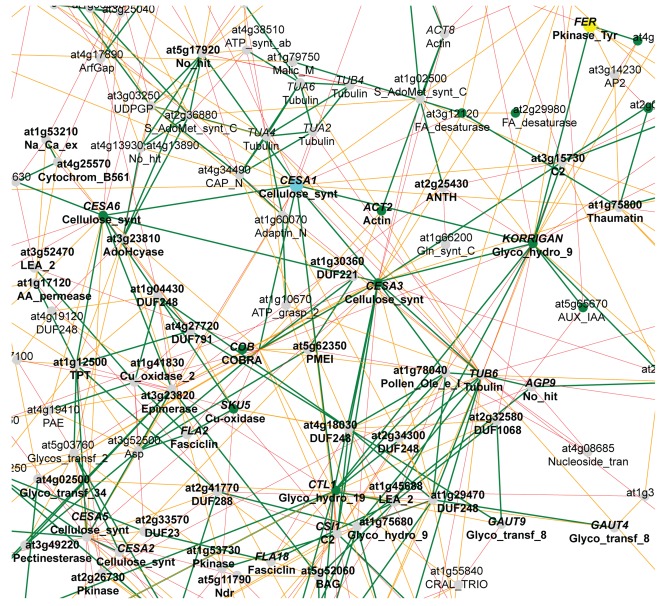
Figure 1 shows a truncated node-vicinity network (NVN) of genes co-expressed with the primary wall-related Arabidopsis CESA1-gene, which was obtained from PlaNet (Mutwil et al., 2011). Several primary wall CESA genes (CESA2, CESA3, CESA5, and CESA6) may be found in close vicinity of CESA1. In addition, many other genes important for cellulose synthesis, such as COBRA (COB), CHITINASE-LIKE (CTL)1, CELLULOSE SYNTHASE INTERACTING (CSI)1/POM-POM2, and KORRIGAN (KOR), are present in this network (Nicol et al., 1998; Zhong et al., 2002; Roudier et al., 2005; Gu et al., 2010; Bringmann et al., 2012). This result confirms previous findings which were based on a smaller number of microarrays (Persson et al., 2005), and is similar to results obtained from other co-expression tools. For example, the many of the genes in this network are also present in the AtCESA1-top 300 list of co-expressed genes in ATTED-II (77 out of 190 genes; see also the genes marked in bold in Figure 1).
FIGURE 1.
Subset of the co-expression gene vicinity network of AtCESA1 (turquoise node). Nodes indicate individual genes, and edges indicate whether two genes are co-expressed above a certain mutual rank threshold. Node colors indicate whether mutations in the gene cause gametophytic lethality (yellow), any biological phenotype (green), or if no mutant phenotype information currently is available (gray) according to TAIR. Green, orange, and red edges indicate a mutual rank relationship <10 (green), 10–19 (orange), and 20–29 (red), respectively, for each connected gene pair. This modified network was obtained from PlaNet (http://aranet.mpimp-golm.mpg.de/aranet). Genes marked in bold are also present in the top 300 co-expression list of AtCESA1 obtained from ATTED-II (http://atted.jp/). The respective gene family according to Pfam annotation is given below the gene names/AGI codes (Finn et al., 2010).
In addition to cellulose synthesis, co-expression approaches have been used to identify genes involved in the synthesis of hemicelluloses. For example, Cocuron et al. (2007) found that the Arabidopsis CSLC4 gene, which presumably is involved in glucan backbone synthesis of xyloglucan, was co-expressed with the xylosyltransferase AtXT1, which has previously been shown to attach xylose residues to a glucan backbone (Faik et al., 2002). Moreover, expression profiling was used to identify IRX15 and an IRX15-like gene, with corresponding single mutants showing a mild irregular xylem phenotype (Jensen et al., 2010; Brown et al., 2011). Both genes belong to a gene family with a domain of unknown function (DUF) 579 and only the corresponding double mutant showed decreased levels of xylan and altered cell wall morphology in stems (Jensen et al., 2010; Brown et al., 2011). Although their exact function is unknown, these results suggest an important role of this DUF579-gene family in xylan biosynthesis (Jensen et al., 2010; Brown et al., 2011).
Interestingly, also many genes that are important for lignin synthesis are co-expressed. Using microarray data from several developmental stages of Arabidopsis stems, Ehlting et al. (2005) showed transcriptional co-ordination of many putative and bona fide genes involved in monolignol biosynthesis, transport, and polymerization. This is consistent with results of Persson et al. (2005), and Brown et al. (2005) that found laccase genes, which are probably involved in polymerization of lignin, in the list of genes that are co-expressed with secondary wall-related CESAs.
COMPARATIVE EXPRESSION ANALYSES OF CELL WALL-RELATED GENES ACROSS SPECIES
The study of Mitchell et al. (2007) was one of the first studies to compare transcript relationships of cell wall-related genes across monocots and dicots. Based on EST data, the authors found that members of the GT43-, GT47-, and GT61-family are enriched in monocots and therefore might be involved in biosynthesis of the grass-specific glucuronoarabinoxylan (Mitchell et al., 2007). Indeed, the same group could recently show that several GT61-members are essential for arabinosylation of xylan (Anders et al., 2012). Another recent study compared the co-expression relationships of xylan-related genes from Arabidopsis and rice (Oikawa et al., 2010). Using the Arabidopsis genes IRX9, IRX10, and IRX14, which have previously been associated with xylan backbone synthesis (Brown et al., 2005; Peña et al., 2007), the tentative rice orthologs were identified based on sequence homology and expression patterns (Oikawa et al., 2010). These genes were then utilized as baits in the ATTED-II tool and revealed many re-occurring homologs in the respective co-expression lists (Oikawa et al., 2010). A similar co-expression approach has been undertaken for cellulose-related genes by comparing the co-expression networks of primary and secondary CESAs from seven species using PlaNet (Ruprecht et al., 2011). Interestingly, many gene families are consistently co-expressed with CESA genes across species. The function of most of these gene families remains unknown; however, their conserved transcriptional co-ordination suggests that they fulfill important functions during cellulose synthesis (Ruprecht et al., 2011). Based on the results, an Arabidopsis mutant corresponding to PINORESINOL REDUCTASE (PRR) 1 was identified that displayed distorted xylem vessels (Ruprecht et al., 2011), probably due to decreased lignin or lignan production.
USING A COMPARATIVE CO-EXPRESSION TOOL TO ANALYZE LIGNIN BIOSYNTHESIS
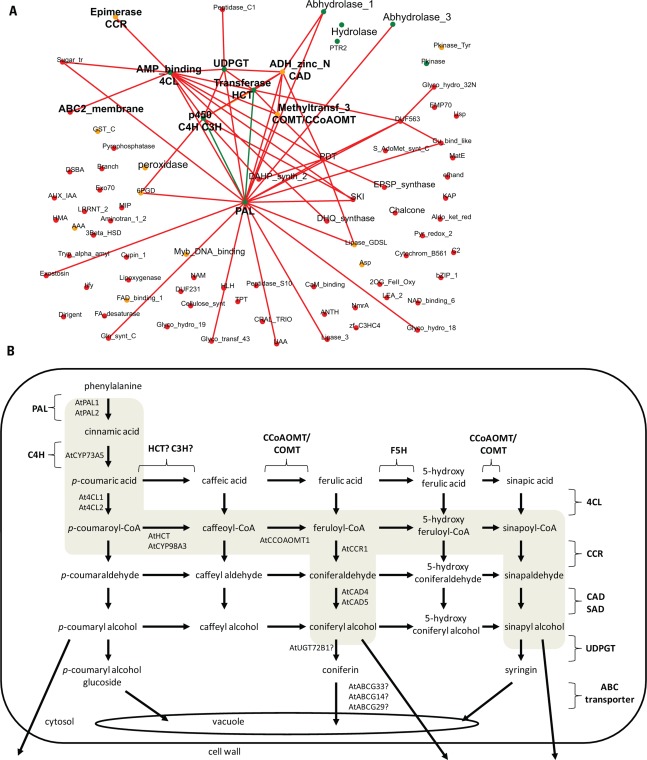
While comparative transcriptional analyses across species have been undertaken for cellulose and hemicelluloses-related genes, conserved co-expression relationships among lignin-related genes have been paid less attention. Similar to previously published results (Ehlting et al., 2005), we found that the co-expression network of Arabidopsis PHENYLALANINE AMMONIA LYASE (PAL) 1, which marks the initial step for monolignol synthesis, comprises many genes involved in lignin formation Figure 2B). Using AtPAL1 as bait in the NetworkComparer-tool of PlaNet, we identified similar co-expression networks for AtPAL1-orthologs in barley, Medicago, poplar, rice, soybean, and wheat. The resulting consensus network indicates that most of the gene families needed for monolignol synthesis are conserved across species (Figure 2). Surrounding the “PAL”-gene family, we found gene families corresponding to the subsequent steps in the lignin pathway such as C4H (which belongs to gene family “p450” according to Pfam annotation, Finn et al., 2010), 4CL (“AMP_binding”), HCT (“Transferase”), C3H (“p450”), CCoAOMT (“Methyltransf_3”), CCR (“Epimerase”), and CAD (“ADH_zinc_N”). In addition, genes functioning upstream of PAL in the phenylalanine synthesis pathway were previously identified as transcriptionally linked with the phenylpropanoid-related genes in Arabidopsis (Tohge and Fernie, 2010), which appeared to be also conserved in other species (i.e., gene families “DAHP_synth_2,” “DHQ_synthase,” “SKI,” “EPSP_synthase,” “PDT”). Interestingly, a gene family denoted as “UDPGT,” corresponding to glycosyltransferases, was present in almost all PAL1-derived co-expression networks across the seven species (Figure 2). Members of this family have been shown to glucosylate various monolignols (Lim et al., 2001) and we therefore hypothesize that the respective co-expressed genes of this family might have a similar function in the different species. Recently, Miao and Liu (2010) showed that ABC-transporters are likely to mediate transport of monolignols, and their glucoconjugates, across plasma and vacuolar membrane, respectively. However, the exact genes involved in this process have not yet been identified. The AtPAL1-network contained three different genes from the ABC-transporter family (“ABC2_membrane”) which might mediate this function in Arabidopsis. While single knock-out mutants for one of them (atabcg33) did not show any obvious phenotypes (Kaneda et al., 2011), it is plausible that the other two members may functionally compensate for the loss of AtABCG33. Interestingly, the consensus network also comprises several highly conserved gene families, whose function in lignin biosynthesis is still elusive, for example, the gene families “Hydrolase,” “Abhydrolase_1,” and “Abhydrolase_3.” One of the co-expressed genes from the “Hydrolase”-family in Arabidopsis is RESPONSIVE-TO-ANTAGONIST (RAN) 1, which encodes for a copper transporter involved in ethylene signaling (Hirayama et al., 1999). Copper is a co-factor of laccases, which are important for polymerization of monolignols to lignin in the cell wall (Boerjan et al., 2003). We therefore speculate that RAN1 has an additional function in providing copper co-factors for enzymatic lignin formation. Moreover, close homologs of the Arabidopsis gene from the “Abhydrolase_1”-family (At3g03990) are methylesterases, suggesting that these genes might function antagonistically to caffeic acid O-methyltransferases (COMTs). Another rather unexpected yet interesting example is the highly conserved “Peroxidase”-family, which one might associate to monolignol polymerization in the cell wall. However, the respective co-expressed gene in Arabidopsis is ASCORBATE PEROXIDASE (APX)1, a cytosolic enzyme that has an important role in scavenging hydrogen peroxide (Davletova et al., 2005). APX1 might therefore function in preventing premature radical coupling of monolignols already in the cytosol. Thus, exemplified by the lignin-related analysis, we propose that comparative co-expression analyses might be useful in the future to reveal novel players for different aspects of cell wall biosynthesis, but also in high-lighting conserved and divergent elements in different biological processes.
FIGURE 2.
Co-expression of genes involved in lignin formation across species. (A) Consensus co-expression network of lignin formation across seven plant species obtained from the NetworkComparer-tool from PlaNet (http://aranet.mpimp-golm.mpg.de/aranet/NetworkComparer). Co-expression network of following genes/probeset IDs were used: Arabidopsis, At2g37040; rice, LOC_Os02g41650; Medicago, Medtr1g076720; barley, Contig1805_s_at; poplar, PtpAffx.1672.3.A1_a_at; wheat, TaAffx.45277.1.S1_x_at. (B) Monolignol biosynthesis pathway (adopted from Boerjan et al., 2003). Arabidopsis genes present in the co-expression network of AtPAL1 were mapped on the pathway. Question marks indicate that the function of the respective genes has not been shown yet. The main routes for monolignol synthesis in dicots are highlighted in gray.
CONCLUSION AND PERSPECTIVE
Co-expression approaches have been especially valuable for identifying new genes involved in secondary cell wall synthesis (Brown et al., 2005,2011; Persson et al., 2005; Jensen et al., 2010; Ruprecht et al., 2011). A comparison of the candidate genes from several different studies showed that similar results were obtained regardless of the underlying microarray datasets (i.e., condition-independent or stem-specific samples) and the bait genes (xylan or cellulose synthesis-related) used (Oikawa et al., 2010). This is probably due to the fact that secondary cell wall formation is a highly coordinated process that is mainly restricted to certain tissue and cell types.
However, many genes that are transcriptionally associated with secondary cell wall formation have already been investigated, and mutant analyses targeting only one gene are likely to only yield mild or no phenotypes (Jensen et al., 2010; Brown et al., 2011; Ruprecht et al., 2011). One likely reason for this is genetic redundancy, and hence, mutant combinations or knock-down approaches that target several homologous genes might be needed in the future to generate informative phenotypes. In addition, detailed comparative transcriptional studies across and within species might be needed to obtain more reliable candidate genes related to cell wall synthesis.
Conflict of Interest Statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Acknowledgments
We thank Marek Mutwil and Takayuki Tohge for helpful comments on the manuscript and the Max-Planck-Gesellschaft for financial support.
Footnotes
REFERENCES
- Anders N., Wilkinson M. D., Lovegrove A., Freeman J., Tryfona T., Pellny T. K., Weimar T., Mortimer J. C., Stott K., Baker J. M., Defoin-Platel M., Shewry P. R., Dupree P., Mitchell R. A. C. (2012). Glycosyl transferases in family 61 mediate arabinofuranosyl transfer onto xylan in grasses. Proc. Natl. Acad. Sci. U.S.A. 109 989–993 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aoki K., Ogata Y., Shibata D. (2007). Approaches for extracting practical information from gene co-expression networks in plant biology. Plant Cell Physiol. 48 381–390 [DOI] [PubMed] [Google Scholar]
- Bassel G. W., Lan H., Glaab E., Gibbs D. J., Gerjets T., Krasnogor N., Bonner A. J., Holdsworth M. J., Provart N. J. (2011). Genome-wide network model capturing seed germination reveals coordinated regulation of plant cellular phase transitions. Proc. Natl. Acad. Sci. U.S.A. 108 9709–9714 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bergmann S., Ihmels J., Barkai N. (2004). Similarities and differences in genome-wide expression data of six organisms. PLoS Biol. 2 e9 10.1371/journal.pbio.0020009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boerjan W., Ralph J., Baucher M. (2003). Lignin biosynthesis. Annu. Rev. Plant Biol. 54 519–546 [DOI] [PubMed] [Google Scholar]
- Bringmann M., Li E., Sampathkumar A., Kocabek T., Hauser M., Persson S. (2012). POM-POM2/CELLULOSE SYNTHASE INTERACTING1 Is essential for the functional association of cellulose synthase and microtubules in Arabidopsis. Plant Cell 24 163–177 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brown D., Wightman R., Zhang Z., Gomez L. D., Atanassov I., Bukowski J., Tryfona T., McQueen-Mason S. J., Dupree P., Turner S. (2011). Arabidopsis genes IRREGULAR XYLEM (IRX15) and IRX15L encode DUF579-containing proteins that are essential for normal xylan deposition in the secondary cell wall. Plant J. 66 401–413 [DOI] [PubMed] [Google Scholar]
- Brown D. M., Zeef L. A. H., Ellis J., Goodacre R., Turner S. R. (2005). Identification of novel genes in Arabidopsis involved in secondary cell wall formation using expression profiling and reverse genetics. Plant Cell 17 2281–2295 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cocuron J., Lerouxel O., Drakakaki G., Alonso A. P., Liepman A. H., Keegstra K., Raikhel N., Wilkerson C. G. (2007). A gene from the cellulose synthase-like C family encodes a beta-1,4 glucan synthase. Proc. Natl. Acad. Sci. U.S.A. 104 8550–8555 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Davletova S., Rizhsky L., Liang H., Shengqiang Z., Oliver D. J., Coutu J., Shulaev V., Schlauch K., Mittler R. (2005). Cytosolic ascorbate peroxidase 1 is a central component of the reactive oxygen gene network of Arabidopsis. Plant Cell 17 268–281 [DOI] [PMC free article] [PubMed] [Google Scholar]
- De Bodt S., Carvajal D., Hollunder J., Van den Cruyce J., Movahedi S., Inzé D. (2010). CORNET: a user-friendly tool for data mining and integration. Plant Physiol. 152 1167–1179 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Desprez T., Juraniec M., Crowell E. F., Jouy H., Pochylova Z., Parcy F., Höfte H., Gonneau M., Vernhettes S. (2007). Organization of cellulose synthase complexes involved in primary cell wall synthesis in Arabidopsis thaliana. Proc. Natl. Acad. Sci. U.S.A. 104 15572–15577 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ehlting J., Mattheus N., Aeschliman D. S., Li E., Hamberger B., Cullis I. F., Zhuang J., Kaneda M., Mansfield S. D., Samuels L., Ritland K., Ellis B. E., Bohlmann J., Douglas C. J. (2005). Global transcript profiling of primary stems from Arabidopsis thaliana identifies candidate genes for missing links in lignin biosynthesis and transcriptional regulators of fiber differentiation. Plant J. 42 618–640 [DOI] [PubMed] [Google Scholar]
- Faik A., Price N. J., Raikhel N. V., Keegstra K. (2002). An Arabidopsis gene encoding an alpha-xylosyltransferase involved in xyloglucan biosynthesis. Proc. Natl. Acad. Sci. U.S.A. 99 7797–7802 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Finn R. D., Mistry J., Tate J., Coggill P., Heger A., Pollington J. E., Gavin O. L., Gunasekaran P., Ceric G., Forslund K., Holm L., Sonnhammer E. L. L., Eddy S. R., Bateman A. (2010). The Pfam protein families database. Nucleic Acids Res. 38 D211–D222 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gardiner J. C., Taylor N. G., Turner S. R. (2003). Control of cellulose synthase complex localization in developing xylem. Plant Cell 15 1740–1748 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gu Y., Kaplinsky N., Bringmann M., Cobb A., Carroll A., Sampathkumar A., Baskin T. I., Persson S., Somerville C. R. (2010). Identification of a cellulose synthase-associated protein required for cellulose biosynthesis. Proc. Natl. Acad. Sci. U.S.A. 107 12866–12871 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hirai M. Y., Sugiyama K., Sawada Y., Tohge T., Obayashi T., Suzuki A., Araki R., Sakurai N., Suzuki H., Aoki K., Goda H., Nishizawa O. I., Shibata D., Saito K. (2007). Omics-based identification of Arabidopsis Myb transcription factors regulating aliphatic glucosinolate biosynthesis. Proc. Natl. Acad. Sci. U.S.A. 104 6478–6483 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hirayama T., Kieber J. J., Hirayama N., Kogan M., Guzman P., Nourizadeh S., Alonso J. M., Dailey W. P., Dancis A., Ecker J. R. (1999). RESPONSIVE-TO-ANTAGONIST1, a Menkes/Wilson disease-related copper transporter, is required for ethylene signaling in Arabidopsis. Cell 97 383–393 [DOI] [PubMed] [Google Scholar]
- Ihmels J., Levy R., Barkai N. (2004). Principles of transcriptional control in the metabolic network of Saccharomyces cerevisiae. Nat. Biotechnol. 22 86–92 [DOI] [PubMed] [Google Scholar]
- Jensen J. K., Kim H., Cocuron J., Orler R., Ralph J., Wilkerson C. G. (2010). The DUF579 domain containing proteins IRX15 and IRX15-L affect xylan synthesis in Arabidopsis. Plant J. 66 387–400 [DOI] [PubMed] [Google Scholar]
- Jupiter D., Chen H., VanBuren V. (2009). STARNET 2: a web-based tool for accelerating discovery of gene regulatory networks using microarray co-expression data. BMC Bioinformatics 10 332 10.1186/1471-2105-10-332 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaneda M., Schuetz M., Lin B. S. P., Chanis C., Hamberger B., Western T. L., Ehlting J., Samuels A. L. (2011). ABC transporters coordinately expressed during lignification of Arabidopsis stems include a set of ABCBs associated with auxin transport. J. Exp. Bot. 62 2063–2077 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee H. K., Hsu A. K., Sajdak J., Qin J., Pavlidis P. (2004). Coexpression analysis of human genes across many microarray data sets. Genome Res. 14 1085–1094 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee I., Ambaru B., Thakkar P., Marcotte E. M., Rhee S. Y. (2010). Rational association of genes with traits using a genome-scale gene network for Arabidopsis thaliana. Nat. Biotechnol. 28 149–156 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee T., Kim Y., Pham T. T. M., Song S. I., Kim J., Kang K. Y., An G., Jung K., Galbraith D. W., Kim M., Yoon U., Nahm B. H. (2009). RiceArrayNet: a database for correlating gene expression from transcriptome profiling, and its application to the analysis of coexpressed genes in rice. Plant Physiol. 151 16–33 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lim E. K., Li Y., Parr A., Jackson R., Ashford D. A., Bowles D. J. (2001). Identification of glucosyltransferase genes involved in sinapate metabolism and lignin synthesis in Arabidopsis. J. Biol. Chem. 276 4344–4349 [DOI] [PubMed] [Google Scholar]
- Manfield I. W., Jen C., Pinney J. W., Michalopoulos I., Bradford J. R., Gilmartin P. M., Westhead D. R. (2006). Arabidopsis Co-expression Tool (ACT): web server tools for microarray-based gene expression analysis. Nucleic Acids Res. 34 W504–W509 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mao L., Van Hemert J. L., Dash S., Dickerson J. A. (2009). Arabidopsis gene co-expression network and its functional modules. BMC Bioinformatics 10 346 10.1186/1471-2105-10-346 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miao Y., Liu C. (2010). ATP-binding cassette-like transporters are involved in the transport of lignin precursors across plasma and vacuolar membranes. Proc. Natl. Acad. Sci. U.S.A. 107 22728–22733 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mitchell R. A. C., Dupree P., Shewry P. R. (2007). A novel bioinformatics approach identifies candidate genes for the synthesis and feruloylation of arabinoxylan. Plant Physiol. 144 43–53 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mutwil M., Klie S., Tohge T., Giorgi F. M., Wilkins O., Campbell M. M., Fernie A. R., Usadel B., Nikoloski Z., Persson S. (2011). PlaNet: combined sequence and expression comparisons across plant networks derived from seven species. Plant Cell 23 895–910 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mutwil M., Obro J., Willats W. G. T., Persson S. (2008). GeneCAT – novel webtools that combine BLAST and co-expression analyses. Nucleic Acids Res. 36 W320–W326 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mutwil M., Usadel B., Schütte M., Loraine A., Ebenhöh O., Persson S. (2010). Assembly of an interactive correlation network for the Arabidopsis genome using a novel heuristic clustering algorithm. Plant Physiol. 152 29–43 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nicol F., His I., Jauneau A., Vernhettes S., Canut H., Höfte H. (1998). A plasma membrane-bound putative endo-1,4-beta-D-glucanase is required for normal wall assembly and cell elongation in Arabidopsis. EMBO J. 17 5563–5576 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Obayashi T., Hayashi S., Saeki M., Ohta H., Kinoshita K. (2009). ATTED-II provides coexpressed gene networks for Arabidopsis. Nucleic Acids Res. 37 D987–D991 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Obayashi T., Kinoshita K. (2009). Rank of correlation coefficient as a comparable measure for biological significance of gene coexpression. DNA Res. 16 249–260 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Obayashi T., Nishida K., Kasahara K., Kinoshita K. (2011). ATTED-II updates: condition-specific gene coexpression to extend coexpression analyses and applications to a broad range of flowering plants. Plant Cell Physiol. 52 213–219 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ogata Y., Sakurai N., Suzuki H., Aoki K., Saito K., Shibata D. (2009). The prediction of local modular structures in a co-expression network based on gene expression datasets. Genome Inform. 23 117–127 [PubMed] [Google Scholar]
- Oikawa A., Joshi H. J., Rennie E. A., Ebert B., Manisseri C., Heazlewood J. L., Scheller H. V. (2010). An integrative approach to the identification of Arabidopsis and rice genes involved in xylan and secondary wall development. PLoS ONE 5 e15481 10.1371/journal.pone.0015481 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oldham M. C., Horvath S., Geschwind D. H. (2006). Conservation and evolution of gene coexpression networks in human and chimpanzee brains. Proc. Natl. Acad. Sci. U.S.A. 103 17973–17978 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peña M. J., Zhong R., Zhou G., Richardson E. A., O’Neill M. A., Darvill A. G., York W. S., Ye Z. (2007). Arabidopsis irregular xylem8 and irregular xylem9: implications for the complexity of glucuronoxylan biosynthesis. Plant Cell 19 549–563 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Persson S., Paredez A., Carroll A., Palsdottir H., Doblin M., Poindexter P., Khitrov N., Auer M., Somerville C. R. (2007). Genetic evidence for three unique components in primary cell-wall cellulose synthase complexes in Arabidopsis. Proc. Natl. Acad. Sci. U.S.A. 104 15566–15571 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Persson S., Wei H., Milne J., Page G. P., Somerville C. R. (2005). Identification of genes required for cellulose synthesis by regression analysis of public microarray data sets. Proc. Natl. Acad. Sci. U.S.A. 102 8633–8638 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rawat A., Seifert G. J., Deng Y. (2008). Novel implementation of conditional co-regulation by graph theory to derive co-expressed genes from microarray data. BMC Bioinformatics 9(Suppl. 9) S7 10.1186/1471-2105-9-S9-S7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roudier F., Fernandez A. G., Fujita M., Himmelspach R., Borner G. H. H., Schindelman G., Song S., Baskin T. I., Dupree P., Wasteneys G. O., Benfey P. N. (2005). COBRA, an Arabidopsis extracellular glycosyl-phosphatidyl inositol-anchored protein, specifically controls highly anisotropic expansion through its involvement in cellulose microfibril orientation. Plant Cell 17 1749–1763 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ruan J., Dean A. K., Zhang W. (2010). A general co-expression network-based approach to gene expression analysis: comparison and applications. BMC Syst. Biol. 4 8 10.1186/1752-0509-4-8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ruprecht C., Mutwil M., Saxe F., Eder M., Nikoloski Z., Persson S. (2011). Large-scale co-expression approach to dissect secondary cell wall formation across plant species. Front. Plant Sci. 2:23 10.3389/fpls.2011.00023 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Srinivasasainagendra V., Page G. P., Mehta T., Coulibaly I., Loraine A. E. (2008). CressExpress: a tool for large-scale mining of expression data from Arabidopsis. Plant Physiol. 147 1004–1016 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Steinhauser D., Usadel B., Luedemann A., Thimm O., Kopka J. (2004). CSB.DB: a comprehensive systems-biology database. Bioinformatics 20 3647–3651 [DOI] [PubMed] [Google Scholar]
- Stuart J. M., Segal E., Koller D., Kim S. K. (2003). A gene-coexpression network for global discovery of conserved genetic modules. Science 302 249–255 [DOI] [PubMed] [Google Scholar]
- Tohge T., Fernie A. R. (2010). Combining genetic diversity, informatics and metabolomics to facilitate annotation of plant gene function. Nat. Protoc. 5 1210–1227 [DOI] [PubMed] [Google Scholar]
- Usadel B., Obayashi T., Mutwil M., Giorgi F. M., Bassel G. W., Tanimoto M., Chow A., Steinhauser D., Persson S., Provart N. J. (2009). Co-expression tools for plant biology: opportunities for hypothesis generation and caveats. Plant Cell Environ. 32 1633–1651 [DOI] [PubMed] [Google Scholar]
- Warde-Farley D., Donaldson S. L., Comes O., Zuberi K., Badrawi R., Chao P., Franz M., Grouios C., Kazi F., Lopes C. T., Maitland A., Mostafavi S., Montojo J., Shao Q., Wright G., Bader G. D., Morris Q. (2010). The GeneMANIA prediction server: biological network integration for gene prioritization and predicting gene function. Nucleic Acids Res. 38 W214–W220 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wei H., Persson S., Mehta T., Srinivasasainagendra V., Chen L., Page G. P., Somerville C., Loraine A. (2006). Transcriptional coordination of the metabolic network in Arabidopsis. Plant Physiol. 142 762–774 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wille A., Zimmermann P., Vranová E., Fürholz A., Laule O., Bleuler S., Hennig L., Prelic A., von Rohr P., Thiele L., Zitzler E., Gruissem W., Bühlmann P. (2004). Sparse graphical Gaussian modeling of the isoprenoid gene network in Arabidopsis thaliana. Genome Biol. 5 R92 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhong R., Kays S. J., Schroeder B. P., Ye Z. (2002). Mutation of a chitinase-like gene causes ectopic deposition of lignin, aberrant cell shapes, and overproduction of ethylene. Plant Cell 14 165–179 [DOI] [PMC free article] [PubMed] [Google Scholar]