Abstract
Nuclear localized inositol phospholipids and inositol phosphates are important for regulating many essential processes in animal and yeast cells such as DNA replication, recombination, RNA processing, mRNA export and cell cycle progression. An overview of the current literature indicates the presence of a plant nuclear phosphoinositide (PI) pathway. Inositol phospholipids, inositol phosphates, and enzymes of the PI pathway have been identified in plant nuclei and are implicated in DNA replication, chromatin remodeling, stress responses and hormone signaling. In this review, the potential functions of the nuclear PI pathway in plants are discussed within the context of the animal and yeast literature. It is anticipated that future research will help shed light on the functional significance of the nuclear PI pathway in plants.
Keywords: nucleus, phosphoinositide, inositol phosphate, phosphatidylinositol, plant, signaling, chromatin remodeling
Introduction
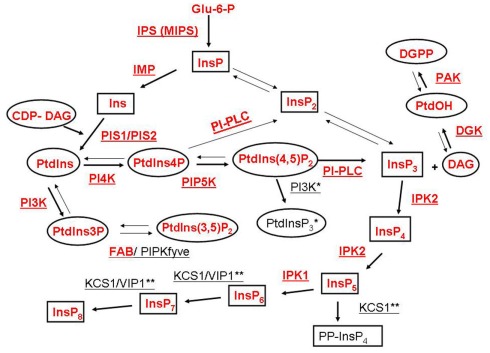
The phosphoinositide (PI) pathway, including the inositol phospholipids and inositol phosphates, is an important regulator of cellular functions in animals and plants (Meijer and Munnik, 2003; Balla et al., 2009). The PI pathway requires the production of myo-inositol-3-phosphate (InsP) from glucose 6-phosphate (Glu-6-P) by myo-inositol phosphate synthase (IPS or MIPS). InsP can be converted into Ins(1,4,5)P3 by two different routes. In a lipid-dependent route, inositol monophosphatase (IMP) produces inositol (Ins) that is converted to phosphatidylinositol (PtdIns) by phosphatidylinositol synthase (PIS). PtdIns is the first lipid of the PI pathway. PtdIns can be phosphorylated to phosphatidylinositol phosphate (PtdIns3P or PtdIns4P and in animals PtdIns5P), which can be further phosphorylated to phosphatidylinositol bisphosphate [PtdIns(3,4)P2, PtdIns(3,5)P2 or PtdIns(4,5)P2], and in animals, to phosphatidylinositol trisphosphate [PtdIns(3,4,5)P3]. Phospholipase C (PLC) hydrolyzes the lipid PtdIns(4,5)P2 to produce the second messengers, inositol trisphosphate (InsP3) and diacylglycerol (DAG). A PtdInsP2 5-phosphatase can decrease PtdInsP2 and increase PtdIns4P. The alternative lipid independent route (also known as the inositol phosphate pathway) can use myo-inositol phosphate (InsP) and inositol phosphate kinases (IPK) to generate inositol bisphosphate (InsP2) and inositol trisphosphate (InsP3). Once formed by either the lipid-dependent or independent pathways, InsP3 can be converted to higher polyphosphorylated inositol phosphates (including InsP4, InsP5, InsP6, InsP7, and InsP8) by IPKs. A schematic of the PI pathway including the enzymes and products is shown in Figure 1. For in depth reviews of the plant PI pathway the reader is directed to: Im et al. (2011), Heilmann (2009), Gillaspy (2011), Valluru and Van den Ende (2011).
Figure 1.
Schematic representation of inositol phospholipids and inositol phosphate metabolism associated with the phosphoinositide (PI) pathway. A generalized PI pathway is shown. Lipid metabolites are represented by ovals, inositol and inositol phosphate derivatives are depicted by boxes and enzymes are underlined. Well characterized pathways are indicated with bold arrows. Enzymes and metabolites that have been found in plants to date are shown in red. Enzyme abbreviations are as follows: IPS, myo-inositol phosphate synthase; IMP, inositol monophosphatase; PIS, phosphatidylinositol synthase; PI3K, PtdIns 3-kinase; PI4K, PtdIns 4-kinase; PIP5K, PtdIns4P 5-kinase; FAB/PIPKfyve, PtdIns3P 5-kinase; PI-PLC, PtdIns/PtdInsP-Phospholipase C; IPK, inositol phosphate kinase; DGK, diacylglycerol kinase; PAK, PtdOH kinase; KCS1, kinase C suppressor 1 (inositol hexakisphosphate (InsP6) kinase); VIP1, InsP6, and inositol heptakisphosphate (InsP7) kinase. Animal Type I PtdIns3K enzyme activity that produces PtdIns(3,4,5)P3 has not been identified in plants and is marked with one (*) asterisk. KCS1 and VIP1 activities have been identified in yeast and animals and are marked with two (**) asterisks.
In animal cells, the nuclear PI pathway is regulated independently from the plasma membrane (PM) PI pathway and plays a critical signaling role in nuclear functions (Irvine, 2003; Bunce et al., 2006; Di Paolo and De Camilli, 2006; Gonzales and Anderson, 2006; York, 2006; Seeds et al., 2007; Barker et al., 2009; Mellman and Anderson, 2009; Barlow et al., 2010; Monserrate and York, 2010; Tsui and York, 2010; Keune et al., 2011; Ramazzotti et al., 2011). It is well established that the nuclear phosphoinositides (PIs) regulate many processes including: nuclear size, nuclear membrane reassembly, chromatin structure, DNA replication, localization of transcription factors, transcription, RNA splicing, mRNA export, and cell cycle progression in animal and yeast cells (as depicted in Figure 2).
Figure 2.
Phosphoinositide pathway functions reported in animal and yeast nuclei. In animals and yeast, phospholipids and inositol phosphates are associated with many critical nuclear processes such as DNA repair, chromatin remodeling and RNA editing and export as described in the text. Nuclear enzymes affected by the PI pathway include: polyadenylate polymerase (PAP), topoisomerase, DNA polymerase and RNA polymerase. Nuclear proteins include: transcription factors (TF), inositol phosphate kinase 2 (IPK2, with transcription factor activity), inositol trisphosphate receptor (InsP3R), histone H1 (H1), histones 2A, 2B, 3 and 4 (Histone Octomer). Nuclear complexes include: DNA repair complex with Non-homologous end joining (NHEJ), RNA editing, mRNA export, Spliceosome, Remodeling complexes, Telomerase complex and the NPC – nuclear pore complex. Data are from both in vivo and in vitro assays. Small circles represent different phospholipids and small squares represent different inositol phosphates as indicated.
Plants and animals both have a complex nuclear structure (reviewed in Cheung and Reddy, 2012) and each structural component of the nucleus is potentially a site for diverse PIs and PI pathway-mediated signaling. The structural components include outer and inner nuclear envelopes (which create a double membrane between the cytoplasm and nucleoplasm generating an intramembrane space around the nucleus, reviewed in Meier and Brkljacic, 2009; Boruc et al., 2012), nuclear pore complexes (the site of active transport between the nucleoplasm and the cytoplasm, reviewed in Boruc et al., 2012), nucleoplasm (similar to cytoplasm), nuclear matrix (structural scaffold for nuclear content, reviewed in Hancock, 2000), chromatin (DNA, protein and metabolites that hold together the tertiary structure of DNA), and nucleoplasmic reticulum (invaginations of the endoplasmic reticulum in the nucleus, reviewed in Malhas et al., 2011). Current studies of the nuclear membrane in plants and animals suggest a similar mechanism for breakdown and reformation during nuclear division (reviewed in Rose et al., 2004; Lloyd and Chan, 2006), suggesting a common origin of the nuclear division. Considering the conservation in nuclear structure, it is not surprising to find that many plant nuclear functions are potential targets of PI pathway regulation, and evidence is mounting for a nuclear PI pathway in plants similar to animal and yeast systems.
In the following sections, we present evidence from the current literature, for the existence of nuclear PIs in plants and discuss potential roles of the plant nuclear PI pathway in cell cycle regulation, DNA synthesis, transcription and RNA processing, stress and hormone responses. However, since our knowledge and understanding of the plant nuclear PI pathway is still limited, we are including relevant examples from the animal and yeast literature in order to highlight the functional significance of these findings and to draw the readers’ attention to promising areas for further investigation.
The PI Pathway Enzymes are Present in Plant Nuclei
Plant PI pathway enzymes have multiple isoforms and their expression, subcellular localization or activities may differ between isoforms in different developmental stages, cell types or environmental conditions (Mueller-Roeber and Pical, 2002; Heilmann, 2009). Nevertheless, many of the enzymes and lipids of the animal nuclear PI pathway have been identified in plant nuclei and these are listed in Table 1.
Table 1.
Phosphoinositide pathway enzymes reported in plant nuclei.
| Enzyme name | Abbreviation | Copy no. | Nuclear isoform | Nuclear activity | Citations |
|---|---|---|---|---|---|
| Myo-inositol synthase | MIPS, IPS | 3 | IPS1 | Nd | Meng et al. (2009) |
| Phosphatidylinositol-3-kinase | PI3K | 1 | PI3K | Yes | Bunney et al. (2000) |
| Phosphatidylinositol-4-kinase | PI4Kα, PI4Kβ | 4 | PI4Kβ1 | Yes | Koroleva et al. (2005), Bunney et al. (2000), Hendrix et al. (1989) |
| Phosphatidylinositol-4-phosphate 5-kinase | PIP5K | 11 | AtPIP5K9 | Yes | Lou et al. (2007), Hendrix et al. (1989) |
| Phosphatidylinositol – phospholipase C | PI-PLC | 9 | Nd | Yes (PtdIns only) | Pfaffmann et al. (1987) |
| Diacylglycerol kinase | DAGK, DGK | 7 | AtDGK1, AtDGK2 | Yes | Vaultier et al. (2008), Wissing and Wagner (1992), Hendrix et al. (1989) |
| Phosphatic acid kinase | PAK | Nd | Nd | Yes | Bunney et al. (2000) |
| Inositol phosphate kinase | IPK, IMPK | 2 | AtIPK2α, AtIPK2β | Nd | Xu et al. (2005), Xia et al. (2003) |
| Inositol (1,3,4) P3 5/6-kinase | ITPK | 4 | AtITPK2, AtITPK1-1 | Nd | Koroleva et al. (2005), Qin et al. (2005) |
| Inositol 5-phosphatase | 5Ptase | 15 | At5Ptase13, At5Ptase7 | Nd | Ananieva et al. (2008), Kaye et al. (2011) |
Many of the plant PI pathway enzymes exhibit dynamic and specific patterns of subcellular localization. The Arabidopsis PIP5K isoforms are the best studied example. In silico prediction places several of the AtPIP5K proteins in the nucleus with 1, 4, 5, 6, 9, and 11 containing possible NLS sequences (WoLF PSORT 2011, Horton et al., 2007). To date only GFP-tagged AtPIP5K9 has been shown to localize to both the nucleus and the PM in transient expression assays in onion cells (Lou et al., 2007), but many of the other AtPIP5K isoforms have been found localized to the PM (Kusano et al., 2008; Stenzel et al., 2008; Camacho et al., 2009; Ischebeck et al., 2011). Transient GFP expression studies can provide results that are difficult to interpret and PIP5Ks turnover rapidly and are difficult to detect with antibodies (Camacho et al., 2009). Biochemical studies have shown PIP5K activity associated with isolated nuclei (Hendrix et al., 1989; Dieck et al., unpublished) suggesting the presence of a nuclear localized PIP5K, therefore it will be important to determine whether the PIP5Ks transiently localize to the nucleus.
Nuclear PI-PLC activity has also been demonstrated in plants using PtdIns as a substrate (Pfaffmann et al., 1987); however the nuclear PI-PLC isoforms have not been identified. Of the nine AtPLC proteins in silico prediction suggests that AtPLC6 (AT2G40116), AtPLC4 (AT5G58700) and AtPLC1 (AT5G58670) may be nuclear targeted (WoLF PSORT 2011, Horton et al., 2007). More genetic, biochemical and in vivo and data are needed to delineate the localization and effects of the nuclear versus PM localized PIP5Ks, PI-PLCs and other PI pathway enzymes on plant growth and development.
The PI Pathway Lipids are Present in Plant Nuclei
The nuclear envelope encloses the nucleus and creates a barrier between the nucleoplasm and the cytoplasm, and is the first point for signaling in the nucleus (reviewed Boruc et al., 2012). The ratios of phospholipids in plant nuclei have been determined, e.g., PtdCho > PtdEtn > PtdIns > PtdSer (Philipp et al., 1976). PtdIns constitutes between 8 to 15% of the total phospholipids in different nuclear fractions (onions roots and stems, Philipp et al., 1976; wheat seedlings, Minasbekyan et al., 2004; tobacco cells grown in suspension culture, Dieck et al., unpublished results). Sub-nuclear localization of the lipids is also important. In wheat embryos 3 days after germination, PtdIns was found in both the nuclear membrane and chromatin, while PtdOH was found in the nuclear membrane, but was not associated with chromatin. While only the dry embryos had PtdOH associated with the chromatin (Minasbekyan et al., 2004), the nuclear membrane and chromatin had similar ratios of most lipids. Additionally, multiple labs have demonstrated the presence of PtdInsP2 in the plant nucleus either directly (König et al., 2008) or through YFP-PH domain constructs that bind PtdInsP2 (Mishkind et al., 2009) and phosphatidylinositol-4-phosphate 5-kinase (PIP5K) activity has been identified in the plant nucleus (Hendrix et al., 1989; Dieck et al., unpublished results) highlighting the localization of PtdInsP2 and at least one plant PIP5K isoform to the nucleus. Importantly, the percentage of polyunsaturated fatty acids (PUFAs) in nuclear-enriched fractions of PtdIns, PtdInsP, and PtdInsP2 are different from those found in the PM (König et al., 2008), suggesting subcellular specificity of lipid fatty acid composition.
The phospholipid composition of plant and animal nuclear membranes is conserved. The amount and location of phospholipids in animal nuclei and their effects on nuclear processes have been well studied (reviewed Irvine, 2003). Animal nuclei and nuclear membranes have a phospholipid profile similar to plant nuclei of PtdCho > PtdEtn > PtdIns > PtdSer (Kleinig, 1970). This general profile is also very similar to lipids in the PM (Kleinig, 1970). PtdIns comprises about 10% of the phospholipids in the nucleus, comparable to the amount in the PM, and many of the PI pathway enzymes and lipids have been identified in the nucleus (reviewed in Irvine, 2003; Bunce et al., 2006; Di Paolo and De Camilli, 2006; Gonzales and Anderson, 2006; York, 2006; Barker et al., 2009; Mellman and Anderson, 2009; Barlow et al., 2010; Monserrate and York, 2010; Tsui and York, 2010; Gillaspy, 2011; Keune et al., 2011; Ramazzotti et al., 2011). Although most lipids in animal and plant nuclei identified so far are similar, it is likely that species-specific inositol lipids and other uncharacterized inositol lipids will be found in the nucleus of plants, animals and yeast. For example, in yeast, a very-long-chain fatty acid PtdIns lipid species with 26 carbons (26:0) is present in both the nuclear membrane and PM. The PtdIns 26:0 lipid species could help stabilize highly curved membrane domains and may be important for the formation of the nuclear pore complex (Schneiter et al., 2004).
Animal nuclei have similar inositol phospholipids to plant nuclei including PtdIns, PtdInsP and PtdInsP2, as well as the animal-specific lipid PtdInsP3 (Garnier-Lhomme et al., 2009). Data from animal cells suggests that the polyphosphorylated inositol phospholipids may be involved in nuclear membrane fusion. Analysis of nuclear envelope remnants from 0.1% non-ionic detergent extraction using mass spectrometry (HPLC-ESI-MS/MS) revealed high levels of phosphorylated inositol phospholipids (12% PtdInsP and PtdInsP2, and 9% PtdInsP3) compared to total nuclear lipids (Garnier-Lhomme et al., 2009). Isolated vesicles associated with nuclear membrane fusion also contain high inositol phospholipid levels as determined by lipid mass spectrometry (20% PtdIns, 20% PtdInsP, 10% PtdInsP2, and 10% PtdInsP3; Byrne et al., 2007). These vesicles have a higher propensity for fusion (Zhendre et al., 2011) as well as enhanced PLCγ association (Byrne et al., 2007). Nuclear membrane assembly normally requires GTP for Ran-mediated activity; however, the addition of PI-PLC can overcome this requirement, and cause nuclear membrane fusion without added GTP. The DAG formed by PI-PLC has been shown to induce nuclear membrane vesicle fusion (Barona et al., 2005). Current studies of the nuclear membrane in plants and animals suggest a similar mechanism for breakdown and reformation during nuclear division (reviewed in Rose et al., 2004; Lloyd and Chan, 2006), which suggests the PI pathway may also be important in plant nuclear membrane fusion/reassembly.
The outer nuclear membrane, which is contiguous with the endoplasmic reticulum, allows for crosstalk between the nucleus and other cellular compartments like the cytosol and ER in plants and animals (reviewed in Raben et al., 1994; Boruc et al., 2012). In animals, sites for synthesis of PtdIns, PtdInsP, and PtdInsP2 are found in the outer nuclear envelope and the flow of PI lipids between the outer and inner nuclear envelope gives evidence that changes in the PI lipids between envelopes are connected (Slomiany and Slomiany, 2011). Nuclear lipids and inositol phosphates can also affect signal transduction via ion channels and membrane bound receptors which regulate the amount of calcium and other ion transport from the intramembrane space across either the outer or inner nuclear membrane (Matzke et al., 2010). It is hypothesized that nuclear calcium signaling can be independent of cytosolic signaling both in animals and plants (Fedorenko et al., 2010; reviewed in Malviya and Rogue, 1998; Pauly et al., 2000). One of the receptors found in animals that can control calcium flux is the inositol trisphosphate receptor (InsP3R). InsP3-mediated nuclear calcium signals have been measured even when cytosolic InsP3 signals are inhibited by heparin, which competes with the cytosolic InsP3-binding site but which can not cross the nuclear pore (Chamero et al., 2008). In animal cells, the InsP3R has been identified in the nucleoplasm (Huh and Yoo, 2003) and on the inner nuclear membrane (Malviya, 1994) providing further evidence for an InsP3 response in the nucleus. The nucleoplasmic reticulum connects the endoplasmic reticulum and outer nuclear envelope to the intranuclear space (reviewed in Bootman et al., 2009). Nuclear calcium signals have been localized to a nucleoplasmic reticulum in animals (Echevarría et al., 2003) and it is possible the InsP3R receptors localize to the nucleoplasmic reticulum. In plants nuclear invaginations similar to the nucleoplasmic reticulum have been observed (Collings et al., 2000), which provides a structural context for localized nuclear signaling in plants. Although a homolog for the animal InsP3R has yet to be identified in plants (Krinke et al., 2007) a number of studies have shown that InsP3 or other inositol phosphates play signaling roles in plants (reviewed in Im et al., 2010; Munnik and Nielsen, 2011), leaving the possibility for a non-canonical InsP3 receptor in plants.
The nuclear matrix is a protein scaffold that supports the nucleus and creates structure for chromatin localization. Animal nuclear matrix PI pathway enzyme activities revealed phosphatidylinositol 4-kinase (PI4K) activity associated with the nuclear periphery and PIP5K, PLC, and diacylglycerol kinase (DAGK) activities associated with the inner nuclear matrix (Payrastre et al., 1992). Intranuclear as well as nuclear matrix associated enzyme activities (as reviewed in Irvine, 2003; Cocco et al., 2009) suggest that PI pathway enzymes in the nucleus are not just nuclear membrane contamination, but could be specifically localized to particular sub-nuclear areas for specific nuclear functions. The presence of PI lipids and enzymes (Table 1) in plant nuclei makes a compelling argument that there is a plant nuclear PI pathway as well (Hendrix et al., 1989; Bunney et al., 2000; Minasbekyan et al., 2004, 2008; König et al., 2008; Mishkind et al., 2009).
The PI Pathway and Cell Cycle Control
Changes in phosphoinositides during the cell cycle have been documented in plants. In coffee plants induction of somatic embryogenesis with hormones causes a transient increase in PI pathway activity followed by decreased activity as measured by [γ32P]ATP labeling of endogenous PtdIns3P and PtdIns4P (Ek-Ramos et al., 2003). Treatment with the PI3/4K inhibitor wortmannin (5 μM) delayed development during somatic embryogenesis, specifically the differentiation from heart to torpedo stages. Treatment was time sensitive, 5 μM wortmannin for 8 days resulted in accumulation of globular and torpedo stages while 38 days of treatment resulted in accumulation of globular and heart stages (Ek-Ramos et al., 2003). This shift suggests that blocking the PI pathway inhibits differentiation; however, because blocking the lipid kinases can have pleiotropic effects on membrane biogenesis and cytoskeletal structure, the results of these treatments are difficult to interpret.
Additional inhibitor studies suggest a requirement for inositol for cell cycle progression in plants. Treatment of rose cells with lithium chloride (an inositol biosynthesis inhibitor) inhibited cell growth, and the addition of inositol overcame this inhibition (Das et al., 1987). Inhibition of myo-inositol production with deoxyglucose also inhibited cell cycle progression which could be overcome with exogenous myo-inositol (Biffen and Hanke, 1990). These data emphasize the importance of inositol for plant cell growth, but do not demonstrate a direct role for the PI pathway. It is essential to recall that inositol is an important plant metabolite essential for cell wall biosynthesis, ascorbic acid biosynthesis and can regulate carbon flux through glycolysis (Loewus and Murthy, 2000). The multiple functions of inositol in plant cells make it particularly difficult to interpret studies that alter de novo inositol synthesis.
A variety of experiments connect the animal nuclear PI pathway with cell cycle progression. Inhibition of PLCβ1 by expression of an antisense PLCβ1 transcript caused inhibition of cell cycle progression induced by the growth factor IGF-I. Conversely, when PLCβ1 was overexpressed increased cell cycle progression was detected (Manzoli et al., 1997). Overexpression of a nuclear localized PLCβ1 increased cell cycle progression even in serum starved cells that would not have progressed through the cell cycle (Faenza et al., 2000). Nuclear localization of overexpressed PLCβ1 was critical for inhibition of differentiation of erythroleukemia cells, while a mutant cytosolic PLCβ1 did not inhibit differentiation (Matteucci et al., 1998). In addition, PI-PLC specific inhibitors led to G2 cell cycle arrest (Sun et al., 1997), while a PC-PLC specific inhibitor did not. Inhibition of nuclear PLC is a current target of cancer therapy (reviewed Cocco et al., 2009).
After synchronization of animal cells, in the G1/S phase radioactive analysis of isolated nuclei showed increased activity of PIP5K, PI4K and DAGK resulting in increased production of [32P] PtdInsP2, [32P] PtdInsP and [32P] PtdOH, respectively (Clarke et al., 2001). Steady state levels of the nuclear phosphoinositides PtdIns4P and PtdIns5P were found to increase while a trend toward decreased PtdInsP2 was observed (Clarke et al., 2001). These results suggest that there is an increase in PtdInsP2 turnover probably caused by the increased nuclear PLCβ1 during cell cycle progression even though PIP5K activity is increased. Localization of the increased PIP5K activity could also be at specific sub-nuclear sites.
Changes in the fatty acid profile of nuclear PtdInsP2 have also been measured during cell cycle progression in animal cells. Decrease of nuclear PtdInsP2 during S phase progression can be specific for certain lipid molecular species (LMS), for example, HeLa cell nuclei showed decreased PtdInsP2 with a fatty acid composition of 34:0 during S phase progression (Ogiso et al., 2010). Surprisingly, nuclei contained a much lower amount of arachidonic acid containing lipids (for example 38:4) compared with whole cell lipids (Ogiso et al., 2010). This suggests that the fatty acid composition of the nuclear PI lipids may be important for their regulation and localization, independent of the cytoplasm and PM.
Cell cycle regulation can culminate in cellular death or apoptosis. In animal cells, Type I PI3K enzymes phosphorylate PtdIns(4,5)P2 to PtdIns(3,4,5)P3 which is a well known regulator of apoptosis. The control of the amount of nuclear PtdInsP3 by specific PI3K and the PtdInsP3 specific phosphatase, phosphatase and tensin homolog deleted on chromosome 10 (PTEN), regulates cellular survival with more PtdInsP3 leading to proliferation and less PtdInsP3 leading to cellular death and cell cycle arrest (Reviewed by Gil et al., 2007). Additional roles for the PTEN and PI3K proteins that do not require their inherent lipid phosphatase or lipid kinase activity have been discovered (Bondeva et al., 1998; Song et al., 2011). Interestingly, Pribat et al. (2012) recently characterized two novel plant PTEN proteins in A. thaliana, AtPTEN2a and AtPTEN2b, which utilize PtdIns3P instead of PtdIns(3,4,5)P3. This reveals a potential plant-specific nuclear signaling pathway since plants have a nuclear PI3K which could produce PtdIns3P (Bunney et al., 2000) as a possible substrate for a PTEN2 protein.
The PI Pathway and DNA Synthesis
In plants, components of the PI pathway may regulate DNA synthesis. In soybean cells, inhibition of PLC activity with the chemical inhibitor U-73122 (5 μM concentration) decreased InsP3 and concomitantly decreased DNA synthesis by >30%. When MS media was added with the PLC inhibitor U-73122, soybean cells did not show significantly decreased DNA synthesis. PLC activity, measured by InsP3 produced, was higher under MS salt conditions compared with a water control, suggesting that an U-73122 resistant PLC activity could still produce enough InsP3 and/or InsP2 to activate DNA synthesis (Shigaki and Bhattacharyya, 2002) and raises the intriguing possibility that inhibitor action on enzymes may be affected by the subcellular localization of enzymes. Human DNA polymerase α has been shown to be activated by InsP2 in vitro (Sylvia et al., 1988), but the affect of inositol phosphates on plant DNA polymerases has not been studied. As previously discussed, PI-PLC activity with PtdIns substrate has been detected in plant nuclei (Pfaffmann et al., 1987) although the specific nuclear localized isoform has not been identified. It is likely that cell cycle or cellular developmental stages will control the nuclear localization of these PLCs as is found in animal cells.
In animals, lipids in the PI pathway have been shown to interact with DNA and may regulate DNA replication (reviewed in Koiv et al., 1995; Kuvichkin, 2002). Studies have shown PtdInsP to decrease α, δ, and ε DNA polymerase activity (Shoji-Kawaguchi et al., 1995). The inhibition of polymerase activity was not confined to PtdInsP as PtdIns inhibited DNA polymerase ε activity and PtdOH inhibited DNA polymerase α and ε activity. The fatty acid composition of PtdIns was a critical factor (Shoji-Kawaguchi et al., 1995) as plant PtdIns (enriched in linoleic (18:2) and palmitic acids (16:0)) inhibited both DNA polymerase α and ε, while animal PtdIns (enriched with arachidonic acid) only inhibited DNA polymerase ε. Furthermore, Ishimaru et al. (2010) showed that animal topoisomerases I and II (involved in DNA synthesis) could be inhibited by PtdOH. Topoisomerase II activity was also inhibited by PtdIns (Ishimaru et al., 2010) and by PtdInsP2 (Lewis et al., 2011). The results indicate that specific isoforms of DNA polymerase and topoisomerase can be regulated by selective lipids in the PI pathway. It will be interesting to see if plant DNA polymerases and topoisomerases are regulated by inositol phospholipids and inositol phosphates as suggested by research from Shigaki and Bhattacharyya (2002).
The PI Pathway and Regulation of Transcription and RNA Processing
The PI pathway may play a role in transcription in plants. Exogenously added PtdIns4P and PtdIns5P caused differential gene expression in A. thaliana (Alvarez-Venegas et al., 2006b), although the mechanisms of the phospholipid effects are unknown. Detergent resistant PI3K and PI4K activities were detected in soybean nuclei, and the PI3K protein showed intranuclear localization that coincided with sites of BrUTP incorporation (Bunney et al., 2000). However, an increase in PI3K activity is associated with root nodule formation (Hong and Verma, 1994) and a YFP-2xFYVE construct that recognizes PtdIns3P (the product of PI3K) co-localizes with endocytic vesicles in vivo (Vermeer et al., 2006). These results indicate PtdIns3P has a prominent role in vesicle trafficking in plants. Specific functions of PtdIns3P, PtdIns4P, and PtdIns5P in plant nuclei have yet to be determined.
Some insights into the function of the PI pathway intermediates in transcription and RNA processing can be gleaned from the animal and yeast literature. Inositol phospholipids and/or inositol phosphates affect RNA processing from transcription to mRNA export. Early studies tested the affect of lipids on transcriptional activity by adding lipid vesicles to isolated nuclei. PtdIns decreased total RNA synthesis, while vesicles of another phospholipid, phosphatidylserine, could increase RNA synthesis (Manzoli et al., 1982). A later study examined the effect of PtdIns, PtdIns4P, PtdInsP2, and PtdIns(3,4,5)P3 on transcription in vitro in the presence of chromatin and histone H1 (Yu et al., 1998). PtdInsP2 decreased the inhibition of RNA polymerase activity by H1 by 62% (Yu et al., 1998). This effect is specific for PtdInsP2 as PtdIns4P and PtdIns(3,4,5)P3 decreased RNA polymerase inhibition by less than 15% and PtdIns had no effect. PtdInsP2 transcription activation may be indirect through the regulation of histone H1; however, antibodies to PtdInsP2 immunoprecipitated RNA polymerase and transcription factors from nuclear extracts suggesting a more direct interaction between PtdInsP2 and the transcriptional machinery including RNA polymerase (Osborne et al., 2001; Lewis et al., 2011). PtdInsP2 also appears to be important for transcription in vivo; the mutant in the nuclear PIP5K skittles in D. melanogaster had decreased transcriptionally active chromatin compared with wild type (Cheng and Shearn, 2004).
RNA polymerase activity is also affected by transcription factors that bind other lipid and inositol phosphate components of the PI pathway. For example, Gozani et al. (2003) showed that the plant homeodomain (PHD) finger of tumor suppressor protein INhibitor of Growth 2 (ING2) bound specifically to PtdIns5P, and that increases in PtdIns5P in vivo affected the localization of endogenous ING2 visualized by immunohistochemistry. Transcription factors with novel phosphoinositide binding domains continue to be identified like the human transcription factors LRH-1 and SF-1, which are NR5 orphan nuclear receptors. The ligands for these receptors had not been identified, but a crystal structure suggested lipid binding and in vitro assays showed specific binding to PtdIns(3,5)P2 and PtdIns(3,4,5)P3 (Krylova et al., 2005) suggesting that these NR5 receptors are inositol phospholipid receptors similar to ING2. Additionally, PtdIns5P labeled with Bodipy-TR, PtdIns3P-Bodipy-TR, and PtdIns4P-Bodipy-TR showed qualitatively distinct nuclear fluorescence patterns (Gozani et al., 2005). Arabidopsis contains 83 PHD finger containing proteins with many similar to ING2 that may bind inositol phospholipids or inositol phosphates (Lee et al., 2009). These results suggest that each plant nuclear PI lipid may have specific sub-nuclear localization and potential effects on unidentified lipid binding transcription factors.
In addition to phospholipids in the PI pathway, IPK and the inositol phosphates have been shown to regulate transcription. First discovered as Arg82, a protein component of a transcriptional complex needed for arginine specific responses in yeast, IPK2 has been identified as a regulator of transcriptional complexes (Odom et al., 2000). The inactive IPK2 enzyme did not recover an IPK2 mutant phenotype, suggesting that the inositol phosphate products of IPK2, InsP4 or InsP5, are important for transcriptional control. Further research is needed in order to understand the role of the PI pathway in transcriptional regulation as IPK2 mutants also have defects in chromatin remodeling (Steger et al., 2003), which can mimic a transcription factor defect.
mRNA processing is an important step to ensure the correct template for translation prior to mRNA export. PtdInsP2 has been identified in nuclear speckles associated with RNA processing and depletion of nuclear PtdInsP2 decreased mRNA splicing (Osborne et al., 2001). Polyadenylation of mRNA transcripts is an important step in mRNA processing and a non-canonical poly-A polymerase, Star-PAP, has been identified that is activated by PtdInsP2 in vitro and in vivo. Star-PAP binds the human HsPIP5K1α and during hyperosmotic stress HsPIP5K1α and Star-PAP regulate a number of specific mRNAs through PtdInsP2 dependent poly-A tail elongation (Mellman et al., 2008). Independent of the hyperosmotic stress response, HsPIP5K1α and Star-PAP are recruited by protein kinase C δ (PKCδ) to regulate the DNA damage/apoptosis response gene BIK by increasing BIK transcript poly-A tail length and subsequently increasing BIK protein expression (Li et al., 2012).
Along with mRNA polyadenylation, some mRNA and tRNA transcripts undergo further processing known as RNA editing where nucleotides are replaced in the RNA transcript. The enzymes for both mRNA editing (ADAR2) and tRNA editing (ADAT1) bind InsP6 and require InsP6 for enzyme stability as well as for ADAR2 activity (Macbeth et al., 2005). The requirement for InsP6 in mRNA editing highlights the need for the study of the inositol phosphate pathway in the nucleus as select nuclear processes can be important for producing the correct mRNA sequence, which will affect the final protein sequence and expression. Efficient export of mRNA can also require InsP6 and the PI pathway (York et al., 1999; Alcázar-Román et al., 2006; Weirich et al., 2006).
The combinatorial regulation and complexity of the animal nuclear PI pathway is highlighted by the existence of cell signaling pathways regulated by HsPIP5K1α, the necessity of PtdInsP2 for mRNA splicing and inositol phosphate requirements in RNA editing. Roles of the plant nuclear PI pathway in mRNA splicing, polyadenylation and stability have yet to be described.
The PI Pathway and Chromatin Remodeling in Plants and Animals
Chromatin is made up of DNA and proteins. The structure of the chromatin controls replication, transcription and localization of the DNA. Basic chromatin structure can be divided into two functional states: repressed or non-active chromatin (heterochromatin) and open or active chromatin (euchromatin). The affect of the PI pathway on chromatin remodeling in plants is still unclear, while a number of studies have shown the PI pathway is important for histone modification and chromatin remodeling in animals.
At least one plant chromatin remodeling enzyme that is sensitive to PI pathway lipids has been described. The chromatin remodeling protein ATX1, a histone trimethyltransferase, has a PHD that binds PtdIns5P (Alvarez-Venegas et al., 2006a). Localization of ATX1 is affected by the lipids in the cell. Exogenously added PtdIns5P results in the relocalization of ATX1 from the nucleus to the PM and subcellular vesicles, and the ATX1-dependent H3K4 trimethylation is decreased (Alvarez-Venegas et al., 2006a). In vivo, overexpression of the PM localized phosphoinositide 3-phosphatase myotubularin (MTM), that hydrolyzes PtdIns(3,5)P2 to increase PM PtdIns5P, causes ATX1 localization to the PM (Ndamukong et al., 2010). It will be interesting to investigate the effect of increased nuclear PtdIns5P on ATX1 localization. We anticipate that further investigations will identify other chromatin remodeling and transcription factors that bind to the PI pathway components in plants.
Histone modification and chromatin remodeling are affected by PI pathway lipids and inositol phosphates in animals. Lipid analysis of isolated animal chromatin revealed that different amounts of lipids are associated with heterochromatin and euchromatin. In addition, higher turnover rates of lipids are associated with euchromatin (Rose and Frenster, 1965). Furthermore, hydrolysis of nuclear phospholipids via PLC changes the chromatin structure (Maraldi et al., 1984).
The skittles mutant in D. melanogaster, which lacks the nuclear localized PIP5K, has hyperphosphorylated histone H1, a biochemical marker of hyper-compacted heterochromatin (Cheng and Shearn, 2004). Yu et al. (1998) showed that both histones H3 and H1 could bind to PtdIns(4,5)P2 and that a specific binding site for PtdIns(4,5)P2 in the C terminal tail of histone H1 overlapped with a phosphorylation site, suggesting that lipid binding could affect histone phosphorylation. In addition to the affect of PtdInsP2 on histone H1 function, remodeling complexes are also affected by the PI pathway. The Brg-associated factor (BAF) remodeling complex in animals binds PtdInsP2 (Rando et al., 2002), and PtdInsP2 activates/stabilizes BAF chromatin binding and activity (Zhao et al., 1998). Small metabolites, like the inositol phosphates, can also affect remodeling complex activities (reviewed in Burgio et al., 2010). Particular inositol phosphates have specific activities, for example InsP6 has an inhibitory effect on remodeling complexes, while InsP4 and InsP5 can activate similar complexes (Shen et al., 2003). Changes in the inositol phosphates were also shown to affect chromatin remodeling in vivo (Steger et al., 2003).
The PI Pathway and Plant Stress Responses
A variety of PI pathway enzymes are important for the regulation of stress responses, and some of these proteins localize to the nucleus, suggesting a direct nuclear signaling capacity (Gillaspy, 2011). Both 5Ptase13 and 5Ptase7, members of a large family of 5Ptase enzymes in plants, have been localized to the nucleus and are required for different stress responses. 5Ptase13 interacts with SnRK1 and regulates responses to sugar and nutrient stress by stabilizing SnRK1 activity (Ananieva et al., 2008). The 5Ptase7 is both nuclear, and PM localized and may help modulate the response to salt stress as 5Ptase7 mutants are hypersensitive to salt. Salt stress induced reactive oxygen species (ROS) in the nucleus (as measured by H2DCFDA fluorescence) and stress responsive transcripts are significantly reduced in 5Ptase7 mutants compared with wild type plants (Kaye et al., 2011).
Plant phosphate stress is an important agronomic concern, and phosphate sensing has been linked to inositol phosphates in yeast. The phenotype of the IPK1 mutation in A. thaliana suggests that the PI pathway also is important for phosphate sensing in plants. When IPK1 is mutated, the IPK1 transcript decreased 70% and the total InsP6 decreased by 82.5% in seeds and over 90% in seedlings (Stevenson-Paulik et al., 2005). ipk1 plants exposed to normal levels of phosphate continue to accumulate phosphate as if they were lacking phosphate and ultimately suffer from phosphate toxicity suggesting that IPK1 (or its inositol phosphate product InsP6) is important for normal phosphate sensing. ipk1 mutant plants are also more susceptible to pathogens; however, the fact that not all InsP6-deficient plants are pathogen sensitive suggests that a selective pool of InsP6 or IPK1-sensitive responses control pathogen response (Murphy et al., 2008). Decreased IPK1 activity in the ipk1 mutant also leads to an accumulation of InsP5, which has a role in increasing jasmonic acid (JA) sensing, and can affect the pathogen response as discussed below.
Cellular PtdInsP2 production was increased sixfold with a heat stress treatment of 40°C for 30 min, and heat stress also caused accumulation of the YFP-PH domain (capable of binding PtdInsP2) in the nucleus of Arabidopsis cells (Mishkind et al., 2009). Increased nuclear PtdInsP2 could result from increased nuclear localization or activation of PIP5K or decreased PtdInsP2 catabolism. The biological relevance of increased YFP-PH accumulation in the nucleus is not yet clear since the high temperature may affect membrane dynamics. Membrane-associated PtdInsP2 has been shown to increase with senescence (Borochov et al., 1994).
Similar to the previously discussed fatty acid composition changes in the animal nuclear PI pathway during the cell cycle (Ogiso et al., 2010), dynamic and transient changes in the fatty acid composition of PI pathway lipids can occur during stress in plants (König et al., 2007). In response to hyperosmotic stress there was a transient increase in PUFAs in total cellular PtdInsP and PtdInsP2 and a decrease in the PUFAs in PtdIns. When cell membranes were fractionated, the transient (15 min) changes in the PM PtdIns and PtdInsP2 were consistent with the whole cell data; however, the lipids in the nuclear-enriched fraction from stressed plants did not show a similar PUFA profile (König et al., 2008). These data are consistent with a separate and dynamic nuclear PI signaling pool.
The PI Pathway and Plant Development
There is evidence that in plants the expression of PI pathway enzymes and lipid content change with developmental progression revealing further similarity to the animal PI pathway. For example, during embryo development and seedling germination, a variety of environmental hormonal and developmental cues are transmitted to the nucleus. As previously discussed, during somatic embryogenesis different stages of embryogenesis showed differences in the activity of PI3K and PI4K, suggesting that a particular point of development requires the PI pathway (Ek-Ramos et al., 2003). Direct changes in the nuclear PtdIns content and distribution can be seen in nuclei isolated during wheat germination (Minasbekyan et al., 2004, 2008). In germinating wheat, changes were observed in both the nuclear membrane and soluble nuclear PI pathway lipids (Minasbekyan et al., 2008). PtdIns was continuously present in the nuclear membrane at day 3 and day 4, but PtdIns in the nucleoplasmic fraction was only observed on day 4. Gibberellin (GA) treatment increased nucleoplasmic PtdIns suggesting that nuclear PtdIns might be a by-product of increased lipid trafficking or might be part of a novel GA regulatory loop that uses PtdIns for nuclear signaling.
Heterologous expression and mutation of enzymes in the PI pathway have revealed potential effects of the PI pathway on the development and differentiation of plants. For example, expression of PI-PLC2 from Brassica napus in canola increased the speed of development by 1 week, resulting in early flowering and an increased number of branches (Georges et al., 2009). The subcellular localization of BnPI-PLC2 in canola was not reported; however, increases in animal nuclear PI-PLC proteins have similar affects in that there is increased cell cycle progression and changes in cell determination (reviewed Cocco et al., 2009). While it is possible that BnPI-PLC2 directly affects nuclear function in plants, indirect effects through regulation of transcription or phytohormones cannot be ruled out.
The A. thaliana inositol kinase IPK2β is a homolog of the yeast IPK2 InsP3/InsP4 dual-specificity 6-/3-/5-kinase, which phosphorylates InsP3 to InsP4, and InsP5 (Stevenson-Paulik et al., 2002). Expression of AtIPK2β in tobacco resulted in similar phenotypes to expression of BnPI-PLC2 in canola (Georges et al., 2009). A slight increase in growth rate and an increased tolerance to abiotic stresses was observed in AtIPK2β expressing plants (Yang et al., 2008). As previously mentioned, IPK2 the yeast homolog of AtIPK2β, can act as a transcription factor (Odom et al., 2000). AtIPK2β also localizes to the nucleus and, in yeast, complemented the ipk2 mutation (Xia et al., 2003; Yang et al., 2008) suggesting an additional role for AtIPK2β as a transcription factor. Constitutive expression of AtIPK2β in tobacco increased expression of stress regulated genes under normal growth conditions, providing evidence that AtIPK2β and/or it is inositol phosphate products can affect transcription in plants (Yang et al., 2008).
A second isoform of AtIPK2, AtIPK2α, was localized to the PM and nucleus by GFP fluorescence. Reduction of AtIPK2α protein and transcript by expression of an antisense construct increased root growth and pollen germination (Xu et al., 2005). The increase in root growth with decreased AtIPK2α expression and increased growth rate with heterologous expression of AtIPK2β demonstrate that different isoforms of the same enzyme from A. thaliana can have different effects on growth and development.
Another IPK, A. thaliana inositol (3,4,5) trisphosphate 5/6-kinase (AtITPK1), has been localized to the plant nucleus by GFP fluorescence (Qin et al., 2005). Moreover, AtITPK1 was shown to interact with the nuclear protein complex the COP9 signalosome (CNS) that controls response to various stresses and developmental cues in plants. Mutants in AtITPK1 also showed decreased hypocotyl growth in red light, suggesting a possible need for the protein kinase or inositol kinase function of the AtITPK1 in photomorphogenesis.
Further evidence that the PI pathway is involved in developmental patterning comes from studies of PtdIns4P-specific regulation of the stem cell factor Poltergeist (Gagne and Clark, 2010) and from studies of A. thaliana PIP5K isoforms on root development (Mei et al., 2011). Whether these phenotypes are related primarily to membrane trafficking, PM signaling or to direct effects on nuclear regulation requires further investigation.
The PI Pathway and Plant Hormone Regulation
Growth, development and stress responses are controlled by many different factors including the growth regulators, auxin, and JA. Auxin and JA both have nuclear receptors (TIR1 and COI1, respectively) that bind their respective hormones and regulate their respective transcription factors. Intriguingly, an InsP6 binding site has been identified in TIR1 and an InsP5 binding site has been identified in COI1. There are no reports thus far that distinguish a role for the nuclear PI pathway versus the cytosolic pathway in generating these inositol phosphates.
Tan et al. (2007) crystallized and solved the structure of TIR1 and found that InsP6 was crystallized with auxin in the binding pocket. The current hypothesis is that InsP6 is a cofactor for auxin perception that allows auxin to bind tighter to TIR1. Computer simulations by Hao and Yang (2010) suggest a structural shift in the TIR1 receptor that requires InsP6 to stabilize a binding pocket for auxin. This stabilization also creates a conformational change in the TIR1 receptor that is favorable for Aux/IAA protein binding, supporting the hypothesized role for InsP6 in auxin perception of the TIR1 protein.
The JA pathway-mediated by the COI1 receptor is also regulated by the inositol phosphates. Sheard et al. (2010) identified InsP4 and InsP5 as cofactors that co-purified with the COI1 and JAZ9 JA sensing complex. In yeast two hybrid analyses, increased levels of InsP5 correlated with an increased binding between COI1 to JAZ9. Further experiments tested the effects of InsP5 in planta. ipk1-1 mutants, which have increased InsP5, showed increased JA responses to wounding and decreased herbivory (Mosblech et al., 2011). In contrast, plants expressing human InsP5Tase with decreased InsP3, InsP5, and InsP6 (Perera et al., 2008), show decreased JA responses and increased herbivory (Mosblech et al., 2008). These experiments suggest that in planta, the cellular content of InsP5 affects the JA pathway through modulation of the COI1 and JAZ9 interaction. It is interesting that Sheard et al. (2010) also found activity with InsP4, suggesting a role for InsP4 in vivo, and highlighting the complexity of the inositol phosphate nuclear code in plants.
Potential Functions of the PI Pathway in Plant Nuclei
While studies of the plant PI pathway and DNA repair, recombination and telomere length have not been reported, we can learn much from data from other organisms. Inositol phosphates and inositol pyrophosphates have been identified as major components of DNA maintenance pathways including repair, recombination and telomere length. InsP6 binds and regulates the DNA repair protein complex protein Ku70 (Cheung et al., 2008), which in turn regulates a non-homologous end joining (NHEJ) complex (Hanakahi and West, 2002; Ma and Lieber, 2002). Additionally a direct connection between amount of inositol phosphates and NHEJ activity was demonstrated (Byrum et al., 2004). Inositol pyrophosphates have been identified to affect recombination in yeast. The yeast inositol hexakisphosphate kinase (InsP6K) that produces InsP7 and InsP8 was first identified as a repressor of the hyper-recombination phenotype of protein kinase C (kinase C suppressor 1, KCS1; Huang and Symington, 1994). A functional KCS1 enzyme was shown to be essential for repression of the hyper-recombination phenotype (Luo et al., 2002) suggesting that an inositol phosphate product (InsP7, InsP8, Luo et al., 2002; PP-InsP4 and others reviewed in York, 2006) or another KCS1 enzymatic activity is needed for the repression.
Regulation of telomere length is a crucial part of DNA processing in eukaryotes (reviewed in d’ Adda di Fagagna et al., 2004; reviewed in Smogorzewska and de Lange, 2004; Schmitt et al., 2007). An elegant set of experiments showed that increasing PP-InsP4 resulted in decreased telomere length, and decreasing PP-InsP4 increased telomere length in yeast. These experiments also showed that when the DNA-PK-like protein Tel1 was mutated, no inositol phosphate-dependent telomere regulation was observed (York et al., 2005). Like Te1, human ataxia telangiectasia mutated (ATM) is also homologous to DNA-PK (Greenwell et al., 1995) and cells with mutated atm show decreased myo-inositol metabolism (Yorek et al., 1999). Like yeast and mammals, plants have DNA-PK homologs, ATM and ATR, which regulate telomere length and response to NHEJ (Vespa et al., 2005). The DNA-PK family of proteins share sequence homology with the PI3K enzymes (Poltoratsky et al., 1995), suggesting an ancient regulatory role. Given the conservation of function of ATM homologs in plants, animals, and yeast, it would not be surprising if plant ATM is regulated by the nuclear inositol phosphates.
Conclusion
Studies in plants make a compelling argument for a developmental and regulatory role for the PI pathway in the nucleus. We have enumerated the PI pathway enzymes and lipid activities associated with plant nuclei. More sensitive detection methods are needed for detailed analyses of plant PI pathway lipids and enzymes in both the cytosolic and nuclear fractions during different developmental stages and stress responses. Attention to the crosstalk and movement of lipids and enzymes between the discrete subcellular PI pathways will be critical for future understanding of the role of the nuclear PI pathway in plants. As demonstrated by the interactions between inositol phosphates and the nuclear auxin and JA receptors, it is anticipated that future research will uncover additional plant-specific processes mediated by the nuclear PI pathway.
Conflict of Interest Statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Acknowledgments
This work was supported in part by grants from the National Science Foundation (MCB-0718452 and MCB-1052034), by the North Carolina Agricultural Research Service, by an Initiative for Future Agriculture and Food Systems (IFAFS) Research Fellowship through the USDA and by a NIH/NCSU Molecular Biotechnology Training Program (MBTP) Fellowship.
References
- Alcázar-Román A. R., Tran E. J., Guo S., Wente S. R. (2006). Inositol hexakisphosphate and Gle1 activate the DEAD-box protein Dbp5 for nuclear mRNA export. Nat. Cell Biol. 8, 711–716 10.1038/ncb1427 [DOI] [PubMed] [Google Scholar]
- Alvarez-Venegas R., Sadder M., Hlavacka A., Baluška F., Xia Y., Lu G., Firsov A., Sarath G., Moriyama H., Dubrovsky J. G., Avramova Z. (2006a). The Arabidopsis homolog of trithorax, ATX1, binds phosphatidylinositol 5-phosphate, and the two regulate a common set of target genes. Proc. Natl. Acad. Sci. U.S.A. 103, 6049–6054 10.1073/pnas.0600944103 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alvarez-Venegas R., Xia Y., Lu G., Avramova Z. (2006b). Phosphoinositide 5-phosphate and phosphoinositide 4-phosphate trigger distinct specific responses of Arabidopsis genes. Plant Signal. Behav. 1, 140–151 10.4161/psb.1.3.2997 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ananieva E. A., Gillaspy G. E., Ely A., Burnette R. N., Les Erickson F. (2008). Interaction of the WD40 domain of a myoinositol polyphosphate 5-phosphatase with SnRK1 links inositol, sugar, and stress signaling. Plant Physiol. 148, 1868–1882 10.1104/pp.108.130575 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Balla T., Szentpetery Z., Kim Y. J. (2009). Phosphoinositide signaling: new tools and insights. Physiology (Bethesda) 24, 231–244 10.1152/physiol.00014.2009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barker C. J., Illies C., Gaboardi G. C., Berggren P.-O. (2009). Inositol pyrophosphates: structure, enzymology and function. Cell. Mol. Life Sci. 66, 3851–3871 10.1007/s00018-009-0115-2 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barlow C. A., Laishram R. S., Anderson R. A. (2010). Nuclear phosphoinositides: a signaling enigma wrapped in a compartmental conundrum. Trends Cell Biol. 20, 25–35 10.1016/j.tcb.2009.09.009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barona T., Byrne R. D., Pettitt T. R., Wakelam M. J. O., Larijani B., Poccia D. L. (2005). Diacylglycerol induces fusion of nuclear envelope membrane precursor vesicles. J. Biol. Chem. 280, 41171–41177 10.1074/jbc.M412863200 [DOI] [PubMed] [Google Scholar]
- Biffen M., Hanke D. E. (1990). Reduction in the level of intracellular myo-inositol in cultured soybean (Glycine max) cells inhibits cell division. Biochem. J. 265, 809. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bondeva T., Pirola L., Bulgarelli-Leva G., Rubio I., Wetzker R., Wymann M. P. (1998). Bifurcation of lipid and protein kinase signals of PI3Kγ to the protein kinases PKB and MAPK. Science 282, 293–296 10.1126/science.282.5387.293 [DOI] [PubMed] [Google Scholar]
- Bootman M. D., Fearnley C., Smyrnias I., MacDonald F., Roderick H. L. (2009). An update on nuclear calcium signalling. J. Cell. Sci. 122, 2337–2350 10.1242/jcs.028100 [DOI] [PubMed] [Google Scholar]
- Borochov A., Cho M. H., Boss W. F. (1994). Plasma membrane lipid metabolism of petunia petals during senescence. Physiol. Plant 90, 279–284 10.1034/j.1399-3054.1994.900206.x [DOI] [Google Scholar]
- Boruc J., Zhou X., Meier I. (2012). Dynamics of the plant nuclear envelope and nuclear pore. Plant Physiol. 158, 78–86 10.1104/pp.111.185256 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bunce M. W., Gonzales M. L., Anderson R. A. (2006). Stress-ING out: phosphoinositides mediate the cellular stress response. Sci. STKE 2006, pe46. 10.1126/stke.3602006pe46 [DOI] [PubMed] [Google Scholar]
- Bunney T. D., Watkins P. A. C., Beven A. F., Shaw P. J., Hernandez L. E., Lomonossoff G. P., Shanks M., Peart J., Drobak B. K. (2000). Association of phosphatidylinositol 3-kinase with nuclear transcription sites in higher plants. Plant Cell 12, 1679–1688 10.2307/387118211006340 [DOI] [Google Scholar]
- Burgio G., Onorati M. C., Corona D. F. V. (2010). Chromatin remodeling regulation by small molecules and metabolites. Biochim. Biophys. Acta 1799, 671–680 [DOI] [PubMed] [Google Scholar]
- Byrne R. D., Garnier-Lhomme M., Han K., Dowicki M., Michael N., Totty N., Zhendre V., Cho A., Pettitt T. R., Wakelam M. J., Poccia D. L., Larijani B. (2007). PLCγ is enriched on poly-phosphoinositide-rich vesicles to control nuclear envelope assembly. Cell. Signal. 19, 913–922 10.1016/j.cellsig.2006.10.011 [DOI] [PubMed] [Google Scholar]
- Byrum J., Jordan S., Safrany S. T., Rodgers W. (2004). Visualization of inositol phosphate-dependent mobility of Ku: depletion of the DNA–PK cofactor InsP6 inhibits Ku mobility. Nucleic Acids Res. 32, 2776–2784 10.1093/nar/gkh592 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Camacho L., Smertenko A. P., Pérez-Gómez J., Hussey P. J., Moore I. (2009). Arabidopsis Rab-E GTPases exhibit a novel interaction with a plasma-membrane phosphatidylinositol-4-phosphate 5-kinase. J. Cell. Sci. 122, 4383. 10.1242/jcs.053488 [DOI] [PubMed] [Google Scholar]
- Chamero P., Manjarres I. M., García-Verdugo J. M., Villalobos C., Alonso M. T., García-Sancho J. (2008). Nuclear calcium signaling by inositol trisphosphate in GH3 pituitary cells. Cell Calcium 43, 205–214 10.1016/j.ceca.2007.05.005 [DOI] [PubMed] [Google Scholar]
- Cheng M. K., Shearn A. (2004). The direct interaction between ASH2, a drosophila trithorax group protein, and SKTL, a nuclear phosphatidylinositol 4-phosphate 5-kinase, implies a role for phosphatidylinositol 4,5-bisphosphate in maintaining transcriptionally active chromatin. Genetics 167, 1213–1223 10.1534/genetics.103.018721 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cheung A. Y., Reddy A. S. N. (2012). Nuclear architecture and dynamics: territories, nuclear bodies, and nucleocytoplasmic trafficking. Plant Physiol. 158, 23–25 10.1104/pp.111.900426 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cheung J. C. Y., Salerno B., Hanakahi L. A. (2008). Evidence for an inositol hexakisphosphate-dependent role for Ku in mammalian nonhomologous end joining that is independent of its role in the DNA-dependent protein kinase. Nucleic Acids Res. 36, 5713–5726 10.1093/nar/gkn572 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clarke J. H., Letcher A. J., D’Santos C. S., Halstead J. R., Irvine R. F., Divecha N. (2001). Inositol lipids are regulated during cell cycle progression in the nuclei of murine erythroleukemia cells. Biochem. J. 357, 905. 10.1042/0264-6021:3570905 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cocco L., Faenza I., Follo M. Y., Billi A. M., Ramazzotti G., Papa V., Martelli A. M., Manzoli L. (2009). Nuclear inositides: PI-PLC signaling in cell growth, differentiation and pathology. Adv. Enzyme Regul. 49, 2–10 10.1016/j.advenzreg.2008.12.001 [DOI] [PubMed] [Google Scholar]
- Collings D. A., Carter C. N., Rink J. C., Scott A. C., Wyatt S. E., Allen N. S. (2000). Plant nuclei can contain extensive grooves and invaginations. Plant Cell 12, 2425–2440 10.1105/tpc.12.12.2425 [DOI] [PMC free article] [PubMed] [Google Scholar]
- d’ Adda di Fagagna F., Teo S.-H., Jackson S. P. (2004). Functional links between telomeres and proteins of the DNA-damage response. Genes Dev. 18, 1781–1799 10.1101/gad.1214504 [DOI] [PubMed] [Google Scholar]
- Das R., Bagga S., Sopory S. K. (1987). Involvement of phosphoinositides, calmodulin and glyoxalase-I in cell proliferation in callus cultures of Amaranthus paniculatus. Plant Sci. 53, 45–51 10.1016/0168-9452(87)90177-4 [DOI] [Google Scholar]
- Di Paolo G., De Camilli P. (2006). Phosphoinositides in cell regulation and membrane dynamics. Nature 443, 651–657 10.1038/nature05185 [DOI] [PubMed] [Google Scholar]
- Echevarría W., Leite M. F., Guerra M. T., Zipfel W. R., Nathanson M. H. (2003). Regulation of calcium signals in the nucleus by a nucleoplasmic reticulum. Nat. Cell Biol. 5, 440–446 10.1038/ncb980 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ek-Ramos M. J., Racagni-Di Palma G., Hernández-Sotomayor S. (2003). Changes in phosphatidylinositol and phosphatidylinositol monophosphate kinase activities during the induction of somatic embryogenesis in Coffea arabica. Physiol. Plant 119, 270–277 10.1034/j.1399-3054.2003.00171.x [DOI] [Google Scholar]
- Faenza I., Matteucci A., Manzoli L., Billi A. M., Aluigi M., Peruzzi D., Vitale M., Castorina S., Suh P.-G., Cocco L. (2000). A role for nuclear phospholipase Cβ1 in cell cycle control. J. Biol. Chem. 275, 30520–30524 10.1074/jbc.M004630200 [DOI] [PubMed] [Google Scholar]
- Fedorenko O., Yarotskyy V., Duzhyy D., Marchenko S. (2010). The large-conductance ion channels in the nuclear envelope of central neurons. Pflügers Arch. Eur. J. Physiol. 460, 1045–1050 10.1007/s00424-010-0882-5 [DOI] [PubMed] [Google Scholar]
- Gagne J. M., Clark S. E. (2010). The Arabidopsis stem cell factor POLTERGEIST is membrane localized and phospholipid stimulated. Plant Cell 22, 729–743 10.1105/tpc.109.068734 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garnier-Lhomme M., Byrne R. D., Hobday T. M. C., Gschmeissner S., Woscholski R., Poccia D. L., Dufourc E. J., Larijani B. (2009). Nuclear envelope remnants: fluid membranes enriched in STEROLS and polyphosphoinositides. PLoS ONE 4, e4255. 10.1371/journal.pone.0004255 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Georges F., Das S., Ray H., Bock C., Nokhrina K., Kolla V. A., Keller W. (2009). Over-expression of Brassica napus phosphatidylinositol-phospholipase C2 in canola induces significant changes in gene expression and phytohormone distribution patterns, enhances drought tolerance and promotes early flowering and maturation. Plant Cell Environ. 32, 1664–1681 10.1111/j.1365-3040.2009.02027.x [DOI] [PubMed] [Google Scholar]
- Gil A., Andrés-Pons A., Pulido R. (2007). Nuclear PTEN: a tale of many tails. Cell Death Differ. 14, 395–399 10.1038/sj.cdd.4402073 [DOI] [PubMed] [Google Scholar]
- Gillaspy G. E. (2011). The cellular language of myo-inositol signaling. New Phytol. 192, 823–839 10.1111/j.1469-8137.2011.03939.x [DOI] [PubMed] [Google Scholar]
- Gonzales M. L., Anderson R. A. (2006). Nuclear phosphoinositide kinases and inositol phospholipids. J. Cell. Biochem. 97, 252–260 10.1002/jcb.20655 [DOI] [PubMed] [Google Scholar]
- Gozani O., Field S. J., Ferguson C. G., Ewalt M., Mahlke C., Cantley L. C., Prestwich G. D., Yuan J. (2005). Modification of protein sub-nuclear localization by synthetic phosphoinositides: evidence for nuclear phosphoinositide signaling mechanisms. Adv. Enzyme Regul. 45, 171–185 10.1016/j.advenzreg.2005.02.010 [DOI] [PubMed] [Google Scholar]
- Gozani O., Karuman P., Jones D. R., Ivanov D., Cha J., Lugovskoy A. A., Baird C. L., Zhu H., Field S. J., Lessnick S. L., Villasenor J., Mehrotra B., Chen J., Rao V. R., Brugge J. S., Ferguson C. G., Payrastre B., Myszka D. G., Cantley L. C., Wagner G., Divecha N., Prestwich G. D., Yuan J. (2003). The PHD finger of the chromatin-associated protein ING2 functions as a nuclear phosphoinositide receptor. Cell 114, 99–111 10.1016/S0092-8674(03)00480-X [DOI] [PubMed] [Google Scholar]
- Greenwell P. W., Kronmal S. L., Porter S. E., Gassenhuber J., Obermaier B., Petes T. D. (1995). TEL1, a gene involved in controlling telomere length in S. cerevisiae, is homologous to the human ataxia telangiectasia gene. Cell 82, 823–829 10.1016/0092-8674(95)90479-4 [DOI] [PubMed] [Google Scholar]
- Hanakahi L. A., West S. C. (2002). Specific interaction of IP6 with human Ku70/80, the DNA-binding subunit of DNA-PK. EMBO J. 21, 2038–2044 10.1093/emboj/21.8.2038 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hancock R. (2000). A new look at the nuclear matrix. Chromosoma 109, 219–225 10.1007/s004120000077 [DOI] [PubMed] [Google Scholar]
- Hao G.-F., Yang G.-F. (2010). The role of Phe82 and Phe351 in auxin-induced substrate perception by TIR1 ubiquitin ligase: a novel insight from molecular dynamics simulations. PLoS ONE 5, e10742. 10.1371/journal.pone.0010742 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heilmann I. (2009). Using genetic tools to understand plant phosphoinositide signalling. Trends Plant Sci. 14, 171–179 10.1016/j.tplants.2008.12.002 [DOI] [PubMed] [Google Scholar]
- Hendrix K. W., Assefa H., Boss W. F. (1989). The polyphosphoinositides, phosphatidylinositol monophosphate and phosphatidylinositol bisphosphate, are present in nuclei isolated from carrot protoplast. Protoplasma 151, 62–72 10.1007/BF01403302 [DOI] [Google Scholar]
- Hong Z., Verma D. P. (1994). A phosphatidylinositol 3-kinase is induced during soybean nodule organogenesis and is associated with membrane proliferation. Proc. Natl. Acad. Sci. U.S.A. 91, 9617–9621 10.1073/pnas.91.20.9617 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Horton P., Park K.-J., Obayashi T., Fujita N., Harada H., Adams-Collier C. J., Nakai K. (2007). WoLF PSORT: protein localization predictor. Nucleic Acids Res. 35, W585–W587 10.1093/nar/gkm259 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang K. N., Symington L. S. (1994). Mutation of the gene encoding protein kinase C 1 stimulates mitotic recombination in Saccharomyces cerevisiae. Mol. Cell. Biol. 14, 6039–6045 10.1128/MCB.14.9.6039 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huh Y. H., Yoo S. H. (2003). Presence of the inositol 1,4,5-triphosphate receptor isoforms in the nucleoplasm. FEBS Lett. 555, 411–418 10.1016/S0014-5793(03)01273-0 [DOI] [PubMed] [Google Scholar]
- Im Y., Phillippy B., Perera I. (2010). “InsP3 in plant cells,” in Lipid Signaling in Plants Plant Cell Monographs, ed. Munnik T. (Berlin: Springer; ), 145–160 [Google Scholar]
- Im Y. J., Heilmann I., Perera I. Y. (2011). “The hull of fame: lipid signaling in the plasma membrane,” in The Plant Plasma Membrane, eds Murphy A. S., Schulz B., Peer W. (Berlin: Springer Berlin Heidelberg; ), 437–455 [Google Scholar]
- Irvine R. F. (2003). Nuclear lipid signalling. Nat. Rev. Mol. Cell Biol. 4, 349–360 10.1038/nrm1152 [DOI] [PubMed] [Google Scholar]
- Ischebeck T., Stenzel I., Hempel F., Jin X., Mosblech A., Heilmann I. (2011). Phosphatidylinositol-4,5-bisphosphate influences Nt-Rac5-mediated cell expansion in pollen tubes of Nicotiana tabacum. Plant J. 65, 453–468 10.1111/j.1365-313X.2010.04435.x [DOI] [PubMed] [Google Scholar]
- Ishimaru C., Takeuchi T., Sugawara F., Yoshida H., Mizushina Y. (2010). Inhibitory effects of diacylglyceride phospholipids on DNA polymerase and topoisomerase activities, and human cancer cell growth. Med. Chem. 6, 114–122 [DOI] [PubMed] [Google Scholar]
- Kaye Y., Golani Y., Singer Y., Leshem Y., Cohen G., Ercetin M., Gillaspy G., Levine A. (2011). Inositol polyphosphate 5-phosphatase7 regulates the production of reactive oxygen species and salt tolerance in Arabidopsis. Plant Physiol. 157, 229–241 10.1104/pp.111.176883 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Keune W. J., Bultsma Y., Sommer L., Jones D., Divecha N. (2011). Phosphoinositide signalling in the nucleus. Adv. Enzyme Regul. 51, 91–99 10.1016/j.advenzreg.2010.09.009 [DOI] [PubMed] [Google Scholar]
- Kleinig H. (1970). Nuclear membranes from mammalian liver. J. Cell Biol. 46, 396–402 10.1083/jcb.46.2.396 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koiv A., Palvimo J., Kinnunen P. K. J. (1995). Evidence for ternary complex formation by histone H1, DNA, and liposomes. Biochemistry 34, 8018–8027 10.1021/bi00025a007 [DOI] [PubMed] [Google Scholar]
- König S., Ischebeck T., Lerche J., Stenzel I., Heilmann I. (2008). Salt-stress-induced association of phosphatidylinositol 4,5-bisphosphate with clathrin-coated vesicles in plants. Biochem. J. 415, 387–399 10.1042/BJ20081306 [DOI] [PubMed] [Google Scholar]
- König S., Mosblech A., Heilmann I. (2007). Stress-inducible and constitutive phosphoinositide pools have distinctive fatty acid patterns in Arabidopsis thaliana. FASEB J. 21, 1958. 10.1096/fj.06-7887com [DOI] [PubMed] [Google Scholar]
- Koroleva O. A., Tomlinson M. L., Leader D., Shaw P., Doonan J. H. (2005). High-throughput protein localization in Arabidopsis using Agrobacterium-mediated transient expression of GFP-ORF fusions. Plant J. 41, 162–174 10.1111/j.1365-313X.2004.02281.x [DOI] [PubMed] [Google Scholar]
- Krinke O., Novotná Z., Valentová O., Martinec J. (2007). Inositol trisphosphate receptor in higher plants: is it real? J. Exp. Bot. 58, 361–376 10.1093/jxb/erl220 [DOI] [PubMed] [Google Scholar]
- Krylova I. N., Sablin E. P., Moore J., Xu R. X., Waitt G. M., MacKay J. A., Juzumiene D., Bynum J. M., Madauss K., Montana V., Lebedeva L., Suzawa M., Williams J. D., Williams S. P., Guy R. K., Thornton J. W., Fletterick R. J., Willson T. M., Ingraham H. A. (2005). Structural analyses reveal phosphatidyl inositols as ligands for the NR5 orphan receptors SF-1 and LRH-1. Cell 120, 343–355 10.1016/j.cell.2005.01.024 [DOI] [PubMed] [Google Scholar]
- Kusano H., Testerink C., Vermeer J. E. M., Tsuge T., Shimada H., Oka A., Munnik T., Aoyama T. (2008). The Arabidopsis phosphatidylinositol phosphate 5-kinase PIP5K3 is a key regulator of root hair tip growth. Plant Cell 20, 367–380 10.1105/tpc.107.056119 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kuvichkin V. (2002). DNA-lipid interactions in vitro and in vivo. Bioelectrochemistry 58, 3–12 10.1016/S1567-5394(02)00123-8 [DOI] [PubMed] [Google Scholar]
- Lee W. Y., Lee D., Chung W.-I., Kwon C. S. (2009). Arabidopsis ING and Alfin1-like protein families localize to the nucleus and bind to H3K4me3/2 via plant homeodomain fingers. Plant J. 58, 511–524 10.1111/j.1365-313X.2009.03795.x [DOI] [PubMed] [Google Scholar]
- Lewis A. E., Sommer L., Arntzen M. Ø., Strahm Y., Morrice N. A., Divecha N., D’Santos C. S. (2011). Identification of nuclear phosphatidylinositol 4,5-bisphosphate-interacting proteins by neomycin extraction. Mol. Cell Proteomics 10, M110.003376. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li W., Laishram R. S., Ji Z., Barlow C. A., Tian B., Anderson R. A. (2012). Star-PAP Control of BIK expression and apoptosis is regulated by nuclear PIPKIα and PKCδ signaling. Mol. Cell 45, 25–37 10.1016/j.molcel.2011.11.017 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lloyd C., Chan J. (2006). Not so divided: the common basis of plant and animal cell division. Nat. Rev. Mol. Cell Biol. 7, 147–152 10.1038/nrm1831 [DOI] [PubMed] [Google Scholar]
- Loewus F. A., Murthy P. P. N. (2000). Myo-inositol metabolism in plants. Plant Sci. 150, 1–19 10.1016/S0168-9452(99)00150-8 [DOI] [Google Scholar]
- Lou Y., Gou J.-Y., Xue H.-W. (2007). PIP5K9, an Arabidopsis phosphatidylinositol monophosphate kinase, interacts with a cytosolic invertase to negatively regulate sugar-mediated root growth. Plant Cell 19, 163–181 10.1105/tpc.106.045658 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luo H. R., Saiardi A., Yu H., Nagata E., Ye K., Snyder S. H. (2002). Inositol pyrophosphates are required for DNA hyperrecombination in protein kinase C1 mutant yeast. Biochemistry 41, 2509–2515 10.1021/bi0118153 [DOI] [PubMed] [Google Scholar]
- Ma Y., Lieber M. R. (2002). Binding of inositol hexakisphosphate (IP6) to Ku but not to DNA-PKcs. J. Biol. Chem. 277, 10756–10759 10.1074/jbc.M106685200 [DOI] [PubMed] [Google Scholar]
- Macbeth M. R., Schubert H. L., VanDemark A. P., Lingam A. T., Hill C. P., Bass B. L. (2005). Inositol hexakisphosphate is bound in the ADAR2 core and required for RNA editing. Science 309, 1534–1539 10.1126/science.1113150 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Malhas A., Goulbourne C., Vaux D. J. (2011). The nucleoplasmic reticulum: form and function. Trends Cell Biol. 21, 362–373 10.1016/j.tcb.2011.03.008 [DOI] [PubMed] [Google Scholar]
- Malviya A. N. (1994). The nuclear inositol 1,4,5-trisphosphate and inositol 1,3,4,5-tetrakisphosphate receptors. Cell Calcium 16, 301–313 10.1016/0143-4160(94)90094-9 [DOI] [PubMed] [Google Scholar]
- Malviya A. N., Rogue P. J. (1998). “Tell me where is calcium bred”: clarifying the roles of nuclear calcium. Cell 92, 17–23 10.1016/S0092-8674(00)80895-8 [DOI] [PubMed] [Google Scholar]
- Manzoli F. A., Capitani S., Mazzotti G., Barnabei O., Maraldi N. M. (1982). Role of chromatin phospholipids on template availability and ultrastructure of isolated nuclei. Adv. Enzyme Regul. 20, 247–262 10.1016/0065-2571(82)90019-X [DOI] [PubMed] [Google Scholar]
- Manzoli L., Billi A. M., Rubbini S., Bavelloni A., Faenza I., Gilmour R. S., Rhee S. G., Cocco L. (1997). Essential role for nuclear phospholipase C β1 in insulin-like growth factor I-induced mitogenesis. Cancer Res. 57, 2137. [PubMed] [Google Scholar]
- Maraldi N. M., Capitani S., Caramelli E., Cocco L., Barnabei O., Manzoli F. A. (1984). Conformational changes of nuclear chromatin related to phospholipid induced modifications of the template availability. Adv. Enzyme Regul. 22, 447–464 10.1016/0065-2571(84)90025-6 [DOI] [PubMed] [Google Scholar]
- Matteucci A., Faenza I., Gilmour R. S., Manzoli L., Billi A. M., Peruzzi D., Bavelloni A., Rhee S.-G., Cocco L. (1998). Nuclear but not cytoplasmic phospholipase C β1 inhibits differentiation of erythroleukemia cells. Cancer Res. 58, 5057–5060 [PubMed] [Google Scholar]
- Matzke A. J. M., Weiger T. M., Matzke M. (2010). Ion channels at the nucleus: electrophysiology meets the genome. Mol. Plant 3, 642–652 10.1093/mp/ssq013 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mei Y., Jia W.-J., Chu Y.-J., Xue H.-W. (2011). Arabidopsis phosphatidylinositol monophosphate 5-kinase 2 is involved in root gravitropism through regulation of polar auxin transport by affecting the cycling of PIN proteins. Cell Res. 22, 581–597 10.1038/cr.2011.150 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meier I., Brkljacic J. (2009). Adding pieces to the puzzling plant nuclear envelope. Curr. Opin. Plant Biol. 12, 752–759 10.1016/j.pbi.2008.09.001 [DOI] [PubMed] [Google Scholar]
- Meijer H. J. G., Munnik T. (2003). Phospholipid-based signaling in plants. Annu. Rev. Plant Biol. 54, 265–306 10.1146/annurev.arplant.54.031902.134748 [DOI] [PubMed] [Google Scholar]
- Mellman D. L., Anderson R. A. (2009). A novel gene expression pathway regulated by nuclear phosphoinositides. Adv. Enzyme Regul. 49, 11–28 10.1016/j.advenzreg.2009.01.007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mellman D. L., Gonzales M. L., Song C., Barlow C. A., Wang P., Kendziorski C., Anderson R. A. (2008). A PtdIns4,5P2-regulated nuclear poly(A) polymerase controls expression of select mRNAs. Nature 451, 1013–1017 10.1038/nature06666 [DOI] [PubMed] [Google Scholar]
- Meng P. H., Raynaud C., Tcherkez G., Blanchet S., Massoud K., Domenichini S., Henry Y., Soubigou-Taconnat L., Lelarge-Trouverie C., Saindrenan P., Renou J. P., Bergounioux C. (2009). Crosstalks between myo-inositol metabolism, programmed cell death and basal immunity in Arabidopsis. PLoS ONE 4, e7364 10.1371/journal.pone.0007364 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Minasbekyan L. A., Yavroyan Z. V., Darbinyan M. R., Vardevanyan P. O. (2004). The Phospholipid composition of nuclear subfractions from germinating wheat embryos. Russ. J. Plant Physiol. 51, 708–712 10.1023/B:RUPP.0000040760.29704.db [DOI] [Google Scholar]
- Minasbekyan L. A., Yavroyan Z. V., Darbinyan M. R., Vardevanyan P. O. (2008). Changes in the composition of phospholipids in nuclear subfractions of wheat seedlings treated with gibberellin. Russ. J. Plant Physiol. 55, 372–377 10.1134/S1021443708030138 [DOI] [Google Scholar]
- Mishkind M., Vermeer J. E. M., Darwish E., Munnik T. (2009). Heat stress activates phospholipase D and triggers PIP accumulation at the plasma membrane and nucleus. Plant J. 60, 10–21 10.1111/j.1365-313X.2009.03933.x [DOI] [PubMed] [Google Scholar]
- Monserrate J. P., York J. D. (2010). Inositol phosphate synthesis and the nuclear processes they affect. Curr. Opin. Cell Biol. 22, 365–373 10.1016/j.ceb.2010.03.006 [DOI] [PubMed] [Google Scholar]
- Mosblech A., König S., Stenzel I., Grzeganek P., Feussner I., Heilmann I. (2008). Phosphoinositide and inositolpolyphosphate signalling in defense responses of Arabidopsis thaliana challenged by mechanical wounding. Mol. Plant 1, 249–261 10.1093/mp/ssm028 [DOI] [PubMed] [Google Scholar]
- Mosblech A., Thurow C., Gatz C., Feussner I., Heilmann I. (2011). Jasmonic acid perception by COI1 involves inositol polyphosphates in Arabidopsis thaliana. Plant J. 65, 949–957 10.1111/j.1365-313X.2011.04480.x [DOI] [PubMed] [Google Scholar]
- Mueller-Roeber B., Pical C. (2002). Inositol phospholipid metabolism in Arabidopsis. Characterized and putative isoforms of inositol phospholipid kinase and phosphoinositide-specific phospholipase C. Plant Physiol. 130, 22–46 10.1104/pp.004770 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Munnik T., Nielsen E. (2011). Green light for polyphosphoinositide signals in plants. Curr. Opin. Plant Biol. 14, 489–497 10.1016/j.pbi.2011.06.007 [DOI] [PubMed] [Google Scholar]
- Murphy A. M., Otto B., Brearley C. A., Carr J. P., Hanke D. E. (2008). A role for inositol hexakisphosphate in the maintenance of basal resistance to plant pathogens. Plant J. 56, 638–652 10.1111/j.1365-313X.2008.03629.x [DOI] [PubMed] [Google Scholar]
- Ndamukong I., Jones D. R., Lapko H., Divecha N., Avramova Z. (2010). Phosphatidylinositol 5-phosphate links dehydration stress to the activity of ARABIDOPSIS TRITHORAX-LIKE factor ATX1. PLoS ONE 5, e13396. 10.1371/journal.pone.0013396 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Odom A. R., Stahlberg A., Wente S. R., York J. D. (2000). A role for nuclear inositol 1,4,5-trisphosphate kinase in transcriptional control. Science 287, 2026–2029 10.1126/science.287.5460.2026 [DOI] [PubMed] [Google Scholar]
- Ogiso H., Nakamura K., Yatomi Y., Shimizu T., Taguchi R. (2010). Liquid chromatography/mass spectrometry analysis revealing preferential occurrence of non-arachidonate-containing phosphatidylinositol bisphosphate species in nuclei and changes in their levels during cell cycle. Rapid Commun. Mass Spectrom. 24, 436–442 10.1002/rcm.4415 [DOI] [PubMed] [Google Scholar]
- Osborne S. L., Thomas C. L., Gschmeissner S., Schiavo G. (2001). Nuclear PtdIns(4,5)P2 assembles in a mitotically regulated particle involved in pre-mRNA splicing. J. Cell. Sci. 114, 2501–2511 [DOI] [PubMed] [Google Scholar]
- Pauly N., Knight M. R., Thuleau P., van der Luit A. H., Moreau M., Trewavas A. J., Ranjeva R., Mazars C. (2000). Cell signalling: control of free calcium in plant cell nuclei. Nature 405, 754–755 10.1038/35015671 [DOI] [PubMed] [Google Scholar]
- Payrastre B., Nievers M., Boonstra J., Breton M., Verkleij A. J., Van Bergen en Henegouwen P. M. (1992). A differential location of phosphoinositide kinases, diacylglycerol kinase, and phospholipase C in the nuclear matrix. J. Biol. Chem. 267, 5078–5084 [PubMed] [Google Scholar]
- Perera I. Y., Hung C.-Y., Moore C. D., Stevenson-Paulik J., Boss W. F. (2008). Transgenic Arabidopsis plants expressing the type 1 inositol 5-phosphatase exhibit increased drought tolerance and altered abscisic acid signaling. Plant Cell 20, 2876–2893 10.1105/tpc.108.061374 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pfaffmann H., Hartmann E., Brightman A. O., Morré D. J. (1987). Phosphatidylinositol specific phospholipase C of plant stems 1. Plant Physiol. 85, 1151–1155 10.1104/pp.85.4.1151 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Philipp E. I., Franke W. W., Keenan T. W., Stadler J., Jarasch E. D. (1976). Characterization of nuclear membranes and endoplasmic reticulum isolated from plant tissue. J. Cell Biol. 68, 11–29 10.1083/jcb.68.1.11 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Poltoratsky V., Shi X., York J., Lieber M., Carter T. (1995). Human DNA-activated protein kinase (DNA-PK) is homologous to phosphatidylinositol kinases. J. Immunol. 155, 4529–4533 [PubMed] [Google Scholar]
- Pribat A., Sormani R., Rousseau-Gueutin M., Julkowska M. M., Testerink C., Joubès J., Castroviejo M., Laguerre M., Meyer C., Germain V., Rothan C. (2012). A novel class of PTEN protein in Arabidopsis displays unusual phosphoinositide phosphatase activity and efficiently binds phosphatidic acid. Biochem. J. 441, 161–171 10.1042/BJ20110776 [DOI] [PubMed] [Google Scholar]
- Qin Z.-X., Chen Q.-J., Tong Z., Wang X.-C. (2005). The Arabidopsis inositol 1,3,4-trisphosphate 5/6 kinase, AtItpk-1, is involved in plant photomorphogenesis under red light conditions, possibly via interaction with COP9 signalosome. Plant Physiol. Biochem. 43, 947–954 10.1016/j.plaphy.2005.08.006 [DOI] [PubMed] [Google Scholar]
- Raben D. M., Jarpe M. B., Leach K. L. (1994). Nuclear lipid metabolism in NEST: nuclear envelope signal transduction. J. Membr. Biol. 142, 1–7 [DOI] [PubMed] [Google Scholar]
- Ramazzotti G., Faenza I., Fiume R., Matteucci A., Piazzi M., Follo M. Y., Cocco L. (2011). The physiology and pathology of inositide signaling in the nucleus. J. Cell. Physiol. 226, 14–20 10.1002/jcp.22334 [DOI] [PubMed] [Google Scholar]
- Rando O. J., Zhao K., Janmey P., Crabtree G. R. (2002). Phosphatidylinositol-dependent actin filament binding by the SWI/SNF-like BAF chromatin remodeling complex. Proc. Natl. Acad. Sci. U.S.A. 99, 2824–2829 10.1073/pnas.032662899 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rose A., Patel S., Meier I. (2004). The plant nuclear envelope. Planta 218, 327–336 10.1007/s00425-003-1162-9 [DOI] [PubMed] [Google Scholar]
- Rose H. G., Frenster J. H. (1965). Composition and metabolism of lipids within repressed and active chromatin of interphase lymphocytes. Biochim. Biophys. Acta 106, 577–591 [DOI] [PubMed] [Google Scholar]
- Schmitt J., Benavente R., Hodzic D., Höög C., Stewart C. L., Alsheimer M. (2007). Transmembrane protein Sun2 is involved in tethering mammalian meiotic telomeres to the nuclear envelope. Proc. Natl. Acad. Sci. U.S.A. 104, 7426–7431 10.1073/pnas.0609198104 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schneiter R., BrüGger B., Amann C. M., Prestwich G. D., Epand R. F., Zellnig G., Wieland F. T., Epand R. M. (2004). Identification and biophysical characterization of a very-long-chain-fatty-acid-substituted phosphatidylinositol in yeast subcellular membranes. Biochem. J. 381, 941. 10.1042/BJ20040320 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seeds A., Frederick J., Tsui M., York J. (2007). Roles for inositol polyphosphate kinases in the regulation of nuclear processes and developmental biology. Adv. Enzyme Regul. 47, 10–25 10.1016/j.advenzreg.2006.12.019 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sheard L. B., Tan X., Mao H., Withers J., Ben-Nissan G., Hinds T. R., Kobayashi Y., Hsu F.-F., Sharon M., Browse J., He S. Y., Rizo J., Howe G. A., Zheng N. (2010). Jasmonate perception by inositol-phosphate-potentiated COI1-JAZ co-receptor. Nature 468, 400–405 10.1038/nature09430 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shen X., Xiao H., Ranallo R., Wu W.-H., Wu C. (2003). Modulation of ATP-dependent chromatin-remodeling complexes by inositol polyphosphates. Science 299, 112–114 10.1126/science.1078068 [DOI] [PubMed] [Google Scholar]
- Shigaki T., Bhattacharyya M. K. (2002). Nutrients induce an increase in inositol 1,4,5-trisphosphate in soybean cells: implication for the involvement of phosphoinositide-specific phospholipase C in DNA synthesis. Plant Biol. 4, 53–61 10.1055/s-2002-20436 [DOI] [Google Scholar]
- Shoji-Kawaguchi M., Izuta S., Tamiya-Koizumi K., Suzuki M., Yoshida S. (1995). Selective inhibition of DNA polymerase ε by phosphatidylinositol. J. Biochem. 117, 1095–1099 [DOI] [PubMed] [Google Scholar]
- Slomiany A., Slomiany B. L. (2011). Transformations of Phosphatidylinositol Phosphates in the Outer and Inner Nuclear Membrane are Linked to Synthesis and Restitution of Cellular Membranes. (Report). Health. Available at: http://www.highbeam.com/doc/1G1-265195831.html [accessed December 20, 2011].
- Smogorzewska A., de Lange T. (2004). Regulation of telomerase by telomeric proteins. Annu. Rev. Biochem. 73, 177–208 10.1146/annurev.biochem.73.071403.160049 [DOI] [PubMed] [Google Scholar]
- Song M. S., Carracedo A., Salmena L., Song S. J., Egia A., Malumbres M., Pandolfi P. P. (2011). Nuclear PTEN regulates the APC-CDH1 tumor-suppressive complex in a phosphatase-independent manner. Cell 144, 187–199 10.1016/j.cell.2010.12.020 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Steger D. J., Haswell E. S., Miller A. L., Wente S. R., O’Shea E. K. (2003). Regulation of chromatin remodeling by inositol polyphosphates. Science 299, 114–116 10.1126/science.1078062 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stenzel I., Ischebeck T., König S., Holubowska A., Sporysz M., Hause B., Heilmann I. (2008). The type B phosphatidylinositol-4-phosphate 5-kinase 3 is essential for root hair formation in Arabidopsis thaliana. Plant Cell 20, 124–141 10.1105/tpc.107.052852 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stevenson-Paulik J., Bastidas R. J., Chiou S.-T., Frye R. A., York J. D. (2005). Generation of phytate-free seeds in Arabidopsis through disruption of inositol polyphosphate kinases. Proc. Natl. Acad. Sci. U.S.A. 102, 12612–12617 10.1073/pnas.0504172102 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stevenson-Paulik J., Odom A. R., York J. D. (2002). Molecular and biochemical characterization of two plant inositol polyphosphate 6-/3-/5-kinases. J. Biol. Chem. 277, 42711–42718 10.1074/jbc.M209112200 [DOI] [PubMed] [Google Scholar]
- Sun B., Murray N. R., Fields A. P. (1997). A role for nuclear phosphatidylinositol-specific phospholipase C in the G2/M phase transition. J. Biol. Chem. 272, 26313–26317 10.1074/jbc.272.50.31845 [DOI] [PubMed] [Google Scholar]
- Sylvia V., Curtin G., Norman J., Stec J., Busbee D. (1988). Activation of a low specific activity form of DNA polymerase α by inositol-1,4-bisphosphate. Cell 54, 651–658 10.1016/S0092-8674(88)80009-6 [DOI] [PubMed] [Google Scholar]
- Tan X., Calderon-Villalobos L. I. A., Sharon M., Zheng C., Robinson C. V., Estelle M., Zheng N. (2007). Mechanism of auxin perception by the TIR1 ubiquitin ligase. Nature 446, 640–645 10.1038/nature05731 [DOI] [PubMed] [Google Scholar]
- Tsui M. M., York J. D. (2010). Roles of inositol phosphates and inositol pyrophosphates in development, cell signaling and nuclear processes. Adv. Enzyme Regul. 50, 324–337 10.1016/j.advenzreg.2009.12.002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Valluru R., Van den Ende W. (2011). Myo-inositol and beyond – emerging networks under stress. Plant Sci. 181, 387–400 10.1016/j.plantsci.2011.07.009 [DOI] [PubMed] [Google Scholar]
- Vaultier M.-N., Cantrel C., Guerbette F., Boutté Y., Vergnolle C., Çiçek D., Bolte S., Zachowski A., Ruelland E. (2008). The hydrophobic segment of Arabidopsis thaliana cluster I diacylglycerol kinases is sufficient to target the proteins to cell membranes. FEBS Lett. 582, 1743–1748 10.1016/j.febslet.2008.04.042 [DOI] [PubMed] [Google Scholar]
- Vermeer J. E. M., van Leeuwen W., Tobeña-Santamaria R., Laxalt A. M., Jones D. R., Divecha N., Gadella T. W. J., Jr., Munnik T. (2006). Visualization of PtdIns3P dynamics in living plant cells. Plant J. 47, 687–700 10.1111/j.1365-313X.2006.02830.x [DOI] [PubMed] [Google Scholar]
- Vespa L., Couvillion M., Spangler E., Shippen D. E. (2005). ATM and ATR make distinct contributions to chromosome end protection and the maintenance of telomeric DNA in Arabidopsis. Genes Dev. 19, 2111–2115 10.1101/gad.1333805 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weirich C. S., Erzberger J. P., Flick J. S., Berger J. M., Thorner J., Weis K. (2006). Activation of the DExD/H-box protein Dbp5 by the nuclear-pore protein Gle1 and its coactivator InsP6 is required for mRNA export. Nat. Cell Biol. 8, 668–676 10.1038/ncb1424 [DOI] [PubMed] [Google Scholar]
- Wissing J. B., Wagner K. G. (1992). Diacylglycerol kinase from suspension cultured plant cells. Plant Physiol. 98, 1148–1153 10.1104/pp.98.3.1148 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xia H.-J., Brearley C., Elge S., Kaplan B., Fromm H., Mueller-Roeber B. (2003). Arabidopsis inositol polyphosphate 6-/3-kinase is a nuclear protein that complements a yeast mutant lacking a functional ArgR-Mcm1 transcription complex. Plant Cell 15, 449–463 10.1105/tpc.006676 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu J., Brearley C. A., Lin W.-H., Wang Y., Ye R., Mueller-Roeber B., Xu Z.-H., Xue H.-W. (2005). A role of Arabidopsis inositol polyphosphate kinase, AtIPK2α, in pollen germination and root growth. Plant Physiol. 137, 94–103 10.1104/pp.104.045427 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang L., Tang R., Zhu J., Liu H., Mueller-Roeber B., Xia H., Zhang H. (2008). Enhancement of stress tolerance in transgenic tobacco plants constitutively expressing AtIpk2beta, an inositol polyphosphate 6-/3-kinase from Arabidopsis thaliana. Plant Mol. Biol. 66, 329–343 10.1007/s11103-007-9267-3 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yorek M. A., Dunlap J. A., Manzo-Fontes A., Bianchi R., Berry G. T., Eichberg J. (1999). Abnormal myo-inositol and phospholipid metabolism in cultured fibroblasts from patients with ataxia telangiectasia. Biochim. Biophys. Acta 1437, 287–300 [DOI] [PubMed] [Google Scholar]
- York J. D. (2006). Regulation of nuclear processes by inositol polyphosphates. Biochim. Biophys. Acta 1761, 552–559 [DOI] [PubMed] [Google Scholar]
- York J. D., Odom A. R., Murphy R., Ives E. B., Wente S. R. (1999). A phospholipase C-dependent inositol polyphosphate kinase pathway required for efficient messenger RNA export. Science 285, 96–100 10.1126/science.285.5424.96 [DOI] [PubMed] [Google Scholar]
- York S. J., Armbruster B. N., Greenwell P., Petes T. D., York J. D. (2005). Inositol diphosphate signaling regulates telomere length. J. Biol. Chem. 280, 4264–4269 10.1074/jbc.M412070200 [DOI] [PubMed] [Google Scholar]
- Yu H., Fukami K., Watanabe Y., Ozaki C., Takenawa T. (1998). Phosphatidylinositol 4,5-bisphosphate reverses the inhibition of RNA transcription caused by histone H1. Eur. J. Biochem. 251, 281–287 10.1046/j.1432-1327.1998.2510281.x [DOI] [PubMed] [Google Scholar]
- Zhao K., Wang W., Rando O. J., Xue Y., Swiderek K., Kuo A., Crabtree G. R. (1998). Rapid and phosphoinositol-dependent binding of the SWI/SNF-like BAF complex to chromatin after T lymphocyte receptor signaling. Cell 95, 625–636 10.1016/S0092-8674(00)81633-5 [DOI] [PubMed] [Google Scholar]
- Zhendre V., Grélard A., Garnier-Lhomme M., Buchoux S., Larijani B., Dufourc E. J. (2011). Key role of polyphosphoinositides in dynamics of fusogenic nuclear membrane vesicles. PLoS ONE 6, e23859. 10.1371/journal.pone.0023859 [DOI] [PMC free article] [PubMed] [Google Scholar]