Abstract
Mutations in cancer-causing genes induce changes in gene expression programs critical for malignant cell transformation. Publicly available gene expression profiles produced by modulating the expression of distinct cancer genes may therefore represent a rich resource for the identification of gene signatures common to seemingly unrelated cancer genes. We combined automatic retrieval with manual validation to obtain a data set of high-quality gene microarray profiles. This data set was used to create logical models of the signaling events underlying the observed expression changes produced by various cancer genes and allowed to uncover unknown and verifiable interactions. Data clustering revealed novel sets of gene expression profiles commonly regulated by distinct cancer genes. Our method allows retrieval of significant new information and testable hypotheses from a pool of deposited cancer gene expression experiments that are otherwise not apparent or appear insignificant from single measurements. The complete results are available through a web-application at http://biodata.ethz.ch/cgi-bin/geologic.
Keywords: cancer genes, gene microarray database analysis, gene expression signatures, meta-analysis, network interactions, clustering
Introduction
The development of cancer requires multiple genetic alterations perturbing distinct cellular pathways. In human cancers, these alterations often arise owing to mutations in tumor suppressor genes and protooncogenes, which in turn trigger uncontrolled cell proliferation, survival and genomic instability. Consequently, the study of tumor suppressor proteins and protooncogenes and the cellular signaling networks deregulated by the corresponding mutant proteins has become a centerpiece of contemporary cancer research. In fact, investigations of their mode of action have pinpointed key mechanisms that protect humans against tumor development and thus provided rational foundations for preventing, detecting, and treating cancer.
Inactivation of tumor suppressor genes or the activation of oncogenes invariably trigger changes in gene expression programs. DNA microarrays1 are in wide use as a method to quantify changes in global expression levels. Public microarray databases contain measurements of transcription programs in cells under thousands of different biological conditions and/or perturbations. One of the most prominent is NCBI Gene Expression Omnibus (GEO),2 a curated repository containing microarray data in a standardized format.3 This database therefore offers a rich resource of quantitative data on the behavior of gene expression changes in response to cancer gene mutations.
By specifically analyzing gene expression program changes associated with cancer gene activation or inactivation, we sought for signatures shared among distinct cancer genes listed in the census of cancer genes4 and integrated the resultant data sets into logical networks of interactions.
Meta-analysis of cancer microarray data has been successfully applied by Rhodes et al to find a common gene-expression signature5 in independent data sets from different cancer types. Ramaswamy and colleagues discovered a predictive signature6 for the metastatic status of tumors from diverse origins and Creighton reported coordinate expression patterns of multiple oncogenic pathway signatures7 in human prostate tumors. Our approach used measurements from cell culture experiments in which the expression of specific cancer-causing genes has been either induced or downregulated. This allowed us to unveil gene expression signatures common to cancer genes that have not been linked previously.
Methods
Selection and acquisition
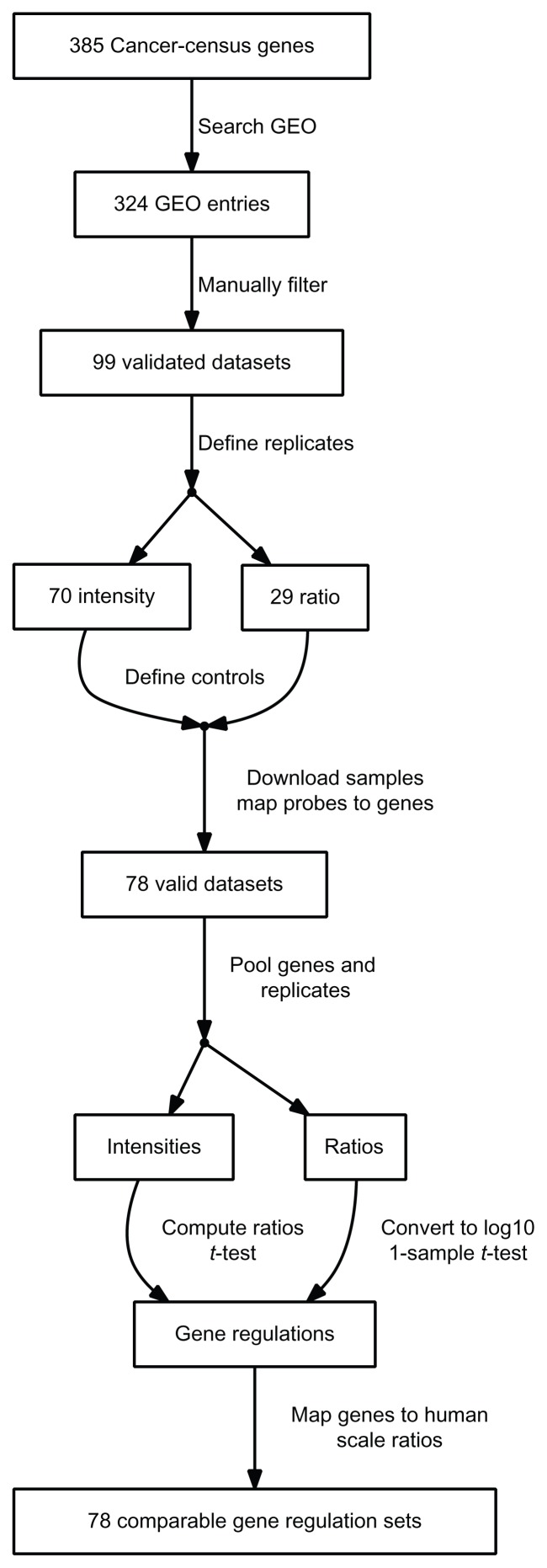
As outlined in Figure 1, the first step in data acquisition was searching the 385 genes present in the cancer gene census in the NCBI GEO repository. 324 GEO entries contained one of these gene names in the title or abstract. The descriptive fields of the entries were duplicated into a local database. False positives were made apparent through an appropriate visualization of the entry description and subsequently removed. We selected experiments in which cancer genes were over-expressed, depleted by the application of small interfering RNAs or genetically eliminated by virtue of gene knock-out in mice. In addition, we included experiments in which dominant-negative forms of cancer genes were expressed. All of these 99 experiments were performed in either human, mouse or rat cell lines as described in the publication accompanying the experiment. If the measured values were given as raw intensities, samples for induced and control as well as their replicates were selected. Replicates and type of logarithm were selected for entries providing ratios. The definition database was made accessible from network-edges in the web-interface.
Figure 1.
Flowchart of the data acquisition process.
Expression values were retrieved for the 607 defined samples. The probe identifiers were mapped to Entrez gene identifiers using the microarray platform description provided by GEO and the UniGene database.8 78 data sets were successfully mapped, the remaining 21 did not contain valid identifiers. NCBI HomoloGene provided human homologues of mouse and rat genes. Measurements were available for 18885 genes, while there was a total of 19978 human genes in HomoloGene. The probe-level measurements (N = 9981226) were grouped by genes and replicates. Intensity values were grouped into induced/control to compute the log(ratio) as well as the P-value of a t-test. In the case of entries containing ratios, they were converted to base 10 logarithm and a one-sample t-test was performed. In order to be able to filter sets of comparably regulated genes, the log-ratios were scaled by subtracting the experiment mean and dividing by the standard deviation. These computations were performed using the Python programming language. The final data matrix is available for download from http://biodata.ethz.ch/cgi-bin/geologic at the bottom of the page.
Clustering
Automatic classification and multiscale bootstrap resampling was performed using the R9 package pvclust.10 A matrix consisting of experiments as columns, genes as rows and gene-expression changes for each pair was used as input. Because not every gene was present in all of the microarray experiments, there are missing values in this matrix. An iterative method was used to reduce the fraction of missing values. In each step, the row or column containing the largest fraction of missing values was removed from the matrix. This procedure was repeated until no more rows or columns contained a fraction of missing values larger than the desired threshold.
The matrices ranging from 1% to 33% missing value thresholds (termed “namax” in the web-application) were computed. Each of them was separately clustered and the resulting dendrograms were made available through the web-application. Negative correlation distance of pairwise complete observation was used as the distance measure. The average linkage method was used in hierarchical clustering. The relative size of the bootstrap sample was increased from 50% to 140% in 10 steps. At each step, 1000 bootstrap replications were performed.
Interaction network analysis
In order to find the significant gene expression changes in our data sets, thresholds were set on the P-value as well as the magnitude of the change between induced and control. As previously described, each mircoarray data set originates from an experiment in which the expression of a gene has been altered by genetic methods. Therefore, a set of 56 “altered genes” is defined. If in any of the data sets one of the other “altered genes” is significantly changed, a connection between this and the one altered in the data set is created. The collectivity of these connection define a network (directed graph) which was plotted using the Graphviz graph visualization software11 and displayed as Scalable Vector Graphics (SVG) or in the ZGRViewer Java applet.12 The P-value and expression change threshold was made variable in the web-application. A default threshold of P-value below 0.01 and abs[log10(ratio)] above 1.5 provided an appropriate number of interactions. The network was searched for sub-networks, in which the direction of gene expression change matched the perturbation in an experiment studying this gene.
The novelty of the gene interactions was determined by searching PubMed. Edges in the web-application link to the manually confirmed PubMed source sentences. The web-application can be controlled through a menu at the top, where the first item (“HELP”) provides information on usage.
Transcription factor analysis
The TRANSFAC database of transcription factors and their binding sites13 was aquired from BIOBASE. All human genes annotated with known binding sites were extracted from the “GENE” table. The gene-factor pairs found by in-vivo ChIP-chip and ChIP-Seq experiments were extracted from the “FRAGMENT” table. Promoter sequences of all human genes from −5000 to +500 relative to the transcriptions start site were retrieved from the ENSEMBL database (version GRCh37) using the Perl API. The “match” program provided by BIOBASE was used to detect further binding sites in these sequences, using the non-redundant vertebrate profile, where matrices from the “MATRIX” table were selected with respect to minimize the rate of false positives. Matches exhibiting a matrix similarity scores above 0.9 were used. Finally, these three data-sets were pooled into one table containing 1458399 gene to binding-site pairs. Over-representation of transcription factor targets in gene sets was detected by a Fisher exact test.
Results and Discussion
Clustered experiment groups
Searching for microarray measurements based on the reported census of genes causally linked to cancer progression4 resulted in 78 individual studies, performed on 56 cancer genes. Table 1 shows which cancer genes were present in which type of perturbation. The processed data matrix of these 78 experiments with expression levels for an average of 12517 genes contained 34% missing values. In order to find groups of experiments in which genes are similarly regulated, this data matrix was clustered. Groups of cancer genes along known classical pathways were detected with high significance in bootstrapped clustering, corroborating the methodology. The dendrograms in the web-application shows clusters above the 95% significance level highlighted by red rectangles. The numbers in red next to the dendrogram branches are the “Approximately Unbiased” (AU) values provided by pvclust, from which the P-value can be deduced by 1 − AU/100.
Table 1.
Cancer genes.
| Increased | Decreased |
|---|---|
| AKT1, BCL2, BRAF, EBF1, EGFR, ERBB2, ERG, FGFR2, FGFR3, GATA6, HOXC13, HRAS, KIT, KRAS, MAP2K1, MKL1, NOTCH1, PPARG, RAF1, RB1, RET, WT1 | APC, ATM, BCL11A, BCL11B, BRCA1, CARD11, CDX2, CREB1, FH, FUS, HNF1A, HOXA11, HOXA9, IRF4, JAK2, MET, MYC, NF1, NOTCH1, NRAS, PARP1, PAX5, PBX1, POU5F1, PPARG, PTEN, RAG1, RBM15, RELA, RING1, RUNX1, SDHB, TFRC, TP53, VHL, WRN, WT1 |
The largest group of genes repeatedly detected in clustering analysis (P-value = 0.05) were based on the following experiments and brought about by cancer genes previously not known to induce common gene expression changes. These include murine sarcoma viral oncogene homolog B1 (BRAF) elevated (+), breast cancer 1 (BRCA1) reduced/absent (−), Notch homolog 1 (NOTCH1) (−), homeobox A9 (HOXA9) (−) and erythroblastosis virus E26 oncogene homolog (ERG) (+). As these perturbations lead to a common gene expression signature, it can by hypothesized that the signaling pathways that these distinct cancer genes affect converge to change a common program in gene expression. Therefore, it is conceivable that deregulated expression of this set of genes contributes to one or more general aspects of the cancer phenotype. Complete results are available in the web-application through the “Experiment Clusters” link.
In-depth analysis
The most significant cancer gene group that produced a common gene expression signature identified here included BRAF+, BRCA1−, NOTCH1−, HOXA9− and ERG+ and has been studied by different authors in different cell lines of which four were of human and one of mouse origin (Table 2). Moreover, three different kinds of microarray chips were used for the measurements. Therefore, the similarity observed is likely not based on experimental variations but rather reflects relevant changes in gene expression and commonalities following perturbations of the above-noted cancer genes. We note that there were 10866 genes common to all five experiments. Genes with average ratios below the mean of up-regulated genes or above the mean of down-regulated genes were excluded to obtain a set of 1296 regulated genes.
Table 2.
GEO Series in the most significant group of five experiments from clustering.
| Target | Type | Author | Cells | GSE | Array | N |
|---|---|---|---|---|---|---|
| BRAF | OE | Ryu | Melanocyte | 13827 | A1 | 17256 |
| BRCA1 | KD | Lee | MCF10A | 4750 | A1 | 17256 |
| HOXA9 | KD | Faber | Leukemia | 13714 | A2 | 12186 |
| ERG | OE | Carver | HEK-293 | 14595 | A2 | 12186 |
| NOTCH1 | KO | Kopan | Hair follicle | 6867 | A3 | 15125 |
Abbreviations: GSE, GEO Series identifier; N, number of probes mapped to human genes; Experiment types: OE, Over-expression; KD, Knock-down; KO, Knock-out; Array names: A1, Affymetrix Human Genome U133 Plus 2.0; A2, Affymetrix Human Genome U133A 2.0; A3, Affymetrix Mouse Genome 430 2.0.
Table 3 lists the 20 genes exhibiting comparably small standard deviation along with their function and ratios from each of the five measurements. Most of these genes control growth directly or through signaling events. The three genes most highly up-regulated were previously shown to contribute to tumor development and include ubiquitin specific peptidase 2 (USP2),14 duffy blood group chemokine receptor (DARC)15 and C-C motif chemokine receptor 3 (CCR3).16 The EED gene is a member of the Polycomb-group (PcG) family, which form multimeric protein complexes involved in maintaining the transcriptional repressive state of genes. The down-regulation of the EED gene on activation of the oncogene BRAF as well as on inactivation of the tumor suppressor BRCA1 leads to inactivation of gene silencing, pointing to a possible mechanism of neoplastic transformation mediated by these distinct cancer genes.
Table 3.
Top 20 most similar regulated genes in the group of five experiments.
| Gene | StdDev | Function | bra | brc | hox | ERG | not | C+ | C− |
|---|---|---|---|---|---|---|---|---|---|
| DDX50 | 0.048 | RNA helicase | −0.15 | −0.27 | −0.15 | −0.18 | −0.14 | 1 | 1 |
| USP2 | 0.058 | Peptidase | 0.27 | 0.38 | 0.22 | 0.35 | 0.29 | 1 | 26 |
| NCAPD2 | 0.059 | Protein binding | −0.22 | −0.22 | −0.14 | −0.09 | −0.08 | 21 | 3 |
| DARC | 0.067 | Cytokine binding | 0.18 | 0.27 | 0.15 | 0.33 | 0.21 | 4 | 46 |
| CCR3 | 0.069 | Cytokine binding | 0.4 | 0.49 | 0.38 | 0.33 | 0.51 | 0 | 0 |
| NUP37 | 0.07 | Nuclear pore | −0.25 | −0.14 | −0.07 | −0.21 | −0.08 | 17 | 1 |
| PSMG1 | 0.071 | Chaperone | −0.2 | −0.07 | −0.19 | −0.22 | −0.04 | 17 | 6 |
| IDI1 | 0.072 | Isomerase | −0.16 | −0.18 | −0.19 | −0.21 | −0.01 | 1 | 6 |
| LRIT1 | 0.072 | Protein binding | 0.22 | 0.17 | 0.29 | 0.39 | 0.28 | 0 | 1 |
| NOL4 | 0.075 | DNA binding | 0.28 | 0.38 | 0.31 | 0.46 | 0.25 | 5 | 10 |
| RPA2 | 0.075 | DNA replication | −0.28 | −0.12 | −0.15 | −0.06 | −0.11 | 2 | 1 |
| CHST1 | 0.076 | Transferase | 0.31 | 0.29 | 0.12 | 0.21 | 0.14 | 5 | 3 |
| PCBP1 | 0.078 | Translation regulator | −0.2 | −0.1 | −0.18 | −0.25 | −0.03 | 2 | 2 |
| HAT1 | 0.079 | Transferase | −0.17 | −0.04 | −0.18 | −0.26 | −0.07 | 18 | 1 |
| ADNP2 | 0.079 | Transcription regulator | −0.28 | −0.1 | −0.15 | −0.04 | −0.18 | 3 | 1 |
| CD7 | 0.079 | Receptor | 0.29 | 0.19 | 0.14 | 0.36 | 0.28 | 2 | 8 |
| DEK | 0.08 | Transcription regulator | −0.23 | −0.08 | −0.08 | −0.25 | −0.07 | 17 | 3 |
| EED | 0.081 | Transferase | −0.26 | −0.16 | −0.2 | −0.15 | −0.02 | 1 | 2 |
| KDR | 0.081 | Growth factor receptor | 0.25 | 0.34 | 0.2 | 0.39 | 0.17 | 1 | 11 |
| EDNRA | 0.081 | Peptide receptor | 0.23 | 0.22 | 0.18 | 0.33 | 0.08 | 31 | 17 |
Notes: Values are log10 ratios between induced and control. C+/C−: number significant unique analyses of cancer tissue in the Oncomine database showing up/down-regulation in the 10% rank percentile, P-value below 1e-4 and fold-change above 2.0.
Abbreviations: StdDev, standard deviation; bra, BRAF+; brc, BRCA1−; hox, HOXA9−; ERG, ERG+; not, NOTCH1−.
The Oncomine database17 was used to check for consistent differential expression in actual cancer tissue. Table 3 contains the number significant unique analyses showing up- or down-regulation in the 10% rank percentile, P-value below 1e-4 and fold-change above 2.0.
From this list, five genes (IDI1, CHST1, ADNP2, EED, EDNRA) showed consistent differential expression in a majority of analyses represented in Oncomine (see Table 3). Thirteen genes (DDX50, USP2, NCAPD2, DARC, NUP37, PSMG1, NOL4, RPA2, PCBP1, HAT1, CD7, DEK, KDR) showed consistent differential expression in a minority of analyses. Only one gene (LRIT1) was inconsistent and one gene (CCR) did not show differential expression at all.
Checking for over-representation of transcription factors in the TRANSFAC database, we found that with the exception of three genes (CD7, DARC, EDNRA), all genes can be potentially regulated by the transcription factor NF-YA (P-value = 6.79e-04). NF-YA functions as part of a heterotrimeric complex that activates a number of genes involved in cell cycle regulation, cell proliferation and survival.18,19
Networks
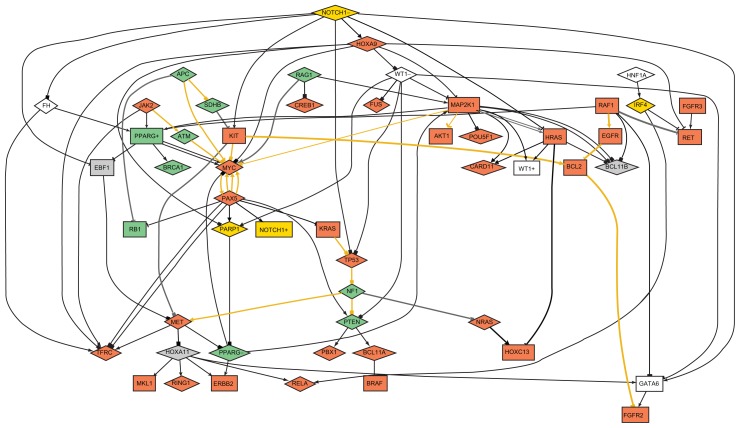
The expression value matrix for the analyzed cancer genes contained 9% missing values, as they were present in essentially all arrays used as a basis for this study. Inspection of the network of protein interactions (Fig. 2) showed that Mitogen-activated protein kinase kinase 1 (MAP2K1), Notch homolog 1 (NOTCH1), V-myc myelocytomatosis viral oncogene homolog (MYC), and Paired box 5 (PAX5) were the most highly interconnected genes, connecting each to more than six other nodes. This observation was constant at all combination of thresholds. For MYC and MAP2K1, PubMed articles confirmed many of the interactions, but only one was found for NOTCH1 and PAX5. This indicates novel links to PAX5 and NOTCH1, outlining a previous unrecognized importance for these potential cancer genes.
Figure 2.
Logical network at P-value below 0.01 and abs[log(ratio)] above 1.5.
Notes: Rectangle: increased expression; Diamond: decreased expression; Pointy arrow: up-regulation; Curved: up-regulation linked to up-expression; Tee: down-regulation; Block: down-regulation linked to down-expression; Golden edges: confirmed by PubMed evidence; Gray: false positive evidence; Black: no evidence; red node background: tumor promoting gene; green node background: tumor suppressive gene; yellow node background: tumor promoting and suppressive gene.
The expression value matrix was randomized and the resulting network compared to the one created from experimental data. In the randomized data network, node connectivity was decreased to 2.2 from 3.8 in experimental data. The number of predicted connections confirmed by publications in PubMed dropped from 16 in experimental to 4 in randomized data.
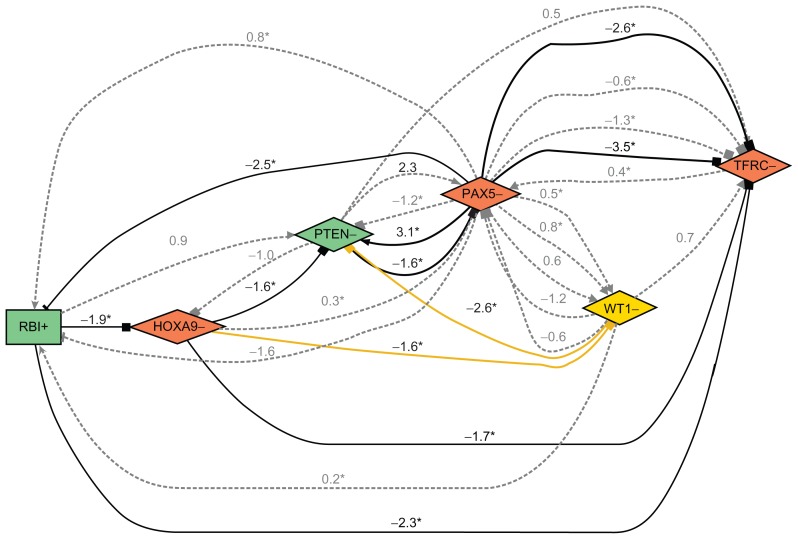
Looking at sub-networks revealed verifiable sequences of interactions between genes, as shown in Figure 3. This sub-network contained retinoblastoma 1 (RB1) (+), homeobox A9 (HOXA9) (−), Phosphatase and tensin homolog (PTEN) (−), PAX5 (−), and Transferrin receptor (TFRC) (−). These results therefore predict that RB1, a key regulator of entry into cell division, can be connected to TFRC, which regulates cellular uptake of iron, an element known to be required for cell division. This is confirmed by the measurement of a down-regulation of TFRC in the RB1 up-regulation experiment and can therefore serve as an internal validation of the prediction.
Figure 3.
Sub-network of the above network.
Notes: Number beside edge: gene expression change in target gene as log(ratio). The asterisk symbol (*) marks significant changes resulting in t-test P-values below the threshold of 0.01.
Conclusions
Through analysis of a large set of gene expression data publicly available, we were able to identify a select class of cancer-causing genes whose activities converge to deregulate a common transcription program. This finding is surprising, as the products of the identified cancer-causing genes are known to function in distinct signaling pathways. However, tumor cell evolution proceeds via a process in which genetic changes confer one or another type of growth advantage. Therefore, it is conceivable that changes in the gene expression signature described here mediate a specific aspect of tumor cell evolution. Clearly, the methodology reported here allows to extract from the inundation of gene expression information key patterns of gene expression and to connect them to the deregulation of specific cancer-causing gene products.
Acknowledgments
We thank Dr. Rehrauer (FGCZ, Zurich) and Dr. Joachim Buhmann (Laboratory of Machine Learning, ETH Zurich) for helpful comments on the manuscript.
Footnotes
Author Contributions
NF, IC and WK conceived the study. NF wrote the programs for the methodology. IC and PW annotated the cancer studies. NF and WK principally wrote the paper, with revisions and contributions from IC. All authors read and approved the final manuscript.
Disclosures and Ethics
As a requirement of publication author(s) have provided to the publisher signed confirmation of compliance with legal and ethical obligations including but not limited to the following: authorship and contributorship, conflicts of interest, privacy and confidentiality and (where applicable) protection of human and animal research subjects. The authors have read and confirmed their agreement with the ICMJE authorship and conflict of interest criteria. The authors have also confirmed that this article is unique and not under consideration or published in any other publication, and that they have permission from rights holders to reproduce any copyrighted material. Any disclosures are made in this section. The external blind peer reviewers report no conflicts of interest.
Funding
This work was supported in part by the Competence Center for Systems Physiology and Metabolic Disease.
References
- 1.Kulesh DA, Clive DR, Zarlenga DS, Greene JJ. Identification of interferon-modulated proliferation-related cDNA sequences. Proc Natl Acad Sci U S A. 1987;84:8453–7. doi: 10.1073/pnas.84.23.8453. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Barrett T, Troup DB, Wilhite SE, et al. NCBI GEO: archive for high-throughput functional genomic data. Nucleic Acids Res. 2009;37:D885–90. doi: 10.1093/nar/gkn764. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Brazma A, Hingamp P, Quackenbush J, et al. Minimum information about a microarray experiment (MIAME)-toward standards for microarray data. Nat Genet. 2001;29:365–71. doi: 10.1038/ng1201-365. [DOI] [PubMed] [Google Scholar]
- 4.Futreal PA, Coin L, Marshall M, et al. A census of human cancer genes. Nat Rev Cancer. 2004;4:177–83. doi: 10.1038/nrc1299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Rhodes D, Chinnaiyan A. Integrative analysis of the cancer transcriptome. Nat Genet. 2005;37:S31–7. doi: 10.1038/ng1570. [DOI] [PubMed] [Google Scholar]
- 6.Ramaswamy S, Ross KN, Lander ES, Golub TR. A molecular signature of metastasis in primary solid tumors. Nat Genet. 2003;33:49–54. doi: 10.1038/ng1060. [DOI] [PubMed] [Google Scholar]
- 7.Creighton CJ. Multiple oncogenic pathway signatures show coordinate expression patterns in human prostate tumors. PloS One. 2008;3:1–8. doi: 10.1371/journal.pone.0001816. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Sayers EW, Barrett T, Benson DA, et al. Database resources of the National Center for Biotechnology Information. Nucleic Acids Res. 2009;37:5–15. doi: 10.1093/nar/gkn741. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.R Development Core Team. R: A Language and Environment for Statistical Computing. R Foundation for Statistical Computing; Vienna, Austria: 2008. [Google Scholar]
- 10.Suzuki R, Shimodaira H. Pvclust: an R package for assessing the uncertainty in hierarchical clustering. Bioinformatics. 2006;22:1540–2. doi: 10.1093/bioinformatics/btl117. [DOI] [PubMed] [Google Scholar]
- 11.Ellson J, Gansner E, Koutsofios E, North S, Woodhull G. Graphviz and dynagraph—static and dynamic graph drawing tools. In: Junger M, Mutzel P, editors. Graph Drawing Software. Springer-Verlag; 2003. pp. 127–48. [Google Scholar]
- 12.Pietriga E. A toolkit for addressing hci issues in visual language environments; IEEE Symposium on Visual Languages and Human-Centric Computing (VL/HCC ‘05); Dallas, TX, USA. September 2005; pp. 145–52. [Google Scholar]
- 13.Matys V, Fricke E, Geffers R, et al. TRANSFAC: transcriptional regulation, from patterns to profiles. Nucleic Acids Res. 2003;31:374–8. doi: 10.1093/nar/gkg108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Priolo C, Tang D, Brahamandan M, et al. The isopeptidase USP2a protects human prostate cancer from apoptosis. Cancer Res. 2006;66:8625–32. doi: 10.1158/0008-5472.CAN-06-1374. [DOI] [PubMed] [Google Scholar]
- 15.Lentsch AB. The Duffy antigen/receptor for chemokines (DARC) and prostate cancer. A role as clear as black and white? FASEB J. 2002;16:1093–5. doi: 10.1096/fj.02-0066hyp. [DOI] [PubMed] [Google Scholar]
- 16.Jaehrer K, Zelle-Rieser C, Perathoner A, et al. Up-regulation of functional chemokine receptor CCR3 in human renal cell carcinoma. Clin Cancer Res. 2005;11:2459–65. doi: 10.1158/1078-0432.CCR-04-0405. [DOI] [PubMed] [Google Scholar]
- 17.Rhodes DR, Yu J, Shanker K, et al. ONCOMINE: a cancer microarray database and integrated data-mining platform. Neoplasia. 2004;6:1–6. doi: 10.1016/s1476-5586(04)80047-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Bhattacharya A, Deng JM, Zhang Z, Behringer R, de Crombrugghe B, Maity SN. The B subunit of the CCAAT box binding transcription factor complex (CBF/NF-Y) is essential for early mouse development and cell proliferation. Cancer Res. 2003;63:8167–72. [PubMed] [Google Scholar]
- 19.Caretti G, Salsi V, Vecchi C, Imbriano C, Mantovani R. Dynamic recruitment of NF-Y and histone acetyltransferases on cell-cycle promoters. J Biol Chem. 2003;278:30435–40. doi: 10.1074/jbc.M304606200. [DOI] [PubMed] [Google Scholar]