Abstract
The Arabidopsis thaliana SFD1 (suppressor of fatty acid desaturase deficiency1) gene (also known as GLY1) is required for accumulation of 34:6 (i.e., 18:3–16:3) monogalactosyldiacylglycerol (MGDG) and for the activation of systemic acquired resistance (SAR), an inducible defense mechanism that confers resistance against a broad spectrum of pathogens. SFD1, which has been suggested to be involved in lipid-based signaling in SAR, contains a putative chloroplast transit peptide and has glycerol-3-phosphate synthesizing dihydroxyacetone phosphate (DHAP) reductase (also referred as glycerol-3-phosphate dehydrogenase) activity. The goals of this study were to determine if the DHAP reductase activity and chloroplast localization are required for SFD1’s involvement in galactolipid metabolism and SAR signaling. The crystal structure of a Leishmania mexicana glycerol-3-phosphate dehydrogenase was used to model SFD1 structure and identify Lys194, Lys279, and Asp332 as potential catalytic site residues in SFD1. Mutational analysis of SFD1 confirmed that Lys194, Lys279, and Asp332 are critical for SFD1’s DHAP reductase activity, and its involvement in SAR. SFD1 proteins with these residues individually substituted by Ala lacked DHAP reductase activity and were unable to complement the SAR defect of the sfd1 mutant. The SFD1–Ala279 protein was also unable to restore 34:6-MGDG content when expressed in the sfd1 mutant. In vivo imaging of a green fluorescent protein-tagged SFD1 protein demonstrated that SFD1 is targeted to the chloroplast. The N-terminal 43 amino acids, which are required for proper targeting of SFD1 to the chloroplast, are also required for SFD1’s function in lipid metabolism and SAR. Taken together, these results demonstrate that SFD1’s DHAP reductase activity is required in the chloroplast for lipid metabolism and defense signaling.
Keywords: lipid signaling, plant defense, systemic acquired resistance
Introduction
Glycerol-3-phosphate (G3P) feeds into glycolysis and gluconeogenesis, and serves as a precursor for the synthesis of membrane lipids and storage lipids. In addition, G3P participates in a mitochondrial shuttle to channel reducing equivalents from the cytosol into the mitochondria for respiration (Shen et al., 2006). Plants have multiple mechanisms for the synthesis of G3P. For example, glycerol kinase catalyzes the phosphorylation of glycerol to yield G3P. In Arabidopsis thaliana, the NHO1 (also known as GLI1)-encoded glycerol kinase synthesizes G3P in the cytosol (Kang et al., 2003). Another G3P biosynthesis mechanism involves the NADH-dependent reduction of dihydroxyacetone phosphate (DHAP) by DHAP reductases (Gee et al., 1988; Kirsch et al., 1992). DHAP reductases are generally grouped with and referred to as G3P dehydrogenase (GPDH). In animals, GPDH catalyze the oxidation of G3P to DHAP. However, in plants, at the physiological pH, most of these enzymes are essentially inactive as GPDHs, functioning only as DHAP reductases (Gee et al., 1988; Kirsch et al., 1992; Wei et al., 2001; Shen et al., 2006).
Four DHAP reductases/GPDHs have been studied in Arabidopsis. The At2g41540-encoded GPDHc1, which is located in the cytosol, functions along with the At3g10370-encoded mitochondrial FAD-dependent GPDH (FAD–GPDH) to shuttle reducing equivalents into the mitochondria for respiration (Shen et al., 2003, 2006). In contrast, the At5g40610-encoded DHAP reductase contains a plastid transit peptide at its N-terminus and is suggested to function in the plastids (Wei et al., 2001). The SFD1 (also known as GLY1) gene (At2g40690) encodes another DHAP reductase, which was shown to be required for glycerolipid synthesis in the chloroplasts (Miquel et al., 1998; Kachroo et al., 2004; Nandi et al., 2004). Loss-of-function mutations in this gene impact the acyl composition of monogalactosyldiacylglycerol (MGDG; Miquel et al., 1998; Nandi et al., 2004), resulting in a substantial reduction in levels of the chloroplast synthesized 18:3–16:3 acyl chain containing 34:6-MGDG species and a parallel increase in the levels of 18:3–18:3 acyl chain containing 36:6-MGDG, which is presumably derived from diacylglycerol that has been shunted from the cytosol into the chloroplast as a compensatory mechanism.
In Arabidopsis, enzymes involved in the synthesis of G3P also function in stress response. For example, GPDHc1 expression was induced in response to stress imposed by high salt and dehydration (Shen et al., 2006). GPDHc1 expression was also induced in response to treatment with abscisic acid, an important stress hormone. Plants lacking GPDHc1 function were impaired in maintaining cellular redox homeostasis, which resulted in a higher steady state level of reactive oxygen species in the gpdhc1 mutant plant and a corresponding increase in ROS scavenging enzymes (Shen et al., 2006). In line with a role for GPDHc1 in plant stress response, the mutant plant exhibited heightened sensitivity to salt stress and abscisic acid (Shen et al., 2006). The NHO1 gene is also involved in plant stress response. NHO1 is required for resistance against non-host and avirulent strains of Pseudomonas spp. (Kang et al., 2003). NHO1 expression is induced in plants inoculated with these pathogens. Furthermore, loss of NHO1 function resulted in increased growth of these pathogens in the nho1 mutant. In contrast, NHO1-mediated resistance is overcome by virulent strains of Pseudomonas syringae, which suppress NHO1 expression. NHO1 is also required for resistance against the fungal pathogen Colletotrichum higginsianum (Chanda et al., 2008).
Like NHO1, SFD1 is also involved in plant defense against pathogens. Although not required for basal resistance to P. syringae (Nandi et al., 2004; Chaturvedi et al., 2008), SFD1 is required for Arabidopsis resistance against C. higginsianum (Chanda et al., 2008). SFD1 is also required for systemic acquired resistance (SAR; Nandi et al., 2004; Chaturvedi et al., 2008; Chanda et al., 2011), which is an inducible defense mechanism that is activated systemically throughout the foliage in response to a prior exposure to pathogen elsewhere on the foliage (Métraux et al., 2002; Durrant and Dong, 2004; Vlot et al., 2008; Shah, 2009). The activation of SAR requires the translocation of a signal from the pathogen-inoculated organ to rest of the foliage, where it stimulates salicylic acid accumulation and enhances resistance against subsequent infections. The vasculature is the conduit for the long-distance translocation of this signal (Heil and Ton, 2008; Shah, 2009). sfd1 Mutant plants are defective in this long-distance signaling (Chaturvedi et al., 2008). Unlike vascular sap-enriched petiole exudates collected from wild type (WT) leaves treated with a SAR-inducing pathogen, comparable exudates collected from the sfd1 mutant leaves were unable to activate SAR when applied to leaves of the WT plant (Chaturvedi et al., 2008). By contrast, the sfd1 mutant leaves were sensitive to the SAR-inducing activity contained in WT petiole exudates. Considering that mutations in sfd1 impact lipid metabolism (Miquel et al., 1998; Kachroo et al., 2004; Nandi et al., 2004), it was suggested that a lipid or a lipid-derived factor was required for the accumulation in the vasculature and/or long-distance translocation of a SAR-inducing activity (Chaturvedi et al., 2008). Alternatively, G3P, which has been shown to impact plant defenses (Kachroo and Kachroo, 2009; Chanda et al., 2011), could have a more direct role in SAR. However, the molecular mechanism of SFD1’s involvement in SAR signaling remains unclear.
SFD1 encodes a 420 amino acid protein (Figure 1A). The sfd1-1 and gly1-2 alleles contain G → A transition mutations within the first 500 nucleotides of the SFD1 coding region that likely impact splicing of the SFD1 RNA (Kachroo et al., 2004; Nandi et al., 2004). In contrast, the gly1-3 allele contains a nonsense mutation that is predicted to result in a truncated protein lacking the C-terminal 109 amino acids (Kachroo et al., 2004), which includes part of the predicted DHAP-binding domain (Figure 1A), and the sfd1-2 allele contains a mis-sense mutation that results in the replacement of Ala at amino acid position 381 with Thr (Nandi et al., 2004). Although this alteration in sfd1-2 is within the DHAP-binding domain, it is not within the predicted active site region of the protein. Thus, the existing data does not exclude the possibility that the involvement of SFD1 in SAR is independent of its DHAP reductase activity. Indeed, moonlighting, that is having more than one biochemical function, is not uncommon for proteins (Jeffery, 1999; Moore, 2004). For example, in humans, the glycolytic enzyme glyceraldehyde-3-phosphate dehydrogenase also possesses a protein kinase activity and a uracil glycosylase activity that is involved in DNA repair (Meyer-Siegler et al., 1991). Likewise, a plant ferredoxin-dependent glutamate synthase was shown to form a complex with UDP-sulfoquinovose synthase and thus participate in sulfolipid biosynthesis, in addition to its role in amino acid metabolism (Shimojima et al., 2005). Thus, it is important to determine if SFD1’s DHAP reductase activity is required for the activation of SAR.
Figure 1.
Amino acid sequence and functional domains of SFD1 protein and their alignment with the Leishmania mexicana GPDH. (A) InterProScan predicted NAD-dependent glycerol-3-phosphate dehydrogenase domain (residues 88–250) indicated in red, and a NAD-dependent glycerol-3-phosphate dehydrogenase C-terminal domain (residues 267–411) indicated in blue. The putative N-terminal chloroplast transit peptide as predicted by iPSORT is overlined. The Gly residues in the GxGxxG NAD-binding motif that is conserved amongst most GPDHs, and the active site residues Lys194, Lys279, and Asp332, which were chosen for site-directed mutagenesis, are in bold black and underlined. (B) Alignment of SFD1 protein to Leishmania mexicana GPDH. ClustalW software was used to obtain multiple sequence alignment between SFD1 and the 1EVYA (GenBank accession: MT5G51560) GPDH from Leishmania mexicana. Active site residues in LmGPDH (Lys125, Ser155, Ala157, Lys210, Asn211, Asp263, Thr267, Ser273, Arg274, and Asn275) are indicated in bold red and underlined, and the corresponding residues of SFD1 indicated in bold red. Other residues that are identical between LmGPDH and SFD1 are in bold blue. The NAD-dependent glycerol-3-phosphate dehydrogenase N-terminal domain is overlined with a dashed line, while a solid overline denotes the C-terminal NAD-dependent glycerol-3-phosphate dehydrogenase domain.
Since the major lipid defect in the sfd1 mutant is the reduction in levels of the plastid synthesized 34:6-MGDG, we had suggested that SFD1 likely functions in the plastids (Nandi et al., 2004). However, there is a lack of agreement between the different subcellular prediction programs in predicting the intracellular location of SFD1. For example, ChloroP 1.11 predicts that SFD1 does not have a chloroplast transit peptide. In contrast, iPSORT2 predicts that SFD1 has, at its N-terminus, an apparent chloroplast transit peptide (Figure 1A). Here we show that the N-terminal 43 amino acids, which are not essential for SFD1’s DHAP reductase activity, are required for proper targeting of SFD1 to chloroplast. Furthermore, we demonstrate that the N-terminal 43 amino acids and DHAP reductase activity of SFD1 are critical for lipid metabolism and its involvement in SAR signaling.
Results
Homology of SFD1 to leishmania mexicana GPDH
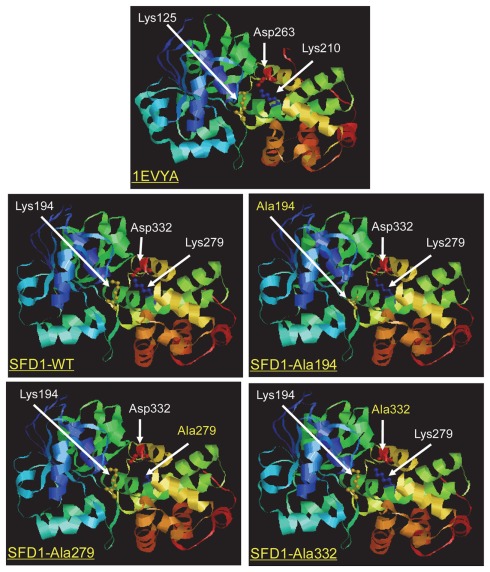
Sequence alignment of the SFD1 protein to other known GPDHs predicted an NAD-dependent GPDH domain corresponding to amino acid residues 267–411 at the C-terminal half of SFD1, and an NAD-binding domain (residues 88–250) in the N-terminal half of the protein (Figure 1A; Nandi et al., 2004). A highly conserved NAD+/NADH-binding GxGxxG motif is present beginning at amino acid 94 (Figure 1A). The crystal structure of the Leishmania mexicana GPDH (LmGPDH) holo enzyme in complex with NADH revealed that amino acids Lys125, Lys210, and Asp263 of LmGPDH are in the vicinity of the nicotinamide moiety of NADH (Suresh et al., 2000). Further study of a ternary complex involving LmGPDH with the substrate DHAP and the product NAD+ confirmed that amino acids Lys125, Ser155, Ala157, Lys210, Asn211, Asp263, Thr267, Ser273, Arg274, and Asn275 are part of the active site region (Suresh et al., 2000; Choe et al., 2003). Alignment of the LmGPDH sequence with the SFD1 amino acid sequence revealed that these amino acids are conserved in SFD1 and correspond to amino acids Lys194, Ser226, Ala228, Lys279, Asn280, Asp332, Thr336, Ser342, Arg343, and Asn344 in SFD1 (Figure 1B). In LmGPDH, Lys125, and Lys210 have been shown to form H-bonds with the O-2 atom of DHAP, from which a proton is removed during the oxidation of G3P to DHAP. Furthermore, replacement of Lys125 and Lys210 with Ala or Met in LmGPDH disrupted its enzyme activity (Choe et al., 2003). To further determine if Lys194 and Lys279 in SFD1 correspond to Lys125 and Lys210 of LmGPDH, a homology model was generated using SWISS-MODEL3, a protein modeling server (Arnold et al., 2006), and the RasMol molecular graphics visualization tool4. Analysis of the derived model of the SFD1-WT protein indicated that the overall structure of SFD1 is similar to that of LmGPDH (Figure 2) and includes 17 helices, 12 strands, and 29 turns. Based on this model (Figure 2), SFD1 amino acids Lys194 and Lys279 are located in the same position as amino acids Lys125 and Lys210 in LmGPDH. This model also predicts that Asp332 in SFD1, which corresponds to Asp263 in LmGPDH, is adjacent to Lys194 and Lys279 (Figure 2) and thus is likely involved in SFD1’s DHAP reductase activity.
Figure 2.
Proposed structure of wild type and mutated SFD1 proteins based on the structure of a Leishmania mexicana G3P dehydrogenase. A ribbon model of the Leishmania mexicana 1EVYA GPDH enzyme based on its crystal structure is shown. The structure of the wild type SFD1 protein (SFD1-WT) and the SFD1–Ala194, SFD1–Ala279, and SFD1–Ala332 proteins in which residues Lys194, Lys279, and Asp332 are replaced by Ala (denoted as beads), were modeled based on the structure of 1EVYA with the SWISS-MODEL and RasMol 2.7.4.2 www.rasmol.org softwares.
Biochemical characterization of wild type and mutant SFD1 proteins
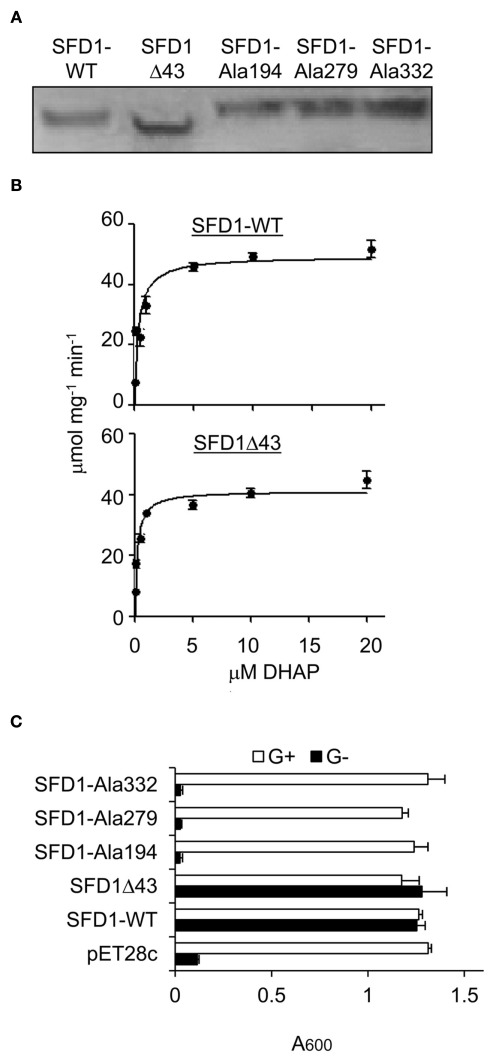
To determine if Lys194, Lys279, and Asp332 are critical for SFD1’s enzymatic activity, recombinant wild type, and mutant SFD1 proteins in which these amino acids were replaced by Ala were produced in Escherichia coli. The recombinant proteins had a His-tag at the C-terminal end, which facilitated their purification over a Ni-column. As shown in Figure 3A, the WT and mutant SFD1 proteins exhibited the expected molecular mass of ∼45 kDa. SFD1-WT demonstrated DHAP reductase activity (Figure 3B) with Michaelis–Menten kinetics with an apparent Km of 0.33 μM for DHAP (Table 1). Apparent Vmax for SFD1-WT was 49 μmol mg−1 min−1 (Table 1). Replacement of any one of the three residues, Lys194, Lys279, and Asp332 with Ala in the SFD1–Ala194, SFD1–Ala279, and SFD1–Ala332 recombinant constructs resulted in complete loss of SFD1’s DHAP reductase activity (Table 1).
Figure 3.
DHAP reductase activity of WT and mutant SFD1 protein. (A) Immunodetection of the recombinant SFD1 proteins with anti-His-tag antibody. (B) DHAP reductase activity of the purified recombinant SFD1-WT and SDF1Δ43 proteins. DHAP reductase activity was measured as the rate of decrease in A340 nm due to oxidation of NADH to NAD+. No activity was detected with the recombinant SFD1–Ala194, SFD1–Ala279, and SFD1–Ala332 proteins. (C) Functional complementation of an E. coli G3P auxotrophic mutant by WT and mutant SFD1 proteins. Growth of the E. coli DHAP reductase-deficient strain BB20-14 expressing the WT and mutant SFD1 constructs on minimal liquid M9 medium with or without 0.1% glycerol. The empty pET28c vector-transformed E. coli provided the negative control. Cell growth was determined by measuring the optical density at A600. G+, supplemented with glycerol; G−, without glycerol.
Table 1.
DHAP reductase activity of WT and mutant SFD1 proteins.
| SFD1 construct | Km (μM) | Vmax (μmol mg−1 min−1) |
|---|---|---|
| SFD1-WT | 0.33 ± 0.16 | 49.24 ± 4.58 |
| SFD1Δ43 | 0.20 ± 0.05 | 41.11 ± 1.93 |
| SFD1–Ala194 | nd | nd |
| SFD1–Ala279 | nd | nd |
| SFD1–Ala332 | nd | nd |
nd, No activity detected.
Further confirmation of the importance of Lys194, Lys279, and Asp332 for SFD1’s enzymatic activity was obtained by expressing the Ala-substituted mutant proteins and the SFD1-WT protein in the E. coli gpsA mutant strain BB20-14. This E. coli mutant lacks a GPDH activity resulting in G3P auxotrophy, which can be complemented by glycerol (Cronan and Bell, 1974) and by the expression of SFD1 (Kachroo et al., 2004; Nandi et al., 2004). As shown in Figure 3C, unlike the SFD-WT construct, the SFD1–Ala194, SFD1–Ala279, and SFD1–Ala332 mutant constructs were unable to complement the G3P auxotrophy of the E. coli gpsA mutant strain BB20-14. The Ala substitutions in SFD1–Ala194, SFD1–Ala279, and SFD1–Ala332 are predicted to not have any significant impact on the structure of SFD1 (Figure 2). Taken together the above results confirm that Lys194, Lys279, and Asp332 are critical for SFD1’s DHAP reductase activity.
SFD1’s DHAP reductase activity is critical for its involvement in SAR and lipid metabolism
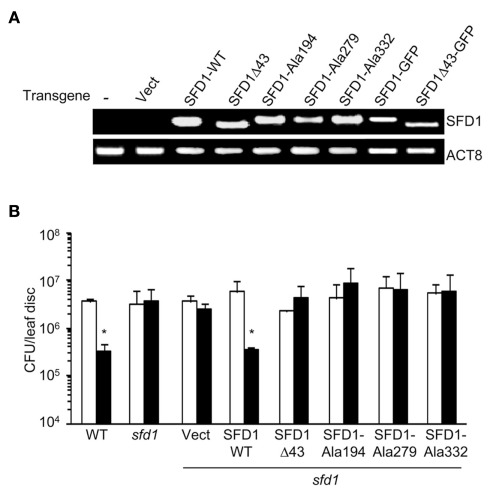
To determine if SFD1’s DHAP reductase activity is required for its involvement in SAR, the SFD1–Ala194, SFD1–Ala279, and SFD1–Ala332 mutant constructs were expressed from the Cauliflower mosaic virus 35S gene promoter in the sfd1-1 mutant background. Plants transformed with the SFD1-WT and empty vector provided the positive and negative controls, respectively, for these experiments. The WT accession Nössen and sfd1-1 mutant plant provided additional controls. Reverse-transcription polymerase chain reactions (RT-PCR) analysis confirmed that compared to the sfd1-1 mutant and sfd1-1 mutant plants transformed with the pMDC32 empty vector (Vect), the SFD1 transcript accumulated at high levels in transgenic plants with the wild type SFD1 (SFD1-WT) and SFD1–Ala194, SFD1–Ala279, and SFD1–Ala332 mutant constructs (Figure 4A). To monitor SAR in these transgenics, a mix of plants derived from three independent transgenic lines for each construct were used. SAR was induced by inoculating (1° inoculation) three fully expanded lower leaves of each plant with a suspension (in 10 mM MgCl2) of an avirulent strain of P. syringae pv. tomato DC3000 (Pto–avrRpt2). Three days later, four upper leaves were challenged (2° inoculation) with P. syringae pv. maculicola (Pma), a virulent pathogen, and bacterial numbers in these Pma-inoculated leaves monitored 3 days post inoculation (3 dpi). Plants that received a 1° inoculation of 10 mM MgCl2 provided the negative control for comparing the extent of SAR-enhanced resistance to Pma. As shown in Figure 4B, in the WT plant Pma numbers were significantly lower in the plants that received a 1° inoculation of Pto–avrRpt2 than in those that were treated with 10 mM MgCl2, indicating that SAR was activated. In contrast, as previously demonstrated (Nandi et al., 2004; Chaturvedi et al., 2008), SAR was not activated in the sfd1-1 mutant. Pma numbers were comparable in the upper leaves of the sfd1-1 mutant plants that were previously treated with 10 mM MgCl2 or Pto–avrRpt2. Like the sfd1-1 mutant and the empty vector (Vect) transformed sfd1-1 mutant plants, SAR was also not activated in transgenic plants transformed with the SFD1–Ala194, SFD1–Ala279, and SFD1–Ala332 mutant constructs, confirming that these amino acids, which are critical for SFD1’s DHAP reductase activity are also required for SFD1’s involvement in SAR.
Figure 4.
Systemic acquired resistance in sfd1-1 transgenic plants expressing the wild type and mutant SFD1. (A) RT-PCR analysis of the SFD1 transgene expression in leaves of the sfd1-1 mutant plants transformed with the empty pMDC32 vector (Vect), the wild type (SFD1-WT) and the SFD1Δ43, SFD1–Ala194, SFD1–Ala279, and SFD1–Ala332 mutant constructs, and the SFD1–GFP and SFD1Δ43–GFP protein fusion expressing constructs. RNA extracted from non-transformed sfd1-1 mutant provided the negative control (−). The attR1-F plus a reverse primer (SFD-RCt) specific for the C-terminus of SFD1 were used for the PCR reaction to monitor expression of the SFD1 transgenes. This primer pair does not detect expression of the genomic SFD1. Expression of the ACTIN gene, ACT8, provided the control for RT-PCR. (B) SAR conferred resistance against P. syringae pv. maculicola in the wild type (WT) Arabidopsis accession Nössen plant, the sfd1-1 mutant, and sfd1-1 mutant plants transformed with the indicated constructs. SAR was induced by prior inoculation of a lower leaf with P. syringae pv. tomato DC3000 carrying the avrRpt2 avirulence gene (black bars). Plants similarly treated with 10 mM MgCl2 (white bars) provided the mock control for SAR. Three days after the primary inoculation the distal leaves were challenged with P. syringae pv. maculicola (Pma). Pma numbers were monitored 3 days post inoculation. Each bar represents the average colony forming units (CFU) of Pma ± SD in three samples each containing five leaf disks. An *indicates significant (P < 0.05) differences in bacterial numbers relative to the corresponding mock control.
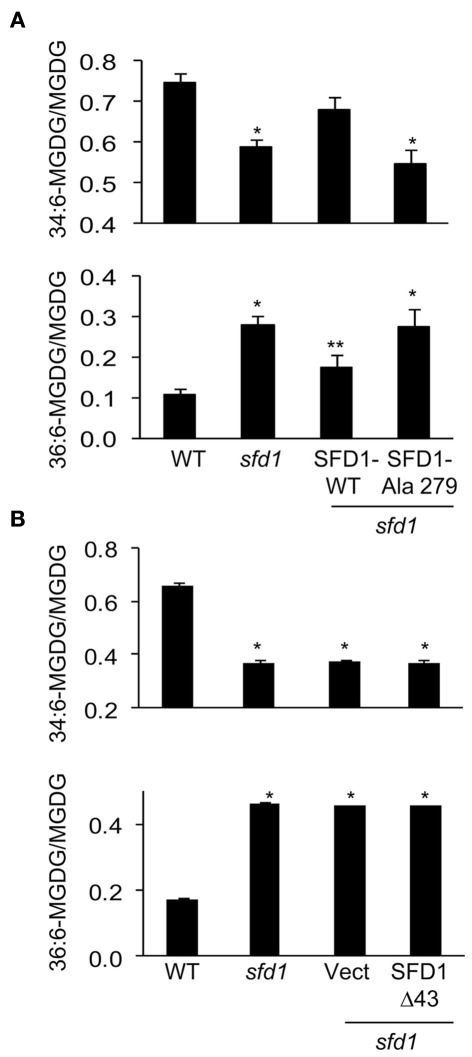
Expression of SFD1-WT also complemented the galactolipid defect of the sfd1-1 allele. As previously demonstrated and shown in Figures 5A,B, the sfd1-1 mutation resulted in a reduction in the content of the plastid synthesized 34:6-MGDG. Furthermore, a corresponding increase in 36:6-MGDG, which is synthesized from DAG derived from the ER pathway, was also observed in the sfd1-1 mutant. To determine if the DHAP reductase activity of SFD1 is essential for its involvement in lipid metabolism, 34:6- and 36:6-MGDG content were compared between the SFD1-WT and SFD1–Ala279 transgenic plants. The WT and the sfd1-1 mutant provided additional controls. As shown in Figure 5A, in comparison to the SFD1-WT plants, 34:6-MGDG content as a fraction of total MGDG was lower in the SFD1–Ala279 plant and 36:6-MGDG content was higher. The 34:6- and 36:6-MGDG contents in the SFD1–Ala279 plant were comparable to those in the sfd1-1 mutant plant, thus confirming that Lys279, and hence SFD1’s DHAP reductase activity is critical for its involvement in Arabidopsis lipid metabolism.
Figure 5.
34:6- and 36:6-MGDG content in sfd1-1 transgenic plants expressing wild type and mutant SFD1. (A) 34:6- and 36:6-MGDG content in the wild type accession Nössen (WT), sfd1-1, and sfd1-1 transformed with the SFD1-WT and SFD1–Ala279 mis-sense mutant. (B) 34:6- and 36:6-MGDG content in the WT, sfd1-1, and sfd1-1 transformed with the empty pMDC32 vector (Vect) and the SFD1Δ43 construct. In (A,B), all values are the average (±SD) of five independent samples given as the fraction of 34:6- and 36:6-MGDG species relative to the total MGDG content. An *denotes significant (P < 0.05) differences relative to the WT. A **denotes significant (P < 0.05) differences relative to all other genotypes.
SFD1 is localized to the chloroplast
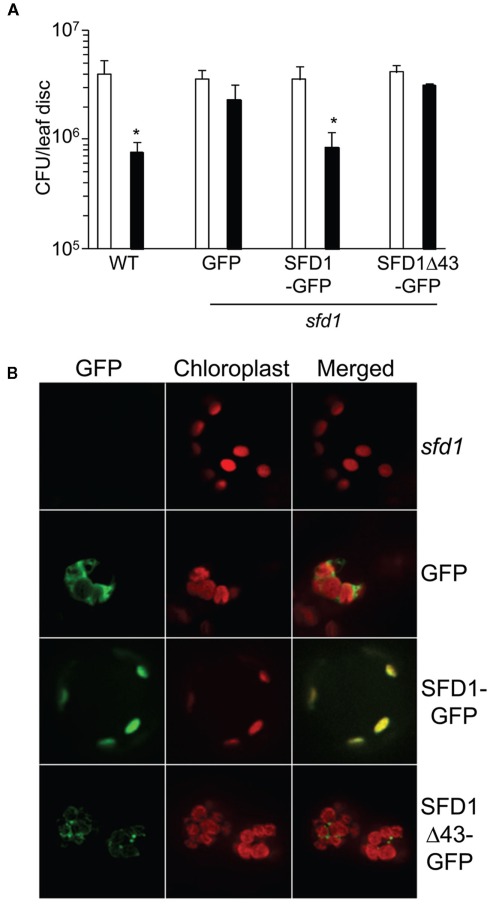
As mentioned above and indicated in Figure 1A, iPSORT predicts that SFD1 has at its N-terminus a chloroplast targeting sequence. Furthermore, proteomics studies have identified SFD1 in the envelope fraction of chloroplasts (Joyard et al., 2010). We tested chloroplast localization of SFD1 in transgenic plants that expressed SFD1 fused to green fluorescent protein (GFP). GFP was fused to the last (carboxyl terminal) amino acid of SFD1. RT-PCR analysis confirmed expression of the transgene in the SFD1–GFP plants (Figure 4A). Expression of the SFD1–GFP fusion also complemented the SAR defect of the sfd1-1 mutant (Figure 6A), confirming that the fusion protein was biologically functional. By comparison, expression of GFP alone was unable to complement the SAR defect of the sfd1-1 mutant. The GFP fluorescence (green) and red autofluorescence of chloroplasts was monitored in protoplasts isolated from the SFD1–GFP transformed sfd1-1 plants, and, as controls in protoplasts isolated from sfd1-1 plants expressing GFP and the non-transformed sfd1-1 plants. As shown in Figure 6B, protoplasts derived from the SFD1–GFP transformed line exhibited a GFP fluorescence pattern that overlapped very well with the red autofluorescence of chloroplasts, confirming that SFD1–GFP fusion is localized to the chloroplast. By contrast, in protoplasts expressing GFP alone, strong green fluorescence was observed outside the chloroplasts. As expected, green fluorescence was not observed in protoplasts derived from the non-transformed sfd1-1 plants.
Figure 6.
The N-terminal 43 amino acids of SFD1 are required for targeting SFD1 to the chloroplast. (A) SAR conferred resistance against P. syringae pv. maculicola (Pma) in the wild type (WT) Arabidopsis accession Nössen plant, and in transgenic sfd1-1 mutant plants that express GFP or the SFD1–GFP and SFD1Δ43–GFP constructs in which the GFP coding sequence is fused in frame to the C-terminus of the SFD1 protein. Refer to legend to Figure 4B on details for the induction of SAR. An *indicates significant (P < 0.05) differences in Pma numbers in plants that were previously treated with an avirulent pathogen (black bars) relative to plants that received a prior mock treatment (white bars). (B) The green fluorescence of GFP and red autofluorescence of chloroplasts were observed in protoplasts isolated from leaves of 4-week-old sfd1-1 mutant, and transgenic sfd1-1 mutant plants expressing GFP, SFD1–GFP, and SFD1Δ43–GFP. Merged images of green fluorescence of GFP and red autofluorescence of chloroplasts are also depicted.
To determine if the N-terminus of SFD1, which contains the predicted chloroplast targeting sequence, is required for targeting SFD1 to the chloroplast, a SFD1Δ43–GFP construct was generated in which the N-terminal 43 amino acids of SFD1 were deleted and GFP was fused to the last (carboxyl terminal) amino acid of SFD1. RT-PCR confirmed that the SFD1Δ43–GFP construct was expressed in the transgenic plants (Figure 4A). Protoplasts derived from transgenic sfd1-1 plants expressing the SFDΔ43–GFP construct were observed for GFP fluorescence pattern. As shown in Figure 6B, the GFP fluorescence pattern of SFD1Δ43–GFP showed a punctate pattern that did not overlap with the red autofluorescence of chloroplasts, thus indicating that the N-terminal 43 amino acids are required for proper targeting of SFD1 to chloroplasts.
N-terminal 43 amino acids are critical for SFD1’s involvement in lipid metabolism and defense signaling
The SFD1Δ43 protein expressed and purified from E. coli (Figure 3A) exhibited DHAP reductase activity with an apparent Km of 0.2 μM for DHAP and Vmax of 41 μmol mg−1 min−1 (Figure 3B; Table 1). In addition, this construct efficiently complemented the G3P auxotrophy of the E. coli gpsA mutant strain BB20-14 (Figure 3C), thus indicating that the N-terminal 43 amino acids of SFD1 are not critical for its DHAP reductase activity. However, expression of SFD1Δ43 in transgenic plants did not complement the MGDG defect of the sfd1-1 mutant (Figure 5B). Furthermore, SFD1Δ43 was unable to restore SAR in the sfd1-1 mutant background (Figure 4B). Similarly, the SFD1Δ43–GFP fusion was unable to complement the SAR defect of sfd1-1 (Figure 6A). Taken together, these results indicate that the N-terminal 43 amino acids, which are required for targeting SFD1 to chloroplasts, are critical for SFD1’s involvement in SAR and lipid metabolism.
Discussion
The Arabidopsis SFD1 gene was previously demonstrated to be required for long-distance signaling associated with the activation of SAR and the accumulation of 34:6-MGDG, a major glycerolipid that is synthesized in the plastid (Nandi et al., 2004; Chaturvedi et al., 2008). Evidence provided here demonstrates that SFD1’s DHAP reductase activity is essential for its involvement in SAR and the accumulation of 34:6-MGDG. Ala substitution of Lys194, Lys279, and Asp332 in SFD1, which correspond to the critical active site residues in the L. mexicana LmGPDH, resulted in loss of SFD1’s DHAP reductase activity (Table 1), and in its ability to complement the SAR and the 34:6-MGDG composition defect of the sfd1-1 mutant (Figures 4B and 5A). Fluorescence microscopy of cells expressing GFP-tagged SFD1 revealed that the SFD1 protein is localized to the chloroplasts (Figure 6B). These results are in agreement with a recent proteomic study, in which SFD1 was recovered with the envelope fraction of chloroplasts (Joyard et al., 2010). The N-terminal 43 amino acids were required for the proper localization of SFD1 to chloroplasts (Figure 6B). The SFD1Δ43 protein, which lacks these 43 amino acids, was unable to complement the SAR and 34:6-MGDG deficiency of the sfd1-1 mutant allele (Figures 4B and 5B). Similarly, the SFD1Δ43–GFP fusion protein was also unable to restore SAR in the sfd1-1 mutant background (Figure 6A). Since the SFD1Δ43 protein possesses DHAP reductase activity, these results suggest that the proper targeting of SFD1 to chloroplasts is critical for its involvement in SAR signaling and lipid metabolism.
The SFD1Δ43 protein retains DHAP reductase activity and is as active as the WT SFD1 protein in vitro (Figure 3B; Table 1). However, since SFD1Δ43 does not exhibit chloroplast localization, any G3P synthesized outside the chloroplast by this protein is not sufficient to compensate for the deficiency of the sfd1-1 mutant, thus suggesting that G3P synthesized within the chloroplast is critical for SAR and galactolipid metabolism. The At5g40610-encoded AtGPDHp is another chloroplastic DHAP reductase from Arabidopsis that has been biochemically characterized (Wei et al., 2001). Many features of AtGPDHp are similar to that of SFD1. Both proteins are of comparable size (40–45 kDa) and have Km’s for DHAP that are in the low micro molar range (AtGPDHp Km = 13 μM; SFD1 Km = 0.3 μM). In addition, AtGPDHp has a pH optimum of 7.5 that is comparable to that of recombinant SFD1 (optimum pH 7.2), and both proteins are localized to the chloroplast. AtGPDHp exhibits 29% (101 of 343 amino acids) identity and 46% (158 of 343 amino acids) similarity with SFD1 (Krothapalli, 2008). However, unlike SFD1, knock-out of AtGPDHp did not have a major impact on the content and composition of galactolipid and other glycerolipids in the atgphdp mutant (Krothapalli, 2008). Furthermore, Krothapalli (2008) found that the atgphdp mutant, unlike the sfd1 mutant, was SAR-competent and the severity of SAR-deficiency was comparable between sfd1 single mutant and the sfd1 atgpdhp double mutant, suggesting that unlike SFD1, the AtGPDHp protein does not have a major role in leaf glycerolipid synthesis and SAR signaling. However, while this manuscript was being prepared, another study showed that SAR was compromised in an atgpdhp mutant (Chanda et al., 2011). Although the reason for the discrepancy between these two independent studies (Krothapalli, 2008; Chanda et al., 2011) is not clear, it is quite possible that the involvement of the atgpdhp gene in SAR is conditioned by other factors and is required under certain conditions, but not in others. However, in concordance with a lack of effect of the AtGPDHp gene in defense, AtGPDHp is not required for Arabidopsis resistance to C. higginsianum as SFD1 is (Chanda et al., 2008). AtGPDHp’s lack of effects or low-level effects on SAR and lipid metabolism may be due to its weak expression in leaf tissue, compared to its expression in floral tissues, siliques, and young seedlings (Wei et al., 2001). It is also plausible that SFD1 and AtGPDHp are expressed in different cell types. Thus, G3P synthesized by these two enzymes has different functions in Arabidopsis. Alternatively, Arabidopsis leaf cells may contain different pools of G3P in the plastid, with the SFD1-derived G3P pool contributing to galactolipid metabolism and SAR and the AtGPDHp-derived G3P pool presumably contributing to other biochemical processes.
In agreement with the results presented here demonstrating that the DHAP reductase activity of SFD1 is required for SAR, Chanda et al. (2011) demonstrated that applying G3P, along with vascular sap from uninfected wild type plants, complements the SAR defect in a gly1 (sfd1) mutant. However, in the absence of vascular sap, G3P was not capable of restoring SAR in the gly1 mutant, suggesting that an additional factor that is present in vascular sap is required for the G3P-induced systemic resistance. Furthermore, G3P, when applied with vascular sap from uninfected plants was unable to induce the systemic accumulation of salicylic acid that is observed in biologically induced SAR. Rather, G3P-induced accumulation of azelaic acid (Chanda et al., 2011), a defense priming metabolite, which like G3P can enhance the strength of SAR (Jung et al., 2009). The sfd1 mutant is also less sensitive to dehydroabietinal, a C20 abietane diterpenoid that was recently identified as a vascular translocated factor, and the most potent activator of SAR (Shah, 2009; R. Chaturvedi and J. Shah, unpublished). Unlike, G3P and azelaic acid, local application of dehydroabietinal leads to a systemic induction in salicylic acid accumulation and enhanced disease resistance. Taken together, these results suggest that SFD1 has an additional function in SAR that promotes the effect of dehydroabietinal in SAR.
In summary, results presented here demonstrate that SFD1’s DHAP reductase activity is required in the chloroplast for its participation in lipid metabolism and defense signaling.
Materials and Methods
Plant and pathogen cultivation
Arabidopsis thaliana plants were grown at 22°C in growth chambers/rooms programmed for 14 h light (100 μE m−2 s−1) and 10 h dark cycle in autoclaved peat-based planting mixture (Premier Pro Mix-PGX, Premier Tech Horticulture5).
Pathogen inoculations
A suspension (107 cfu ml−1 in 10 mM MgCl2) of an avirulent strain of Pseudomonas syringae pv. tomato DC3000 carrying the avrRpt2 avirulence gene (Pto–avrRpt2) was infiltrated (1° inoculation) into three fully expanded leaves to activate SAR, as previously described (Chaturvedi et al., 2008). 10 mM MgCl2-infiltrated plants provided the mock controls. Three days later, four upper leaves were challenged (2° inoculation) with a suspension (105 cfu ml−1) of the virulent pathogen P. syringae pv maculicola ES4326 (Pma). Three days after the 2° inoculation the Pma-inoculated leaves were harvested and bacterial counts in leaf disks (area = 0.28 cm2) determined as previously described (Chaturvedi et al., 2008). A total of 15 leaf disks (three replications of five leaf disks in each sample) were analyzed for each treatment.
Plasmids for in planta expression of recombinant SFD1 protein
DNA containing wild type, truncated and mutant versions of the SFD1 open reading frames were cloned in the pET28c vector (Novagen6) to generate SFD1-WT (full length), SDF1Δ43 (lacks DNA encoding the N-terminal 43 amino acids of SFD1), and SFD1–Ala194, SFD1–Ala297, and SFD1–Ala332 in which Lys at amino acids 194 and 279, and Asp at 332 were replaced by Ala, respectively. The pET28c− SFD1-WT clone was generated by ligating the SFD1 cDNA from the plasmid U10925 (obtained from the Arabidopsis Biological Resource Center, Ohio State University7) into the EcoRI and SalI sites of pET28c. Two sets of PCR reactions were set up in which the wild type SFD1 clone in pET28c (SFD1-WT) was used as a template to generate the SFD1–Ala194, SFD1–Ala297, and SFD1–Ala332 mutant constructs. For construction of SFD1–Ala194 the first set of PCR reactions utilized the forward mutagenic primer SFD1-a1 and the T7-R primer (Table 2) to amplify SFD1 sequences 3′ to the region spanning amino acid 194. The second set of reactions involved the T7-F primer and the reverse mutagenic primer SFD1-a2 to amplify SFD1 sequences 5′ to the region spanning amino acid 194. Both reaction products were subsequently combined, diluted, and used as a template in a PCR reaction with the T7-F and T7-R primers. The resultant PCR product was treated with EcoRI and SalI (New England Biolabs8) and the digested amplicon cloned between the EcoRI and SalI sites of pET28c to yield pET28c− SFD1–Ala194. The SFD1–Ala297 and SFD1–Ala332 constructs were similarly made using the SFD1-b1 and SFD1-b2, and SFD1-c1 and SFD1-c2 mutant primers, respectively. To generate the pET28c− SDF1Δ43 construct, the PCR product derived with the SDF1Δ43-F1 and SDF1Δ43-R1 primers was first ligated into pGEM-T Easy vector (Promega9). The insert was subsequently excised with BamHI and SalI and cloned into the BamHI and SalI linearized pET28a vector. All constructs were sequenced to confirm the presence of the desired mutations and lack of any other unwanted alterations. The sequences of primers used in this study are provided in Table 2.
Table 2.
Primers used in this study.
| Primer name | Sequence 5′ → 3′ |
|---|---|
| SFD1-a1 | CCTTTTATATCTCTTAGCGCGGGTCTGGAGCTTAATACTC |
| SFD1-a2 | GAGTATTAAGCTCCAGACCCGCGCTAAGAGATATAAAAGG |
| SFD1-b1 | GAAATCGCCGGTGCCCTGGCGAATGTTCTAGCAATAG |
| SFD1-b2 | CTATTGCTAGAACATTCGCCAGGGCACCGGCGATTTC |
| SFD1-c1 | GTTTATCAGGAACTGGGGCGATAATGCTTACGTGTTTTG |
| SFD1-c2 | CAAAACACGTAAGCATTATCGCCCCAGTTCCTGATAAAC |
| SDF1ΔNt43-F1 | GGATCCGAATTCATCTCTGGTCCGCCTG |
| SDF1ΔNt43-R1 | AAGCTTGTCGACTCATACTTCTTCAATCTGAGG |
| SDF1ΔNt43-F2start | ATCTCTGGTCCGCCTGATATC |
| SDF1ΔNt43-R2stop | TCATACTTCTTCAATCTGAGGA |
| SDF1ΔNt43-R2Ct | TACTTCTTCAATCTGAGGAAG |
| SFD1-Fstart | ATGGCGGCTTCGGTGCAACCTGC |
| SFD1-Rstop | TCATACTTCTTCAATCTGAGGA |
| SFD1-RCt | TACTTCTTCAATCTGAGGAAG |
| T7-F | TAATACGACTCACTATAGGG |
| T7-R | TAGTTATTGCTCAGCGGTGG |
| Hyg-F | GATGTTGGCGACCTCGTATT |
| Hyg-R | GATGTAGGAGGGCGTGGATA |
| attR1-F | TTGGAGAGGACCTCGACTCT |
To generate clones for expressing wild type and mutant versions of SFD1 in planta, the pET28c− SFD1-WT, pET28c− SFD1–Ala194, pET28c− SFD1–Ala279, and pET28c− SFD1–Ala332 vectors were used as templates in PCR reactions to amplify the SFD1-WT, SFD1–Ala194, SFD1–Ala297, and SFD1–Ala332 DNA with the SFD1-F start and SFD1-R stop primers. To generate a clone for expressing SFD1Δ43 in planta, the pET28c− SFD1Δ43 plasmid was used as a template in PCR with the primers SFD1Δ43-F2 and SFD1Δ43-R2stop. The amplified products were cloned into pCR®8/GW/TOPO® vector (Invitrogen). The inserts in the resultant plasmids were fully sequenced and used for the LR recombination reaction with the destination vector pMDC32 (Invitrogen; Curtis and Grossniklaus, 2003), which contains the Cauliflower mosaic virus 35S gene promoter. To generate constructs expressing SFD1–GFP and SFD1Δ43–GFP in Arabidopsis, the pET28c− SFD1-WT and pET28c− SFD1Δ43 plasmids were used as templates in PCR reactions with the primer pairs SFD1-Fstart plus SFD1-RCt, and SFD1Δ43-F2 plus SFD1Δ43-R2Ct, respectively. The amplified products were cloned into the pCR®8/GW/TOPO® vector. The resultant plasmids were used in the LR recombination reaction with destination vector pMDC83, such that the GFP coding sequence in pMDC83 was fused in frame to the C-terminal amino acid of the SFD1 protein (Curtis and Grossniklaus, 2003). The pMDC32 empty vector and pMDC83 empty vectors were obtained through LR reaction of pCR®8/GW/TOPO® empty vector with pMDC32 and pMDC83, respectively.
Plant transformation
pMDC32 and pMDC83 plasmids containing the desired inserts were electroporated into the Agrobacterium tumefaciens strain GV3101. Presence of the corresponding plasmids in the GV3101 transformants was confirmed by PCR. GV3101 transformants containing pMDC32 or pMDC83 with inserts of interest were used to transform sfd1-1 mutant plants by the floral dip method (Clough and Bent, 1998). Hygromycin resistant seeds were selected on MS agar plates supplemented with hygromycin (20 μg ml−1). Expression of the transgene was confirmed by RT-PCR analysis. Plants from three independently derived transgenic lines for each construct were mixed and used for SAR bioassays and lipid analysis.
RNA extraction and RT-PCR analysis
Leaf tissue was ground in liquid nitrogen and RNA was extracted using an acid guanidinium thiocyanate/phenol/chloroform mix (Chomczynski and Sacchi, 1987). The isolated RNA was purified and used in the RT-PCR. The PCR primers for the ACT8 gene (At1g49240) were as previously described (Pegadaraju et al., 2005). To confirm expression of the hygromycin resistance selection marker (data not shown) and the chimeric SFD1 transgenes, the RT step was followed by PCR with Hyg-F plus Hyg-R, and the attR1-F plus a reverse primer (SFD1-RCt) specific for the C-terminus of SFD1, respectively. Primer sequences can be found in Table 2. The PCR with the Hyg-F plus Hyg-R primers was performed with the following conditions: 95°C for 5 min followed by 25 cycles of 95°C for 30 s, 50°C for 45 s, 72°C for 1 min, with final extension at 72°C for 10 min. The PCR with the attR1-F plus the SFD1-RCt primers was performed with the following conditions: 95°C for 5 min followed by 35 cycles of 95°C for 30 s, 55°C for 45 s, 72°C for 1 min, with final extension at 72°C for 10 min.
Subcellular localization of SFD1–GFP protein
Leaves were cut from the 4-week-old transgenic plants and used for enzymatic protoplasts isolation. Protoplasts were prepared by incubating leaves at room temperature for 2 h in a solution containing 1% cellulase, 0.3% macerozyme, 0.4 M mannitol, 20 mM KCl, and 20 mM MES (pH 6.0). The protoplasts were washed with the protoplast storage buffer (0.5 M mannitol, 20 mM KCl, 4 mM MES pH 6.0) before analysis. GFP fluorescence and chloroplast autofluorescence were monitored using a Zeiss Axiovert 200 M optical microscope connected with a Hamamatsu camera and Yokogawa Confocal Scanner Unit CSU10. Excitation wavelengths/emission filters were 488 nm/band-pass 505–535 nm for GFP and 488 nm/band-pass 672–712 nm for chloroplast autofluorescence. The entire system was controlled with Simple PCI software from Hamamatsu Corporation.
Recombinant protein purification and enzyme assays
A recombinant SFD1-WT, SDF1Δ43, and mis-sense mutant fusion proteins (SFD1–Ala194, SFD1–Ala297, and SFD1–Ala332) containing a 6× His-tag were expressed from the pET28c vector in E. coli and then purified under native conditions over an affinity Ni-NTA column (Qiagen). E. coli strains BL21 containing (WT) and mutant SFD1 plasmids were cultured in LB medium supplemented with 50 μg ml−1 kanamycin at 37°C. Induction of SFD1 and mutant proteins expression was achieved by adding 1 mM IPTG. Cells were harvested and broken down through sonication. Induction of protein and further purification were performed according to suggested protocols (The QIAexpressionistTM, QIAGEN). The protein concentration was determined using Bio-Rad protein assay kit (Bio-Rad), with BSA used as a standard. The BL21 strain harboring the pET28c empty vector was cultured and treated in parallel as a control. To determine the pH optimum for SFD1, recombinant SFD1 protein was incubated at 30°C in a reaction mixture containing 0.16 mM NADH, 1 mM DTT, 10 mM DHAP in 100 mM HEPES buffer of varying pH. The final volume of the reaction mix was 500 μl. The pH optimum for SFD1’s DHAP reductase activity was determined to be 7.2. The assay was based on the DHAP-dependent oxidation of NADH to NAD+ and consisted of two stages. The background of NADH oxidation was first determined in reaction mixtures without DHAP, after which DHAP was added at 5–200 μM and the reductase activity measured. The activity measured was the rate of decrease in A340 due to oxidation of NADH to NAD+. For measuring the kinetic constants the reaction was conducted at pH 7.2. The GraphPad Prism 5 program was used to determine kinetic constants. The WT SFD1 protein retained DHAP reductase activity when kept overnight at room temperature. The enzyme was unstable if left at 4°C or lower temperature. Stability of the SFD1 protein was achieved by storage in 2 M glycerol and addition of 1 mM DTT. To reactivate the enzyme it was heated to 30°C for 10 min.
Complementation of bacterial strain BB20-14
Escherichia coli gpsA mutant strain BB20-14 (Cronan and Bell, 1974) was obtained from the E. coli Genetic Stock Center at Yale University. BB20-14 was maintained in M9 medium (Sambrook et al., 1989) supplemented with 0.4% glucose, 1 mM MgSO4, 100 μM CaCl2, and 1% glycerol (v/v). BB20-14 was transformed with empty pET28c vector and pET28c vectors containing cDNA of SFD1-WT, SDF1Δ43 and different mis-sense SFD1 versions. The transformation mix was plated on M9 agar supplemented with kanamycin (50 μg mL−1), 0.4% glucose, 1 mM MgSO4, 100 μM CaCl2 and 0.1% glycerol (v/v). Kanamycin-resistant colonies were inoculated into liquid M9 medium containing kanamycin (50 μg mL−1), 1 mM IPTG, 1 mM MgSO4, 100 μM CaCl2 with or without 0.1% glycerol. Bacteria were grown at 37°C for 2 days and the absorbance at A600 was determined.
Lipid extraction and ESI-MS/MS analysis
Lipids were extracted using the protocol available at the Kansas Lipidomics Research Center website10 and analyzed by MS as previously described (Xiao et al., 2010). Unfractionated lipid extracts were introduced by continuous infusion into the ESI source on a triple quadrupole MS/MS (API 4000, Applied Biosystems, Foster City, CA, USA) at 30 μl/min. MGDG was detected as [M + NH4]+ in positive ion mode with Neutral Loss of 179.1. The lipids in each class were quantified in comparison to 2.01 nmol 16:0–18:0-MGDG and 0.39 nmol di18:0-MGDG internal standards.
Conflict of Interest Statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Acknowledgments
We thank Lon Turnbull for assistance with fluorescence confocal microscopy. This work was supported by grants from the National Science Foundation (IOS-0827200 and MCB-0920600). Kansas Lipidomics Research Center instrument acquisition and method development was supported by National Science Foundation (EPS-0236913, MCB-0455318, MCB-0920663, DBI-0521587), Kansas Technology Enterprise Corporation, K-IDeA Networks of Biomedical Research Excellence (INBRE) of National Institute of Health (P20RR16475), and Kansas State University.
Footnotes
Erratum
On page 7, last paragraph, right column of this article, it was mistakenly stated “However, in concordance with a lack of effect of the AtGPDHp gene in defense, AtGPDHp is not required for Arabidopsis resistance to C. higginsianum as SFD1 is (Chanda et al., 2008).” The authors would like to rectify that Chanda et al. (2008) showed that mutation in another GPDH encoding gene, At2g41540, did not impact resistance against this fungus. They did not report the effect of AtGPDHp encoded by At5g40610 on Arabidopsis resistance to Colletotrichum higginsianum. In addition, on page 8, the fourth sentence of the first full paragraph, left column, should read “Similarly, G3P was unable to induce accumulation of azelaic acid (Chanda et al., 2011), a defense priming metabolite, which can enhance the strength of SAR (Jung et al., 2009).” and not “Rather, G3P-induced accumulation of azelaic acid (Chanda et al., 2011), a defense priming metabolite, which like G3P can enhance the strength of SAR (Jung et al., 2009)”.
References
- Arnold K., Bordoli L., Kopp J., Schwede T. (2006). The SWISS-MODEL Workspace: a web-based environment for protein structure homology modelling. Bioinformatics 22, 195–201 10.1093/bioinformatics/bti770 [DOI] [PubMed] [Google Scholar]
- Chanda B., Venugopal S. C., Kulshreshtha S., Navarre D. A., Downie B., Vaillancourt L., Kachroo A., Kachroo P. (2008). Glycerol-3-phosphate levels are associated with basal resistance to the hemibiotrophic fungus Colletotrichum higginsianum in Arabidopsis. Plant Physiol. 147, 2017–2029 10.1104/pp.108.121335 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chanda B., Xia Y., Mandal M. K., Yu K., Sekine K.-T., Gao Q.-M., Selote D., Hu Y., Stromberg A., Navarre D. A., Kachroo A., Kachroo P. (2011). Glycerol-3-phosphate is a critical mobile inducer of systemic immunity in plants. Nat. Genet. 43, 421–427 10.1038/ng.798 [DOI] [PubMed] [Google Scholar]
- Chaturvedi R., Krothapalli K., Makandar R., Nandi A., Sparks A. A., Roth M. R., Welti R., Shah J. (2008). Plastid omega3-fatty acid desaturase-dependent accumulation of a systemic acquired resistance inducing activity in petiole exudates of Arabidopsis thaliana is independent of jasmonic acid. Plant J. 54, 106–117 10.1111/j.1365-313X.2007.03400.x [DOI] [PubMed] [Google Scholar]
- Choe J., Guerra D., Michels P. A. M., Hol W. G. J. (2003). Leishmania mexicana glycerol-3-phosphate dehydrogenase showed conformational changes upon binding a bi-substrate adduct. J. Mol. Biol. 329, 335–349 10.1016/S0022-2836(03)00421-2 [DOI] [PubMed] [Google Scholar]
- Chomczynski P., Sacchi N. (1987). Single-step method of RNA isolation by acid guanidinium thiocyanate-phenol-chloroform extraction. Anal. Biochem. 162, 156–159 10.1016/0003-2697(87)90021-2 [DOI] [PubMed] [Google Scholar]
- Clough S. J., Bent A. F. (1998). Floral dip: a simplified method for Agrobacterium-mediated transformation of Arabidopsis thaliana. Plant J. 16, 735–743 10.1046/j.1365-313x.1998.00343.x [DOI] [PubMed] [Google Scholar]
- Cronan J. E., Bell R. M. (1974). Mutants of Escherichia coli defective in membrane phospholipid synthesis: mapping of the structural gene for L-glycerol 3-phosphate dehydrogenase. J. Bacteriol. 118, 598–605 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Curtis M. D., Grossniklaus U. A. (2003). Gateway cloning vector set for high-throughput functional analysis of genes in planta. Plant Physiol. 133, 462–469 10.1104/pp.103.027979 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Durrant W. E., Dong X. (2004). Systemic acquired resistance. Annu. Rev. Phytopathol. 42, 185–209 10.1146/annurev.phyto.42.040803.140421 [DOI] [PubMed] [Google Scholar]
- Gee R. E., Byerrum R. U., Gerber D. W., Tolbert N. E. (1988). Dihydroxyacetone phosphate reductases in plants. Plant Physiol. 86, 98–103 10.1104/pp.86.1.98 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heil M., Ton J. (2008). Long-distance signalling in plant defence. Trends Plant Sci. 13, 264–272 10.1016/j.tplants.2008.03.005 [DOI] [PubMed] [Google Scholar]
- Jeffery C. J. (1999). Moonlighting proteins. Trends Biochem. Sci. 24, 8–11 10.1016/S0968-0004(98)01335-8 [DOI] [PubMed] [Google Scholar]
- Joyard J., Ferro M., Masselon C. H., Seigneurin-Berny D., Salvi D., Garin J., Rolland N. (2010). Chloroplast proteomics highlights the subcellular compartmentation of lipid metabolism. Prog. Lipid Res. 49, 128–158 10.1016/j.plipres.2009.10.003 [DOI] [PubMed] [Google Scholar]
- Jung H. W., Tschaplinski T. J., Wang L., Glazebrook J., Greenberg J. T. (2009). Priming in systemic plant immunity. Science 324, 89–91 10.1126/science.1170025 [DOI] [PubMed] [Google Scholar]
- Kachroo A., Kachroo P. (2009). Fatty acid-derived signals in plant defense. Annu. Rev. Phytopathol. 47, 153–176 10.1146/annurev-phyto-080508-081820 [DOI] [PubMed] [Google Scholar]
- Kachroo A., Srivathsa C. V., Lapchyk L., Falcone D., Hildebrand D., Kachroo P. (2004). Oleic acid levels regulated by glycerolipid metabolism modulate defense gene expression in Arabidopsis. Proc. Natl. Acad. Sci. U.S.A. 101, 5152–5157 10.1073/pnas.0401315101 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kang L., Li J., Zhao T., Xiao F., Tang X., Thilmony R., He S.-Y., Zhou J.-M. (2003). Interplay of the Arabidopsis non-host resistance gene NHO1 with bacterial virulence. Proc. Natl. Acad. Sci. U.S.A. 100, 3519–3524 10.1073/pnas.0637377100 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kirsch T., Gerber D. W., Byerrum R. U., Tolbert N. E. (1992). Plant dihydroxyacetone phosphate reductase. Plant Physiol. 100, 352–359 10.1104/pp.100.1.352 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krothapalli K. (2008). Association of Plastid Lipid Metabolism with the Activation of Systemic Acquired Resistance in Arabidopsis thaliana. Ph.D. thesis, Kansas State University, Manhattan [Google Scholar]
- Métraux J. P., Nawrath C., Genoud T. (2002). Systemic acquired resistance. Euphytica 124, 237–243 10.1023/A:1015690702313 [DOI] [Google Scholar]
- Meyer-Siegler K., Mauro D., Seal G., Wurzer J., DeRiel J., Sirover M. (1991). A human nuclear uracil DNA glycosylase is the 37-kDa subunit of glyceraldehyde-3-phosphate dehydrogenase. Proc. Natl. Acad. Sci. U.S.A. 88, 8460–8464 10.1073/pnas.88.19.8460 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miquel M., Cassagne C., Browse J. (1998). A new class of Arabidopsis mutants with reduced hexadecatrienoic acid fatty acid levels. Plant Physiol. 117, 923–930 10.1104/pp.117.3.923 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moore B. D. (2004). Bifunctional and moonlighting enzymes: lighting the way to regulatory control. Trends Plant Sci. 9, 221–228 10.1016/j.tplants.2004.03.005 [DOI] [PubMed] [Google Scholar]
- Nandi A., Welti R., Shah J. (2004). The Arabidopsis thaliana dihydroxyacetone phosphate reductase gene SUPPRESSOR OF FATTY ACID DESATURASE DEFICIENCY1 is required for glycerolipid metabolism and for the activation of systemic acquired resistance. Plant Cell 16, 465–477 10.1105/tpc.016907 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pegadaraju V., Knepper C., Reese J., Shah J. (2005). Premature leaf senescence modulated by the Arabidopsis thaliana PAD4 gene is associated with defense against the phloem-feeding green peach aphid. Plant Physiol. 139, 1927–1934 10.1104/pp.105.070433 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sambrook J., Fritsch E. F., Maniatis T. (1989). Molecular Cloning: A Laboratory Manual, 2nd Edn Cold Spring Harbor, NY: Cold Spring Harbor Laboratory Press [Google Scholar]
- Shah J. (2009). Plants under attack: systemic signals in defense. Curr. Opin. Plant Biol. 12, 459–464 10.1016/j.pbi.2009.05.011 [DOI] [PubMed] [Google Scholar]
- Shen W., Wei Y., Dauk M., Tan Y., Taylor D. C., Selvaraj G., Zou J. (2006). Involvement of a glycerol-3-phosphate dehydrogenase in modulating the NADH/NAD+ ratio provides evidence of a mitochondrial glycerol-3-phosphate shuttle in Arabidopsis. Plant Cell 18, 422–441 10.1105/tpc.105.039750 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shen W., Wei Y., Dauk M., Zheng Z., Zou J. (2003). Identification of a mitochondrial glycerol-3-phosphate dehydrogenase from Arabidopsis thaliana: evidence for a mitochondrial glycerol-3-phosphate shuttle in plants. FEBS Lett. 536, 92–96 10.1016/S0014-5793(03)00033-4 [DOI] [PubMed] [Google Scholar]
- Shimojima M., Hoffmann-Benning S., Graravito R. M., Benning C. (2005). Ferredoxin-dependent glutamate synthase moonlights in plant sulfolipid biosynthesis forming a complex with SQD1. Arch. Biochem. Biophys. 436, 206–214 10.1016/j.abb.2005.02.005 [DOI] [PubMed] [Google Scholar]
- Suresh S., Turley S., Opperdoes F. R., Michels P. A. M., Hol W. G. J. (2000). A potential target enzyme for trypanocidal drugs revealed by the crystal structure of NAD-dependent glycerol-3-phosphate dehydrogenase from Leishmania mexicana. Structure 8, 541–552 10.1016/S0969-2126(00)00135-0 [DOI] [PubMed] [Google Scholar]
- Vlot A. C., Klessig D. F., Park S. W. (2008). Systemic acquired resistance: the elusive signal(s). Curr. Opin. Plant Biol. 11, 436–442 10.1016/j.pbi.2008.05.003 [DOI] [PubMed] [Google Scholar]
- Wei Y., Periappuram C., Datla R., Selvaraj G., Zou J. (2001). Molecular and biochemical characterizations of a plastidic glycerol-3-phosphate dehydrogenase from Arabidopsis. Plant Physiol. Biochem. 39, 841–848 10.1016/S0981-9428(01)01308-0 [DOI] [Google Scholar]
- Xiao S., Gao W., Chen Q.-F., Chan S.-W., Zheng S.-X., Ma J., Wang M., Welti R., Chen M.-L. (2010). Overexpression of Arabidopsis acyl-CoA binding protein ACBP3 promotes starvation-induced and age-dependent leaf senescence. Plant Cell 22, 1–20 10.1105/tpc.110.075333 [DOI] [PMC free article] [PubMed] [Google Scholar]