Abstract
In land plants, peroxisomes play key roles in various metabolic pathways, including the most prominent examples, that is lipid mobilization and photorespiration. Given the large number of substrates that are exchanged across the peroxisomal membrane, a wide spectrum of metabolite and cofactor transporters is required and needs to be efficiently coordinated. These peroxisomal transport proteins are a prerequisite for metabolic reactions inside plant peroxisomes. The entire peroxisomal “permeome” is closely linked to the adaption of photosynthetic organisms during land plant evolution to fulfill and optimize their new metabolic demands in cells, tissues, and organs. This review assesses for the first time the distribution of these peroxisomal transporters within the algal and plant species underlining their evolutionary relevance. Despite the importance of peroxisomal transporters, the majority of these proteins, however, are still unknown at the molecular level in plants as well as in other eukaryotic organisms. Four transport proteins have been recently identified and functionally characterized in Arabidopsis so far: one transporter for the import of fatty acids and three carrier proteins for the uptake of the cofactors ATP and NAD into plant peroxisomes. The transport of the three substrates across the peroxisomal membrane is essential for the degradation of fatty acids and fatty acids-related compounds via β-oxidation. This metabolic pathway plays multiple functions for growth and development in plants that have been crucial in land plant evolution. In this review, we describe the current state of their physiological roles in Arabidopsis and discuss novel features in their putative transport mechanisms.
Keywords: plant, peroxisomes, transport proteins, metabolites
Introduction
Peroxisomes are eukaryotic organelles, which have been identified in various photosynthetic organisms, from single algal cells to land plants (Igamberdiev and Lea, 2002). Overall our knowledge of how peroxisomes are distributed across the plant kingdom and their versatile roles is limited (Igamberdiev and Lea, 2002; Gabaldon, 2010). Peroxisomes adapted coincidently as land plants derived from the freshwater green algal ancestor that is closely related to the modern Charophytes. Going from water to terrestrial habits led to the development of a complex plant body composed of specialized organs, such as leaves, roots, flowers, and seeds (Langdale, 2008). Plant peroxisomes underwent specialization during land plant evolution and thus contain highly variable and dynamic enzymatic content depending on the specific organ function (Igamberdiev and Lea, 2002).
In vascular plants, peroxisomes are assigned to many processes, including storage oil mobilization via β-oxidation of fatty acids coupled with glyoxylate cycle, photorespiration, membrane lipids turnover, and branched amino acid breakdown during leaf senescence, purine catabolism for nitrogen remobilization, biosynthesis of plant hormones, photomorphogenesis, and pathogen defense (Kaur et al., 2009; Reumann, 2011). Most of these pathways are shared between peroxisomes and other organelles. Peroxisomes are required to scavenge oxidative reactions catalyzed by flavin-containing oxidoreductases (oxidases) that produce highly toxic hydrogen peroxide (del Rio et al., 2006; Nyathi and Baker, 2006). Peroxisomes compartmentalize these lethal steps of metabolism, because they contain efficient ROS-scavenging systems, like catalases, to prevent poisoning of the cell (del Rio et al., 2006; Nyathi and Baker, 2006). Thus, peroxisomes have been described as organelles at the crossroad (Erdmann et al., 1997). Most of our current knowledge is based on detailed studies of fatty acid oxidation and photorespiration, which are both present in algae as well in plants.
During land plant evolution a major innovation was the development of seeds allowing spermatophytes to proliferate and spread to drier areas (Linkies et al., 2010). In seed oil storing dicotyledonous plants (dicots), peroxisomes are involved in storage reserve mobilization to support seedling growth and development until the seedlings become photoautotrophic (Graham, 2008). Upon germination, the fatty acids are released from the seed oil triacylglycerols (TAGs) and degraded via β-oxidation to acetyl-CoA, which is subsequently condensated via glyoxylate cycle to 4-carbon compounds (Figure 1). The resulting dicarbonic acids are further converted in mitochondria to malate, which is then either exported for sucrose synthesis or used as a substrate for respiration (Graham, 2008).
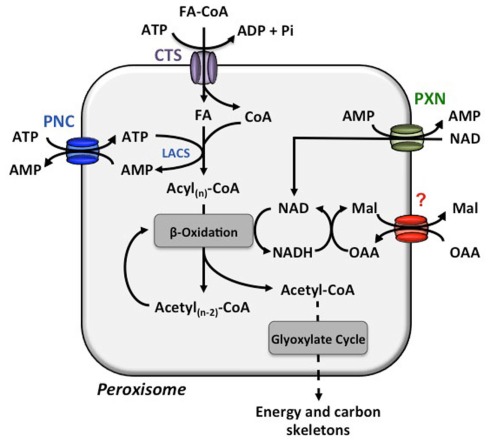
Figure 1.
Transport processes across the peroxisome membrane involved in β-oxidation. The COMATOSE (CTS) transporter facilitates the import of fatty acids from the cytosol to the peroxisomal matrix. The peroxisomal long-chain acyl-coenzyme A synthetase (LACS) activates the free fatty acid (FA) to enter β-oxidation for a stepwise release of acetyl-CoA. For the energy consuming esterification of FAs with coenzyme A (CoA), ATP is required and provided by the import via the PCN proteins. The released AMP is the counter-substrate for the next round of ATP import. The reduction equivalent NAD is reduced during β-oxidation. The PXN transport most likely imports NAD to set-up a sufficient NAD pool in the peroxisomes and the main route for NADH regeneration is enabled via the not yet molecularly identified oxaloacetate (OAA)–malate (Mal) shuttle system.
Many monocotyledonous plants (monocots), such as cereal grasses, store starch in the endosperm of their seeds to fuel seedling establishment. In this case, β-oxidation coupled with the glyoxylate cycle plays another critical role (Igamberdiev and Lea, 2002). Both pathways together metabolize TAGs that are present in considerable amounts in the aleurone layer and in the scutellum, not only to provide energy or carbon skeletons for the synthesis of starch-hydrolyzing enzymes secreted into the endosperm, but also to allow the export of dicarbonic acids, which causes an acidification of the endosperm required for α-amylase activities (Drozdowicz and Jones, 1995).
Beside the breakdown of fatty acids during seedling development, β-oxidation enzymes are also present in other plant organs, such as seeds, leaves, roots, and flowers (Pracharoenwattana and Smith, 2008). Fatty acid oxidation and the glyoxylate cycle contribute to multiple processes, including seed development, seed dormancy, flower fertility, turnover of membrane lipids, and branched amino acids during senescence, the generation of jasmonic acid in response to wounding, and auxin biosynthesis to drive root hair and cotyledon cell expansion during seedling development (Baker et al., 2006; Poirier et al., 2006; Strader et al., 2010).
Many algae that mainly store starch as their storage reserve are capable of accumulating TAGs, particularly under abiotic stress conditions, such as nutrient deprivation or high-light exposure (Thompson, 1996; Hu et al., 2008). The enzymes involved in the TAG degradation have not been intensively studied at the biochemical and molecular levels in algae (Thompson, 1996). It was demonstrated that the charophyte Mougeotia metabolizes fatty acids via the β-oxidation pathway located in the peroxisomes as it is in embryophytes (Stabenau et al., 1984). It is proposed that storage oil turnover in algae contributes primarily to the assembly of membrane lipids to drive rapid cell division after the cessation of nutrient limitations (Thompson, 1996).
Leaves, which are the major photosynthetic parts of the plant, developed after the conquest of land by leafless plants (Dolan, 2009). When land plants appeared, atmospheric oxygen had risen and carbon dioxide (CO2) fallen substantially. In such a high-oxygen containing environment the CO2-fixing enzyme Ribulose-1,5-bisphosphate (RuBP) carboxylase/oxygenase (Rubisco) favors the oxygenation of RuBP, producing the toxic compound phosphoglycolate (2PG; Bauwe et al., 2010). Plants scavenge 2PG to the Calvin cycle intermediate 3-phoshoglycerate (3PGA) by a sequence of reactions called the photorespiratory C2 cycle. Thus a high photorespiration rate is a necessary precondition for plants to invade terrestrial habitats (Bauwe et al., 2010).
The metabolic steps are distributed between chloroplasts, peroxisomes, and mitochondria (Bauwe et al., 2010). Leaf-type peroxisomes contain many key enzymes of the photorespiratory pathway, most notably glycolate oxidase and hydroxypyruvate reductase (Reumann and Weber, 2006). A defect in one of these peroxisomal enzymes causes lethality under low CO2 conditions (Reumann and Weber, 2006). A complete C2 photorespiratory cycle is also found in C4 species, although they recently innovated a sophisticated CO2 concentration mechanism (CCM; Bauwe et al., 2010). It seems that despite the existence of an active CCM, photorespiration is still essential for C4 metabolism.
Since the photorespiratory pathway was transferred from cyanobacteria via endosymbiosis, a complete C2 cycle has been demonstrated in certain green algae (Bauwe et al., 2010). In algae, photorespiration proceeds with lower rates. Due to their ability to concentrate CO2 around Rubisco via carbonic anhydrase, the glycolate production is suppressed (Stabenau and Winkler, 2005). Few algae metabolize glycolate via the mitochondrial glycolate (d-lactate) dehydrogenase. This reaction is coupled with electron transport in mitochondria (Paul and Volcani, 1976). In case of high photorespiration rates, however, most algae excrete glycolate into the surrounding medium (Stabenau and Winkler, 2005).
Both pathways, the breakdown of fatty acids and the photorespiratory C2 cycle, necessitate an efficient flux of metabolites and cofactors between peroxisomes and other cellular compartments. These transport processes across the single bilayer are important, because they connect and control the peroxisomal metabolism with that of the other organelles (Theodoulou et al., 2011). A “two-channel” concept describes the permeability of the peroxisome membrane that consists of channel-forming proteins or “porins” along with specific transport proteins (Antonenkov and Hiltunen, 2006; Visser et al., 2007).
Peroxisomal channels allow free diffusion of small compounds with molecular masses less than 300 kDa, as it known for the outer membrane from mitochondria and plastids (Antonenkov and Hiltunen, 2006; Visser et al., 2007). In addition, selective transport proteins are responsible for the exchange of “bulky” substrates, such as cofactors (ATP, NAD) or fatty acids and their derivatives (Antonenkov and Hiltunen, 2006; Visser et al., 2007). Despite the importance of these porins and transporters, our knowledge is limited in plants, as well as in other eukaryotic organisms. The molecular identity of the channel-forming pore and most of the transporters has not been discovered so far (Theodoulou et al., 2011).
The present review describes the current state of knowledge of the peroxisomal transport processes in plants. Considerable progress has recently been made in the identification of peroxisomal transporters in Arabidopsis (Theodoulou et al., 2011). To date, the proteins of two transporter families are known to reside in the peroxisomal membrane: The peroxisomal ATP-binding cassette (ABC) transporter involved in the import of substrates for β-oxidation and three members of the mitochondrial carrier (MC) family required for the influx of the cofactors ATP or NAD, respectively. This review summarizes their biochemical transport properties and their metabolic role for peroxisomes. In the context of land plant evolution, we analyzed the presence of these peroxisomal transporters in algae and other plant species.
The Peroxisomal ABC Transporter
The peroxisomal ABC transporter imports the substrates for β-oxidation, such as fatty acids or related derivatives, into plant peroxisomes (Theodoulou et al., 2006, 2011). Several independent forward genetic screens identified the peroxisomal ABC transporter in Arabidopsis, here referred to as COMATOSE (CTS, At4g39850; Theodoulou et al., 2006, 2011). Pro- and eukaryotic ABC transporters (TC 3.A.1), in general, are composed of two transmembrane domains and two ABC. The energy from the ATP hydrolysis drives the transport of various molecules across the membrane, often against concentration gradients (Higgins, 1992).
The Arabidopsis CTS encoded by a single gene is functional as a full-length ABC protein with two dissimilar halves. Interestingly, the non-plant peroxisomal ABC transporter from human and yeast are half-transporters (Wanders et al., 2007). Among the Arabidopsis ABC superfamily, that consists of more than 100 members, CTS belongs to the subgroup D (Verrier et al., 2008). In contrast, the second protein of this subgroup (At1g54350) represents a half-size transporter, but its function and putative plastidic localization remain to be analyzed (Verrier et al., 2008).
Function of the peroxisomal ABC transporter in plants
Analysis of Arabidopsis cts null mutants demonstrated that CTS is a transporter with relatively broad spectrum of substrates for β-oxidation. It mediates the uptake of several biologically important molecules into peroxisomes, including fatty acids or fatty acid-derived signaling molecules, such as precursors of auxin and jasmonic acid (Baker et al., 2006; Theodoulou et al., 2006). The loss-of-function led to variable phenotypes, reflecting different roles of β-oxidation in a number of developmental processes in Arabidopsis (Baker et al., 2006; Theodoulou et al., 2006).
The cts mutant alleles exhibit a classical sucrose-dependent phenotype, as mutants defective in β-oxidation. The mutant seedlings are arrested in growth and development due to a block in storage oil mobilization (Zolman et al., 2001; Footitt et al., 2002; Hayashi et al., 2002). High levels of fatty acids in these cts seedlings suggest that CTS transports TAG-derived fatty acids for peroxisomal β-oxidation to fuel seedling establishment. This seedling growth phenotype can be rescued in the presence of an exogenous carbon source, such as sucrose (Zolman et al., 2001; Footitt et al., 2002; Hayashi et al., 2002).
Besides mobilization of storage oil during early seedling growth, CTS provides peroxisomal β-oxidation with fatty acids that are hydrolyzed from membrane lipids during lipid turnover (Baker et al., 2006). This process has not only house-keeping function; it plays a crucial role under conditions, when carbon and energy status are low (Kunz et al., 2009; Slocombe et al., 2009). For instance, under extended darkness conditions, β-oxidation respires fatty acids as an energy source. In case of the cts mutant, free fatty acids, most likely derived from plastidial lipids, dramatically accumulate, which causes a rapid lethal phenotype compared to wildtype (Kunz et al., 2009; Slocombe et al., 2009).
The root growth of the cts mutants is resistant against the protoauxin indole-3-butyric acid (IBA) or the proherbicide 2,4-dichlorophenoxybutyric acid (2,4-DB; Zolman et al., 2001; Hayashi et al., 2002). Due to the loss of CTS both compounds cannot be taken up into peroxisomes, where they are converted into the active auxin indole-3-acetic acid (IAA) and the herbicide 2,4-dichlorophenoxyacetic acid (2,4-D), respectively, which both severely inhibit primary root elongation and cotyledon cell expansion (Zolman et al., 2001; Hayashi et al., 2002; Strader et al., 2010). Since several other pathways contribute to auxin biosynthesis, β-oxidation-dependent conversion of IBA to IAA is not essential for other auxin-dependent cellular responses. Notably, in the cts mutant the elongation of stamen filaments is inhibited, which could be restored by auxin application. This observation indicates an involvement of peroxisomal β-oxidation to supply auxin (Footitt et al., 2007).
COMATOSE also plays a role in jasmonic acid (JA) biosynthesis, demonstrated by the fact that the levels of both basal and wound-inducible JA are reduced in the cts mutants (Theodoulou et al., 2005). It is assumed that CTS imports 12-oxo-phytodienoic acid (OPDA), an intermediate of the JA biosynthesis, into the peroxisomes, where it is further converted by three rounds of β-oxidation to JA (Theodoulou et al., 2005). Because the cts mutant still contains residual JA levels, it suggests an alternative route for the peroxisomal OPDA uptake (Theodoulou et al., 2005). Possible transport mechanisms could be either the existence of an unidentified transporter or a passive transport by anion trapping (Theodoulou et al., 2005).
Unlike other mutants involved in JA biosynthesis, cts plants are not male-sterile, implying that they have sufficient residual JA to produce fertile pollen (Theodoulou et al., 2005). However, the reduced fertility, observed for cts mutants, is caused by an inability to mobilize reserve lipids in both pollen and female gametophytic tissues (Footitt et al., 2007). Therefore, during fertilization, CTS is required to import fatty acids released from stored oil into peroxisomes, where they are broken down via β-oxidation to support energy for pollen germination and tube growth (Footitt et al., 2007).
Mutations in the CTS locus results in seeds that fail to germinate, even in the presence of sucrose, which led to the name COMATOSE (Russell et al., 2000; Footitt et al., 2002; Pinfield-Wells et al., 2005). Only when the seed coat was nicked could cts seeds germinate. cts embryos have the potential to germinate, but they are unable to rupture the seed coat (Kanai et al., 2010). Transcriptome analysis of the cts seeds revealed the molecular mechanism for this phenomenon. The transcription factor ABSCISIC ACID-INSENSITIVE 5 (ABI5) is up-regulated in the mutant and as a consequence, cts seeds contain high transcript levels of polygalacturonase inhibiting proteins (Kanai et al., 2010). These proteins inhibit the degradation of pectin in the seed coat, and thus prevent seed coat rupture during germination (Kanai et al., 2010).
Since other β-oxidation mutants also showed a seed dormant phenotype, CTS might import a molecule into the peroxisomes, which is processed via β-oxidation to generate an as-yet unidentified signal molecule for ABI5 gene repression (Kanai et al., 2010). Recently, it has been reported that the cts seeds contain elevated levels of 12-oxo-phytodienoic acid (OPDA; Dave et al., 2011). Given the fact that CTS imports OPDA into peroxisomes (Theodoulou et al., 2005), OPDA might be the regulator that triggers, along with ABA, ABI5 protein abundance in cts seeds (Dave et al., 2011).
Transport function of the peroxisomal ABC transporter
Based on the phenotypic mutant analysis, CTS imports various β-oxidation substrates into peroxisomes; but it is still a matter for debate whether the accepted substrates are free fatty acids or activated acyl-CoA esters (Theodoulou et al., 2006, 2011). Supportive evidence for the uptake of free acids via CTS is the presence of several acyl-activating enzymes (AAEs) in peroxisomes from Arabidopsis (Fulda et al., 2004; Koo et al., 2006; Wiszniewski et al., 2009). These proteins differ in their substrate specificity and catalyze the esterification of fatty acids or other related molecules, such as the JA precursor or IBA, with coenzyme A (CoA). Such an ATP-dependent activation is essential prior to entering β-oxidation (Fulda et al., 2004; Koo et al., 2006; Wiszniewski et al., 2009). Accordingly, a mutation in these activating enzymes inhibits peroxisomal β-oxidation, resulting in arrested seedling growth, reduced jasmonic acids levels, or IBA resistance (Fulda et al., 2004; Koo et al., 2006; Wiszniewski et al., 2009). Since the cts mutant displays the same defects, this implies that CTS and activation reaction operate in the same – rather than parallel – pathways. Thus, CTS delivers the unesterified fatty acids to the peroxisomal matrix, where they are subsequently activated.
Alternatively, it cannot be excluded that CTS imports the activated CoA esters, as it is known for the yeast homolog. In yeast, the peroxisomal ABC transporter, which consists of two heterodimers named PXA1 and PXA2, is involved in the transport of long-chain acyl-CoA esters (Hettema et al., 1996). Expression of CTS in the pxa1Δ pxa2Δ double mutant rescued the growth phenotype on oleic acid (Nyathi et al., 2010). This complementation assay indicates that CTS is able to transport acyl-CoA esters across the peroxisomal membrane, which is required to metabolize oleic acids as the sole carbon source via β-oxidation. Another implication for activated fatty acids as potential substrates is that acyl-CoA esters, but not fatty acids, stimulate the basal ATPase activity of CTS (Nyathi et al., 2010). In case CTS transports acyl-CoA esters, the β-oxidation substrates have to be activated in the cytosol. However, the corresponding cytosolic enzymes have not been identified so far.
A third model combines the opposite assumptions: CTS imports CoA esters into peroxisomes, the CoA moiety is cleaved off either by CTS or by peroxisomal thioesterases inside the peroxisome to drive the import, and the peroxisomal acyl-CoA activating enzymes re-esterify the β-oxidation substrates (Fulda et al., 2004). At this point the key experiment will be the in vitro transport studies using reconstituted CTS protein to determine the biochemical identity of its substrates. In summary, CTS is the entry point for the substrates of the peroxisomal β-oxidation, which is a crucial pathway for plant growth and development.
The Peroxisomal MC-Type Carrier
Transport proteins that belong to the large MC family (TC 2.A.29) reside in the peroxisomal membrane (Picault et al., 2004; Haferkamp, 2007; Palmieri et al., 2011). This family is named after its prominent member the mitochondrial ATP/ADP carrier (AAC), which is highly abundant in the inner mitochondrial membrane. AAC was one of the first transporter identified at molecular levels and up to now the best biochemically characterized transporter. Members of this family are present in all eukaryotes. The MC members have common structural features. They arose by tandem intragenic triplication. Each replicate contains two a-helical TMDs and a characteristic signature motif (PFAM PF00153; Picault et al., 2004; Haferkamp, 2007; Palmieri et al., 2011). Recently, the three dimensional structure of AAC has been solved, suggesting that MCs are functional as a monomer (Kunji and Crichton, 2010). Despite their related structure, MCF members cover a wide range of transported substrates, exhibit different transport mechanisms, and are distributed to various subcellular membranes.
The Arabidopsis genome encodes for 58 MCs, while three members are localized into peroxisomes (Picault et al., 2004; Haferkamp, 2007; Palmieri et al., 2011). These peroxisomal MC-type transporters are involved in the import of cofactors, such as ATP and NAD (Arai et al., 2008; Linka et al., 2008; Bernhardt et al., 2012). These striking findings for specific ATP and NAD uptake systems refute early biochemical data that the peroxisomal membrane is impermeable for these cofactors (Antonenkov et al., 2004). These transport proteins allow plant peroxisomes to share a common pool of cofactors with the cytosol.
The Peroxisomal ATP Transporters
The peroxisomal adenine nucleotide carrier (PNC) transports ATP into the matrix of plant peroxisomes to fuel ATP-dependent reactions. Arabidopsis peroxisomes contain two PNC proteins, PNC1 (At3g05290) and PNC2 (At5g27520; Arai et al., 2008; Linka et al., 2008). A candidate gene approach identified the two related Arabidopsis PNC proteins based on sequence similarity to the soybean and yeast homolog, which both are prominent membrane proteins in peroxisomes from Glycine max and Saccharomyces cerevisiae (Arai et al., 2008; Linka et al., 2008).
Functional complementation studies verified the ATP transport function of the PNC proteins using a yeast mutant lacking the endogenous peroxisomal ATP carrier Ant1p (Palmieri et al., 2001). This yeast mutant is unable to use medium-chain fatty acids as sole carbon source. Yeast metabolizes medium-chain fatty acids exclusively via peroxisomal β-oxidation. A prerequisite is the import of medium-chain fatty acids and their activation in the peroxisomal matrix. Thus, a lack of the Ant1p inhibits fatty acid breakdown (Palmieri et al., 2001) and expression of PNC1 or PNC2 rescued the growth phenotype of the yeast mutant, demonstrating that both are able to supply yeast peroxisomes with ATP (Linka et al., 2008).
Further, in vitro ATP uptake experiments using recombinant proteins provide a direct proof that PNCs indeed facilitate ATP import (Arai et al., 2008; Linka et al., 2008). Expression and functional integration of PNC2 into the Escherichia coli membrane revealed ATP and ADP uptake activities into intact bacterial cells, while PNC1 could not be expressed (Arai et al., 2008). Interestingly, the presence of AMP did not inhibit the uptake of external ATP (Arai et al., 2008). On the other hand, reconstitution of the two Arabidopsis carriers expressed in yeast led to high ATP uptake activities into liposomes, but only when lipid vesicles were preloaded with another adenine nucleotide (Linka et al., 2008). This clearly shows that PNC1 and PNC2 function as an antiporter that catalyzes the strict counter-exchange of adenine nucleotides (Linka et al., 2008). In contrast to the E. coli uptake studies, the reconstitution systems strongly suggest that PNC proteins accept all adenine nucleotides, including AMP (Linka et al., 2008).
Function of the peroxisomal ATP transporter in plants
Plant peroxisomes need ATP to drive their energy consuming reactions. For instance, the activation of β-oxidation substrates depends on peroxisomal ATP, before they are introduced into this cycle and in Arabidopsis, PNC1 and PNC2 supply peroxisomes with cytosolic ATP. The action of β-oxidation is severely impaired when both PNC genes are repressed via RNA interference (RNAi; Arai et al., 2008; Linka et al., 2008).
The RNAi lines were compromised in seedling establishment in the absence of sucrose (Arai et al., 2008; Linka et al., 2008). The seed oil-derived fatty acids cannot enter the β-oxidation pathway and dramatically accumulate in the iPNC1/2 lines (Arai et al., 2008; Linka et al., 2008). The apparent seedling phenotype is consistent with that of the double knockout mutant for the two acyl-activating enzymes LACS6 and LACS7 in Arabidopsis (Fulda et al., 2004).
The PNC-mediated ATP import is also crucial for peroxisomal enzymes that activate IBA and 2,4-DB (Arai et al., 2008; Linka et al., 2008). An impaired ATP uptake into peroxisomes results in partial resistance against these compounds, which inhibit root growth when converted by a full functional β-oxidation (Arai et al., 2008; Linka et al., 2008). Whether additional ATP-dependent activation steps for β-oxidation are affected in the PNC1/2i plants, such as JA precursor, is currently under investigation (Linka, personal communication).
In sum, the phenotypic analysis of the iPNC1/2 plants shows that (i) there are no other ATP-generating systems in plant peroxisomes, such as substrate-level phosphorylation, and that (ii) the PNC-mediated transport pathway is the only source for intraperoxisomal ATP. Interestingly, the sucrose-dependent phenotype is only partially rescued by exogenous sucrose, indicating a further function of the peroxisomal ATP pool beyond β-oxidation. Latest peroxisomal proteomic approaches detected kinases and other ATP-consuming enzymes, e.g., chaperones, in Arabidopsis, but their functions remains to be determined (Reumann et al., 2007, 2009). A future aim will be to elucidate whether PNC proteins are required to supply these yet-to-be defined steps with external ATP.
Transport function of the peroxisomal ATP transporter
Consistent with the substrate specificities obtained by in vitro uptake studies and their in planta role for β-oxidation, PNC proteins most likely facilitate exchange of cytosolic ATP for peroxisomal AMP under physiological conditions: PNCs import ATP into peroxisomes, which is then hydrolyzed to AMP by the acyl-CoA synthetases. In turn, PNCs export the resulting AMP back to the cytosol, where it is recycled to ATP (Linka et al., 2008). Although PNC catalyze the ATP/AMP exchange in vivo, the following important physiological questions regarding the transport mechanism are not yet clarified.
-
(i)
How is the nucleotide pool in plant peroxisomes loaded in the first place? In yeast, the peroxisomal ATP transporter Ant1p catalyzes in vitro a unidirectional net import of ATP, in addition to the antiport. Such a uniport mechanism allows a net influx of ATP into yeast peroxisomes early in their genesis (Lasorsa et al., 2004). Both Arabidopsis ATP transporters, however, facilitate under the experimental conditions only the exchange of adenine nucleotides (Linka et al., 2008). So it is tempting to speculate that an unidentified uniporter for adenine nucleotides resides in the membrane of plant peroxisomes. Alternatively, PNCs are able in vivo to mediate unidirectional transport since they rescued the loss of the yeast carrier without any obvious remaining phenotype.
-
(ii)
Is the transport of ATP and AMP mediated by PNCs electrogenic or electroneutral? An exchange of ATP4− versus AMP2− would result in a net transfer of two negative charges. In case of the AAC, the electrogenic nature of ADP/ATP transport is crucial for its physiological role. The membrane potential of the inner mitochondrial membrane generated by the respiration chain favors the export of ATP synthesized in the mitochondrial matrix and the entry of cytosolic ADP (Traba et al., 2010). For peroxisomes, there are no reports describing that an ATPase or electron transport chain energized the membranes. The uptake of ATP into peroxisomes might be driven by a concentration gradient. Especially during postgerminative growth, the LACS proteins directly consume ATP and produce AMP in the peroxisomal matrix and favor AMP export to the cytosol (Stitt et al., 1982). Under these conditions, the ATP import catalyzed by PNCs is favored. This is also supported by the kinetic properties of the PNC proteins. The apparent inhibition constant (Ki) of AMP is three times higher than for ATP, indicating that AMP inhibits ATP import only at high concentrations of AMP in the cytosol (Linka et al., 2008).
If the PNCs catalyze an electroneutral transport, counter-ions are required to compensate the charge difference of the ATP/AMP exchange (Figure 2). PNCs might perform a proton-coupled transport, like it was demonstrated for the yeast carrier ANT1p (Lasorsa et al., 2004). Consequently, PNCs transfer protons into peroxisomes and as a result generate a pH gradient across the peroxisomal membrane. To validate this scenario, the pH of plant peroxisomes has to be determined. For human fibroblasts, Chinese hamster ovary cells, and yeast reports about the peroxisomal pH are controversial describing a basic, acidic or neutral lumen for peroxisomes (Dansen et al., 2000; Jankowski et al., 2001; Lasorsa et al., 2004; van Roermund et al., 2004). Notably, calculated isoelectric points of the peroxisomal proteins found by proteomic approaches suggest that the peroxisomal lumen is basic (Reumann et al., 2004). To avoid acidification due to the proton-compensated ATP/AMP exchange, maintenance of the peroxisomal pH is mandatory to ensure a constant internal environment for optimal enzyme activities. Despite the extensive studies of the peroxisomal proteome, there is no evidence for a proton-pumping ATPase at the peroxisomal membrane in any eukaryotic organisms. Alternatively, the “Donnan equilibrium” might maintain the alkaline pH in peroxisomes. The positively net charge of proteins inside peroxisomes drives the free penetration of hydroxyl ions across the peroxisomal membrane. As a result the pH inside peroxisomes is kept more basic than outside (Antonenkov and Hiltunen, 2006; Rokka et al., 2009).
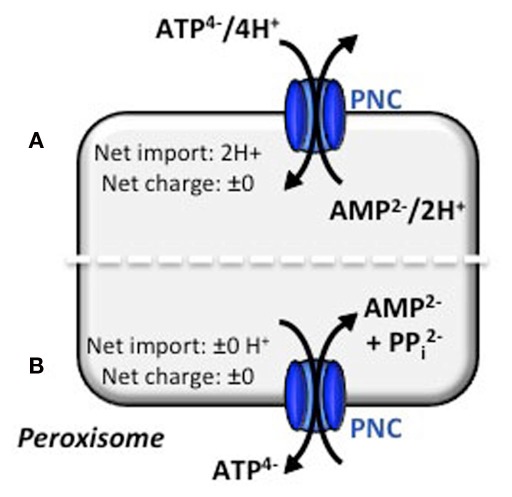
Figure 2.
Electroneutral transport mechanisms for the peroxisomal ATP carrier. Based on the literature concerning similar transport mechanisms, two modes of transport actions for ATP can be hypothesized. (A) Electroneutral ATP/AMP counter-exchange is accomplished by co-transport of protons. The import of 2H+ must be compensated by the Donnan equilibrium. (B) Electroneutral ATP/AMP counter-exchange is maintained by a three substrate-model. AMP and PPi are exchanged for ATP and no proton net import is needed. Both models are under investigation (N. Linka, personal communication).
Conversely, an export of pyrophosphate (PPi) from the peroxisomal matrix could balance the electrogenic ATP/AMP exchange. Beside the electroneutrality, this co-transport also prevents the accumulation of PPi inside peroxisomes, which is produced during the intraperoxisomal ATP-dependent activation of β-oxidation substrates (Fulda et al., 2004). The plastidial ATP transporter, for example, mediates such a phosphate-coupled transport. It imports ATP from the cytosol into plastids in exchange with stromal ADP and inorganic phosphate (Pi) and hence maintains the cellular phosphate homeostasis (Trentmann et al., 2008). For the PNCs, Pi as a third efflux substrate can be excluded, because none of the Arabidopsis encoded pyrophosphatases are targeted to peroxisomes, which could decompose PPi into two Pi molecules. Thus the export of PPi from the peroxisomal matrix is essential. The reconstitution of peroxisomal membranes from mammals revealed phosphate/pyrophosphate transport activities (Visser et al., 2005). The existence of a phosphate carrier would reject the hypothesis that PNCs catalyze an ATP/AMP + PPi antiport. However, the corresponding gene for this postulated transport process has not yet been assigned (Visser et al., 2005).
Concisely stated, additional transport studies are required to understand in more detail how the PNC proteins transport ATP across the peroxisomal membrane, e.g., influence of protonophores on ATP/AMP exchange or stimulation of internal PPi on ATP uptake against AMP.
The Peroxisomal Nad Carrier in Arabidopsis
The transport of NAD into plant peroxisomes is mediated via the peroxisomal NAD carrier PXN (At2g39970; Bernhardt et al., 2012). The existence of a transport protein that imports cytosolic NAD into peroxisomes has been controversially discussed for decades (Rottensteiner and Theodoulou, 2006). The biosynthesis of NAD in the cytosol necessitates the import of NAD into peroxisomes for numerous reduction/oxidation (redox) reactions (Noctor et al., 2006; Hashida et al., 2009).
Proteomic studies detected an abundant membrane protein with an apparent mass of 38 kDa in the purified peroxisomal membrane fraction of etiolated pumpkin cotyledons by Edman degradation (Fukao et al., 2001). The peptide sequence obtained showed a high sequence similarity to the PXN. Since its discovery 10 years ago, it has been assumed that PXN represents the peroxisomal ATP transport due to its close relationship to the mitochondrial AAC (Fukao et al., 2001). This hypothesis was recently disproved, considering that PXN was unable to complement the growth phenotype of the ant1Δ yeast mutant that is impaired in the peroxisomal ATP uptake (Linka et al., 2008). Further phylogenetic analysis exhibited that At2g39970 clusters with recently identified NAD carrier from plastids and mitochondria (Palmieri et al., 2009), proposing a NAD transport function for PXN (Bernhardt et al., 2012).
The in vitro synthesized PXN protein demonstrated the uptake of NAD into liposomes, which were preloaded with NAD (Bernhardt et al., 2012). No significant NAD uptake activities were detectable in the absence of a counter-exchange substrate. This compelling result points out that PXN transports NAD in an antiport mechanism, like the plastidial and mitochondrial NAD transporters (Bernhardt et al., 2012). Further analysis identified NADH, AMP, and ADP as suitable efflux substrates for the NAD import (Bernhardt et al., 2012). The highest NAD uptake rates against AMP indicated that PXN preferentially mediates the NAD/AMP antiport, like the other previously characterized NAD carriers from Arabidopsis and yeast (Todisco et al., 2006; Palmieri et al., 2009). However, PXN is unique in terms of its capability to transport in vitro NAD in its reduced form (Bernhardt et al., 2012).
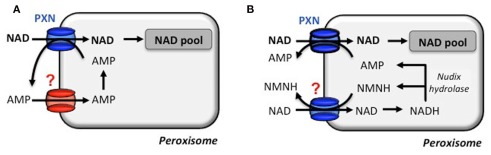
Based on the in vitro transport data, PXN imports NAD against AMP. But how does PXN achieve a net influx of NAD to fuel NAD-dependent reactions? To balance the loss of peroxisomal AMP, two scenarios can be proposed for a net NAD influx (Bernhardt et al., 2012). An unknown peroxisomal carrier might be involved in the unidirectional re-import of cytosolic AMP into peroxisomes. The presence of an adenylate uniporter could refill the peroxisomal adenine nucleotide pool (Figure 3A). In Arabidopsis a peroxisomal Nudix hydrolase with a typical peroxisomal targeting signal might provide the efflux substrates for PXN (Reumann et al., 2004). This enzyme favors in vitro the hydrolysis of NADH to AMP and NMNH (Ge et al., 2007; Ge and Xia, 2008). The net import of one NAD molecule occurs, when PXN imports two NAD molecules against one AMP molecule and one NMNH molecule. Both efflux substrates derived form the hydrolysis of one NADH molecule (Figure 3B). In the cytosol they can be fed in the salvage pathway to regenerate NAD (Noctor et al., 2006; Hashida et al., 2009).
Figure 3.
Transport modes for a net NAD influx via the peroxisomal NAD carrier. The proposed net import of NAD by the transport protein PXN is strictly accompanied by an export of AMP. (A) NAD import is hypothesized to be supported by an AMP uniporter to provide a peroxisomal AMP counter-substrate pool in the first place. (B) Alternatively, an in silico predicted peroxisomal Nudix hydrolase is postulated to degrade NADH to AMP and reduced nicotinamide mononucleotide (NMNH). Both function as a counter-substrate and deliver a net import of NAD. A NMNH/NAD carrier has not been described and NMNH has not been tested for PXN (N. Linka, personal communication).
Function of the peroxisomal NAD carrier in plants
Redox reactions play fundamental roles in plant peroxisomes (Cousins et al., 2008; Pracharoenwattana and Smith, 2008). For instances, the multifunctional protein strictly requires NAD as a cofactor for the oxidative degradation of fatty acids (Graham, 2008; Quettier and Eastmond, 2009). The peroxisomal NAD carrier might be involved in the uptake of NAD to fuel β-oxidation.
A link between the function of PXN and fatty acid breakdown is indicated by the publicly available microarray database Arabidopsis eFP Browser (Winter et al., 2007). PXN is ubiquitously expressed in all tissues, but dry mature seeds store high amounts of PXN mRNA, indicating a possibly involvement in early developmental stages upon germination (Nakabayashi et al., 2005). The steady state transcript level of PXN is also up-regulated in senescent leaves and thus might be required for the process of senescence (van der Graaff et al., 2006).
Consistent with the gene expression on the transcript level are the results of the immunoblot analysis using the related pumpkin protein of PXN (PMP38). Like the enzymes of the β-oxidation, PMP38 is highly abundant in young etiolated pumpkin seedlings (Fukao et al., 2001). Once the transition to photoautotrophic metabolism occurred, the protein amount was markedly decreased. In addition, an accumulation of PMP38 protein was observed during in vivo senescence when green pumpkin cotyledons were kept in extended darkness (Fukao et al., 2001). For both processes, NAD-dependent β-oxidation plays a pivotal role in mobilization of storage or membrane lipids (Baker et al., 2006; Graham, 2008).
Surprisingly, Arabidopsis loss-of-function mutants did not display an apparent sucrose-dependent phenotype, indicating that the storage oil mobilization was not completely blocked during seedling establishment (Bernhardt et al., 2012). A detailed analysis, revealed that the fatty acid breakdown was delayed, because fatty acids accumulated and oil bodies were still present in the pxn seedlings (Bernhardt et al., 2012). Nevertheless, the pxn mutants were able to metabolize a sufficient portion of the seed storage oil that enabled normal seedling growth. Taken together, PXN delivers the peroxisomal β-oxidation with NAD to ensure its optimal operation during storage oil mobilization (Bernhardt et al., 2012).
The intermediate seedling phenotype suggests, for instances, the existence of a second NAD import system in the peroxisomal membrane that builds up the peroxisomal NAD pool together with PXN. Alternatively, plant peroxisomes already contain a “catalytic” amount of NAD for their metabolism, when they derive from the ER (Hoepfner et al., 2005). Since peroxisomes are highly dynamic organelles and they multiply via fission according to their metabolic demand (Kaur and Hu, 2009), each division would dilute the intraperoxisomal NAD content. Hence, PXN might replenish a certain NAD level in peroxisomes instead of setting it up. Thirdly, NAD is taken up together with its NAD-dependent enzymes from the cytosol via the peroxisomal protein importomer. This large pore allows the passage of fully folded and assembled proteins across the membrane (Meinecke et al., 2010). In yeast, it has been shown that the FAD-dependent acyl-CoA oxidase binds its cofactor in the cytosol, where it is synthesized, and is then targeted to the peroxisomes (Titorenko et al., 2002).
In contrast to its proposed function as a net NAD importer, PXN might catalyze in vivo the import of NAD in exchange with NADH. This unique transport feature of PXN would lead in vivo to the transfer reducing equivalents across the peroxisomal membrane (Bernhardt et al., 2012). It is widely accepted that the peroxisomal malate/oxaloacetate shuttle is the exclusive route for the exchange of the oxidized and reduced forms of NAD with the cytosol (Mettler and Beevers, 1980). In the course of β-oxidation NAD is reduced to NADH, which is re-oxidized by the peroxisomal malate dehydrogenase (PMDH) via the reversible reduction of oxaloacetate to malate (Pracharoenwattana et al., 2007, 2010). This enzyme is part of this redox shuttle together with the so far unidentified transporters exchanging these two dicarbonic acids. Since PXN and the peroxisomal redox shuttle are redundant in their function, it would explain the partial inhibition of β-oxidation in the pxn mutants as well as in plants deficient in PMDH activities (Pracharoenwattana et al., 2007, 2010; Bernhardt et al., 2012).
Future analyses are required to elucidate the in vivo role of the peroxisomal NAD carrier in plants. For example, further investigations will address whether other peroxisomal NAD-dependent processes, such as photorespiration, depend on PXN. The pxn plants that were grown under photorespiratory conditions did not exhibit an obvious phenotype (Linka, personal communication); however, a detailed metabolic analysis remains to be done.
Distribution of the Peroxisomal Transporter Proteins within the Plant Kingdom
To date, members of the ABC transporter and MC families represent carrier proteins of the peroxisome membrane in plants. Their function and peroxisomal localization have so far been investigated only in Arabidopsis. We conducted a comprehensive BLASTP analysis (Altschul et al., 1997) using protein database of 22 fully sequenced plant genomes from PLAZA 2.0 (http://bioinformatics.psb.ugent.be/plaza), including 5 algae, 1 moss, and 17 land plants. To identify homologs for CTS, PNCs, and PXN, the reciprocal best hit was chosen if it was in both directions among the three best hits in the other genome (Figure 4). This finding indicates that the evolution of these peroxisomal transporters was an early event before plants evolved. The four plant-specific peroxisomal transporters were already present in the eukaryotic host cells before the cyanobacterial endosymbiont was engulfed. This is consistent with the fact that peroxisomes are eukaryotic organelles and present in animals, plants, and fungi. The peroxisomal transport proteins from plants most likely derived from already existing host membrane proteins and are conserved in green plants.
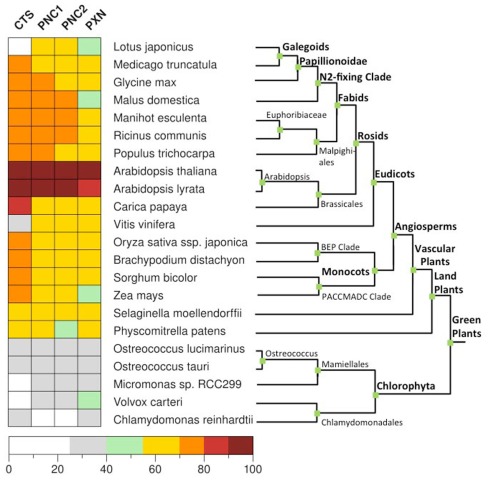
Figure 4.
Distribution of the peroxisomal transporter involved in the uptake of fatty acids, ATP, and NAD in green plants. Homologs genes for CTS (At4g39850), PNC1 (At3g05290), PNC2 (At5g27520), and PXN (At2g39970) were identified in 22 fully sequenced plant genomes from PLAZA 2.0 (http://bioinformatics.psb.ugent.be/plaza) using BLASTP (Altschul et al., 1997). For rice only Oryza sativa ssp. japonica was taken into account. The reciprocal hit was chosen if it was in both directions among the three best hits in the other genome. As a cut-off value we used a normalized sequence identity greater than 25%.
It is not surprisingly that members of the ABC transporter and MC families reside in the peroxisomal membrane. Both families have extensively expanded during evolution. The members are highly abundant in various eukaryotes, display a broad substrate spectrum, and are widely distributed in all types of cellular membranes, such as mitochondria, plastids, peroxisomes. The diversity within a species is driven by duplication of genes that allow for the acquisition of new functions. This contributes to plant complexity and diversification, allowing the plant peroxisomes to adapt during land plant evolution.
Among most of the analyzed land plants, the putative CTS genes encode for a full size ABC protein. This gene structure appears to be conserved in land plants with a few exceptions. In algae the candidates for the peroxisomal ABC transporter represent a half-size transporter, like the human and yeast homologs (Table 1). The presence of CTS-related proteins in algae is contradictory to earlier reports, stating that no peroxisomal ABC transporter genes were found in the algal genomes (Schulze and Kolukisaoglu, 2006).
Table 1.
Plant homologs of the peroxisomal ABC transporter from Arabidopsis.
| Organism | CTS (PLAZA gene ID) | Identity (%) | Length (AA) | Molecular weight (kDa) | Sub-group |
|---|---|---|---|---|---|
| L. japonicus | No hit | – | – | – | – |
| M. trunculata | MT3G36770 | 77 | 1335 | 150.15 | FS |
| G. max | GM11G38160 | 77 | 1287 | 144.79 | FS |
| M. domestica | MD07G005530 | 76 | 1952 | 220.18 | FS |
| M. esculenta | ME05923G00060 | 76 | 796 | 89.49 | HS |
| R. communis | RC30186G00190 | 75 | 1339 | 151.05 | FS |
| P. trichocarpa | PT05G07590 | 78 | 987 | 110.4 | HS |
| A. thaliana | AT4G39850 | 100 | 1337 | 149.6 | FS |
| A. lyrata | AL7G02070 | 98 | 1337 | 149.55 | FS |
| C. papaya | CP00213G00160 | 81 | 465 | 51.76 | HS |
| V. vinifera | VV19G09290 | 26 | 697 | 78.83 | HS |
| O. sativa | OS01G73530 | 72 | 1326 | 147.95 | FS |
| B. distachyon | BD2G61780 | 74 | 1251 | 140.38 | FS |
| S. bicolor | SB03G047010 | 72 | 1324 | 148.03 | FS |
| Z. mays | ZM08G17060 | 72 | 1326 | 148.26 | FS |
| S. moellendorffii | SM00029G00480 | 62 | 1306 | 145.81 | FS |
| P. patens | PP00150G00750 | 65 | 1271 | 141.49 | FS |
| O. lucimarinus | OL02G00720 | 32 | 583 | 64.69 | HS |
| O. tauri | OT02G06480 | 26 | 722 | 79.43 | HS |
| Micromonas. sp. RCC299 | No hit | – | – | – | – |
| V. carteri | No hit | – | – | – | – |
| C. reinhardtii | CR15G00650 | 26 | 753 | 81.34 | HS |
The table summarizes the information of the genes used for the distribution analysis of the CTS gene (Figure 4). The following information is listed: PLAZA gene identity (ID) number of the reciprocal hit in the individual genome, relative identity of the genes to the Arabidopsis homolog, molecular weight, amino acid (AA) length, and molecular structure half or full size (HS or FS). The pairwise normalized identities were calculated by dividing the number of identical positions by the length of the shorter protein. As a cut-off value we used a normalized sequence identity greater than 25%. In case for A. thaliana the best match was the query sequence itself and has been highlighted in bold.
Regarding the peroxisomal MC-type proteins, at least two different candidates have been identified within the green plants that might be putative ATP or NAD transporters (Table 2). One exception is the single-cell chlorophyte Chlamydomonas, the genome of which encodes only for one peroxisomal MCF-type gene that is related to PXN. In contrast, two paralogs for the peroxisomal ATP carrier are found in Arabidopsis, the apple, and the moss Physcomitrella. In case of Arabidopsis, PNC1 and PNC2 are derived from the recent chromosome re-arrangement in the genome (Palmieri et al., 2011).
Table 2.
Plant homologs of the peroxisomal MC-type transporter from Arabidopsis.
| Organism | PNC1 (PLAZA gene ID) | Identity (%) | PNC2 (PLAZA gene ID) | Identity (%) | PXN (PLAZA gene ID) | Identity (%) |
|---|---|---|---|---|---|---|
| L. japonicus | LJ5G027490 | 63 | LJ5G027490 | 63 | LJ0G084980 | 55 |
| M. trunculata | MT5G33690 | 70 | MT5G33690 | 64 | MT5G64320 | 66 |
| G. max | GM02G11800 | 72 | GM02G11800 | 67 | GM18G03400 | 66 |
| M. domestica | MD00G459280 | 71 | MD00G150490 | 72 | MD01G023270 | 51 |
| M. esculenta | ME03905G00190 | 72 | ME03905G00190 | 71 | ME04268G00320 | 70 |
| R. communis | RC30128G01600 | 76 | RC30128G01600 | 71 | RC29669G00050 | 69 |
| P. trichocarpa | PT13G02220 | 76 | PT13G02220 | 70 | PT10G18860 | 69 |
| A. thaliana | AT3G05290 | 100 | AT5G27520 | 100 | AT2G39970 | 100 |
| A. lyrata | AL3G05130 | 98 | AL6G27740 | 99 | AL4G26590 | 90 |
| C. papaya | CP00994G00020 | 69 | CP00994G00020 | 68 | CP00091G00750 | 60 |
| V. vinifera | VV05G04420 | 68 | VV05G04420 | 64 | VV13G03870 | 69 |
| O. sativa | OS05G32630 | 69 | OS05G32630 | 67 | OS03G15860 | 65 |
| B. distachyon | BD2G26480 | 68 | BD2G26480 | 65 | BD1G67180 | 63 |
| S. bicolor | SB09G019430 | 68 | SB09G019430 | 68 | SB01G040130 | 63 |
| Z. mays | ZM08G13190 | 67 | ZM08G13190 | 66 | ZM08G15920 | 47 |
| S. moellendorffii | SM00005G05600 | 57 | SM00005G05600 | 59 | SM00006G03610 | 58 |
| P. patens | PP00086G00350 | 58 | PP00022G01420 | 55 | PP00090G00790 | 57 |
| O. lucimarinus | OL01G02450 | 39 | OL01G02450 | 37 | OL20G02530 | 39 |
| O. tauri | OT01G02440 | 35 | OT01G02440 | 33 | OT14G02730 | 37 |
| Micromonas. sp. RCC299 | MRCC299_16G02440 | 27 | MRCC299_16G02440 | 26 | MRCC299_13G00830 | 38 |
| V. carteri | VC00102G00130 | 27 | VC00102G00130 | 26 | VC00025G00570 | 42 |
| C. reinhardtii | No hit | – | No hit | 0 | CR07G08520 | 30 |
The table summarizes the information of the genes used for the distribution analysis of the MC-type transporters PNC1, PNC2, and PXN (Figure 4). Following information is listed: PLAZA gene identity (ID) number of the reciprocal best hit in the individual genome, relative identity of the genes to the Arabidopsis homolog. The pairwise normalized identities were calculated by dividing the number of identical positions by the length of the shorter protein. As a cut-off value we used a normalized sequence identity greater than 25%. In case for A. thaliana the best matches were the query sequences itself and have been highlighted in bold.
The conducted candidate gene approach was based on protein sequence similarity. None of the predicted candidates have been experimentally investigated up to now. Future efforts are required to confirm the predicted function of the MC- or ABC proteins from algae, moss, and land plants, including measurements of substrate specificity using the recombinant proteins and verification of the peroxisomal localization by GFP fusion.
Conclusion
Future efforts will discover additional peroxisomal transport proteins. Recent proteome studies using purified peroxisomes revealed only the identification of highly abundant membrane proteins that have been already known, such as CTS, PXN, PNC, and a number of biogenesis proteins. This indicates that the other peroxisomal transporters are underrepresented in the peroxisome membrane. A challenge will be to enrich sufficient amounts of peroxisomal membrane proteins for upcoming proteomic approaches. Complementary strategies bear good prospects of elucidating the peroxisomal permeome, including co-expression analyses, mutant studies in combination with metabolic profiling, and genome-wide localization studies of putative transport proteins tagged with GFP.
For decades, several researchers have searched for the postulated transporter that transfers the intermediates of the glyoxylate cycle and photorespiration in plants. This missing link is important to understanding how the flux through the photorespiration is mediated. Recently, it has been hypothesized that the peroxisomal membrane protein with a molecular mass of 22 kDa (PMP22) might be a candidate for peroxisomal porin in plants (Reumann, 2011). To address this, the channel activities have to be determined using electrophysiological techniques in the near future.
Transporters connect the highly compartmentalized metabolic network in plant cells, tissues, and organs and unite the separate parts. A deeper knowledge of the organism-wide permeome will have a major impact on our understanding and designing of metabolic fluxes. This is also true for the subset of transporters in the peroxisomal membrane. Important pathways such as β-oxidation, glyoxylate cycle, photorespiration, branched amino acid breakdown, purine catabolism, biosynthesis of plant hormones, and pathogen defense resides completely or in part in peroxisomes. Fine-tuning the flux via transporters is a promising approach for applied sciences. Peroxisomal research should be intensified in order to show the biotechnological potential and also to close the knowledge gap compared to other organelles. There is still a lot to learn about peroxisomal transporters and a comprehensive understanding of the transport mechanism is required in model organisms prior the transfer into crop plants.
Conflict of Interest Statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Acknowledgments
This work was supported by the DFG-grant 1781/1-1 and 1781/1-2 (to Nicole Linka) and DFG-iGRAD plant GRK1525 (to Christian Esser). We thank Andreas P. M. Weber for helpful discussions. We are also thankful that Kristin Bernhardt, Martin Schroers, Sarah K. Vigelius, Jan Wiese, and Thomas J. Wrobel committed to elucidating of the peroxisomal permeome in plants.
References
- Altschul S. F., Madden T. L., Schäffer A. A., Zhang J., Zhang Z., Miller W., Lipman D. J. (1997). Gapped BLAST and PSI-BLAST: a new generation of protein database search programs. Nucleic Acids Res. 25, 3389–3402 10.1093/nar/25.17.3389 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Antonenkov V. D., Hiltunen J. K. (2006). Peroxisomal membrane permeability and solute transfer. Biochim. Biophys. Acta 1763, 1697–1706 10.1016/j.bbamcr.2006.08.044 [DOI] [PubMed] [Google Scholar]
- Antonenkov V. D., Sormunen R. T., Hiltunen J. K. (2004). The rat liver peroxisomal membrane forms a permeability barrier for cofactors but not for small metabolites in vitro. J. Cell Sci. 117, 5633–5642 10.1242/jcs.01485 [DOI] [PubMed] [Google Scholar]
- Arai Y., Hayashi M., Nishimura M. (2008). Proteomic identification and characterization of a novel peroxisomal adenine nucleotide transporter supplying ATP for fatty acid β-oxidation in soybean and Arabidopsis. Plant Cell 20, 3227–3240 10.1105/tpc.108.062877 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baker A., Graham I. A., Holdsworth M., Smith S. M., Theodoulou F. L. (2006). Chewing the fat: β-oxidation in signalling and development. Trends Plant Sci. 11, 124–132 10.1016/j.tplants.2006.01.005 [DOI] [PubMed] [Google Scholar]
- Bauwe H., Hagemann M., Fernie A. R. (2010). Photorespiration: players, partners and origin. Trends Plant Sci. 15, 330–336 10.1016/j.tplants.2010.03.006 [DOI] [PubMed] [Google Scholar]
- Bernhardt K., Wilkinson S., Weber A. P. M., Linka N. (2012). A peroxisomal carrier delivers NAD+ and contributes to optimal fatty acid degradation during storage oil mobilisation. Plant J. 69, 1–13 10.1111/j.1365-313X.2011.04775.x [DOI] [PubMed] [Google Scholar]
- Cousins A. B., Pracharoenwattana I., Zhou W., Smith S. M., Badger M. R. (2008). Peroxisomal malate dehydrogenase is not essential for photorespiration in Arabidopsis but its absence causes an increase in the stoichiometry of photorespiratory CO2 release. Plant Physiol. 148, 786–795 10.1104/pp.108.122622 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dansen T. B., Wirtz K. W., Wanders R. J., Pap E. H. (2000). Peroxisomes in human fibroblasts have a basic pH. Nat. Cell Biol. 2, 51–53 10.1038/71375 [DOI] [PubMed] [Google Scholar]
- Dave A., Hernandez M. L., He Z., Andriotis V. M., Vaistij F. E., Larson T. R., Graham I. A. (2011). 12-Oxo-phytodienoic acid accumulation during seed development represses seed germination in Arabidopsis. Plant Cell 23, 583–599 10.1105/tpc.110.081489 [DOI] [PMC free article] [PubMed] [Google Scholar]
- del Rio L. A., Sandalio L. M., Corpas F. J., Palma J. M., Barroso J. B. (2006). Reactive oxygen species and reactive nitrogen species in peroxisomes. Production, scavenging, and role in cell signaling. Plant Physiol. 141, 330–335 10.1104/pp.106.078204 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dolan L. (2009). Body building on land: morphological evolution of land plants. Curr. Opin. Plant Biol. 12, 4–8 10.1016/j.pbi.2008.12.001 [DOI] [PubMed] [Google Scholar]
- Drozdowicz Y. M., Jones R. L. (1995). Hormonal regulation of organic and phosphoric acid release by barley aleurone layers and scutella. Plant Physiol. 108, 769–776 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Erdmann R., Veenhuis M., Kunau W. H. (1997). Peroxisomes: organelles at the crossroads. Trends Cell Biol. 7, 400–407 10.1016/S0962-8924(97)01126-4 [DOI] [PubMed] [Google Scholar]
- Footitt S., Dietrich D., Fait A., Fernie A. R., Holdsworth M. J., Baker A., Theodoulou F. L. (2007). The COMATOSE ATP-binding cassette transporter is required for full fertility in Arabidopsis. Plant Physiol. 144, 1467–1480 10.1104/pp.107.099903 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Footitt S., Slocombe S. P., Larner V., Kurup S., Wu Y., Larson T., Graham I., Baker A., Holdsworth M. (2002). Control of germination and lipid mobilization by COMATOSE, the Arabidopsis homologue of human ALDP. EMBO J. 21, 2912–2922 10.1093/emboj/cdf300 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fukao Y., Hayashi Y., Mano S., Hayashi M., Nishimura M. (2001). Developmental analysis of a putative ATP/ADP carrier protein localized on glyoxysomal membranes during the peroxisome transition in pumpkin cotyledons. Plant Cell Physiol. 42, 835–841 10.1093/pcp/pce108 [DOI] [PubMed] [Google Scholar]
- Fulda M., Schnurr J., Abbadi A., Heinz E., Browse J. (2004). Peroxisomal acyl-CoA synthetase activity is essential for seedling development in Arabidopsis thaliana. Plant Cell 16, 394–405 10.1105/tpc.019646 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gabaldon T. (2010). Peroxisome diversity and evolution. Philos. Trans. R. Soc. Lond. B Biol. Sci. 365, 765–773 10.1098/rstb.2009.0240 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ge X., Li G. J., Wang S. B., Zhu H., Zhu T., Wang X., Xia Y. (2007). AtNUDT7, a negative regulator of basal immunity in Arabidopsis, modulates two distinct defense response pathways and is involved in maintaining redox homeostasis. Plant Physiol. 145, 204–215 10.1104/pp.107.103374 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ge X., Xia Y. (2008). The role of AtNUDT7, a Nudix hydrolase, in the plant defense response. Plant Signal. Behav. 3, 119–120 10.4161/psb.3.2.5019 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Graham I. A. (2008). Seed storage oil mobilization. Annu. Rev. Plant Biol. 59, 115–142 10.1146/annurev.arplant.59.032607.092938 [DOI] [PubMed] [Google Scholar]
- Haferkamp I. (2007). The diverse members of the mitochondrial carrier family in plants. FEBS Lett. 581, 2375–2379 10.1016/j.febslet.2007.02.020 [DOI] [PubMed] [Google Scholar]
- Hashida S. N., Takahashi H., Uchimiya H. (2009). The role of NAD biosynthesis in plant development and stress responses. Ann. Bot. 103, 819–824 10.1093/aob/mcp019 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hayashi M., Nito K., Takei-Hoshi R., Yagi M., Kondo M., Suenaga A., Yamaya T., Nishimura M. (2002). Ped3p is a peroxisomal ATP-binding cassette transporter that might supply substrates for fatty acid β-oxidation. Plant Cell Physiol. 43, 1–11 10.1093/pcp/pcf023 [DOI] [PubMed] [Google Scholar]
- Hettema E. H., van Roermund C. W., Distel B., van den Berg M., Vilela C., Rodrigues-Pousada C., Wanders R. J., Tabak H. F. (1996). The ABC transporter proteins Pat1 and Pat2 are required for import of long-chain fatty acids into peroxisomes of Saccharomyces cerevisiae. EMBO J. 15, 3813–3822 [PMC free article] [PubMed] [Google Scholar]
- Higgins C. F. (1992). ABC transporters: from microorganisms to man. Annu. Rev. Cell Biol. 8, 67–113 10.1146/annurev.cellbio.8.1.67 [DOI] [PubMed] [Google Scholar]
- Hoepfner D., Schildknegt D., Braakman I., Philippsen P., Tabak H. F. (2005). Contribution of the endoplasmic reticulum to peroxisome formation. Cell 122, 85–95 10.1016/j.cell.2005.04.025 [DOI] [PubMed] [Google Scholar]
- Hu Q., Sommerfeld M., Jarvis E., Ghirardi M., Posewitz M., Seibert M., Darzins A. (2008). Microalgal triacylglycerols as feedstocks for biofuel production: perspectives and advances. Plant J. 54, 621–639 10.1111/j.1365-313X.2008.03492.x [DOI] [PubMed] [Google Scholar]
- Igamberdiev A. U., Lea P. J. (2002). The role of peroxisomes in the integration of metabolism and evolutionary diversity of photosynthetic organisms. Phytochemistry 60, 651–674 10.1016/S0031-9422(02)00179-6 [DOI] [PubMed] [Google Scholar]
- Jankowski A., Kim J. H., Collins R. F., Daneman R., Walton P., Grinstein S. (2001). In situ measurements of the pH of mammalian peroxisomes using the fluorescent protein pHluorin. J. Biol. Chem. 276, 48748–48763 10.1074/jbc.M009527200 [DOI] [PubMed] [Google Scholar]
- Kanai M., Nishimura M., Hayashi M. (2010). A peroxisomal ABC transporter promotes seed germination by inducing pectin degradation under the control of ABI5. Plant J. 62, 936–947 [DOI] [PubMed] [Google Scholar]
- Kaur N., Hu J. (2009). Dynamics of peroxisome abundance: a tale of division and proliferation. Curr. Opin. Plant Biol. 12, 781–788 10.1016/j.pbi.2009.08.001 [DOI] [PubMed] [Google Scholar]
- Kaur N., Reumann S., Hu J. (2009). “Peroxisome biogenesis and function,” in The Arabidopsis Book, eds Somerville C. R., Meyerowitz E. M. (Rockville: American Society of Plant Biologists; ). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koo A. J., Chung H. S., Kobayashi Y., Howe G. A. (2006). Identification of a peroxisomal acyl-activating enzyme involved in the biosynthesis of jasmonic acid in Arabidopsis. J. Biol. Chem. 281, 33511–33520 10.1074/jbc.M511083200 [DOI] [PubMed] [Google Scholar]
- Kunji E. R. S., Crichton P. G. (2010). Mitochondrial carriers function as monomers. Biochim. Biophys. Acta 1797, 817–831 10.1016/j.bbabio.2010.03.023 [DOI] [PubMed] [Google Scholar]
- Kunz H. H., Scharnewski M., Feussner K., Feussner I., Flügge U. I., Fulda M., Gierth M. (2009). The ABC transporter PXA1 and peroxisomal β-oxidation are vital for metabolism in mature leaves of Arabidopsis during extended darkness. Plant Cell 21, 2733–2749 10.1105/tpc.108.064857 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Langdale J. A. (2008). Evolution of developmental mechanisms in plants. Curr. Opin. Genet. Dev. 18, 368–373 10.1016/j.gde.2008.05.003 [DOI] [PubMed] [Google Scholar]
- Lasorsa F. M., Scarcia P., Erdmann R., Palmieri F., Rottensteiner H., Palmieri L. (2004). The yeast peroxisomal adenine nucleotide transporter: characterization of two transport modes and involvement in DeltapH formation across peroxisomal membranes. Biochem. J. 381, 581–585 10.1042/BJ20040856 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Linka N., Theodoulou F. L., Haslam R. P., Linka M., Napier J. A., Neuhaus H. E., Weber A. P. M. (2008). Peroxisomal ATP import is essential for seedling development in Arabidopsis thaliana. Plant Cell 20, 3241–3257 10.1105/tpc.108.062042 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Linkies A., Graeber K., Knight C., Leubner-Metzger G. (2010). The evolution of seeds. New Phytol. 186, 817–831 10.1111/j.1469-8137.2010.03249.x [DOI] [PubMed] [Google Scholar]
- Meinecke M., Cizmowski C., Schliebs W., Kruger V., Beck S., Wagner R., Erdmann R. (2010). The peroxisomal importomer constitutes a large and highly dynamic pore. Nat. Cell Biol. 12, 273–277 [DOI] [PubMed] [Google Scholar]
- Mettler I. J., Beevers H. (1980). Oxidation of NADH in glyoxysomes by a malate-aspartate shuttle. Plant Physiol. 66, 555–560 10.1104/pp.66.4.555 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakabayashi K., Okamoto M., Koshiba T., Kamiya Y., Nambara E. (2005). Genome-wide profiling of stored mRNA in Arabidopsis thaliana seed germination: epigenetic and genetic regulation of transcription in seed. Plant J. 41, 697–709 10.1111/j.1365-313X.2005.02337.x [DOI] [PubMed] [Google Scholar]
- Noctor G., Queval G., Gakiere B. (2006). NAD(P) synthesis and pyridine nucleotide cycling in plants and their potential importance in stress conditions. J. Exp. Bot. 57, 1603–1620 10.1093/jxb/erj202 [DOI] [PubMed] [Google Scholar]
- Nyathi Y., Baker A. (2006). Plant peroxisomes as a source of signalling molecules. Biochim. Biophys. Acta 1763, 1478–1495 10.1016/j.bbamcr.2006.08.031 [DOI] [PubMed] [Google Scholar]
- Nyathi Y., De Marcos Lousa C., van Roermund C. W., Wanders R. J., Johnson B., Baldwin S. A., Theodoulou F. L., Baker A. (2010). The Arabidopsis peroxisomal ABC transporter, comatose, complements the Saccharomyces cerevisiae pxa1 pxa2 delta mutant for metabolism of long chain fatty acids and exhibits fatty acyl-CoA stimulated ATPase activity. J. Biol. Chem. 285, 29892–29902 10.1074/jbc.M110.151225 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Palmieri F., Pierri C. L., De Grassi A., Nunes-Nesi A., Fernie A. R. (2011). Evolution, structure and function of mitochondrial carriers: a review with new insights. Plant J. 66, 161–181 10.1111/j.1365-313X.2011.04516.x [DOI] [PubMed] [Google Scholar]
- Palmieri F., Rieder B., Ventrella A., Blanco E., Do P. T., Nunes-Nesi A., Trauth A. U., Fiermonte G., Tjaden J., Agrimi G., Kirchberger S., Paradies E., Fernie A. R., Neuhaus H. E. (2009). Molecular identification and functional characterization of Arabidopsis thaliana mitochondrial and chloroplastic NAD carrier proteins. J. Biol. Chem. 284, 31249–31259 10.1074/jbc.M109.041830 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Palmieri L., Rottensteiner H., Girzalsky W., Scarcia P., Palmieri F., Erdmann R. (2001). Identification and functional reconstitution of the yeast peroxisomal adenine nucleotide transporter. EMBO J. 20, 5049–5059 10.1093/emboj/20.18.5049 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Paul J. S., Volcani B. E. (1976). A mitochondrial glycolate: cytochrome C reductase in Chlamydomonas reinhardtii. Planta 129, 59–61 10.1007/BF00390914 [DOI] [PubMed] [Google Scholar]
- Picault N., Hodges M., Palmieri L., Palmieri F. (2004). The growing family of mitochondrial carriers in Arabidopsis. Trends Plant Sci. 9, 138–146 10.1016/j.tplants.2004.01.007 [DOI] [PubMed] [Google Scholar]
- Pinfield-Wells H., Rylott E. L., Gilday A. D., Graham S., Job K., Larson T. R., Graham I. A. (2005). Sucrose rescues seedling establishment but not germination of Arabidopsis mutants disrupted in peroxisomal fatty acid catabolism. Plant J. 43, 861–872 10.1111/j.1365-313X.2005.02498.x [DOI] [PubMed] [Google Scholar]
- Poirier Y., Antonenkov V. D., Glumoff T., Hiltunen J. K. (2006). Peroxisomal β-oxidation–a metabolic pathway with multiple functions. Biochim. Biophys. Acta 1763, 1413–1426 10.1016/j.bbamcr.2006.08.034 [DOI] [PubMed] [Google Scholar]
- Pracharoenwattana I., Cornah J. E., Smith S. M. (2007). Arabidopsis peroxisomal malate dehydrogenase functions in β-oxidation but not in the glyoxylate cycle. Plant J. 50, 381–390 10.1111/j.1365-313X.2007.03055.x [DOI] [PubMed] [Google Scholar]
- Pracharoenwattana I., Smith S. M. (2008). When is a peroxisome not a peroxisome? Trends Plant Sci. 13, 522–525 10.1016/j.tplants.2008.07.003 [DOI] [PubMed] [Google Scholar]
- Pracharoenwattana I., Zhou W., Smith S. M. (2010). Fatty acid β-oxidation in germinating Arabidopsis seeds is supported by peroxisomal hydroxypyruvate reductase when malate dehydrogenase is absent. Plant Mol. Biol. 72, 101–109 10.1007/s11103-009-9554-2 [DOI] [PubMed] [Google Scholar]
- Quettier A. L., Eastmond P. J. (2009). Storage oil hydrolysis during early seedling growth. Plant Physiol. Biochem. 47, 485–490 10.1016/j.plaphy.2008.12.005 [DOI] [PubMed] [Google Scholar]
- Reumann S. (2011). Toward a definition of the complete proteome of plant peroxisomes: where experimental proteomics must be complemented by bioinformatics. Proteomics 11, 1764–1779 10.1002/pmic.201000681 [DOI] [PubMed] [Google Scholar]
- Reumann S., Babujee L., Ma C., Wienkoop S., Siemsen T., Antonicelli G. E., Rasche N., Luder F., Weckwerth W., Jahn O. (2007). Proteome analysis of Arabidopsis leaf peroxisomes reveals novel targeting peptides, metabolic pathways, and defense mechanisms. Plant Cell 19, 3170–3193 10.1105/tpc.107.050989 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reumann S., Ma C., Lemke S., Babujee L. (2004). AraPerox. A database of putative Arabidopsis proteins from plant peroxisomes. Plant Physiol. 136, 2587–2608 10.1104/pp.104.043695 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reumann S., Quan S., Aung K., Yang P., Manandhar-Shrestha K., Holbrook D., Linka N., Switzenberg R., Wilkerson C. G., Weber A. P. M., Olsen L. J., Hu J. (2009). In-depth proteome analysis of Arabidopsis leaf peroxisomes combined with in vivo subcellular targeting verification indicates novel metabolic and regulatory functions of peroxisomes. Plant Physiol. 150, 125–143 10.1104/pp.109.137703 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reumann S., Weber A. P. (2006). Plant peroxisomes respire in the light: some gaps of the photorespiratory C2 cycle have become filled–others remain. Biochim. Biophys. Acta 1763, 1496–1510 10.1016/j.bbamcr.2006.09.008 [DOI] [PubMed] [Google Scholar]
- Rokka A., Antonenkov V. D., Soininen R., Immonen H. L., Pirilä P. L., Bergmann U., Sormunen R. T., Weckström M., Benz R., Hiltunen J. K. (2009). Pxmp2 is a channel-forming protein in mammalian peroxisomal membrane. PLoS ONE 4, e5090. 10.1371/journal.pone.0005090 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rottensteiner H., Theodoulou F. L. (2006). The ins and outs of peroxisomes: co-ordination of membrane transport and peroxisomal metabolism. Biochim. Biophys. Acta 1763, 1527–1540 10.1016/j.bbamcr.2006.08.012 [DOI] [PubMed] [Google Scholar]
- Russell L., Larner V., Kurup S., Bougourd S., Holdsworth M. (2000). The Arabidopsis COMATOSE locus regulates germination potential. Development 127, 3759–3767 [DOI] [PubMed] [Google Scholar]
- Schulz B., Kolukisaoglu H. U. (2006). Genomics of plant ABC transporters: the alphabet of photosynthetic life forms or just holes in membranes? FEBS Lett. 580, 1010–1016 10.1016/j.febslet.2006.01.002 [DOI] [PubMed] [Google Scholar]
- Slocombe S. P., Cornah J., Pinfield-Wells H., Soady K., Zhang Q., Gilday A., Dyer J. M., Graham I. A. (2009). Oil accumulation in leaves directed by modification of fatty acid breakdown and lipid synthesis pathways. Plant Biotechnol. J. 7, 694–703 10.1111/j.1467-7652.2009.00435.x [DOI] [PubMed] [Google Scholar]
- Stabenau H., Winkler U. (2005). Glycolate metabolism in green algae. Physiol. Plant. 123, 235–245 10.1111/j.1399-3054.2005.00442.x [DOI] [Google Scholar]
- Stabenau H., Winkler U., Säftel W. (1984). Enzymes of β-oxidation in different types of algal microbodies. Plant Physiol. 75, 531–533 10.1104/pp.75.3.531 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stitt M., Lilley R. M., Heldt H. W. (1982). Adenine nucleotide levels in the cytosol, chloroplasts, and mitochondria of wheat leaf protoplasts. Plant Physiol. 70, 971–977 10.1104/pp.70.4.971 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Strader L. C., Culler A. H., Cohen J. D., Bartel B. (2010). Conversion of endogenous indole-3-butyric acid to indole-3-acetic acid drives cell expansion in Arabidopsis seedlings. Plant Cell 153, 1577–1586 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Theodoulou F. L., Holdsworth M., Baker A. (2006). Peroxisomal ABC transporters. FEBS Lett. 580, 1139–1155 10.1016/j.febslet.2005.12.095 [DOI] [PubMed] [Google Scholar]
- Theodoulou F. L., Job K., Slocombe S. P., Footitt S., Holdsworth M., Baker A., Larson T. R., Graham I. A. (2005). Jasmonic acid levels are reduced in COMATOSE ATP-binding cassette transporter mutants. Implications for transport of jasmonate precursors into peroxisomes. Plant Physiol. 137, 835–840 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Theodoulou F. L., Zhang X., De Marcos Lousa C., Nyathi Y., Baker A. (2011). “Peroxisomal transport systems: roles in signaling and metabolism,” in Transporters and Pumps in Plant Signaling (Signaling and Communications in Plants), eds Geisler M., Venema K. (Berlin: Springer; ), 327–351 [Google Scholar]
- Thompson G. A. (1996). Lipids and membrane function in green algae. Biochim. Biophys. Acta 1302, 17–45 [DOI] [PubMed] [Google Scholar]
- Titorenko V. I., Nicaud J. M., Wang H., Chan H., Rachubinski R. A. (2002). Acyl-CoA oxidase is imported as a heteropentameric, cofactor-containing complex into peroxisomes of Yarrowia lipolytica. J. Cell Biol. 156, 481–494 10.1083/jcb.200111075 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Todisco S., Agrimi G., Castegna A., Palmieri F. (2006). Identification of the mitochondrial NAD transporter in Saccharomyces cerevisiae. J. Biol. Chem. 281, 1524–1531 10.1074/jbc.M510425200 [DOI] [PubMed] [Google Scholar]
- Traba J., Satrustegui J., del Arco A. (2010). Adenine nucleotide transporters in organelles: novel genes and functions. Cell. Mol. Life Sci. 68, 1183–1206 10.1007/s00018-010-0612-3 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Trentmann O., Jung B., Neuhaus H. E., Haferkamp I. (2008). Non-mitochondrial ATP/ADP transporters accept phosphate as third substrate. J. Biol. Chem. 283, 36486–36493 10.1074/jbc.M806903200 [DOI] [PMC free article] [PubMed] [Google Scholar]
- van der Graaff E., Schwacke R., Schneider A., Desimone M., Flügge U. I., Kunze R. (2006). Transcription analysis of Arabidopsis membrane transporters and hormone pathways during developmental and induced leaf senescence. Plant Physiol. 141, 776–792 10.1104/pp.106.079293 [DOI] [PMC free article] [PubMed] [Google Scholar]
- van Roermund C. W., De Jong M., Ijlst L., van Marie J., Dansen T. B., Wanders R. J., Waterham H. R. (2004). The peroxisomal lumen in Saccharomyces cerevisiae is alkaline. J. Cell Sci. 117, 4231–4237 10.1242/jcs.01305 [DOI] [PubMed] [Google Scholar]
- Verrier P. J., Bird D., Burla B., Dassa E., Forestier C., Geisler M., Klein M., Kolukisaoglu U., Lee Y., Martinoia E., Murphy A., Rea P. A., Samuels L., Schulz B., Spalding E. P., Yazaki K., Theodoulou F. L. (2008). Plant ABC proteins–a unified nomenclature and updated inventory. Trends Plant Sci. 13, 151–159 10.1016/j.tplants.2008.02.001 [DOI] [PubMed] [Google Scholar]
- Visser W. F., van Roermund C. W., Ijlst L., Hellingwerf K. J., Wanders R. J., Waterham H. R. (2005). Demonstration and characterization of phosphate transport in mammalian peroxisomes. Biochem. J. 389, 717–722 10.1042/BJ20041846 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Visser W. F., van Roermund C. W., Ijlst L., Waterham H. R., Wanders R. J. (2007). Metabolite transport across the peroxisomal membrane. Biochem. J. 401, 365–375 10.1042/BJ20061352 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wanders R. J., Visser W. F., van Roermund C. W., Kemp S., Waterham H. R. (2007). The peroxisomal ABC transporter family. Pflugers Arch. 453, 719–734 10.1007/s00424-006-0142-x [DOI] [PubMed] [Google Scholar]
- Winter D., Vinegar B., Nahal H., Ammar R., Wilson G. V., Provart N. J. (2007). An “electronic fluorescent pictograph” browser for exploring and analyzing large-scale biological data sets. PLoS ONE 2, e718. 10.1371/journal.pone.0000718 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wiszniewski A. A. G., Zhou W. X., Smith S. M., Bussell J. D. (2009). Identification of two Arabidopsis genes encoding a peroxisomal oxidoreductase-like protein and an acyl-CoA synthetase-like protein that are required for responses to pro-auxins. Plant Mol. Biol. 69, 503–515 10.1007/s11103-008-9431-4 [DOI] [PubMed] [Google Scholar]
- Zolman B. K., Silva I. D., Bartel B. (2001). The Arabidopsis pxa1 mutant is defective in an ATP-binding cassette transporter-like protein required for peroxisomal fatty acid β-oxidation. Plant Physiol. 127, 1266–1278 10.1104/pp.010550 [DOI] [PMC free article] [PubMed] [Google Scholar]