Abstract
Potato yellow dwarf virus (PYDV) is the type species of the genus Nucleorhabdovirus and, like all members of this genus, replication and morphogenesis occurs inside the nuclei of infected cells. Protein localization prediction algorithms failed to identify a nuclear localization signal (NLS) in PYDV nucleocapsid (N) protein, although PYDV-N has been shown to localize exclusively to the nucleus when expressed as a green fluorescent protein (GFP):N fusion in plant cells. Deletion analysis using fragments of PYDV-N identified a karyophilic region in the carboxy-terminal 122 amino acids. Alanine-scanning mutagenesis was performed across this region in the context of the full-length N protein. Mutants were assayed for their ability to nuclear localize using live-cell imaging and a yeast-based assay. Two amino acid motifs, 419QKR421 and 432KR433 were shown to be essential for nuclear import and interaction with importin-α. Additional bimolecular fluorescence complementation showed that the PYDV-N-NLS mutants cannot be ferried into the nucleus via interaction with PYDV-P or -M. In contrast, interaction with N-NLS mutants appeared to retard the nuclear import of PYDV-P. GFP fused to aa 419–434 established that the PYDV-N-NLS can function outside the context of this protein. Taken together, it was determined that PYDV-N contains the bipartite NLS 419QKRANEEAPPAAQKR433.
Keywords: rhabdovirus, importin-α, plant nucleus, BiFC, plant virus
Introduction
Rhabdoviruses are structurally complex enveloped viruses, the plant-adapted members of which are separated into the genera Cytorhabdovirus, which replicate in the cytoplasm of infected cells, and Nucleorhabdovirus, the viruses considered here, which replicate in nuclei of infected cells. The minimal infectious unit of these viruses is a nucleocapsid composed of viral genomic single-stranded negative-sense RNA encapsidated over its entire length by a nucleocapsid (N) protein and sub-molar amounts of an RNA-dependent RNA polymerase composed of a large (L) protein and a phosphoprotein (P; Goodin et al., 2001; Jackson et al., 2005; Ammar el et al., 2009). The N proteins of viruses belonging to the genus Nucleorhabdovirus contain nuclear localization signals (NLSs) that not only direct the protein itself to the nucleus but also the viral nucleocapsid, which is required to establish sites of replication and viral assembly in nuclei of infected plant cells (Wagner et al., 1996; Wagner and Jackson, 1997; Jackson et al., 2005; Kuzmin et al., 2009). In this regard, the N protein-mediated transport of nucleocapsids parallels the role of VirE2 in the transfer of transfer DNA (T-DNA) from Agrobacterium tumefaciens to nuclei of plant cells to which it is attached. In broader terms, the nuclear transport and subsequent cell-to-cell movement of rhabdoviral nucleocapsids provides insight into the general macromolecular trafficking in plant cells. As such the identification and mapping of NLSs in N proteins is critically required to understand the mechanism by which these proteins, and their ribonucleoprotein complexes are targeted to the nuclei (Citovsky et al., 1992, 2004; Deng et al., 2007).
In this study, we report the characterization of the NLS responsible for nuclear targeting of the nucleocapid protein of Potato yellow dwarf virus (PYDV), which is the type species of the nucleorhabdovirus genus (Hsu and Black, 1973, 1974; Bandyopadhyay et al., 2010). Nucleocapsids of these insect-vectored viruses are transported to the nucleus where they establish sites of replication known as viroplasms. These viroplasms are spatially separated from the sites of virion assembly, which takes place on the inner nuclear membrane, with condensed nucleocapsids accumulating in the perinuclear space of infected cells (Goodin et al., 2001, 2007; Martin et al., 2009). In order to initiate new rounds of the infection cycle, nucleocapsids must exit the nucleus, via a mechanism that is thought to involve multiple host factors, microtubules, and membranes (Min et al., 2010).
We have shown previously that despite the fact that protein localization prediction algorithms failed to identify an NLS in PYDV-N, this protein is fully capable of directing Green fluorescent protein (GFP) exclusively to the nucleus in plant cells expressing GFP:PYDV-N fusions (Bandyopadhyay et al., 2010). Additionally, we have shown that PYDV-N interacts with one of two isoforms of the karyophilic transporter, importin-α, in plant cells (Martin et al., 2009). Finally, we have shown that PYDV-N interacts with the nuclear localized matrix (M) and P proteins of PYDV, thus raising the possibility that these two proteins may in fact play a role in facilitating the nuclear transport of N (Bandyopadhyay et al., 2010). While these studies contribute to an understanding of plant–virus interactions, the identification of novel NLSs aids the categorization of the nuclear transport signals that are able to function in plants. As such, this provides a means to improve predictive algorithms for identifying plant proteins associated with nuclei (Kosugi et al., 2009).
Materials and Methods
Plant materials and virus maintenance
All plants, including transgenic marker lines, were maintained in greenhouse on open benches under ambient conditions (Martin et al., 2009). PYDV (American Type Culture Collection accession PV-234) was maintained by serial passage in Nicotiana benthamiana and N. rustica in insect-proof cages under ambient conditions.
Generation of site-directed mutants
Site-directed mutagenesis was performed using the Change-IT™ Multiple Mutation Site-Directed Mutagenesis Kit (USB) according to protocols provided by the manufacturer. All mutagenesis was conducted using sequence-validated portions of the PYDV-N open reading frame cloned into plasmid pDONR221 (Invitrogen). Mutagenized clones were validated by DNA sequencing to confirm the presence of specific mutations.
Protein expression in plant cells
Expression of autofluorescent protein fusions in plant cells for localization and bimolecular fluorescence complementation (BiFC) assays, were performed as described previously (Chakrabarty et al., 2007; Goodin et al., 2007; Martin et al., 2009). In this study, the vector pSITE-2C (GFP-fusion) was used for localization assays, and pSITE–BiFC–nEYFP and pSITE–BiFC–cEYFP vectors for BiFC assays. Recombinant vectors were transformed into A. tumefaciens strain LBA4404. Agroinfiltration for expression of protein fusions in plant cells was conducted as described previously (Goodin et al., 2002). Each expression construct was examined in 25 mm2 leaf pieces taken from a minimum of two leaves, from a minimum of two independent plants. Multiple micrographs representative of several hundred examined cells were acquired for documentation.
Laser-scanning confocal microscopy
All microscopy was performed on an Olympus FV1000 laser-scanning confocal microscope as described previously (Goodin et al., 2005). Micrographs for dual-color imaging were acquired sequentially, as described in Goodin et al. (2007).
Nuclear import assays in yeast
Sequence-validated clones in pDONR221 (Invitrogen) were recombined into pNIA–DEST plasmids (Bandyopadhyay et al., 2010). Recombinant pNIA–DEST plasmids were transformed into Saccharomyces cerevisiae strain L40 (Zaltsman et al., 2007). Transformed yeast were grown at 30°C for 3 days, on minimal media lacking tryptophan (Trp−). Colonies were then re-streaked onto minimal media lacking both tryptophan and histidine (His−), containing 5 mM 3-amino-1,2,4-triazole (3AT). Growth of yeast cultures on Trp−/His− media indicates a functional NLS in proteins expressed from pNIA–DEST (Zaltsman et al., 2007).
Results
A NLS is present in the C-terminus of PYDV-N
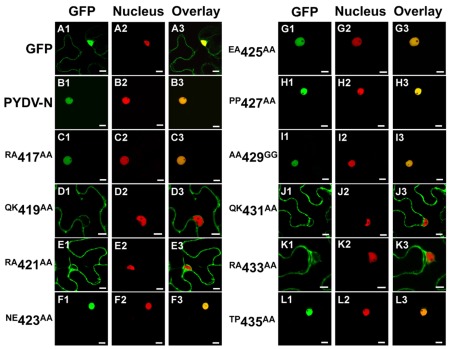
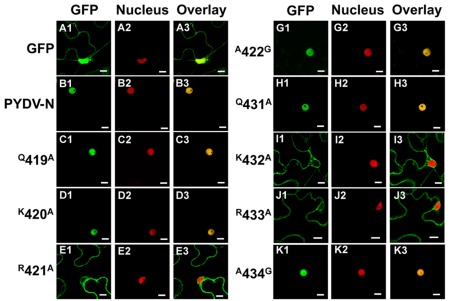
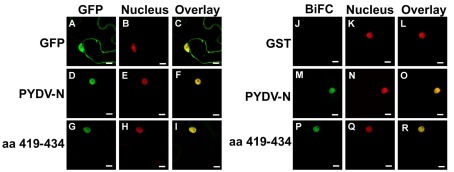
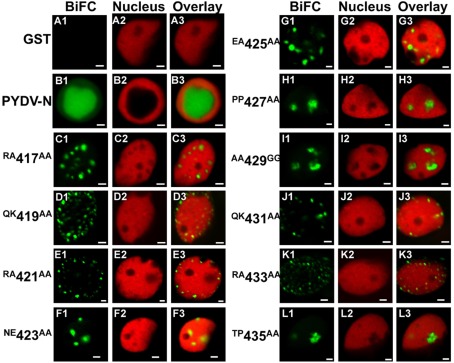
We have reported previously that PYDV-N localizes exclusively to nuclei when expressed as a GFP-fusion in transgenic N. benthamiana, despite the absence of a predictable NLS (Bandyopadhyay et al., 2010). To determine the karyophilic region of PYDV-N we expressed six overlapping fragments of PYDV-N as GFP-fusions, and determined their subcellular localization in planta. Fragments 1–5 were found throughout the cell or excluded from the nucleus, whereas the C-terminal Fragment 6 (aa 350–473) was found to localize exclusively in the nucleus (data not shown). In order to define the location of the NLS in PYDV-N we used alanine-scanning mutagenesis in the context of full-length N protein. A subset of these mutations, shown in Figure 1, demonstrated the location of the PYDV-N-NLS between amino acids 419–434. Scanning mutagenesis upstream or downstream of this region did not influence nuclear localization. In particular, two QKRA repeats (419QKRA422 and 431QKRA434) appeared to be required for NLS function (Figures 1D,E,J,K), whereas the eight amino acid residues separating them could be mutated without affecting nuclear localization (Figures 1F–I). Fine mapping using single amino acid mutations within the QKRA motifs (Figure 2), revealed that amino acids R421, K432, and R433 are all required for NLS function (Figures 2E,I,J).
Figure 1.
Single-plane confocal micrographs of GFP: protein fusions expressed transiently in leaf epidermal cells of N. benthamiana plants transgenic for the nuclear marker, RFP: histone 2B. Shown in panels 1, 2, and 3 are micrographs of GFP (fusions), RFP, and the resultant overlay, respectively. (A1–A3) GFP only. (B1–B3) PYDV-N. (C–L) PYDV-N site-directed double mutants. Numbers indicate the amino acid residue in the full-length N protein where a two-residue change to alanine was introduced. Changes to glycine were made in cases where the residue was alanine in the native protein. Scale bar = 10 μm.
Figure 2.
Single-plane confocal micrographs of protein fusions expressed in leaf epidermal cells of transgenic N. benthamiana plants expressing RFP fused to a nuclear marker, histone 2B. Shown in panels 1, 2, and 3 are micrographs of GFP (fusions), RFP, and the resultant overlay, respectively. (A1–A3) GFP only. (B1–B3) GFP:PYDV-N. (C–K) PYDV-N site-directed single mutants. Numbers indicate the amino acid residue in the full-length N protein where a single residue change to alanine was introduced. In cases where the residue was alanine in the native protein, changes to glycine were made. Scale bar = 10 μm.
Interaction of PYDV-N mutants with importin-α
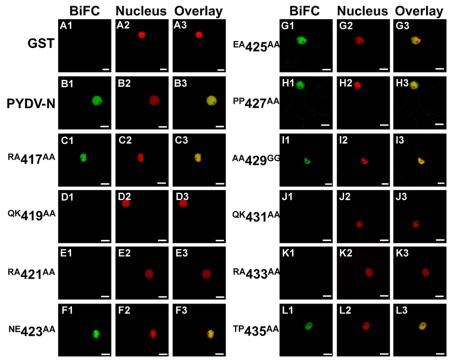
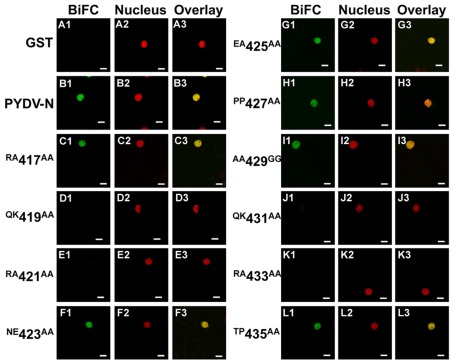
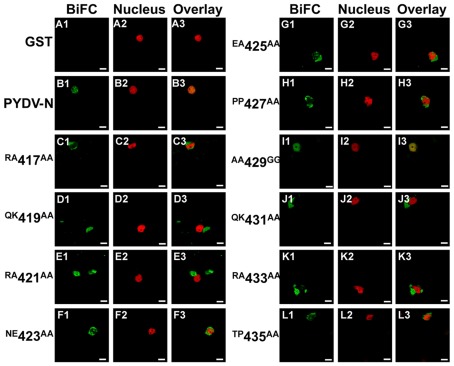
The initial mutagenesis and protein localization revealed two QKRA repeats that appeared to be required for nuclear localization of PYDV-N. This arrangement of amino acids, though not predicted by protein localization algorithms, were reminiscent of bipartite NLSs that interact with the nuclear import receptor importin-α, two isoforms of which are known to exist in N. benthamiana (Kanneganti et al., 2007). We have shown previously that PYDV-N interacts preferentially with N. benthmiana importin-α1 (Nbimportin-α1; Martin et al., 2009). Therefore, to investigate the role of the QKRA motifs on NLS function, we conducted BiFC assays to test the interaction of PYDV-N mutants and Nbimportin-α1. (Figure 3). GST was used as a negative-binding control as it has no interaction with Nbimportin-α1 (Figures 3A1–A3). Conversely, wild-typed PYDV-N interacted strongly with Nbimportin-α1 (Figure 3B). However, PYDV-N mutants that failed to localize to the nucleus (see Figures 1 and 2) also failed to interact with Nbimportin-α1 (Figures 3D,E,J,K). In addition, the mutants that were still localized to the nucleus when expressed in planta as GFP-fusions interacted with Nbimportin-α1 in the nucleus (Figures 3C,F–I,L).
Figure 3.
Confocal micrographs showing results of BiFC assays to determine interactions between importin-α and PYDV-N mutants. Interaction assays were conducted in leaf epidermal cells of transgenic N. benthamiana expressing fluorescent nuclear marker protein fusion. Panels 1, 2, and 3 show micrographs of YFP (BiFC), nuclear marker (Nucleus), and the resultant overlay, respectively. (A1–A3) Non-binding control GST. (B1–B3) BiFC between Importin-α and PYDV-N. (C–L) BiFC between Importin-α and double mutants of PYDV-N, described in Figure 1. Scale bar = 10 μm.
Nuclear import of PYDV-N mutants is confirmed by a yeast-based assay
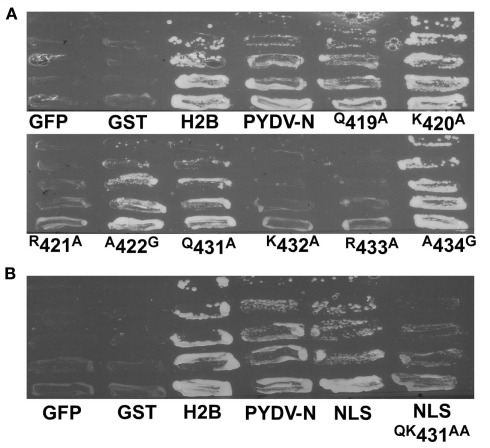
To confirm the in planta localization data for PYDV-N and mutants thereof, a yeast-based nuclear import assay (NIA) was employed (Figure 4). In this assay, only proteins containing a functional NLS will facilitate the nuclear import of a transcriptional activator required for expression of a reporter gene in yeast cells (Zaltsman et al., 2007). Consistent with the in planta localization patterns, PYDV-N, and PYDV-N mutants Q419A, K420A, A422G, Q431A, and A434G, were positive in the NIA. PYDV-N mutants R421A, K432A, and R433A were NIA-negative. In addition, the potential PYDV-N-NLS, aa 419–434, was NIA-positive, whereas the same sequence containing the QK431–432AA double mutant was NIA-negative.
Figure 4.
Yeast-based assay for identification of proteins containing a functional NLS. (A) Positive [histone 2B (H2B)] and negative (GFP and GST) control proteins along with PYDV proteins (N, and site-directed N mutants) were expressed from pNIA–DEST in yeast strain L40. Only proteins containing a functional NLS are able to facilitate yeast growth on media lacking histidine. (B) Localization of the karyophilic domain of PYDV-N (aa 419–434; NLS) fused to NIA transcriptional activator. Introducing the QK431AA mutation into this domain abolished nuclear import activity.
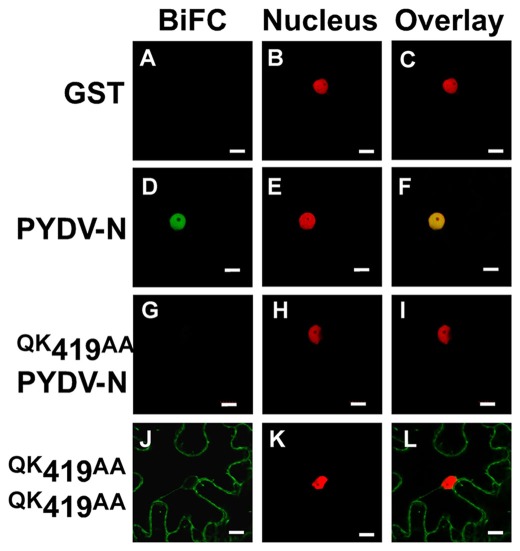
The region spanning aa 419–434 of PYDV-N is sufficient to direct nuclear localization
In order to test function of the PYDV-N-NLS outside the context of the full-length protein, aa 419–434 of PYDV-N were expressed as a fusion to GFP in plant cells. The addition of the PYDV-N aa 419–434 was able to localize GFP exclusively to the nucleus (Figures 5A–I). In addition, when BiFC was performed between PYDV-N aa 419–434 and Nbimportin-α1, they were shown to interact and the interaction was localized to the nucleus (Figures 5J–R).
Figure 5.
Confocal micrographs of protein fusions expressed in leaf epidermal cells of transgenic N. benthamiana plants expressing a fluorescent a nuclear marker (Nucleus). (A–I) Show localization of GFP-fusions while (J–R) show results of BiFC assays to test interactions with importin-α. (A–C) Localization of GFP, nuclear marker, and overlay, respectively of GFP. (D–F) Localization of full-length PYDV-N fused to GFP. (G–I) Localization of the karyophilic domain of PYDV-N (aa 419–434) fused to GFP. (J–L) BiFC interaction of GST and importin-α. (M–O) BiFC interaction of PYDV-N and importin-α. (P–R) BiFC interaction of aa 419–434 PYDV-N and importin-α. Scale bar = 10 μm.
The PYDV-N–N self-interaction may occur after nuclear import
It has previously been shown that PYDV-N interacts with itself, PYDV-P, and PYDV-M in the nucleus (Bandyopadhyay et al., 2010). Such interactions may occur prior to or following nuclear import. To investigate if a PYDV-N containing a functional NLS could interact with and facilitate the nuclear import of NLS mutants, BiFC was performed between PYDV-N and N mutants (Figure 6). It was found that the mutants that retained the ability to be imported into nuclei (Figures 6C,F–I,L) were also capable of interaction with PYDV-N in the nucleus. In contrast, mutants containing a non-functional NLS did not interact with PYDV-N (Figures 6D,E,J,K).
Figure 6.
Confocal micrographs of BiFC assays testing the ability of PYDV-N mutant proteins to associate with the wild-type N protein. Interaction assays were conducted in leaf epidermal cells of transgenic N. benthamiana expressing a fluorescent nuclear marker (Nucleus). Panels 1, 2, and 3 show micrographs of YFP (BiFC), nuclear marker, and the resultant overlay, respectively. Shown here are BiFC assays conducted with wild-type PYDV-N expressed as a fusion to the carboxy-terminal portion of YFP and N mutants expressed as fusions to the amino-terminal fragment of YFP. (A) GST, used as a non-binding control. (B) Wild-type PYDV-N. (C–L) BiFC between PYDV-N and site-directed double mutants of PYDV-N shown in Figure 1. Scale bar = 10 μm.
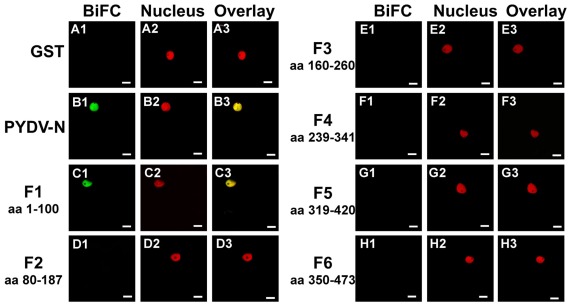
To determine if the absence of interaction between PYDV-N and mutants QK419–420AA, RA421–422AA, QK431–432AA, and RA433–434AA, was due to a disruption of the N-interacting domain, BiFC was performed to test self-interaction of these mutants. This showed that although these mutants no longer interact with wild-type PYDV-N, they are able to self-interact and interact with each other, and this interaction occurs outside of the nucleus (Figure 7). In addition, BiFC between PYDV-N and the overlapping fragments of PYDV-N showed that only the N-terminal fragment, Fragment 1, showed interaction with PYDV-N (Figure 8), suggesting that the N–N interaction domain is contained in this fragment. Taken together, these results suggest that the PYDV-N self-interaction occurs after nuclear import.
Figure 7.
Confocal micrographs showing results of BiFC assays to determine the ability to self-associate. Interaction assays were conducted in leaf epidermal cells of transgenic N. benthamiana expressing a fluorescent nuclear marker (Nucleus). (A–C) GST, as a non-binding control. (D–F) Self-interaction of wild-type PYDV-N. (G–I) BiFC between PYDV-N and an N mutant that is incapable of nuclear import. (J–L) Self-interaction of QK419AA mutant of PYDV-N. All N-NLS mutants were found to retain the ability to self-interact (data not shown). Scale bar = 10 μm.
Figure 8.
Confocal micrographs showing PYDV-N fragment interactions with PYDV-N determined by BiFC. Interaction assays were conducted in leaf epidermal cells of transgenic N. benthamiana expressing a fluorescent nuclear marker (Nucleus). Shown is the ability of fragments of PYDV-N to interact with wild-type N protein in BiFC assays. (A1–A3) GST. (B1–B3) Wild-type PYDV-N. (C–H) BiFC between PYDV-N and PYDV-N fragments F1–F6. The portions of PYDV-N used in the BiFC assays are indicated by the amino acid (aa) designation for each panel. Scale bar = 10 μm.
N-NLS mutants cannot be ferried into the nucleus by the PYDV phosphoprotein or matrix protein
Bimolecular fluorescence complementation assays using wild-type and NLS mutants of PYDV-N suggested that this protein does not enter the nucleus as homo-oligomeric complexes. To address the possibility that other interaction partners may play a role in the nuclear import of N, we conducted BiFC assays with the N mutants and PYDV-P or -M.
Assays with PYDV-P showed, contrary to expectation, that NLS mutants of N were capable of interacting with PYDV-P and in doing so retarded the nuclear import of PYDV-P (Figure 9). Mutants that were no longer localized to the nucleus as GFP-fusions expressed in plant cells (Figures 9D,E,J,K) showed interaction with PYDV-P completely outside of the nucleus.
Figure 9.
Confocal micrographs showing PYDV-N mutant protein interactions with PYDV-P determined by BiFC. Interaction assays were conducted in leaf epidermal cells of transgenic N. benthamiana expressing a fluorescent nuclear marker protein fusion. Panels 1, 2, and 3 show micrographs of YFP (BiFC), nuclear marker (Nucleus), and the resultant overlay, respectively. All interactions were tested against PYDV-P expressed as a fusion to the carboxy-terminal of YFP. (A1–A3) Non-binding control GST. (B1–B3) BiFC between PYDV-P and PYDV-N. (C–L) BiFC between PYDV-P and double mutants of PYDV-N. Scale bar = 10 μm.
In a similar fashion, N-NLS mutants retained their ability to interact with PYDV-M (Figure 10). N proteins that were competent for nuclear import interacted with N on intranuclear sites (Figures 10C,F–I,L). In contrast, N proteins with a non-functional NLS interacted with M at the periphery of nuclei and not in intranuclear loci (Figures 10D,E,J,K).
Figure 10.
Confocal micrographs showing PYDV-N mutant protein interactions with PYDV-M determined by BiFC. Interaction assays were conducted in leaf epidermal cells of transgenic N. benthamiana expressing a fluorescent nuclear marker (Nucleus). Panels 1, 2, and 3 show micrographs of YFP (BiFC), nuclear marker (Nucleus), and the resultant overlay, respectively. All interactions were tested against PYDV-M expressed as a fusion to the carboxy-terminal of YFP. (A1–A3) Non-binding control GST. (B1–B3) BiFC between PYDV-M and PYDV-N. (C–L) BiFC between PYDV-M and double mutants of PYDV-N. Scale bar = 2 μm.
Discussion
At present, determination of the subcellular localization of proteins for organisms whose genomes have been sequenced, is based most frequently on the use of predictive algorithms and, to a far lesser extent, actual protein localization data. The consequence of this is likely to be extreme underestimation in the number of proteins that are targeted to the nucleus as algorithms used to predict nuclear localization are designed primarily to look for basic, arginine/lysine-rich regions, typical of proteins imported into the nucleus by importin α/β-mediated pathways (Cokol et al., 2000). However, many novel non-basic NLSs have been identified, including the M9 and other non-canonical NLSs that are not detected using the most popular predictive algorithms (Mattaj and Englmeier, 1998; Kumar and Raghava, 2009). The situation becomes more complex if we consider the fact that no less than 10 different subnuclear loci, which have markedly different functions, have been identified (Spector, 2006). Localization to these loci is not predicted by publicly available algorithms, which are limited to predicting only nuclear or nucleolar localization (Nakai and Horton, 2007; Kumar and Raghava, 2009).
In the present study we have used protein interaction and in vivo localization to map and characterize the bipartite NLS of PYDV-N. Although this NLS could not be predicted by in silico methods, our in vivo analyses provide comprehensive evidence that the PYDV-N-NLS is located between the amino acids 419–433 of the mature N protein. Mutagenesis scanning employing double mutants revealed that repeated QKRA motifs were essential for nuclear localization. However, fine mapping where single amino acids were mutated showed that the most critical residues were R421, K432, and R433 located within the two QKR repeats. This apparent discrepancy might be due to greater perturbation of NLS function in the double mutants compared to the single mutations, which revealed only those residues that had the greatest impact on nuclear import and on binding to importin-α. Ultimately, the PYDV-N-NLS was determined to be 419QKRANEEAPPAAQKR433, which is generally consistent with the consensus KRX10–12K(K/R)(K/R) found in bipartite NLSs that function in plants (Kosugi et al., 2009). A key difference however is the lack of a third basic residue after the spacer region, suggesting that a basic amino acid at this site is not critical to the function of some NLSs. Importantly, although plants and yeast encode different numbers of importin-α isoforms, both in planta and yeast-based NIAs returned similar data thus further validating the utility of yeast to screen plant nuclear localized proteins (Zaltsman et al., 2007).
Comparison to other predicted or physically mapped NLSs in nucleorhabdoviral N proteins, for which sequence data are available, it is apparent that N-NLSs are found in all cases, thus far, near the carboxy terminus of these proteins (Table 1). The reason for this may be that, where mapped, the N–N interaction domain resides in the amino-terminus of these proteins (Goodin et al., 2001). Additionally, in the majority of cases, the N-NLSs are of the bipartite class. Importantly, the PYDV-N-NLS is not restricted to this virus but is also found in a plethora of Arabidopsis proteins with known or predicted functions in nuclei, suggesting that the QKR motif is an important class in plant NLS (Table 2).
Table 1.
Nuclear localization signals in nucleocapsid proteins of nucleorhabdoviruses.
| Virus | Type | NLS |
|---|---|---|
| MFSV | Bipartite | 436KRSSDGTGNVSKKKRK452 |
| SYNV | Bipartite | 465PSRKRRSDALTTEKPKK481 |
| RYSV | Bipartite | 405KKLGPPRANAHSRRKEP421 |
| MMV | Monopartite | 438PAPKKTR444 |
| IMMV | Monopartite | 432PRLRRGS438 |
| TaroVCV | Monopartite | 422PTKKRTW428 |
| PYDV | Bipartite | 419QKRANEEAPPAAQKR433 |
Only the SYNV and PYDV signals have been mapped physically, others have been predicted using in silico methods. The numbers indicate the amino acid residues in the full-length proteins that mark the NLSs. MFSV, maize fine streak virus; RYSV, rice yellow stunt virus; MMV, maize mosaic virus; IMMV, maize Iranian mosaic virus; TaVCV, taro vein chlorosis virus.
Table 2.
The PYDV-N-NLS was used as a probe in BLASTp searches to identify Arabidopsis proteins with QKR motif NLSs.
| Accession number | Description | Sequence |
|---|---|---|
| EU183122 | PYDV-N | QKRANEEAPPAAQKR |
| dbj|BAD95377.1| | RING finger-like protein | QKRRKCVEGESSGKR |
| dbj|BAF01759.1| | Putative non-LTR reverse transcriptase | QKRINGWTSKFLSKR |
| gb|AEC08614.1| | RAD50 DNA repair protein | QKRAKDEIKMGISKR |
| gb|AEE85106.1| | FtsJ-like methyltransferase family protein | QKRVDRRMKSDDRKR |
| dbj|BAA96899.1| | DNA polymerase III catalytic subunit | QKRVSRYAAWLSLKR |
| gb|AEE30605.1| | Transcription factor MEE8 | QKRSAESRREGKKKR |
| gb|AEE79234.1| | F-Box protein | QKRRKCVEGESSGKR |
| gb|AED90531.1| | E3 ubiquitin-protein ligase UPL4 | QKRMEVVEELPADKR |
| gb|AEE30120.1| | zinc finger CCCH domain-containing protein | QKRNPSDLDSSNLKR |
| ref|NP_188215.1| | UDP-glycosyltransferase-like protein | QKRSGTCDRKSDFKR |
The top 10 hits are shown. Residues in bold correspond to those found in the QKR motifs of the PYDV-N NLS.
Our BiFC data suggest that N-NLS mutants retain the ability to self-associate and that wild-type N proteins could not facilitate nuclear import of mutant proteins. It is likely therefore that wild-type N proteins do not self-associate until they have entered the nucleus. In contrast, the N-NLS mutants were able to inhibit the nuclear import of P proteins, suggesting that the N:P interaction domain overlaps with the P-NLS. This finding could have consequences for engineering resistance to protect plants from rhabdovirus infections. Not only may N proteins incapable of nuclear import serve as dominant-negative mutants of N function, but by interfering with P may provide an additional level of inhibition of critical viral functions. Curiously, the interaction of N-NLS mutants with the M protein occurred at punctate loci on the periphery of nuclei suggestive of the location of nuclear pore complexes (NPCs; Fiserova et al., 2009). In the absence of other viral proteins, the PYDV-M protein induces the intranuclear accumulation of the inner nuclear membrane (Martin et al., 2009; Bandyopadhyay et al., 2010). Why an N-NLS mutant would inhibit this process will likely require identification of additional components of the NPC in N. benthamiana, as has recently been done for Arabidopsis (Meier and Brkljacic, 2009; Tamura et al., 2010; Tamura and Hara-Nishimura, 2011).
Conflict of Interest Statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
References
- Ammar el D., Tsai C. W., Whitfield A. E., Redinbaugh M. G., Hogenhout S. A. (2009). Cellular and molecular aspects of rhabdovirus interactions with insect and plant hosts. Annu. Rev. Entomol. 54, 447–468 10.1146/annurev.ento.54.110807.090454 [DOI] [PubMed] [Google Scholar]
- Bandyopadhyay A., Kopperud K., Anderson G., Martin K., Goodin M. (2010). An integrated protein localization and interaction map for Potato yellow dwarf virus, type species of the genus Nucleorhabdovirus. Virology 402, 61–71 10.1016/j.virol.2010.03.001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chakrabarty R., Banerjee R., Chung S. M., Farman M., Citovsky V., Hogenhout S. A., Tzfira T., Goodin M. (2007). PSITE vectors for stable integration or transient expression of autofluorescent protein fusions in plants: probing Nicotiana benthamiana-virus interactions. Mol. Plant Microbe Interact. 20, 740–750 10.1094/MPMI-20-7-0740 [DOI] [PubMed] [Google Scholar]
- Citovsky V., Kapelnikov A., Oliel S., Zakai N., Rojas M. R., Gilbertson R. L., Tzfira T., Loyter A. (2004). Protein interactions involved in nuclear import of the Agrobacterium VirE2 protein in vivo and in vitro. J. Biol. Chem. 279, 29528–29533 10.1074/jbc.M403159200 [DOI] [PubMed] [Google Scholar]
- Citovsky V., Zupan J., Warnick D., Zambryski P. (1992). Nuclear localization of Agrobacterium VirE2 protein in plant cells. Science 256, 1802–1805 10.1126/science.1615325 [DOI] [PubMed] [Google Scholar]
- Cokol M., Nair R., Rost B. (2000). Finding nuclear localization signals. EMBO Rep. 1, 411–415 10.1093/embo-reports/kvd092 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deng M., Bragg J. N., Ruzin S., Schichnes D., King D., Goodin M. M., Jackson A. O. (2007). Role of the Sonchus yellow net virus N protein in formation of nuclear viroplasms. J. Virol. 81, 5362–5374 10.1128/JVI.00770-07 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fiserova J., Kiseleva E., Goldberg M. W. (2009). Nuclear envelope and nuclear pore complex structure and organization in tobacco BY-2 cells. Plant J. 59, 243–255 10.1111/j.1365-313X.2009.03865.x [DOI] [PubMed] [Google Scholar]
- Goodin M., Yelton S., Ghosh D., Mathews S., Lesnaw J. (2005). Live-cell imaging of rhabdovirus-induced morphological changes in plant nuclear membranes. Mol. Plant Microbe Interact. 18, 703–709 10.1094/MPMI-18-0703 [DOI] [PubMed] [Google Scholar]
- Goodin M. M., Austin J., Tobias R., Fujita M., Morales C., Jackson A. O. (2001). Interactions and nuclear import of the N and P proteins of Sonchus yellow net virus, a plant nucleorhabdovirus. J. Virol. 75, 9393–9406 10.1128/JVI.75.19.9393-9406.2001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goodin M. M., Chakrabarty R., Yelton S., Martin K., Clark A., Brooks R. (2007). Membrane and protein dynamics in live plant nuclei infected with Sonchus yellow net virus, a plant-adapted rhabdovirus. J. Gen. Virol. 88, 1810–1820 10.1099/vir.0.82698-0 [DOI] [PubMed] [Google Scholar]
- Goodin M. M., Dietzgen R. G., Schichnes D., Ruzin S., Jackson A. O. (2002). pGD vectors: versatile tools for the expression of green and red fluorescent protein fusions in agroinfiltrated plant leaves. Plant J. 31, 375–383 10.1046/j.1365-313X.2002.01360.x [DOI] [PubMed] [Google Scholar]
- Hsu H. T., Black L. M. (1973). Inoculation of vector cell monolayers with Potato yellow dwarf virus. Virology 52, 187–198 10.1016/0042-6822(73)90408-X [DOI] [PubMed] [Google Scholar]
- Hsu H. T., Black L. M. (1974). Multiplication of Potato yellow dwarf virus on vector cell monolayers. Virology 59, 331–334 10.1016/0042-6822(74)90232-3 [DOI] [PubMed] [Google Scholar]
- Jackson A. O., Dietzgen R. G., Goodin M. M., Bragg J. N., Deng M. (2005). Biology of plant rhabdoviruses. Annu. Rev. Phytopathol. 43, 623–660 10.1146/annurev.phyto.43.011205.141136 [DOI] [PubMed] [Google Scholar]
- Kanneganti T. D., Bai X., Tsai C. W., Win J., Meulia T., Goodin M., Kamoun S., Hogenhout S. A. (2007). A functional genetic assay for nuclear trafficking in plants. Plant J. 50, 149–158 10.1111/j.1365-313X.2007.03029.x [DOI] [PubMed] [Google Scholar]
- Kosugi S., Hasebe M., Matsumura N., Takashima H., Miyamoto-Sato E., Tomita M., Yanagawa H. (2009). Six classes of nuclear localization signals specific to different binding grooves of importin alpha. J. Biol. Chem. 284, 478–485 10.1074/jbc.M807017200 [DOI] [PubMed] [Google Scholar]
- Kumar M., Raghava G. P. (2009). Prediction of nuclear proteins using SVM and HMM models. BMC Bioinformatics 10, 22. 10.1186/1471-2105-10-22 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kuzmin I. V., Novella I. S., Dietzgen R. G., Padhi A., Rupprecht C. E. (2009). The rhabdoviruses: biodiversity, phylogenetics, and evolution. Infect. Genet. Evol. 9, 541–553 10.1016/j.meegid.2009.02.005 [DOI] [PubMed] [Google Scholar]
- Martin K., Kopperud K., Chakrabarty R., Banerjee R., Brooks R., Goodin M. M. (2009). Transient expression in Nicotiana benthamiana fluorescent marker lines provides enhanced definition of protein localization, movement and interactions in planta. Plant J. 59, 150–162 10.1111/j.1365-313X.2009.03850.x [DOI] [PubMed] [Google Scholar]
- Mattaj I. W., Englmeier L. (1998). Nucleocytoplasmic transport: the soluble phase. Annu. Rev. Biochem. 67, 265–306 10.1146/annurev.biochem.67.1.265 [DOI] [PubMed] [Google Scholar]
- Meier I., Brkljacic J. (2009). Adding pieces to the puzzling plant nuclear envelope. Curr. Opin. Plant Biol. 12, 752–759 10.1016/j.pbi.2008.09.001 [DOI] [PubMed] [Google Scholar]
- Min B. E., Martin K., Wang R., Tafelmeyer P., Bridges M., Goodin M. (2010). A host-factor interaction and localization map for a plant-adapted rhabdovirus implicates cytoplasm-tethered transcription activators in cell-to-cell movement. Mol. Plant Microbe Interact. 23, 1420–1432 10.1094/MPMI-04-10-0097 [DOI] [PubMed] [Google Scholar]
- Nakai K., Horton P. (2007). Computational prediction of subcellular localization. Methods Mol. Biol. 390, 429–466 10.1007/978-1-59745-466-7_29 [DOI] [PubMed] [Google Scholar]
- Spector D. L. (2006). SnapShot: cellular bodies. Cell 127, 1071. 10.1016/j.cell.2006.11.037 [DOI] [PubMed] [Google Scholar]
- Tamura K., Fukao Y., Iwamoto M., Haraguchi T., Hara-Nishimura I. (2010). Identification and characterization of nuclear pore complex components in Arabidopsis thaliana. Plant Cell 22, 4084–4097 10.1105/tpc.110.079947 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tamura K., Hara-Nishimura I. (2011). Involvement of the nuclear pore complex in morphology of the plant nucleus. Nucleus 2, 168–172 10.4161/nucl.2.3.16175 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wagner J. D., Choi T. J., Jackson A. O. (1996). Extraction of nuclei from Sonchus yellow net rhabdovirus-infected plants yields a polymerase that synthesizes viral mRNAs and polyadenylated plus-strand leader RNA. J. Virol. 70, 468–477 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wagner J. D., Jackson A. O. (1997). Characterization of the components and activity of Sonchus yellow net rhabdovirus polymerase. J. Virol. 71, 2371–2382 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zaltsman A., Yi B. Y., Krichevsky A., Gafni Y., Citovsky V. (2007). Yeast-plant coupled vector system for identification of nuclear proteins. Plant Physiol. 145, 1264–1271 10.1104/pp.107.105973 [DOI] [PMC free article] [PubMed] [Google Scholar]