Abstract
RNA binding proteins (RBPs) play an important role not only in nuclear gene expression, but also in cytosolic events, including RNA transport, localization, translation, and stability. Although over 200 RBPs are predicted from the Arabidopsis genome alone, relatively little is known about these proteins in plants as many exhibit no homology to known RBPs in other eukaryotes. Furthermore, RBPs likely have low expression levels making them difficult to identify and study. As part of our continuing efforts to understand plant cytosolic gene expression and the factors involved, we employed a combination of affinity chromatography and proteomic techniques to enrich for low abundance RBPs in developing rice seed. Our results have been compiled into RiceRBP (http://www.bioinformatics2.wsu.edu/RiceRBP), a database that contains 257 experimentally identified proteins, many of which have not previously been predicted to be RBPs. For each of the identified proteins, RiceRBP provides information on transcript and protein sequence, predicted protein domains, details of the experimental identification, and whether antibodies have been generated for public use. In addition, tools are available to analyze expression patterns for the identified genes, view phylogentic relationships and search for orthologous proteins. RiceRBP is a valuable tool for the community in the study of plant RBPs.
Keywords: rice, RNA binding protein, RiceRBP database, proteomics
Introduction
Advances in proteomic technology have allowed researchers to more deeply probe an organism’s proteome resulting in a wealth of information. These methodologies have in part led to a shift from a global approach of assessing gene expression to one that is targeted and aims to identify and characterize proteins involved in a particular biological event (Köcher and Superti-Furga, 2007). Although most highly utilized for studies in yeast and animals, functional proteomic applications in photosynthetic organisms are on the rise, particularly in Arabidopsis and rice, and have been employed to investigate such areas as development, stress response, and post-translational modification (Jorrín-Novo et al., 2009; Wienkoop et al., 2010; Agrawal and Rakwal, 2011). They have also been used to explore protein interactions with other proteins, complexes and particular affinity matrices, which is ultimately important for understanding systems biology (Baginsky, 2009). The use of affinity purification techniques has the added benefit of simplifying a complex mixture of proteins and enriching for those present in physiologically low amounts, facilitating detection.
One area where affinity isolation approaches have had success is the study of RNA binding proteins (RBPs). RBPs are critical components of gene expression events in both the nucleus and cytoplasm and are involved in all facets of RNA metabolism including transcription, pre-mRNA processing, nuclear export, transport, localization, translation, and stability (Wilkinson and Shyu, 2001; Dreyfuss et al., 2002). Well-studied in yeast and metazoans, RBPs are highly diverse and multifunctional (Wilkinson and Shyu, 2001; Dreyfuss et al., 2002) with many characterized by the presence of one or more RNA binding domains, such as the RNA recognition motif (RRM) or K homology (KH) domain (Anantharaman et al., 2002). Less is known about RBPs in plants, however, and even though sequence analysis has predicted over 200 RRM- and KH domain-containing proteins within the Arabidopsis genome alone, few are functionally characterized (Lorkovic and Barta, 2002). Contributing to this lack of knowledge is that many RBPs are plant-specific and lack homologs characterized in yeast and animal models (Lorkovic and Barta, 2002; Lorkovic, 2009). As the importance of RBPs in plant development and stress response becomes increasingly clear, it is imperative that more focus be directed at identifying and elucidating their activity (Fedoroff, 2002; Bailey-Serres et al., 2009; Lorkovic, 2009). A few groups have attempted to do so by combining nucleic acid and ion exchange affinity chromatography with 2D gel electrophoresis (2DE) to enrich for low abundance RBPs in Arabidopsis, spinach, and rice (Baginsky et al., 2007; Xu et al., 2007; Masaki et al., 2008; Ni et al., 2010).
Our laboratory has been particularly interested in identifying plant RBPs involved in cytosolic RNA localization, a process common to eukaryotes that spatially and temporally controls gene expression by targeting RNAs to specific subcellular locations (St. Johnston, 2005; Martin and Ephrussi, 2009). RNA localization is dependent upon cis-localization, or zipcode, sequences within the RNA which are recognized by one or more RBPs (Singer, 1993). Along with other trans-factors, these RBPs form a dynamic ribonucleoprotein complex that regulates nuclear processing and export, cytoskeleton-associated transport, localization, translation, and RNA stability (Wilkinson and Shyu, 2001; Dreyfuss et al., 2002; Martin and Ephrussi, 2009). RNA localization has been well-characterized in yeast and metazoans, but relatively few studies have been reported in plants (Okita and Choi, 2002; Crofts et al., 2004; Bailey-Serres et al., 2009). One of the best models, however, is the localization of storage protein RNAs in developing rice seed. For many years, our lab has been investigating the cytoskeleton-dependent, asymmetric localization of prolamine, and glutelin mRNAs to distinct subdomains of the cortical endoplasmic reticulum (ER), the predominate site of protein synthesis in endosperm cells (Crofts et al., 2004, 2005; Doroshenk et al., 2012). Similar to other eukaryotes, this targeted transport requires cis-localization sequences likely recognized by RBPs (Hamada et al., 2003; Washida et al., 2009). Proper RNA localization is critical to subsequent protein localization, as mis-targeting storage protein mRNAs to the incorrect ER domain results in improper protein deposition (Crofts et al., 2005; Washida et al., 2012). In an attempt to elucidate the trans-acting RBPs involved in RNA transport, we previously identified OsTudor-SN, a cytoskeleton-associated RBP that binds both prolamine and glutelin mRNAs in rice endosperm cells and co-localizes with cytoplasmic prolamine mRNA transport particles (Sami-Subbu et al., 2001; Wang et al., 2008). It is our goal, however, to identify the many other RBPs that are likely involved in this process to better understand the mechanism of transport and ultimately, cytoplasmic plant gene expression.
As previously mentioned, the increasing popularity of plant proteomics has led to a massive amount of information and efforts have been made to catalog these results (Jorrín-Novo et al., 2009). Here, we summarize our own efforts to provide such a resource for plant RBPs identified by multiple affinity purification and proteomic experiments through the creation of the RiceRBP database (Morris et al., 2011; http://www.bioinformatics2.wsu.edu/RiceRBP). To our knowledge, RiceRBP is the first attempt to catalog experimentally identified RBPs for use by the plant biology community.
Proteomic Identification of Rice RBPs
Our interest in cytosolic gene expression in plants and the lack of available data in the literature led us to investigate what nucleic acid binding proteins, and in particular RBPs, are found within the cytoplasm of developing rice endosperm cells. Our ultimate goal was to deposit these findings into a publicly accessible database to share knowledge and resources with other plant scientists also interested in the field. We attempted to identify such proteins using a combination of affinity chromatography and proteomic techniques. As part of a more global approach, we enriched for nucleic acid binding proteins from a cytoskeleton-enriched developing rice seed extract using Poly(U)-Sepharose column chromatography, separated proteins in the bound fraction by 2DE and identified reproducible protein spots by reverse phase liquid chromatography-tandem mass spectrometry (LC-MS/MS; Doroshenk et al., 2009). From these experiments, over 150 distinct proteins were identified including putative RBPs, translation factors, and metabolic enzymes. Of the 20 proteins identified with suggested roles in RNA metabolism, only four were previously characterized in the literature including OsTudor-SN (Sami-Subbu et al., 2001; Wang et al., 2008), confirming the successful application of such enrichment techniques. Many of the uncharacterized RBPs identified were predicted to be involved in RNA processing based on the presence of RNA binding domains such as the RRM and KH domain (Anantharaman et al., 2002). Interestingly, review of the literature revealed that a number of proteins identified from this study categorized as having established roles in polypeptide biosynthesis or carbon metabolism were similar to proteins in other organisms that exhibited dual functionality and actually possessed RNA binding activity (Doroshenk et al., 2009, and references therein). This included protein chaperones and protein turnover enzymes involved in RNA stability as well as membrane-associated transport proteins and carbohydrate metabolic enzymes with suggested roles in RNA localization.
The results of the Poly(U)-Sepharose experiments exposed the complexity of proteins that may be involved in cytosolic gene expression in plants. As our particular interest lies in the mechanism of RNA localization using rice seed storage protein RNA as a model, a more targeted approach was attempted to identify those proteins that interacted specifically with the prolamine RNA cis-localization element, or zipcode. Again taking an affinity chromatography/proteomics approach, the 36 nucleotide zipcode sequence from prolamine mRNA was biotinylated and used as bait to isolate interacting proteins from a cytoskeleton-enriched developing rice seed extract (Crofts et al., 2010). The binding assay was performed in the presence of the competitive binding inhibitor heparin, a highly charged anionic molecule, to ensure specificity. Bound proteins were separated by 1D SDS-PAGE and analyzed by LC-MS/MS. Compared to a control capture experiment using non-zipcode RNA as bait, 15 proteins were identified as having specificity for the prolamine zipcode (Crofts et al., 2010). Ten of these proteins contained at least one predicted RNA binding domain and of those, seven shared significant homology to heterogeneous nuclear ribonucleoproteins (hnRNPs), a class of proteins well-studied in metazoans and yeast with both nuclear and cytoplasmic roles in pre-RNA processing, localization, and translation (Krecic and Swanson, 1999; Dreyfuss et al., 2002). Interestingly, only one of the 15 proteins identified in the prolamine zipcode capture experiment was also identified in the Poly(U)-Sepharose binding fraction detailed above (Doroshenk et al., 2009), likely the result of the low abundance of these RBPs in the cytosol and need for enhanced methods of enrichment. Current efforts include further characterization of the identified prolamine RBPs to establish their role in gene expression.
Because of the highly stringent nature of the prolamine zipcode capture experiment as a result of the addition of heparin in the binding buffer (Crofts et al., 2010), the experiment was repeated without the use of heparin to establish whether an even greater number of RBPs could be identified. This was based partly on results from previous experiments demonstrating the known prolamine RBP OsTudor-SN did not bind its target in the presence of heparin (Sami-Subbu et al., 2001; Crofts et al., 2010). Again using biotinylated prolamine zipcode RNA as bait, bound proteins were analyzed by 1D SDS-PAGE and LC-MS/MS (Morris et al., 2011). 132 putative RBPs were identified including 12 proteins from the more stringent prolamine zipcode capture (Crofts et al., 2010). Importantly, 77 proteins not found in either the Poly(U)-Sepharose affinity chromatography (Doroshenk et al., 2009) or stringent prolamine zipcode capture experiments(Crofts et al., 2010) were identified and include a number of uncharacterized proteins with predicted RNA binding domains (Morris et al., 2011). More study is necessary, though, to determine the nature of their role in plant RNA metabolism.
RiceRBP: A Catalog of Experimentally Identified RBPs
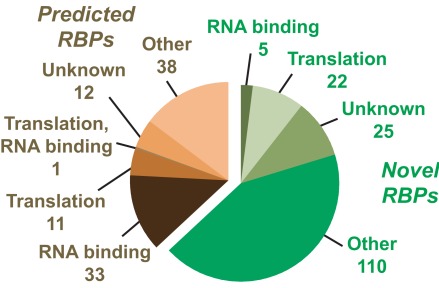
The three proteomic studies described above which sought to investigate RBPs involved in plant cytosolic gene expression led to the combined identification of 257proteins derived from at least 221 distinct rice genes. These results have been compiled into RiceRBP (found at http://www.bioinformatics2.wsu.edu/RiceRBP or bioinformatics1.smb.wsu.edu/RiceRBP), a publicly accessible database for use by the scientific community (Morris et al., 2011). RiceRBP is the only database to our knowledge containing data and analysis tools dedicated to the study of experimentally identified RBPs. In fact, when compared to the POGs/PlantRBP database (Walker et al., 2007) which predicts plant RBPs solely on sequence similarity, only 37% of the experimentally identified rice proteins had been previously annotated as RBPs (Figure 1; Morris et al., 2011). Functional annotations from the Rice Genome Annotation Project (Ouyang et al., 2007) revealed a number of the experimentally identified RBPs had putative roles in RNA processing (RNA binding/translation) as expected (Figure 1). Of interest, however, are the numerous proteins that had other unrelated predicted functions, particularly for the novel RBPs not previously annotated as such by the POGS/PlantRBP database (Walker et al., 2007), highlighting how little is known about this important class of plant proteins and the need for further study.
Figure 1.
Classification of experimentally identified RNA binding proteins (RBPs) cataloged within RiceRBP. Functional annotations were obtained from the Rice Genome Annotation Project (Ouyang et al., 2007) and the number of entries in each grouping is indicated. Proteins predicted to be RBPs by the POGs/PlantRBP database (Walker et al., 2007) are represented in brown, while those newly identified as such are in green. Reproduced with permission from Morris et al. (2011).
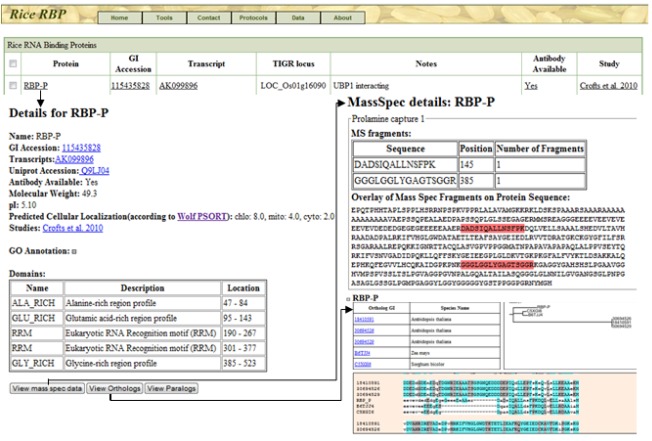
Some of the features of RiceRBP are demonstrated in Figure 2 using RBP-P, a putative oligouridylate binding protein identified from the stringent prolamine zipcode capture experiment (Crofts et al., 2010), as an example. Each unique, identified protein in RiceRBP has been assigned a random identifier (i.e., RBP-A, -B or RBP-1, -2, etc.) and cross-referenced to accession numbers representing gene, cDNA and protein sequences from a variety of sources (when available) including NCBI, the Rice Genome Annotation Project, Rice Annotation Project Database and UniProt (Morris et al., 2011 and references therein). Information regarding predicted molecular weight, protein domains, subcellular localization, and functional annotation are given for each entry, as is whether the identification resulted from the Poly(U)-Sepharose (PolyU capture; Doroshenk et al., 2009), high stringency prolamine zipcode (prolamine capture 1; Crofts et al., 2010), and/or low stringency prolamine zipcode (prolamine capture 2; Morris et al., 2011) affinity chromatography experiments. Detailed experimental methodologies for each of these experiments are provided in printable format as well. For each protein entry, the peptide sequences identified by mass spectrometry and position within the entire polypeptide sequence are highlighted (a sub-set of mass spectrometry data is shown in Figure 2). Separate links are also provided for each entry to view sequence alignments and phylogenetic tress highlighting paralogous family members within rice, as well as prokaryotic and eukaryotic orthologs, including important agricultural species such as barley (Hordeum vulgare), maize (Zea mays), sorghum (Sorghum bicolor), and wheat (Triticum aestivum).
Figure 2.
Illustration of several features of RiceRBP using RBP-P, a prolamine RNA binding protein (Crofts et al., 2010), as an example. Each protein entry contains information regarding accession identifiers, predicted function, antibody availability, and from which RBP capture experiment it was identified. Links to web pages dedicated to each RBP provide additional information on predicted domains and subcellular localization as well as peptide data obtained from mass spectrometry analysis. A list of orthologous and paralogous family members for each entry, visualized by phylogenetic trees and sequence alignments, is also available.
A variety of tools are offered for users of RiceRBP including the ability to search the database for particular entries of interest using keywords or gene, transcript or protein accession identifiers. User supplied transcript or amino acid sequence can be entered to BLAST against rice proteins within the database as well. Another resource includes visualization of transcript expression data from microarray experiments compiled from multiple public sources (Morris et al., 2011 and references therein) for a particular RBP entry if available. These datasets have been generated from a variety of rice tissue samples as well as developmental stages and experimental treatments. For those users interested in identifying proteins related to rice RBPs in a specific species, an ortholog search tool is available with the option of selecting prokaryotic and/or eukaryotic queries and a downloadable results format. For details of the specific methodology for constructing RiceRBP and available analysis tools, readers are directed to Morris et al. (2011).
The Future of RiceRBP
RiceRBP continues to be a work in progress with the aim of providing a current and extensive plant RBP resource to the community. For instance, we hope to update the database with additional RBP orthologs as more plant genome sequences become available. We also plan to incorporate results from current efforts aimed at characterizing the functional roles of specific rice RBPs in cytosolic gene expression, and particularly RNA localization, as this data becomes available. Antibodies have been generated to some of these proteins and their availability to the public has been noted within RiceRBP. These antibodies are being used for RNA binding and immunofluorescence localization studies, as illustrated by RBP-A and RBP-D, two putative hnRNPs identified from the stringent prolamine zipcode RNA capture experiment (Crofts et al., 2010). RNA-immunoprecipitation (RNA-IP) using RBP specific antibodies revealed both RBP-A and RBP-D interact with prolamine and glutelin RNA in vivo (Crofts et al., 2010; Crofts et al., unpublished results). Furthermore, both proteins exist as multiple populations within rice endosperm cells, with RBP-A localized to the nucleus, microtubules, and cortical ER (Crofts et al., 2010) and RBP-D to the nucleus and particulate structures associated with actin filaments (Crofts et al., unpublished results). Dual localization of nuclear assembled ribonucleoprotein complex components has been demonstrated in other organisms as well (Giorgi and Moore, 2007)and for RBP-A and -D may suggest multiple, diverse roles in pre-mRNA processing, nuclear to cytoplasmic shuttling, cytoskeletal-associated RNA transport, translational regulation or anchoring to the ER. Additional rice RBPs will be subjected to similar studies.
RNA binding protein specific antibodies are also being used to identify both RNA and protein targets which may offer further insight into functionality. RNA-IP combined with microarray and next generation sequencing will identify RNA targets specific to each RBP to better understand the global role of that particular RBP within the cell. Based on the interacting RNA sequences identified, prediction software is being utilized to determine whether particular RNA sequence or structural motifs exist, which could be used to elucidate additional RNA targets in rice. This information could also be used to identify targets of orthologous RBPs in other plant species. RBP specific antibodies are also being used in traditional immunoprecipitation experiments to determine what other proteins may be associated. Ribonucleoprotein complexes contain multiple proteins and RNAs (Jansen, 2001; Martin and Ephrussi, 2009) and it would be of great interest to identify other protein components to understand the diverse mechanisms of RNA transport, localization and stability within the cell.
Finally, mutant studies to further our knowledge of proteins involved in RNA metabolism are underway. A number of RNAi lines for particular rice RBPs of interest have been generated while others are in construction. For example, characterization of RBP OsTudor-SN RNAi transgenic plants not only revealed a decrease in OsTudor-SN transcript and protein levels, but also a decrease in prolamine transcript and protein levels, indicating a role in prolamine gene expression (Wang et al., 2008).In addition, rice genetic mutants are being screened using the TILLING method (Suzuki et al., 2008) to identify those lines containing mutations within RBPs of interest for further characterization. Successful screens have identified candidate mutants for OsTudor-SN and RBP-Pand are currently being studied. Specific to our interest in RNA localization, genetic mutants impaired in storage protein synthesis, sorting or processing, which may be an indication of disrupted RNA transport, have also been identified (Crofts et al., 2004, 2005). One such mutant, glup4, has been found to partially mislocalize both glutelin RNA and protein and displays highly irregular membrane architecture within endosperm cells (Doroshenk et al., 2010; Fukuda et al., 2011). The Glup4 gene encodes Rab5 (Satoh-Cruz et al., 2010), a small GTPase with roles in early endosome formation and cytoskeleton-dependent transport, cargo recruitment, and ER structuring (Zerial and McBride, 2001; van der Bliek, 2005; Audhya et al., 2007). Proteomic characterization of glup4 developing rice seed by two-dimensional difference in gel electrophoresis (2D-DIGE) revealed a number of proteins that were differentially expressed in the mutant, including membrane-associated proteins and those involved in the biosynthesis of cell wall components and seed storage reserves (Doroshenk et al., 2010). The combined results of these studies suggest an interesting possibility that RNA transport from the nucleus to the cortical ER in rice endosperm is a membrane-associated process mediated by Rab proteins(Doroshenk et al., 2010, 2012), an idea supported by work in Drosophila oocytes (Ruden et al., 2000; Jankovics et al., 2001; Dollar et al., 2002). As part of its role in membrane trafficking, the rice Rab5 may bean important component of gene expression events within the cell. Current studies include investigating whether Rab5 plays an actual role in RNA transport and if so, what other factors (such as RBPs) may be involved. Similar 2D-DIGE analysis of other genetic and transgenic mutants is also of interest.
We anticipate that future versions of the RiceRBP database will incorporate RNA binding targets of selected RBPs based on RNA-IP microarray and sequencing data. Additional functional and localization studies of RBPs may also be included in the RiceRBP database as such data becomes available. Hence, we expect that the RiceRBP database will continue to provide a centralized resource for biologists interested in this important, yet understudied, class of proteins and their role in plant gene expression.
Conflict of Interest Statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Acknowledgments
This research was generously supported by National Science Foundation Grants IOB-0544469 and DBI-0605016 and USDA grant 2006-35301-17043. We thank Nicholas Lewis, Adam Mitchell, Dale Cox, Ryan Hoffman, and Leanne Whitmore for their help in developing and testing the RiceRBP database and website.
References
- Agrawal G. K., Rakwal R. (2011). Rice proteomics: a move toward expanded proteome coverage to comparative and functional proteomics uncovers the mysteries of rice and plant biology. Proteomics 11, 1630–1649 10.1002/pmic.201000696 [DOI] [PubMed] [Google Scholar]
- Anantharaman V., Koonin E. V., Aravind L. (2002). Comparative genomics and evolution of proteins involved in RNA metabolism. Nucleic Acids Res. 30, 1427–1464 10.1093/nar/30.7.1427 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Audhya A., Desai A., Oegema K. (2007). A role for Rab5 in structuring the endoplasmic reticulum. J. Cell Biol. 178, 43–56 10.1083/jcb.200701139 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baginsky S. (2009). Plant proteomics: concepts, applications, and novel strategies for data interpretation. Mass Spectrom. Rev. 28, 93–120 10.1002/mas.20183 [DOI] [PubMed] [Google Scholar]
- Baginsky S., Grossmann J., Gruissem W. (2007). Proteome analysis of chloroplast mRNA processing and degradation. J. Proteome Res. 6, 809–820 10.1021/pr060473q [DOI] [PubMed] [Google Scholar]
- Bailey-Serres J., Sorenson R., Juntawong P. (2009). Getting the message across: cytoplasmic ribonucleoprotein complexes. Trends Plant Sci. 14, 443–453 10.1016/j.tplants.2009.05.004 [DOI] [PubMed] [Google Scholar]
- Crofts A. J., Crofts N., Whitelegge J. P., Okita T. W. (2010). Isolation and identification of cytoskeleton-associated prolamine mRNA binding proteins from developing rice seeds. Planta 231, 1261–1276 10.1007/s00425-010-1125-x [DOI] [PubMed] [Google Scholar]
- Crofts A. J., Washida H., Okita T. W., Ogawa M., Kumamaru T., Satoh H. (2004). Targeting of proteins to endoplasmic reticulum-derived compartments in plants. The importance of RNA localization. Plant Physiol. 136, 3414–3419 10.1104/pp.104.048934 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Crofts A. J., Washida H., Okita T. W., Ogawa M., Kumamaru T., Satoh H. (2005). The role of mRNA and protein sorting in seed storage protein synthesis, transport and deposition. Biochem. Cell Biol. 83, 728–737 10.1139/o05-156 [DOI] [PubMed] [Google Scholar]
- Dollar G., Struckhoff E., Michaud J., Cohen R. S. (2002). Rab11 polarization of the Drosophila oocyte: a novel link between membrane trafficking, microtubule organization, and oskar mRNA localization and translation. Development 129, 517–526 [DOI] [PubMed] [Google Scholar]
- Doroshenk K. A., Crofts A. J., Morris R. T., Wyrick J. J., Okita T. W. (2009). Proteomic analysis of cytoskeleton-associated RNA binding proteins in developing rice seed. J. Proteome Res. 8, 4641–4653 10.1021/pr900537p [DOI] [PubMed] [Google Scholar]
- Doroshenk K. A., Crofts A. J., Washida H., Satoh-Cruz M., Crofts N., Sugino A., Okita T. W., Morris R. T., Wyrick J. J., Fukuda M., Kumamaru T., Satoh H. (2010). Characterization of the rice glup4 mutant suggests a role for the small GTPase Rab5 in the biosynthesis of carbon and nitrogen storage reserves in developing endosperm. Breed. Sci. 60, 568–574 10.1270/jsbbs.60.568 [DOI] [Google Scholar]
- Doroshenk K. A., Crofts A. J., Washida H., Satoh-Cruz M., Crofts N., Yang Y., Morris R. T., Okita T. W., Fukuda M., Kumamaru T., Satoh H. (2012). “mRNA localization in plants and the role of RNA binding proteins,” in RNA Binding Proteins, ed. Lorkovic Z. (Austin: Landes Bioscience; ). [Google Scholar]
- Dreyfuss G., Kim V. N., Kataoka N. (2002). Messenger-RNA-binding proteins and the messages they carry. Nat. Rev. Mol. Cell Biol. 3, 195–205 10.1038/nrm760 [DOI] [PubMed] [Google Scholar]
- Fedoroff N. V. (2002). RNA-binding proteins in plants: the tip of an iceberg? Curr. Opin. Plant Biol. 5, 452–459 10.1016/S1369-5266(02)00280-7 [DOI] [PubMed] [Google Scholar]
- Fukuda M., Satoh-Cruz M., Crofts A. J., Wen L., Sugino A., Washida H., Okita T. W., Ogawa M., Kawagoe Y., Maeshima M., Kumamaru T. (2011). The small GTPase Rab5a is essential for intracellular transport of glutelin precursor from Golgi apparatus and endosomal membrane organization in developing rice endosperm. Plant Physiol. 157, 632–644 10.1104/pp.111.180505 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Giorgi C., Moore M. J. (2007). The nuclear nurture and cytoplasmic nature of localized mRNPs. Semin. Cell Dev. Biol. 18, 186–193 10.1016/j.semcdb.2007.01.002 [DOI] [PubMed] [Google Scholar]
- Hamada S., Ishiyama K., Sakulsingharoj C., Choi S. B., Wu Y., Wang C., Singh S., Kawai N., Messing J., Okita T. W. (2003). Dual regulated RNA transport pathways to the cortical region in developing rice endosperm. Plant Cell 15, 2265–2272 10.1105/tpc.013466 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jankovics F., Sinka R., Erdélyi M. (2001). An interaction type of genetic screen reveals a role of the Rab11 gene in oskar mRNA localization in the developing Drosophila melanogaster oocyte. Genetics 158, 1177–1188 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jansen R. P. (2001). mRNA localization: message on the move. Nat. Rev. Mol. Cell Biol. 2, 247–256 10.1038/35067016 [DOI] [PubMed] [Google Scholar]
- Jorrín-Novo J. V., Maldonado A. M., Echevarría-Zomeño S., Valledor L., Castillejo M. A., Curto M., Valero J., Sghaier B., Donoso G., Redondo I. (2009). Plant proteomics update (2007–2008): second-generation proteomic techniques, an appropriate experimental design, and data analysis to fulfill MIAPE standards, increase plant proteome coverage and expand biological knowledge. J. Proteomics 72, 285–314 10.1016/j.jprot.2009.01.026 [DOI] [PubMed] [Google Scholar]
- Köcher T., Superti-Furga G. (2007). Mass spectrometry-based functional proteomics: from molecular machines to protein networks. Nat. Methods 4, 807–815 10.1038/nmeth1093 [DOI] [PubMed] [Google Scholar]
- Krecic A. M., Swanson M. S. (1999). hnRNP complexes: composition, structure, and function. Curr. Opin. Cell Biol. 11, 363–371 10.1016/S0955-0674(99)80051-9 [DOI] [PubMed] [Google Scholar]
- Lorkovic Z. (2009). Role of plant RNA-binding proteins in development, stress response and genome organization. Trends Plant Sci. 14, 229–236 10.1016/j.tplants.2009.01.007 [DOI] [PubMed] [Google Scholar]
- Lorkovic Z. J., Barta A. (2002). Genome analysis: RNA recognition motif (RRM) and K homology (KH) domain RNA-binding proteins from the flowering plant Arabidopsis thaliana. Nucleic Acids Res. 30, 623–635 10.1093/nar/30.3.623 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martin K. C., Ephrussi A. (2009). mRNA localization: gene expression in the spatial dimension. Cell 136, 719–730 10.1016/j.cell.2009.01.044 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Masaki S., Yamada T., Hirasawa T., Todaka D., Kanekatsu M. (2008). Proteomic analysis of RNA-binding proteins in dry seeds of rice after fractionation by ssDNA affinity column chromatography. Biotechnol. Lett. 30, 955–960 10.1007/s10529-007-9619-8 [DOI] [PubMed] [Google Scholar]
- Morris R. T., Doroshenk K. A., Crofts A. J., Lewis N., Okita T. W., Wyrick J. J. (2011). RiceRBP: a database of experimentally identified RNA-binding proteins in Oryza sativa L. Plant Sci. 180, 204–211 10.1016/j.plantsci.2010.08.004 [DOI] [PubMed] [Google Scholar]
- Ni R. J., Shen Z., Yang C. P., Wu Y. D., Bi Y. D., Wang B. C. (2010). Identification of low abundance polyA-binding proteins in Arabidopsis chloroplast using polyA-affinity column. Mol. Biol. Rep. 37, 637–641 10.1007/s11033-009-9478-6 [DOI] [PubMed] [Google Scholar]
- Okita T. W., Choi S.-B. (2002). mRNA localization in plants: targeting to the cell’s cortical region and beyond. Curr. Opin. Plant Biol. 5, 553–559 10.1016/S1369-5266(02)00304-7 [DOI] [PubMed] [Google Scholar]
- Ouyang S., Zhu W., Hamilton J., Lin H., Campbell M., Childs K., Thibaud-Nissen F., Malek R., Lee Y., Zheng L., Orvis J., Haas B., Wortman J., Buell C. (2007). The TIGR rice genome annotation resource: improvements and new features. Nucleic Acids Res. 35, D883–D887 10.1093/nar/gkl976 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ruden D. M., Sollars V., Wang X., Mori D., Alterman M., Lu X. (2000). Membrane fusion proteins are required for oskar mRNA localization in the Drosophila egg chamber. Dev. Biol. 218, 314–325 10.1006/dbio.1999.9583 [DOI] [PubMed] [Google Scholar]
- Sami-Subbu R., Choi S.-B., Wu Y., Wang C., Okita T. W. (2001). Identification of a cytoskeleton-associated 120 kDa RNA binding protein in developing seeds. Plant Mol. Biol. 46, 79–88 10.1023/A:1010643209402 [DOI] [PubMed] [Google Scholar]
- Satoh-Cruz M., Fukuda M., Ogawa M., Satoh H., Kumamaru T. (2010). Glup4 gene encodes small GTPase Rab5a in rice. Rice Genet. Newsl. 25, 48–49 [Google Scholar]
- Singer R. H. (1993). RNA zipcodes for cytoplasmic addresses. Curr. Biol. 3, 719–721 10.1016/0960-9822(93)90079-4 [DOI] [PubMed] [Google Scholar]
- St. Johnston D. (2005). Moving messages: the intracellular localization of mRNAs. Nat. Rev. Mol. Cell Biol. 6, 363–375 10.1038/nrm1643 [DOI] [PubMed] [Google Scholar]
- Suzuki T., Euguchi M., Kumamaru T., Satoh H., Matsusaka H., Moriguchi K., Nagato Y., Kurata N. (2008). MNU-induced mutant pools and high performance TILLING enable finding of any gene mutation in rice. Mol. Genet. Genomics 279, 213–223 10.1007/s00438-007-0293-2 [DOI] [PubMed] [Google Scholar]
- van der Bliek A. M. (2005). A sixth sense for Rab5. Nat. Cell Biol. 7, 548–550 10.1038/ncb0605-548 [DOI] [PubMed] [Google Scholar]
- Walker N. S., Stiffler N., Barkan A. (2007). POGs/PlantRBP: a resource for comparative genomics in plants. Nucleic Acids Res. 35, D852–D856 10.1093/nar/gkl795 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang C., Washida H., Crofts A. J., Hamada S., Katsube-Tanaka T., Kim D., Choi S.-B., Modi M., Singh S., Okita T. W. (2008). The cytoplasmic-localized, cytoskeletal-associated RNA binding protein OsTudor-SN: evidence for an essential role in storage protein RNA transport and localization. Plant J. 55, 443–454 10.1111/j.1365-313X.2008.03489.x [DOI] [PubMed] [Google Scholar]
- Washida H., Kaneko S., Crofts N., Sugino A., Wang C., Okita T. W. (2009). Identification of cis-localization elements that target glutelin RNAs to a specific subdomain of the cortical endoplasmic reticulum in rice endosperm cells. Plant Cell Physiol. 50, 635–643 10.1093/pcp/pcp103 [DOI] [PubMed] [Google Scholar]
- Washida H., Sugino A., Doroshenk K. A., Satoh-Cruz M., Nagamine A., Katsube-Tanaka T., Ogawa M., Kumamaru T., Satoh H., Okita T. W. (2012). RNA targeting to a specific ER subdomain is required for efficient transport and packaging of α-globulins to the protein storage vacuole in developing rice endosperm. Plant J. 70, 471–479 10.1111/j.1365-313X.2011.04880.x [DOI] [PubMed] [Google Scholar]
- Wienkoop S., Baginsky S., Weckwerth W. (2010). Arabidopsis thaliana as a model organism for plant proteome research. J. Proteomics 73, 2239–2248 10.1016/j.jprot.2010.07.012 [DOI] [PubMed] [Google Scholar]
- Wilkinson M. F., Shyu A. B. (2001). Multifunctional regulatory proteins that control gene expression in both the nucleus and the cytoplasm. Bioessays 23, 775–787 10.1002/bies.1113 [DOI] [PubMed] [Google Scholar]
- Xu Y., Wang B. C., Zhu Y. X. (2007). Identification of proteins expressed at extremely low level in Arabidopsis leaves. Biochem. Biophys. Res. Commun. 358, 808–812 10.1016/j.bbrc.2007.04.189 [DOI] [PubMed] [Google Scholar]
- Zerial M., McBride H. (2001). Rab proteins as membrane organizers. Nat. Rev. Mol. Cell Biol. 2, 107–117 10.1038/35052055 [DOI] [PubMed] [Google Scholar]