Abstract
To identify preferentially expressed genes in the central endocrine organs of the hypothalamus and pituitary gland, we generated transcriptome-wide mRNA profiles of the hypothalamus, pituitary gland, and parietal cortex in male mice (12–15 weeks old) using serial analysis of gene expression (SAGE). Total counts of SAGE tags for the hypothalamus, pituitary gland, and parietal cortex were 165824, 126688, and 161045 tags, respectively. This represented 59244, 45151, and 55131 distinct tags, respectively. Comparison of these mRNA profiles revealed that 22 mRNA species, including three potential novel transcripts, were preferentially expressed in the hypothalamus. In addition to well-known hypothalamic transcripts, such as hypocretin, several genes involved in hormone function, intracellular transduction, metabolism, protein transport, steroidogenesis, extracellular matrix, and brain disease were identified as preferentially expressed hypothalamic transcripts. In the pituitary gland, 106 mRNA species, including 60 potential novel transcripts, were preferentially expressed. In addition to well-known pituitary genes, such as growth hormone and thyroid stimulating hormone beta, a number of genes classified to function in transport, amino acid metabolism, intracellular transduction, cell adhesion, disulfide bond formation, stress response, transcription, protein synthesis, and turnover, cell differentiation, the cell cycle, and in the cytoskeleton and extracellular matrix were also preferentially expressed. In conclusion, the current study identified not only well-known hypothalamic and pituitary transcripts but also a number of new candidates likely to be involved in endocrine homeostatic systems regulated by the hypothalamus and pituitary gland.
Keywords: transcriptome, endocrine, serial analysis of gene expression
Introduction
Identification of actively transcribed genes is fundamental to understanding the function, molecular biology, and physiology of specific tissues and organs. The serial analysis of gene expression (SAGE) method and complementary DNA (cDNA) microarray or gene chip analysis are efficient strategies which enable gene expression profiling at the transcriptome level. While DNA microarray and gene chip strategies are limited by their ability to analyze only previously known genes, SAGE does not require a priori knowledge of the sequence of mRNA transcripts expressed in the tissues of interest. Thus, SAGE can reveal unexpected but important changes in the expression of a number of genes simultaneously. Moreover, SAGE allows the discovery of potential novel transcripts that are differentially expressed. Indeed, SAGE is a powerful method that can globally characterize gene expression in tissues of interest and that can detect transcripts differentially expressed under various physiological and diseases conditions (Velculescu et al., 1995; St-Amand et al., 2001; Nishida et al., 2006, 2009).
The hypothalamus and pituitary gland are key organs responsible for the maintenance of homeostasis by controlling endocrine systems and instinctive behaviors, such as feeding and drinking, metabolism, reproduction, and the stress response. A previous study using directional tag polymerase chain reaction (PCR) subtraction identified well-known hypothalamus-specific transcripts, such as oxytocin, vasopressin, and proopiomelanocortin (Pomc), as well as several novel mRNA species (Gautvik et al., 1996). A gene expression profiling study using human samples also reported that specific genes, such as proteolipid protein, myelin basic protein, carboxypeptidase E, and selenoprotein P were actively transcribed in the hypothalamus (Hu et al., 2000). Additionally, there are several reports on hypothalamic nuclei specific transcriptome using laser capture microdissection coupled with microarray technology (Segal et al., 2005; Tung et al., 2008; Paulsen et al., 2009; Jovanovic et al., 2010). For instance, an earlier study identified top 12 preferentially expressed genes in ventromedial hypothalamic nucleus relative to two adjacent hypothalamic nuclei (arcuate and dorsomedial hypothalamus), such as steroidogenic factor 1 and cytochrome P450CYP2J9 (Segal et al., 2005). Genes preferentially expressed in the pituitary have also been identified by gene profiling studies (Hu et al., 2000; Tanaka et al., 2002). These include, not only classical pituitary genes, such as growth hormone (Gh), prolactin, and Pomc, but also novel candidates responsible for the unique function of the pituitary gland (Hu et al., 2000; Tanaka et al., 2002). However, it is likely that a large number of genes that are essential for endocrine homeostatic systems of the central endocrine regulatory organs remain to be identified.
To identify preferentially expressed transcripts in the hypothalamus and pituitary gland, we generated comprehensive mRNA profiles using SAGE. In addition to the above mentioned strengths of the SAGE method, this method has a very high reproducibility, with r2 = 0.96 (Dinel et al., 2005). Moreover, no significant differences in expression levels were observed among mRNA species estimated from two SAGE libraries constructed from the same pool of total RNA (Dinel et al., 2005). Thus, the current study provides global characterization of the gene expression in the hypothalamus and pituitary gland. Our study also identifies genes with presently unknown roles, including potential novel transcripts, which may be involved in the key roles these organs essential for endocrine homeostatic systems.
Materials and Methods
Rodents and tissue sampling
Male C57BL/6 mice (12–15 weeks old) were obtained from Charles River Laboratories (St. Constant, QC, Canada). Female mice were not used to avoid variability due to menstrual cycles. The male mice were housed in an air-conditioned room (19–25°C) with controlled lighting from 07:15 to 19:15 h and were given free access to laboratory chow (Lab Rodent Diet No. 5002, Ren’s Feed and Suppliers, ON, Canada) and water. The hypothalamus, pituitary gland, and parietal cortex of 51 mice were pooled and used for expression profiling. After vertebral cervical dislocation under isoflurane anesthesia, the brain was removed from the skull and the three brain regions (hypothalamus, pituitary gland, and parietal cortex) were immediately dissected, snap-frozen in liquid nitrogen and stored at −80°C until further use. All mice were handled in a facility approved by the Canadian Council on Animal Care in accordance with their Guide for Care and Use of Experimental Animals.
SAGE and data analysis
Total RNA was isolated from tissues using an RNA extraction kit (TRIzol Reagent, Invitrogen, ON, Canada). Approximately, 6 μg of mRNA was extracted with Oligotex mRNA Mini Kit (Qiagen, Mississauga, Canada). The SAGE method was performed as previously described (Velculescu et al., 1995; St-Amand et al., 2001). In brief, double strand DNA complementary to RNA (cDNA) was synthesized from the mRNA using a biotinylated (T)18 primer and a cDNA synthesis kit (Gibco BRL, ON, Canada). The cDNA libraries were digested with the restriction enzyme NlaIII, which recognizes the sequence CATG. The 3′-terminal cDNA fragments were captured using streptavidin-coated magnetic beads (Dynal, Oslo, Norway). After ligation of two annealed linker pairs to the NlaIII-compatible sticky ends, the cDNA fragments were digested with the tagging enzyme BsmFI (site present in linkers), thereby releasing cDNA fragments that possessed short 15 base pairs tags. The released tags were ligated to form long concatemers of ditags separated by NlaIII sites. The concatemers were cloned into the SphI site of pUC19. White colonies were screened by PCR and agarose gel electrophoresis to select inserts longer than 500 bp for automated sequencing. The sequence and occurrence of each tag were analyzed by the SAGEana software program, which is a new version of SAGEparser.pl1. Tags corresponding to linker sequences were discarded and duplicate concatemers were counted only once. To identify the transcripts, the sequences of 15 bp SAGE tags (the CATG NlaIII site plus the adjacent 11 bp tags) were matched with public databases. Classification of transcripts was mainly based on the genome directory (Adams et al., 1995) at the TIGR web site2 and at gene ontology3. Gene names of preferentially expressed genes were used to search for their functions in the websites and information from previously published literatures were also used for the classification of transcripts. The SAGE tags which did not match any sequence in the public databases were classified as potential novel transcripts. SAGE method quantifies the gene expression level relatively to total mRNA population (Velculescu et al., 1995; St-Amand et al., 2001; Dinel et al., 2005; Nishida et al., 2005a, 2006, 2009). Therefore, SAGE does not require housekeeping genes as internal standard. Tag number normalized by 100,000 is shown in Tables.
Serial analysis of gene expression data are available at GEO4 under accession nos.: GSM38889, GSM38891, and GSM38893, which correspond to the raw SAGE data of the male mouse hypothalamus, pituitary gland, and parietal cortex, respectively.
Quantitative real-time PCR
For the confirmation of SAGE results, the expression levels of six mRNAs of interest [three mRNAs for preferentially expressed genes in the hypothalamus (growth factor receptor bound protein 10, pregnancy upregulated non-ubiquitously expressed CaM kinase, and huntingtin-associated protein 1) and three mRNAs for preferentially expressed genes in the pituitary gland (RAB25 member RAS oncogene family, guanine nucleotide binding protein beta 3, and serine protease inhibitor Kunitz type 2)] were measured in four male C57BL/6 mice (12–15 weeks old) using Quantitative real-time PCR (Q_RT-PCR). Brain regions (hypothalamus, pituitary gland, and parietal cortex) were harvested as described above, preserved in RNAlater solution (Applied Biosystems, CA, USA), and stored at −80°C until RNA extraction and Q_RT-PCR analysis. Total RNA was isolated from tissues using the RNAqueous®-Micro Kit (Applied Biosystems, CA, USA). First-strand cDNA was synthesized using isolated RNA with High Capacity RNA-to-cDNA kit (Applied Biosystems, CA, USA). The cDNA was analyzed in a Real-Time PCR System (StepOnePlus™, Applied Biosystems, CA, USA) with specific Taq Man probes and PCR master mix (TaqMan® Gene Expression Assays and TaqMan® Gene Expression Master Mix, Applied Biosystems, CA, USA). The relative abundance of mRNAs was calculated by the comparative cycle of threshold method with hypoxanthine guanine phosphoribosyl transferase (Hprt) mRNA as an internal standard. Hprt has stable expression levels from embryonic life through adulthood in various tissues (Warrington et al., 2000). These measurements were performed in triplicate and had <3% of coefficient of variation. The expression levels were expressed as fold changes (mean ± SE) to the parietal cortex values as control.
Statistical analyses
The comparative count display (CCD) program (Lash et al., 2000) was used to identify the transcripts differentially expressed between brain regions with more than a twofold change. The gene expression levels were compared to detect preferentially expressed transcripts, i.e., mRNA expressed at higher levels in a specific region compared to the other two brain regions. For analyzing Q_RT-PCR data, Mann–Whitney’s U test was performed. For all tests, differences were considered statistically significant at p < 0.05.
Results
Using SAGE, we have identified preferentially expressed transcripts in the central endocrine organs of the hypothalamus and pituitary gland. Total counts of SAGE tags from the hypothalamus, pituitary gland, and parietal cortex were 165824, 126688, and 161045 tags, respectively. This represented 59244, 45151, and 55131 distinct tag species, respectively. Comparison of the three transcriptomes revealed 22 and 106 transcripts expressed preferentially in the hypothalamus and pituitary gland, respectively. Among the 22 preferentially expressed transcripts in the hypothalamus, 13 were functionally classified as involved in cell signaling and communication (Table 1) or other functions (Table 2). Of the 106 preferentially expressed transcripts in the pituitary gland, 40 were functionally classified as involved in components of hormone pathways (Table 3), metabolism and cell signaling and communication (Table 4), cell and organism defense, and gene expression (Table 5), and cell structure and division (Table 6). Both the hypothalamus and pituitary gland have six preferentially expressed, functionally unclassified transcripts (Table 7). In the hypothalamus and pituitary gland, 3 and 60 potentially novel transcripts, respectively, were detected (Table 8). In addition, 62 transcripts (29 functionally classified, 8 functionally unclassified, and 25 potential novel transcripts) were preferentially expressed in the parietal cortex (Tables A1– A4 in Appendix).
Table 1.
Components of cell signaling and communication preferentially expressed in the hypothalamus.
| Function and tag sequences | Tag frequency |
Description (UniGene cluster, GenBank accession no.) | Chromosomal location | ||
|---|---|---|---|---|---|
| Parietal cortex | Pituitary gland | Hypothalamus | |||
| NEUROPEPTIDE (FEEDING BEHAVIOR) | |||||
| TGTGACGCTGG | 1 | 0 | 37 | Cocaine and amphetamine regulated transcript (Mm.75498, NM_013732) | 13 |
| GGCCGCCGCGC | 1 | 0 | 33 | Hypocretin (Mm.10096, NM_010410) | 11 |
| HORMONE | |||||
| GAGACAAGGCC | 1 | 0 | 39 | Angiotensinogen (Mm.285467, NM_007428) | 8 |
| GTCAGATGAGG | 0 | 0 | 8 | Thyrotropin releasing hormone (Mm.1363, NM_009426) | 6 |
| INTRACELLULAR TRANSDUCERS | |||||
| TAACCGTTTTC | 2 | 2 | 17 | Growth factor receptor bound protein 10 (Mm.273117, BC016111) | 11 |
| CATTGTGTGGC | 5 | 1 | 39 | Pregnancy upregulated non-ubiquitously expressed CaM kinase (Mm.89564, NM_012040) | X |
| METABOLISM | |||||
| TCTTGGGCTAA | 6 | 2 | 31 | Acetylcholinesterase (Mm.255464, NM_009599) | 5 |
| AGTTCCTTCGC | 1 | 0 | 34 | Calbindin 2 (Mm.2755, NM_007586) | 8 |
| TRANSPORT PROTEINS | |||||
| GGGCTGGGTAG | 4 | 2 | 36 | Gamma-aminobutyric acid GABA-A transporter 4 (Mm.44683, NM_172890) | 6 |
All tags are expressed at higher levels in the hypothalamus compared to other two regions, p < 0.05.
Table 2.
Other transcripts preferentially expressed in the hypothalamus.
| Function and tag sequences | Tag frequency |
Description (UniGene cluster, GenBank accession no.) | Chromosomal location | ||
|---|---|---|---|---|---|
| Parietal cortex | Pituitary gland | Hypothalamus | |||
| STEROIDOGENESIS | |||||
| GCTCTGGGAGT | 0 | 1 | 16 | 3β-Hydroxysteroid dehydrogenase-1 (Mm.140811, NM_008293) | 3 |
| EXTRACELLULAR MATRIX | |||||
| GAACATTGCAC | 14 | 13 | 228 | Secreted acidic cysteine rich glycoprotein (Mm.291442, BC004638) | 11 |
| GENES RELATED TO BRAIN DISEASE | |||||
| TCTGCCAGAAG | 3 | 5 | 43 | Huntingtin-associated protein 1 (Mm.281700, NM_010404) | 11 |
| TAGGAGCAAAG | 11 | 2 | 40 | Megalencephalic leukoencephalopathy with subcortical cysts 1 homolog human (Mm.32780, NM_133241) | 15 |
All tags are expressed at higher levels in the hypothalamus compared to other two regions, p < 0.05.
Table 3.
Hormones preferentially expressed in pituitary gland.
| Tag sequences | Tag frequency |
Description (UniGene cluster, GenBank accession no.) | Chromosomal location | ||
|---|---|---|---|---|---|
| Parietal cortex | Hypothalamus | Pituitary gland | |||
| AAGTGTCGCCG | 0 | 4 | 14963 | Growth hormone (Mm.343934, NM_008117) | 11 |
| AAGTGTTGCCG | 0 | 0 | 28 | EST growth hormone (Mm.343934, AVI 73676) | |
| CGCAGCGACGA | 0 | 0 | 218 | EST growth hormone (Mm.343934, BB749308) | |
| AAAGTGTCGCC | 0 | 0 | 40 | EST growth hormone (Mm.343934, CB236481) | |
| GCACCACGGGC | 0 | 0 | 12 | Growth hormone releasing hormone receptor (Mm.89928, XM_132546) | 6 |
| CTGTGTGGCCA | 2 | 2 | 347 | Glycoprotein hormones, alpha subunit (Mm.l361, NM_009889) | 4 |
| GTCCGAGTACT | 0 | 0 | 75 | Luteinizing hormone beta (Mm.5706l, NM_008497) | 7 |
| CTTGGGTGCAA | 0 | 0 | 1307 | Prolactin (Mm.1270, NM_011164) | 13 |
| CACTGAAAACT | 0 | 0 | 107 | EST prolactin (Mm.1270, BB025395) | |
| AACAGCCAGGG | 0 | 0 | 23 | EST prolactin (Mm.1270, CB231671) | |
| GGCGAGCTGAT | 0 | 0 | 1496 | Proopiomelanocortin-alpha (Mm.153634, BC061215) | 12 |
| GAGCACTTCCG | 0 | 0 | 21 | Proopiomelanocortin-alpha (Mm.277996, M30489) | |
| TTACTCCTTAT | 0 | 0 | 49 | Thyroid stimulating hormone, beta subunit (Mm.110730, NM_009432) | 3 |
All tags are expressed at higher levels in the pituitary gland compared to other two regions, p < 0.05.
Table 4.
Components of the metabolism and cell signaling/communication preferentially expressed in pituitary gland.
| Function and tag sequences | Tag frequency |
Description (UniGene cluster, GenBank accession no.) | Chromosomal location | ||
|---|---|---|---|---|---|
| Parietal cortex | Hypothalamus | Pituitary gland | |||
| TRANSPORT | |||||
| AAGGGTGCTGG | 6 | 7 | 39 | Annexin A5 (Mm.1620, NM_009673) | 3 |
| TGACTAGCGTG | 2 | 0 | 48 | Chromogranin B (Mm.255241, NM_007694) | 2 |
| CAGCCGGCTAT | 4 | 1 | 53 | Chromogranin B (Mm.255241, AK019893) | |
| GCTGTGGCCCT | 1 | 1 | 9 | RAB4B, member RAS oncogene family (Mm.197516, NM_019394) | 7 |
| TGATCTGCCCA | 0 | 0 | 10 | RAB25, member RAS oncogene family (Mm.26994, NM_016899) | 3 |
| AMINO ACID METABOLISM | |||||
| AAGTGTCGCTG | 0 | 0 | 41 | Mus musculus similar to spermidine/spermine Nl-acetyltransferase (LOC231081; Mm.319224, XM_131957) | X |
| INTRACELLULAR TRANSDUCERS | |||||
| ATAATGAATAA | 0 | 0 | 17 | Delta- like 1 homolog Drosophila (Mm.157069, NM_010052) | 12 |
| AGCCCCCTATT | 0 | 1 | 12 | EST dystrophia myotonica kinase, B15 (Mm.6529, BC056615) | 7 |
| ACTATGGATGT | 0 | 0 | 15 | Guanine nucleotide binding protein, beta 3 (Mm.68889, NM_013530) | 6 |
| CAGGGCCTCAC | 0 | 0 | 21 | Tumor-associated calcium signal transducer 1 (Mm.4259, BC005618) | 17 |
| TGCATAATTAA | 0 | 0 | 12 | Tumor-associated calcium signal transducer 1 (Mm.4259, XM_147278) | |
| CELL ADHESION | |||||
| TGACTCCTTGC | 3 | 4 | 29 | CD164 antigen (Mm.269815, NM_016898) | 10 |
All tags are expressed at higher levels in the pituitary gland compared to other two regions, p < 0.05.
Table 5.
Components of the cell and organism defense, and gene expression preferentially expressed in pituitary gland.
| Function and tag sequences | Tag frequency |
Description (UniGene cluster, GenBank accession no.) | Chromosomal location | ||
|---|---|---|---|---|---|
| Parietal cortex | Hypothalamus | Pituitary gland | |||
| DISULFIDE BONDS FORMATION | |||||
| CAGGAGGAGTT | 13 | 21 | 62 | Glucose-regulated protein (Mm.263177, NM_007952) | 2 |
| GCCTGTGGCCT | 7 | 11 | 48 | Prolyl 4-hydroxylase, beta polypeptide (Mm.16660, XM_126743) | 11 |
| STRESS RESPONSE | |||||
| ACAAAACTGAT | 6 | 7 | 28 | EST heat shock 70 kD protein 5 glucose-regulated protein (Mm.330160, CB233866) | 2 |
| TRANSCRIPTION FACTORS | |||||
| TAATGAGATTA | 1 | 1 | 11 | Achaete–scute complex homolog-like 1 Drosophila (Mm.76039, BC055748) | 10 |
| TCGCCGGGCGC | 0 | 0 | 8 | Paired-like homeodomain transcription factor 1 (Mm.135195, BC012696) | 13 |
| ATAGGTTTCTC | 0 | 0 | 13 | Sine oculis-related homeobox 6 homolog Drosophila (Mm.57138, AF050130) | 12 |
| PROTEIN SYNTHESIS AND TURNOVER | |||||
| AAGGAAGACGG | 0 | 0 | 8 | Proteosome (prosome, macropain) subunit, beta type 8 (large multifunctional protease 7; Mm.180191, NM_010724) | 17 |
| GGAAGCCACTT | 28 | 25 | 78 | Ribosomal protein S27a (Mm.l80003, NM_024277) | 9 |
| GTTCTGATCCT | 2 | 7 | 28 | Serine protease inhibitor, Kunitz type 2 (Mm.295230, NM_011464) | 7 |
| CELL DIFFERENTIATION | |||||
| CGCCTGCGAAG | 0 | 0 | 9 | Claudin 9 (Mm.103738, BC058186) | 17 |
| GGGGGAGTGGA | 22 | 203 | 848 | Neuronatin (Mm.233903, NM_010923) | 2 |
All tags are expressed at higher levels in the pituitary gland compared to other two regions, p < 0.05.
Table 6.
Components of cell structure and division preferentially in the pituitary.
| Function and tag sequences | Tag frequency |
Description (UniGene cluster, GenBank accession no.) | Chromosomal location | ||
|---|---|---|---|---|---|
| Parietal cortex | Hypothalamus | Pituitary gland | |||
| CYTOSKELETAL | |||||
| CAAACTGTGCA | 1 | 1 | 12 | Keratin complex 1, acidic, gene 18 (Mm.22479, NM_010664) | 15 |
| EXTRACELLULAR MATRIX | |||||
| CGCCTGCTAGC | 1 | 1 | 21 | Procollagen, type I, alpha 2 (Mm.277792, NM_007743) | 6 |
| CELL CYCLE | |||||
| ACCAAAACGCA | 2 | 2 | 15 | Cyclin D2 (Mm.333406, NM_009829) | 6 |
| TACCCGCCGTC | 1 | 1 | 18 | EST Cyclin D2 (Mm.333406, BQ445605) | |
All tags are expressed at higher levels in the pituitary gland compared to other two regions, p < 0.05.
Table 7.
Unclassified transcripts preferentially expressed in the hypothalamus and pituitary gland.
| Tag sequences | Tag frequency |
Description (UniGene cluster, GenBank accession no.) | Chromosomal location | ||
|---|---|---|---|---|---|
| Hypothalamus | Pituitary gland | Parietal cortex | |||
| HYPOTHALAMUS | |||||
| ACCCTGCTGTG | 55 | 9 | 13 | Abelson helper integration site (Mm.253280, NM_026203) | 10 |
| GAGAACTGGTT | 59 | 10 | 19 | Dispatched homolog 2 Drosophila (Mm.221499, NM_170593) | 2 |
| AAGAACAGGAG | 23 | 2 | 2 | Hypothetical protein 9430073I07 (Mm.l33187, NM_173016) | 8 |
| ACAGCAGTGTC | 45 | 8 | 0 | RIKEN cDNA 0610007P22 gene (Mm.328857, BC022659) | 17 |
| TGTATCCAGTG | 133 | 17 | 35 | RIKEN cDNA 1110020M21 gene (Mm.191892, NM_021432) | 6 |
| GCTGACTCTTG | 31 | 1 | 2 | RIKEN cDNA 2700055K07 gene (Mm.29358, NM_026481) | 8 |
| PITUITARY GLAND | |||||
| TTTCTTTGTTA | 0 | 8 | 0 | cDNA sequence BC031468 (Mm.23713, NM_194055) | 4 |
| TGCTGAGATCA | 2 | 15 | 2 | EST GTL2, imprinted maternally expressed untranslated mRNA (Mm.289645, BM123457) | 12 |
| GTGCTTTCAAT | 1 | 31 | 2 | EST GTL2, imprinted maternally expressed untranslated mRNA (Mm.289645, Y13832) | 12 |
| CTCCTCCACTC | 0 | 9 | 0 | Expressed sequence AI987662 (Mm.206911, NM_178899) | 6 |
| TGAGAGGTGCC | 2 | 36 | 1 | RIKEN cDNA 5330417C22 gene (Mm.316890, BC022655) | 3 |
| GATGGATGGTG | 0 | 15 | 0 | RIKEN cDNA 5330437I02 gene (Mm.183576, NM_177028) | 18 |
Tags are expressed at higher levels in each brain region compared to other two regions, p < 0.05.
Table 8.
Potential novel transcripts in the hypothalamus and pituitary gland.
| Tag sequences | Tag frequency |
||
|---|---|---|---|
| Hypothalamus | Pituitary gland | Parietal cortex | |
| HYPOTHALAMUS | |||
| GAAAATGAGAA | 80 | 13 | 18 |
| ATTTTCAGTTT | 26 | 4 | 2 |
| TGGGTTCTGAC | 10 | 0 | 1 |
| PITUITARY GLAND | |||
| GCGGGAAAAGC | 1 | 1383 | 0 |
| AAGTGTCGCCC | 0 | 646 | 0 |
| AAGTGTCGCCT | 0 | 601 | 0 |
| TTGGCGTCAAA | 0 | 436 | 0 |
| AAGTGTCGCCA | 0 | 408 | 0 |
| TCGGTTCTCTG | 0 | 329 | 0 |
| ACGTACTTCCG | 1 | 222 | 0 |
| GCTGGGGCCCG | 0 | 194 | 0 |
| TATGAAGAGAA | 33 | 156 | 7 |
| ACCCGCAGGTA | 0 | 114 | 0 |
| CCCTTGTCCAG | 0 | 77 | 0 |
| GAGTGTCGCCG | 0 | 62 | 0 |
| CAGGGAGGGGG | 0 | 53 | 0 |
| GAGTAGGAGCG | 1 | 53 | 0 |
| GTGAAGAAACC | 7 | 50 | 9 |
| AGGTGTCGCCG | 0 | 42 | 0 |
| AAGCGTCGCCG | 0 | 40 | 0 |
| ACCTCCGAGAA | 0 | 39 | 0 |
| AAGCCACCGTA | 0 | 37 | 0 |
| AAGTGCCGCCG | 0 | 36 | 0 |
| TCTGGACGGTC | 7 | 36 | 1 |
| TGTGCTGGCTT | 1 | 34 | 0 |
| ACCAGAATGAC | 0 | 30 | 0 |
| GTGACCACGGG | 5 | 28 | 7 |
| CAACCCCATTC | 2 | 28 | 2 |
| GCGGGAAAAGG | 0 | 26 | 0 |
| CTGACACAGCC | 0 | 25 | 0 |
| AAGTGACGCCG | 0 | 24 | 0 |
| AAGAGTCGCCG | 0 | 23 | 0 |
| GCGGGAAAAGT | 0 | 21 | 0 |
| GTAGGATCTTG | 4 | 21 | 1 |
| AAGTGTCACCG | 0 | 21 | 0 |
| GGAAGCTTTCA | 0 | 21 | 0 |
| TGGCCTCCGAG | 0 | 19 | 0 |
| AAGTGTCTCCG | 0 | 17 | 0 |
| GCATTAGCATT | 1 | 16 | 1 |
| GAATTCAATGA | 0 | 16 | 0 |
| AAGTGTCCCCG | 0 | 16 | 0 |
| AAATGTCGCCG | 0 | 15 | 0 |
| GTCACAGAGCT | 1 | 15 | 1 |
| ATGTGTCGCCG | 0 | 14 | 0 |
| GAATTGCTTCG | 0 | 14 | 0 |
| CTTGGGTGCGA | 0 | 13 | 0 |
| AAGTATCGCCG | 0 | 13 | 0 |
| ACTGGATGAGC | 0 | 13 | 1 |
| TGCTTGAGTTT | 1 | 12 | 0 |
| GGTCACTTCCG | 0 | 12 | 0 |
| CAAATCAGGCT | 0 | 11 | 1 |
| GCGGGAAAAGA | 0 | 10 | 0 |
| ACGTACTTCCC | 0 | 10 | 0 |
| ACGTACTTCCT | 0 | 10 | 0 |
| TGGCTAAGAGT | 0 | 10 | 0 |
| AAGTGTGGCCA | 0 | 10 | 0 |
| AAGTGTCGTCG | 0 | 10 | 0 |
| GTGCCCCGGCC | 1 | 10 | 0 |
| AAGACCATAGC | 0 | 9 | 0 |
| GAAGTGTCGCC | 0 | 9 | 0 |
| TTCCTGCATAT | 0 | 8 | 0 |
| TAGTGTCGCCG | 0 | 8 | 0 |
| GGCGAGCAGAA | 0 | 8 | 0 |
Tags are expressed at higher levels in each brain region compared to other two regions, p < 0.05.
Preferentially expressed genes in the hypothalamus
Three well-known hypothalamic transcripts, cocaine and amphetamine regulated transcript (Cart), hypocretin, and thyrotropin releasing hormone (Trh), were preferentially expressed in the hypothalamus (Table 1). Genes involved in hormone pathways (angiotensinogen), intracellular transduction [growth factor receptor bound protein 10 (Grb10) and pregnancy upregulated non-ubiquitously expressed CaM kinase (Pnck)], metabolism (acetylcholinesterase and calbindin 2), and protein transportation [gamma-aminobutyric acid GABA-A transporter 4 (Gat4)] were also preferentially expressed. Other hypothalamic genes identified were genes involved in steroidogenesis [3β-hydroxysteroid dehydrogenase-1 (3β-Hsd-1)], the extracellular matrix [secreted acidic cysteine rich glycoprotein (Sparc)], and mRNAs related to brain disease [huntingtin-associated protein 1 (Hap1) and megalencephalic leukoencephalopathy with subcortical cysts 1 homolog human (Mlc1); Table 2].
Preferentially expressed genes in the pituitary gland
A number of well-known pituitary genes, such as Gh, GH releasing hormone receptor, glycoprotein hormones alpha subunit, luteinizing hormone (Lh) beta, prolactin, proopiomelanocortin (Pomc), and thyroid stimulating hormone (Tsh) beta subunit were preferentially expressed in the pituitary gland (Table 3). Genes involved in transport [annexin A5 (Anxa5), chromogranin B, members of the RAS oncogene family, RAB4B and RAB25], amino acid metabolism [Mus musculus similar to spermidine/spermine N1-acetyltransferase (Sat1)], intracellular transducers [delta-like 1 homolog Drosophila (Dlk1), dystrophia myotonica kinase (Dmpk), guanine nucleotide binding protein beta 3 (Gnb3), and tumor-associated calcium signal transducer 1], and cell adhesion (CD164 antigen) were also preferentially expressed in this organ (Table 4). Other pituitary genes identified were potentially involved in disulfide bond formation [glucose-regulated protein (Grp58) and prolyl 4-hydroxylase beta polypeptide (P4hb)], stress response (heat shock 70 kD protein 5 glucose-regulated protein), transcription [achaete–scute complex homolog-like 1 Drosophila (Ascl1), paired-like homeodomain transcription factor 1 (Pitx1), and sine oculis-related homeobox 6 homolog Drosophila (Six6)], protein synthesis and turnover [proteasome (prosome, macropain) subunit beta type 8 (large multifunctional protease 7), ribosomal protein S27a, and serine protease inhibitor Kunitz type 2 (Spint2)] and cell differentiation (claudin 9 and neuronatin; Table 5). Moreover, three transcripts involved in the cytoskeleton (keratin complex 1 acidic gene 18), the extracellular matrix [procollagen type I alpha 2 (Col1a2)], and cell cycle (cyclin D2) were identified as preferentially expressed pituitary genes (Table 6).
Confirmation of SAGE results with Q_RT-PCR
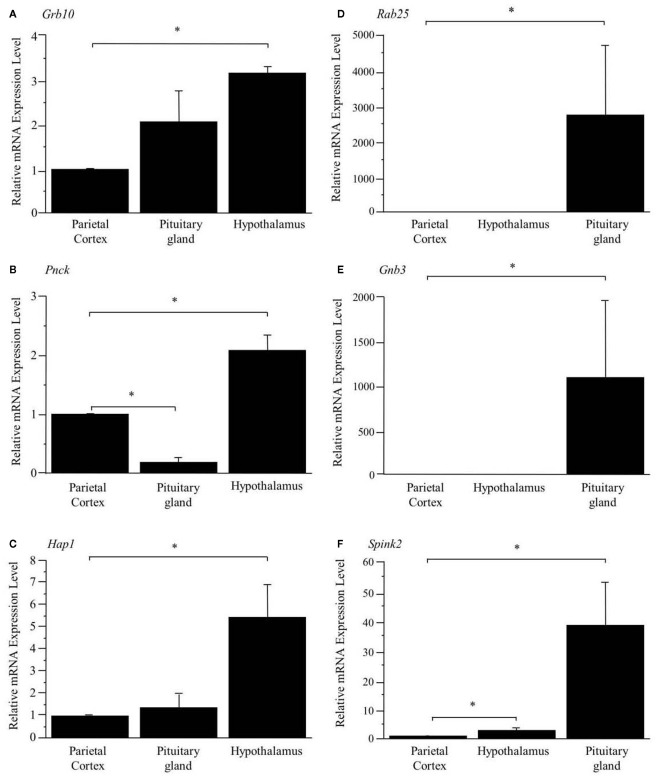
As shown in Figure 1, the preferential expression of three genes in the hypothalamus (A, B, C) and three genes in the pituitary gland (D, E, F) detected by SAGE were confirmed by Q_RT-PCR.
Figure 1.
Confirmation of preferentially expressed genes in the hypothalamus (A–C) and pituitary gland (D–F). The expression levels are normalized to Hprt and are shown as fold difference compared with the parietal cortex. *Represents a significant difference compared to the parietal cortex (p < 0.05). Data are represented as mean ± SE.
Discussion
The 13 preferentially expressed mRNAs in the hypothalamus included neuropeptides, hormones, intracellular transducers and genes involved in metabolism, steroidogenesis, the extracellular matrix, and brain disease (Tables 1 and 2). Among the 13 transcripts, 4 are well-known hypothalamic neuropeptides or hormones (Cart, hypocretin, angiotensinogen, and Trh). For instance, hypocretin and Cart are key hypothalamic genes involved in the regulation of feeding behavior. Hypocretin is known as an orexigenic gene, while Cart is an anorexigenic gene (Kristensen et al., 1998; Sakurai, 2003). Hypocretin has other functions than the regulation of food intake; it regulates sleep and awake states and the hypocretin knockout mouse is a model of human narcolepsy, a hypersomnia disorder (Chemelli et al., 1999).
The preferentially expressed hypothalamic transcripts involved in intracellular transduction were Grb10 and Pnck. GRB10 is a member of a superfamily of adaptor proteins. In mouse, the Grb10 gene is expressed from the maternal allele in almost all organs; however, in the brain this gene shows biallelic expression (Hitchins et al., 2002). GRB10 binds to the tyrosine-phosphorylated insulin receptor via its Src homology 2 domain and inhibits signal transduction (Langlais et al., 2004). GRB10 is known as a potential regulator of GH signaling in hepatoma cells (Moutoussamy et al., 1998), while in brain GRB10 is involved in neuronal insulin signaling and energy metabolism (Lim et al., 2004). Another transcript involved in intracellular transduction was Pnck. Ca2+ is an important intracellular second-messenger in signal transduction pathways. Many of the effects of Ca2+ are mediated through its interaction with the Ca2+-binding protein, calmodulin. The Ca2+/calmodulin complex activates calmodulin-dependent protein kinases, which ultimately regulate cellular processes, such as neurotransmitter release, metabolism, and gene expression (Schulman, 1993; Gardner et al., 2000). Pnck mRNA is expressed in a tissue-specific manner in adult mice with the highest levels of expression in the sex organs, the uterus, ovary, and testis, and in the brain (Gardner et al., 2000). Since the current study showed that Pnck mRNA is preferentially expressed in the hypothalamus, the expression of this gene might be important not only in the peripheral reproductive organs but also in the central hypothalamus, which regulates the secretion of sex hormones from the uterus, ovary, and testis.
The hypothalamus is a central brain region of the autonomic nervous system that uses acetylcholine as a neurotransmitter. Acetylcholinesterase inhibits the actions of acetylcholine by degrading acetylcholine into acetate and choline. The preferential expression of acetylcholinesterase is consistent with a previous report showing neuronal acetylcholinesterase staining in the human hypothalamus (Darvesh and Hopkins, 2003). Acetylcholinesterase gene expression can be critical for controlling the activity of sympathetic and parasympathetic nervous systems; the preferential expression of acetylcholinesterase is a molecular feature of the hypothalamus, which plays an important role as a brain center of the autonomic nervous system. Another transcript classified under metabolism is calbindin 2 (or calretinin). Calbindin 2 has Ca2+ buffering action against cytotoxicity of Ca2+ overload in brain neurons (Persechini et al., 1989). Calbindin 2 is present in oxytocin nerve terminals in the posterior pituitary, whose cell bodies are located in the hypothalamus, and dehydration induced by drinking 2% NaCl and deprivation of drinking water increased the level of calbindin 2 protein in the posterior pituitary (Miyata et al., 2000). Thus, the current data support the view that calbindin 2 is a key hypothalamic molecule involved in the regulation of drinking behavior.
GAT4 is a transporter of gamma-aminobutyric acid (GABA), one of the major inhibitory neurotransmitters in the central nervous system. There are four GABA transporters, GAT1–4 (Nelson, 1998). Gat2 and Gat3 are expressed in non-neural organs, such as liver and kidney, whereas Gat4 and GAT1 are expressed in the brain (Nelson, 1998). They play a role in the regulation of GABA concentration in the synapse, and in its duration of action by removing GABA from the synaptic cleft (Iverson, 1975). The preferential expression of Gat4 in the hypothalamus is in agreement with a previous study using in situ hybridization which showed that Gat4 mRNA was observed in several brain regions, including the hypothalamus, but not the cortex (Durkin et al., 1995). In hypothalamic neurons, neuropeptide Y (NPY) and POMC exert opposing actions on energy balance with NPY increasing food intake. Subpopulations of NPY neuron terminals contain GABA and synapse onto POMC neurons (Cowley et al., 2001). In addition, POMC neurons also release GABA (Hentges et al., 2004). The preferential expression of Gat4 in the hypothalamus may suggest an important role for Gat4 in the regulation of GABA concentration in the hypothalamus.
Transcripts involved in steroidogenesis (3β-Hsd-1) and in the extracellular matrix (Sparc) are also preferentially expressed in the hypothalamus. It has been reported that the brain is a steroidogenic organ that expresses the key steroidogenic enzyme, 3β-Hsd-1 (Weidenfeld et al., 1980; Zwain and Yen, 1999). 3β-Hsd-1 converts pregnenolone to progesterone, which is an important hormone for the normal reproductive behavior in both sexes (Olster and Blaustein, 1988). Thus, preferential expression of 3β-Hsd-1 in the hypothalamus may suggest an involvement of this gene in the regulation of sexual behavior by locally producing progesterone. Sparc was also preferentially expressed in the hypothalamus. We previously showed that Sparc mRNA is one of the most abundant transcripts in the hypothalamus (Nishida et al., 2005b). Interestingly, Sparc-null mice have greater deposits of subcutaneous fat, larger epididymal fat pads and elevated levels of serum leptin compared to wild-type counterparts (Bradshaw et al., 2003). On the other hand, Sparc mRNA levels in adipose tissue have also been reported to be markedly increased (three- to sixfold) in three different rodent models of obesity (Tartare-Deckert et al., 2001). Physiological roles of the preferentially expressed Sparc gene in the hypothalamus remain to be determined.
Two transcripts related to brain disease (Hap1 and Mlc1) were preferentially expressed in the hypothalamus. Huntington’s disease is an inherited autosomal dominant disorder, with progressive dementia and abnormal movements that usually take the form of choreic, rapid dance-like movements, typically of the hands and neck. Huntington’s disease is caused by trinucleotide expansions of the glutamine encoding CAG triplet in the huntingtin gene. The polyglutamine expansion causes huntingtin to interact abnormally with a number of proteins. HAP1 was found as a protein which binds to huntingtin in proportion to the number of glutamines present in the glutamine repeat region (Li et al., 1995). Interestingly, mice lacking Hap1 showed neurodegeneration in the hypothalamic region and decreased feeding behavior and loss of body weight (Li et al., 2003). Another transcript classified as involved in brain disease is Mlc1. The human Mlc1 gene and its murine homolog encode a putative transmembrane protein expressed primarily in brain. Mlc1 is highly expressed in the olfactory bulb and cerebellum (Schmitt et al., 2003). Recessive mutations within this gene cause megalencephalic leukoencephalopathy with subcortical cysts, whose diagnostic features are motor disability in the form of spasticity and ataxia, occasional seizures, mild cognitive decline with slow progression (Singhal et al., 2003). The precise physiological role of Mlc1 in the hypothalamus remains to be clarified.
The pituitary gland is characterized by preferential expression of genes related to hormones, such as Gh, GH releasing hormone receptor, prolactin, Tsh beta subunit, Lh beta, glycoprotein hormone alpha subunit, and Pomc (Table 3). The detection of preferential expression of well-known pituitary and hypothalamic markers validates our application of the SAGE technique.
Many mRNA transcripts involved in metabolism and cell signaling/communication are preferentially expressed in the pituitary (Table 4). Our results showing the preferential expression of chromogranin B is in agreement with a previous report which used human pituitary gland (Tanaka et al., 2002). Another transcript, Anxa5 is classified as a transporter that stimulates Lh beta mRNA expression and secretion of LH and follicle-stimulating hormone (FSH; Kawaminami et al., 2002). In addition, in the current study, RAB4B and RAB25 were preferentially expressed in the pituitary. RAB proteins are low molecular weight GTPases that are master regulators of vesicle transport. Vesicle transport is an essential process for secretion, since newly synthesized secretory proteins are translocated into the endoplasmic reticulum and are then transported to the membrane via the Golgi apparatus by vesicles (Takai et al., 2001). Among the RAB proteins, RAB27B is associated with secretory granules and granuphilin, an effector protein of RAB27A, in a corticotroph cell line, and over-expression of inactive RAB27B mutants significantly inhibits adrenocorticotropic hormone (ACTH) secretion in these cells (Zhao et al., 2002). The current results suggest that RAB25 and RAB4B mRNAs are new candidates for important roles in the secretory pathway in the pituitary gland.
Spermine, spermidine, and putrescine are representative polyamines, which are abundantly present in endocrine organs, such as the prostate and pancreas, as well as in carcinomas. The polyamines function in cell proliferation and growth. The current study showed that M. musculus similar to Sat1 was preferentially expressed in the pituitary. This is a rate-limiting enzyme which catalyzes the N1 acetylation of spermidine and spermine, and converts them to putrescine. Female transgenic mice over-expressing Sat1 are infertile (Pietila et al., 1997). Because the reproductive function of the ovary is regulated by gonadotropin hormones, which are synthesized in the pituitary gland, the current result suggests potential importance of Sat1 expression in the regulation of pituitary gonadotropin hormones.
Heterotrimeric guanine nucleotide binding proteins, known as G proteins, play a key role in signal transduction from seven-transmembrane receptors to intracellular effectors. They are heterodimers composed of three distinct subunits, alpha, beta, and gamma (Lania et al., 2001). Among these subunits, the current study revealed that Gnb3 was preferentially expressed in the pituitary. Previous studies have reported that defects in G protein-coupled signal transduction cause endocrine disorders (Lania et al., 2001). Previous and current data, therefore, suggest that this molecule may play an important role in normal endocrine regulation, by its preferential expression in the pituitary gland.
Dlk1 is an epidermal growth factor (EGF)-like protein, synthesized as a transmembrane protein with six tandem EGF like repeats. Dlk1 expression is restricted to endocrine organs, such as the adrenal gland and placenta (Laborda et al., 1993). The preferential expression of Dlk1 in the pituitary is in agreement with the expression of this gene being restricted to endocrine organs. Myotonic dystrophy, which shows a progressive decrease in muscle mass, is an autosomal dominant disorder caused by CTG repeat expansion in the Dmpk gene. Patients with myotonic dystrophy present hypogonadism and higher basal levels of ACTH and corticosteroid (Brisson et al., 2002). The current result of preferential expression of Dmpk in the pituitary supports the idea that this gene plays an important role in gonadotropin and ACTH secretion.
The current study also revealed two pituitary genes (three mRNAs) classified under metabolism and cell signaling/communication (tumor-associated calcium signal transducer 1 and CD164 antigen). Tumor-associated calcium signal transducer 1 (Epithelial cell adhesion molecule) was first identified as a tumor-specific antigen on several carcinomas. CD164, is expressed in hematopoietic cells in bone marrow and plays a role in hematopoiesis (Zannettino et al., 1998). CD164 protein contains 178 amino acids and is extremely rich in serine and threonine (Zannettino et al., 1998). Thus, these molecules with presently unknown functions in the pituitary are promising targets for future studies of cell signaling in the pituitary gland.
We detected three preferentially expressed transcription factors in the pituitary (Table 5). PITX1 was previously characterized as a mouse transcription factor on the basis of its ability to activate pituitary transcription of the Pomc gene (Lamonerie et al., 1996). Six6 is strongly expressed in the developing pituitary gland (Lopez-Rios et al., 1999) and Ascl1 is a basic helix-loop-helix transcription factor and acts as a developmental regulator in the mammalian central nervous system (Guillemot et al., 1993). In addition, there are two preferentially expressed transcripts involved in cell differentiation. Neuronatin is selectively expressed in the developing brain (Joseph et al., 1994). In adult humans, the anterior pituitary gland was the only place where neuronatin was highly expressed among various tissues (Usui et al., 1997). We previously reported that neuronatin is one of the most abundant transcripts in the mouse pituitary (Nishida et al., 2005b). Claudin 9 belongs to the claudin family of tight junction transmembrane proteins, which is expressed in the developing mouse submandibular gland (Hashizume et al., 2004). The current results suggest that these genes have important roles not only in the developing but also in the adult pituitary gland.
We also identified three preferentially expressed genes involved in protein degradation, ribosomal protein S27a, which encodes ubiquitin, SPINT2, a proteasome (prosome, macropain) subunit, and proteasome subunit beta type 8 (large multifunctional protease 7). Proteasome inhibitors induce apoptosis in GH- and prolactin-secreting rat pituitary tumor cells (Yu et al., 2002). In addition, exposure of pancreatic islets to cytosolic cysteine protease (calpain) inhibitors impairs mitochondrial fuel metabolism and the exocytosis of insulin (Zhou et al., 2003). Northern blot analysis showed that Spint2 mRNA was expressed in endocrine organs, such as pancreas, prostate, and testis (Kawaguchi et al., 1997). Spint2 contains a putative N-terminal signal peptide sequence, a hydrophobic membrane-associated C-terminal region and a central region that encodes the cleaved protein, suggesting that this protein could be produced as an active membrane-associated protein that is proteolytically cleaved and secreted (Kawaguchi et al., 1997). Furthermore, DNA arrays revealed that Spint2 is expressed at significantly lower levels in abnormal testis in humans (Rockett et al., 2004). These previous results and the preferential expression of these three genes in the pituitary suggest important roles for genes related to protein synthesis and degradation in the regulation of pituitary endocrine function.
P4HB is a ubiquitous, abundant protein that is located primarily in the endoplasmic reticulum (Cheng et al., 1987). P4HB has a protein disulfide isomerase activity (Koivu et al., 1987). Disulfide bonds between and within polypeptides stabilize tertiary and quaternary structures. GRP58 also has protein disulfide isomerase activity. Since GH has two disulfide bonds and its mRNA is highly abundant in the adult pituitary gland (Nishida et al., 2005b), it is reasonable that genes involved in forming disulfide bonds are preferentially expressed in the pituitary.
There are four preferentially expressed transcripts classified as components of cell structure and cell division (Table 6). The precise function of these genes in the adult pituitary is unclear at present. One of these genes is cyclin D2. This gene is known to be extensively inactivated in various pituitary tumor cell lines by increased DNA methylation (Qian et al., 2000). Ovaries of FSH-deficient mice demonstrate a modest decrease in cyclin D2 mRNA, without up-regulation of cell cycle inhibitor mRNAs associated with luteinization (Burns et al., 2001). Although components of the FSH-null phenotype may be caused by partial cyclin D2 loss of function, these findings indicate that the mechanisms of granulosa cell cycle arrest in FSH beta knockout mice are distinct from those of cycle withdrawal at luteinization (Burns et al., 2001). The preferential expression of cyclin D2 suggests that this molecule plays a key role for normal cell cycling in the adult pituitary gland.
The present study has identified six unclassified and three potential novel transcripts preferentially expressed in the hypothalamus (Table 7). Moreover, 6 unclassified and 60 potential novel transcripts were detected to be preferentially expressed in the pituitary (Table 8). These mRNAs potentially play important roles in these central endocrine regions; therefore, full-length cloning and further characterization of the potential novel transcripts should be undertaken.
There is a limitation of the current study that should be described. We used whole hypothalamus, which is a heterogeneous region of the brain. Since each of hypothalamic nuclei occupies relatively a small portion of whole hypothalamus, mRNA species concentrated in a specific nucleus might be diluted by other parts of the hypothalamus to an undetectable level. For instance, hypothalamic neuropeptides POMC and NPY are localized in the arcuate nucleus (Cowley et al., 2001). In the current study, we could not detect tags corresponding to Pomc and Npy, probably because of their low abundance in the whole hypothalamus. Additionally, orexigenic Npy is not expected to be abundantly expressed in the hypothalamus of ad libitum fed mice used in the present study. It is assumed that low abundance mRNA species such as Pomc and Npy would be detected with greater number of SAGE tags. Since a large amount of total RNA (100 μg per group) was required to generate each cDNA library in the current SAGE study, we used 51 mice to obtain sufficient amount of RNA from the small brain regions, and to eliminate the individual variation. Thus, for the SAGE study it would be impractical to use specific hypothalamic nuclei in mice, which is exceedingly small. Other researchers have investigated transcriptome from individual hypothalamic nuclei using laser capture microdissection coupled with microarray technology (Segal et al., 2005; Tung et al., 2008; Paulsen et al., 2009; Jovanovic et al., 2010). SAGE method can precisely measure changes in the expression of previously well-studied genes, as well as the less commonly studied, and even novel transcripts. Additionally, although SAGE is time-consuming, this method does not suffer from non-specific hybridization. Thus, further effort to identify preferentially expressed genes in the hypothalamic nuclei specific transcriptome using superior techniques such as laser capture coupled with SAGE is warranted.
In conclusion, the current analysis of mRNA profiles identified a number of genes with preferential expression in the central endocrine organs of the hypothalamus and pituitary gland. In addition to well-known genes, such as Cart and Trh, the current study revealed that genes encoding Grb10, Pnck, Gat4, 3β-Hsd-1, Hap1, and Mlc1 were preferentially expressed in the hypothalamus. In the pituitary gland, not only well-known pituitary genes, such as Gh, but a number of other genes, such as Anxa5, Sat1, Dlk1, CD164 antigen, Grp58, Ascl1, Spint2, claudin 9, and cyclin D2 were also preferentially expressed in the pituitary. Moreover, the current study showed a number of preferentially expressed, uncharacterized transcripts, including potential novel transcripts (3 in the hypothalamus and 60 in the pituitary). Therefore, the current study characterized the global preferential gene expression in the central endocrine organs of the hypothalamus and pituitary gland. In addition to the preferential expression of several well-known hypothalamic and pituitary genes, the current SAGE analysis identified a number of new candidates involved in endocrine homeostatic systems regulated by the hypothalamus and pituitary gland, such as feeding and drinking, metabolism, reproduction, and the stress response.
Conflict of Interest Statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Acknowledgments
We thank Drs. Céline Martel, Claude Labrie, Ei’ichi Iizasa, and Kazutoshi Nishijima for procedures with mice, Marc André Rodrigue and Dr. Vincent Raymond for SAGE tag sequencing, Pascal Belleau and Dr. Jean Morissette for bioinformatic analyses, and Dr. Fernand Labrie, the leader of the ATLAS project. This work was supported by grants from Genome Quebec and Genome Canada, the Japanese Ministry of Education, Culture, Sports, Science, and Technology (No. 19200049, Strategic Research Infrastructure), and the Global FU Program, funded by Fukuoka University. Dr. Mayumi Yoshioka is supported as a postdoctoral fellow by the Heart and Stroke Foundation of Canada (HSFC), the Canadian Institute of Health Research (CIHR), and the Canadian Diabetes Association (CDA). Dr. Jonny St-Amand is supported by the Fonds de la recherche en santé du Québec (FRSQ).
Appendix
Table A1.
Components of cell signaling preferentially expressed in the cortex.
| Function and tag sequences | Tag frequency |
Description (UniGene cluster, GenBank accession no.) | Chromosomal location | ||
|---|---|---|---|---|---|
| Hypothalamus | Pituitary gland | Parietal cortex | |||
| CALCIUM AND CALMODULIN SIGNAL TRANSDUCTION | |||||
| GCTTCCCCACC | 8 | 0 | 82 | Calcium/calmodulin-dependent protein | 18 |
| ACAAACTTAGG | 68 | 32 | 171 | Calmodulin 1 (Mm.285993, M 19381) | 12 |
| GTCCTGCAGGA | 1 | 0 | 14 | Inositol 1,4,5-trisphosphate 3-kinase A | 2 |
| CAGCTCTGCCT | 29 | 0 | 363 | Neurogranin (Mm.335065, AK002933) | 9 |
| TTACCATACTG | 0 | 0 | 9 | Neurogranin (Mm.335065, NM_022029) | |
| AAGGACTGATT | 2 | 2 | 30 | Protein phosphatase 3, catalytic subunit, alpha isoform (Mm.80565, XM_131226) | 3 |
| AGGTTTTGTTT | 1 | 1 | 26 | RAS guanyl releasing protein 1 (Mm.42150, BC057341) | 2 |
| CELL ADHESION | |||||
| GCTGTGGGTCG | 3 | 1 | 17 | Intercellular adhesion molecule 5, telencephalin (Mm.4629, NM_008319) | 9 |
| HORMONE | |||||
| GGCTGGATGGA | 8 | 0 | 122 | Cholecystokinin (Mm.26l9, NM_031161) | 9 |
| RECEPTOR | |||||
| GCTGCCCTGCC | 1 | 1 | 14 | EST cholinergic receptor, muscarinic 1, CNS (Mm.240607, BY652206) | 19 |
| TRANSPORT | |||||
| TAGCTGTAACG | 24 | 41 | 111 | ATPase, Na+/K+ transporting, alpha 1 polypeptide (Mm.280l03, NM_144900) | 3 |
| AATTCGCGGAT | 6 | 0 | 91 | Transthyretin (Mm.2l08, BC024702) | 18 |
All tags are expressed at higher levels in the parietal compared to other two regions, p < 0.05.
Table A2.
Components of metabolism, gene expression, and others preferentially expressed in the cortex.
| Function and tag sequences | Tag frequency |
Description (UniGene cluster, GenBank accession no.) | Chromosomal location | ||
|---|---|---|---|---|---|
| Hypothalamus | Pituitary gland | Parietal cortex | |||
| METABOLISM | |||||
| Energy | |||||
| ATAATACATAA | 71 | 24 | 196 | ATP6 (NC_001569:Pos:8596) | Mitochondrial DNA |
| Lipid | |||||
| GTCTGGGGGGA | 4 | 5 | 32 | Monoglyceride lipase (Mm.272197, BC057965) | 6 |
| TGAGCATCGGG | 8 | 7 | 39 | Monoglyceride lipase (Mm.272197, NM_011844) | |
| TATTGCAAGGT | 3 | 2 | 32 | Oxysterol binding protein-like 1A (Mm.259470, NM_020573) | 18 |
| Sugar and glycolysis | |||||
| TGCTAAGTCCC | 1 | 0 | 14 | UDP-N-acetyl-alpha-d-galactosamine polypeptide N-acetylgalactosaminyltransferase 9 (Mm.275748, XM_132253) | 5 |
| TRANSCRIPTION FACTORS | |||||
| TAAATAGACAA | 2 | 2 | 18 | Myocyte enhancer factor 2C (Mm.24001, NM_025282) | 13 |
| AAGCTTGGATT | 2 | 0 | 17 | EST myocyte enhancer factor 2C (Mm.24001, BE654834) | |
| GAGCTATCTGG | 1 | 0 | 16 | Forkhead box Gl (Mm.4704, NM_008241) | 12 |
| DNA BINDING | |||||
| AAGTGCAGTAC | 8 | 2 | 30 | Purine rich element binding protein B (Mm.296150, AK009365) | 11 |
| CYTOSKELETAL | |||||
| GTGCAGATAGT | 11 | 3 | 43 | Spectrin beta 3 (Mm.2ll852, XM_129130) | 19 |
| NEURITE GROWTH | |||||
| AGGGGCCGGTT | 7 | 1 | 48 | Nenritin 1 (Mm.232930, NM_153529) | 13 |
| HOMEOSTASIS-GENERAL | |||||
| TGGACGGTGGT | 5 | 3 | 26 | Carbonic anhydrase 11 (Mm.27736, NM_009800) | 7 |
All tags are expressed at higher levels in the parietal cortex compared to other two organs, p < 0.05.
Table A3.
Components of synapse preferentially expressed in the cortex.
| Tag sequences | Tag frequency |
Description (UniGene cluster, GenBank accession no.) | Chromosomal location | ||
|---|---|---|---|---|---|
| Hypothalamus | Pituitary gland | Parietal cortex | |||
| GTACTTGGCTG | 1 | 2 | 24 | Synaptic vesicle glycoprotein 2 b (Mm.273082, AK122359) | 7 |
| GGAACAGGAGG | 2 | 0 | 25 | Synaptopodin (Mm.252321, AK129267) | 18 |
| TATATTAAATC | 1 | 0 | 11 | Synaptosomal-associated protein 25 (Mm.45953, XM_130450) | 2 |
| CTGTAATGATA | 6 | 1 | 35 | EST synaptosomal-associated protein 25 (Mm.45953, AV330750) | |
| CAGCGGGAGCT | 16 | 4 | 57 | Syntaxin 1A brain (Mm.6225, NM_016801) | 5 |
All tags are expressed at higher levels in the parietal cortex compared to other two organs, p < 0.05.
Table A4.
Unclassified transcripts preferentially expressed in the parietal cortex.
| Tag sequences | Tag frequency |
Description (UniGene cluster, GenBank accession no.) | Chromosomal location | ||
|---|---|---|---|---|---|
| Hypothalamus | Pituitary gland | Parietal cortex | |||
| ATTCTCCCTGT | 5 | 0 | 20 | Expressed sequence N28178 (Mm.240965, AK122428) | 4 |
| CATTACTGCAG | 1 | 0 | 16 | EST expressed sequence BB075781 (Mm.230702, W39846) | 17 |
| TTCTTTTCCCT | 1 | 0 | 12 | Formin-like (Mm.138913, AF215666) | 11 |
| CAGGGATCAAT | 1 | 0 | 10 | EST Mus musculus 10 days neonate cortex cDNA, RIKEN full-length enriched library, clone A830028E23 product unknown EST, full insert sequence (Mm.192128, AK043754) | 6 |
| ACTGGTGACTG | 3 | 2 | 25 | ProSAPiPl protein (Mm.40621, NM_197945) | 2 |
| GTATTTGCAAA | 21 | 5 | 206 | RIKEN cDNA 1810006K23 gene (Mm.41603, AK007352) | 4 |
| CTGTGTGGCCC | 1 | 2 | 22 | RIKEN cDNA E130012A19 gene (Mm.24506, NM_175332) | 11 |
| CGGTCTAGTCC | 12 | 2 | 136 | EST RIKEN cDNA 1810006K23 gene (Mm.41603, BF323011) | 4 |
| POTENTIAL NOVEL TRANSCRIPTS | |||||
| AGGAACCAAAG | 45 | 11 | 148 | ||
| ACCGACAAGGC | 12 | 8 | 76 | ||
| GCGAGGAAGAA | 1 | 1 | 76 | ||
| TGTGCCCCAGG | 14 | 5 | 75 | ||
| TCACATCCAAG | 9 | 2 | 63 | ||
| CTCCCTGGGGG | 8 | 0 | 52 | ||
| TGATGGAAGGT | 8 | 6 | 47 | ||
| CAAAACAGGCA | 5 | 2 | 42 | ||
| TGGTCTTTTTG | 10 | 3 | 37 | ||
| CACAAAAGAAT | 4 | 0 | 36 | ||
| AAGGTGGATAC | 2 | 1 | 29 | ||
| GCTATTTGCCT | 5 | 2 | 29 | ||
| GTATCATTGGG | 4 | 2 | 22 | ||
| TGCTCTGGAGG | 4 | 2 | 22 | ||
| CTGGGCGCGTG | 2 | 0 | 22 | ||
| GGACGGCAGTT | 1 | 0 | 20 | ||
| TGACAAGACAC | 4 | 1 | 20 | ||
| GTCGCGTCCGG | 4 | 2 | 19 | ||
| GAGCCCCTTGG | 2 | 1 | 18 | ||
| TGGCCCCGGAA | 1 | 0 | 18 | ||
| TAGTGTTTTCC | 2 | 0 | 14 | ||
| GCTGGAACAGA | 2 | 2 | 14 | ||
| GACATCAGTTG | 2 | 0 | 12 | ||
| TAGGGCCCTAG | 1 | 0 | 11 | ||
| AAAACCTCCTC | 1 | 0 | 9 | ||
Tags are expressed at higher levels in the parietal cortex region compared to other two regions, p < 0.05.
Footnotes
Abbreviations
3β-Hsd-1, 3β-hydroxysteroid dehydrogenase-1; ACTH, adrenocorticotropic hormone; Anxa5, annexin A5; Ascl1, achaete–scute complex homolog-like 1 Drosophila; Cart, cocaine and amphetamine regulated transcript; CCD, comparative count display; cDNA, complementary DNA; Col1a2, procollagen, type I alpha 2; Dlk1, delta-like 1 homolog Drosophila; Dmpk, dystrophia myotonica kinase; DNA, deoxyribonucleic acid; EGF, epidermal growth factor; EST, expressed sequence tag; FSH, follicle-stimulating hormone; GABA, gamma-aminobutyric acid; Gat4, gamma-aminobutyric acid GABA-A transporter 4; Gh, growth hormone; Grb10, growth factor receptor bound protein 10; Hap1, huntingtin-associated protein 1; Hprt, hypoxanthine guanine phosphoribosyl transferase; Lh, luteinizing hormone; Mlc1, megalencephalic leukoencephalopathy with subcortical cysts 1 homolog human; NPY, neuropeptide Y; PCR, polymerase chain reaction; P4hb, prolyl 4-hydroxylase beta polypeptide; Pitx1, paired-like homeodomain transcription factor 1; Pnck, pregnancy upregulated non-ubiquitously expressed CaM kinase; Pomc, proopiomelanocortin; SAGE, serial analysis of gene expression; Sat1, spermidine/spermine N1-acetyltransferase; Six6, sine oculis-related homeobox 6 homolog Drosophila; Sparc, secreted acidic cysteine rich glycoprotein; Spint2, serine protease inhibitor Kunitz type 2; Trh, thyrotropin releasing hormone; Tsh, thyroid stimulating hormone.
References
- Adams M. D., Kerlavage A. R., Fleischmann R. D., Fuldner R. A., Bult C. J., Lee N. H., Kirkness E. F., Weinstock K. G., Gocayne J. D., White O. (1995). Initial assessment of human gene diversity and expression patterns based upon 83 million nucleotides of cDNA sequence. Nature 377, 3–174 10.1038/377003a0 [DOI] [PubMed] [Google Scholar]
- Bradshaw A. D., Graves D. C., Motamed K., Sage E. H. (2003). SPARC-null mice exhibit increased adiposity without significant differences in overall body weight. Proc. Natl. Acad. Sci. U.S.A. 100, 6045–6050 10.1073/pnas.1932759100 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brisson D., Houde G., St-Pierre J., Vohl M. C., Mathieu J., Gaudet D. (2002). The pleiotropic expression of the myotonic dystrophy protein kinase gene illustrates the complex relationships between genetic, biological and clinical covariates of male aging. Aging Male 5, 223–232 10.1080/713604716 [DOI] [PubMed] [Google Scholar]
- Burns K. H., Yan C., Kumar T. R., Matzuk M. M. (2001). Analysis of ovarian gene expression in follicle-stimulating hormone beta knockout mice. Endocrinology 142, 2742–2751 10.1210/en.142.7.2742 [DOI] [PubMed] [Google Scholar]
- Chemelli R. M., Willie J. T., Sinton C. M., Elmquist J. K., Scammell T., Lee C., Richardson J. A., Williams S. C., Xiong Y., Kisanuki Y., Fitch T. E., Nakazato M., Hammer R. E., Saper C. B., Yanagisawa M. (1999). Narcolepsy in orexin knockout mice: molecular genetics of sleep regulation. Cell 98, 437–451 10.1016/S0092-8674(00)81973-X [DOI] [PubMed] [Google Scholar]
- Cheng S. Y., Gong Q. H., Parkison C., Robinson E. A., Appella E., Merlino G. T., Pastan I. (1987). The nucleotide sequence of a human cellular thyroid hormone binding protein present in endoplasmic reticulum. J. Biol. Chem. 262, 11221–11227 [PubMed] [Google Scholar]
- Cowley M. A., Smart J. L., Rubinstein M., Cerdan M. G., Diano S., Horvath T. L., Cone R. D., Low M. J. (2001). Leptin activates anorexigenic POMC neurons through a neural network in the arcuate nucleus. Nature 411, 480–484 10.1038/35078085 [DOI] [PubMed] [Google Scholar]
- Darvesh S., Hopkins D. A. (2003). Differential distribution of butyrylcholinesterase and acetylcholinesterase in the human thalamus. J. Comp. Neurol. 463, 25–43 10.1002/cne.10751 [DOI] [PubMed] [Google Scholar]
- Dinel S., Bolduc C., Belleau P., Boivin A., Yoshioka M., Calvo E., Piedboeuf B., Snyder E. E., Labrie F., St-Amand J. (2005). Reproducibility, bioinformatic analysis and power of the SAGE method to evaluate changes in transcriptome. Nucleic Acids Res. 33, e26. 10.1093/nar/gni025 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Durkin M. M., Smith K. E., Borden L. A., Weinshank R. L., Branchek T. A., Gustafson E. L. (1995). Localization of messenger RNAs encoding three GABA transporters in rat brain: an in situ hybridization study. Brain Res. Mol. Brain Res. 33, 7–21 10.1016/0169-328X(95)00101-W [DOI] [PubMed] [Google Scholar]
- Gardner H. P., Rajan J. V., Ha S. I., Copeland N. G., Gilbert D. J., Jenkins N. A., Marquis S. T., Chodosh L. A. (2000). Cloning, characterization, and chromosomal localization of Pnck, a Ca2+/calmodulin-dependent protein kinase. Genomics 63, 279–288 10.1006/geno.1999.6091 [DOI] [PubMed] [Google Scholar]
- Gautvik K. M., de Lecea L., Gautvik V. T., Danielson P. E., Tranque P., Dopazo A., Bloom F. E., Sutcliffe J. G. (1996). Overview of the most prevalent hypothalamus-specific mRNAs, as identified by directional tag PCR subtraction. Proc. Natl. Acad. Sci. U.S.A. 93, 8733–8738 10.1073/pnas.93.16.8733 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guillemot F., Lo L. C., Johnson J. E., Auerbach A., Anderson D. J., Joyner A. L. (1993). Mammalian achaete-scute homolog 1 is required for the early development of olfactory and autonomic neurons. Cell 75, 463–476 10.1016/0092-8674(93)90381-Y [DOI] [PubMed] [Google Scholar]
- Hashizume A., Ueno T., Furuse M., Tsukita S., Nakanishi Y., Hieda Y. (2004). Expression patterns of claudin family of tight junction membrane proteins in developing mouse submandibular gland. Dev. Dyn. 231, 425–431 10.1002/dvdy.20142 [DOI] [PubMed] [Google Scholar]
- Hentges S. T., Nishiyama M., Overstreet L. S., Stenzel-Poore M., Williams J. T., Low M. J. (2004). GABA release from proopiomelanocortin neurons. J. Neurosci. 24, 1578–1583 10.1523/JNEUROSCI.3952-03.2004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hitchins M. P., Bentley L., Monk D., Beechey C., Peters J., Kelsey G., Ishino F., Preece M. A., Stanier P., Moore G. E. (2002). DDC and COBL, flanking the imprinted GRB10 gene on 7p12, are biallelically expressed. Mamm. Genome 13, 686–691 10.1007/s00335-002-3028-z [DOI] [PubMed] [Google Scholar]
- Hu R. M., Han Z. G., Song H. D., Peng Y. D., Huang Q. H., Ren S. X., Gu Y. J., Huang C. H., Li Y. B., Jiang C. L., Fu G., Zhang Q. H., Gu B. W., Dai M., Mao Y. F., Gao G. F., Rong R., Ye M., Zhou J., Xu S. H., Gu J., Shi J. X., Jin W. R., Zhang C. K., Wu T. M., Huang G. Y., Chen Z., Chen M. D., Chen J. L. (2000). Gene expression profiling in the human hypothalamus-pituitary-adrenal axis and full-length cDNA cloning. Proc. Natl. Acad. Sci. U.S.A. 97, 9543–9548 10.1073/pnas.97.4.1661 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Iverson F. (1975). Affinity and carbamylation rate constants of propoxur in reaction with erythrocyte and serum cholinesterase. Biochem. Pharmacol. 24, 1537–1538 10.1016/0006-2952(75)90032-5 [DOI] [PubMed] [Google Scholar]
- Joseph R., Dou D., Tsang W. (1994). Molecular cloning of a novel mRNA (neuronatin) that is highly expressed in neonatal mammalian brain. Biochem. Biophys. Res. Commun. 201, 1227–1234 10.1006/bbrc.1994.1836 [DOI] [PubMed] [Google Scholar]
- Jovanovic Z., Tung Y. C., Lam B. Y., O’Rahilly S., Yeo G. S. (2010). Identification of the global transcriptomic response of the hypothalamic arcuate nucleus to fasting and leptin. J. Neuroendocrinol. 22, 915–925 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kawaguchi T., Qin L., Shimomura T., Kondo J., Matsumoto K., Denda K., Kitamura N. (1997). Purification and cloning of hepatocyte growth factor activator inhibitor type 2, a Kunitz-type serine protease inhibitor. J. Biol. Chem. 272, 27558–27564 10.1074/jbc.272.44.27558 [DOI] [PubMed] [Google Scholar]
- Kawaminami M., Etoh S., Miyaoka H., Sakai M., Nishida M., Kurusu S., Hashimoto I. (2002). Annexin 5 messenger ribonucleic acid expression in pituitary gonadotropes is induced by gonadotropin-releasing hormone (GnRH) and modulates GnRH stimulation of gonadotropin release. Neuroendocrinology 75, 2–11 10.1159/000048216 [DOI] [PubMed] [Google Scholar]
- Koivu J., Myllyla R., Helaakoski T., Pihlajaniemi T., Tasanen K., Kivirikko K. I. (1987). A single polypeptide acts both as the beta subunit of prolyl 4-hydroxylase and as a protein disulfide-isomerase. J. Biol. Chem. 262, 6447–6449 [PubMed] [Google Scholar]
- Kristensen P., Judge M. E., Thim L., Ribel U., Christjansen K. N., Wulff B. S., Clausen J. T., Jensen P. B., Madsen O. D., Vrang N., Larsen P. J., Hastrup S. (1998). Hypothalamic CART is a new anorectic peptide regulated by leptin. Nature 393, 72–76 10.1038/29993 [DOI] [PubMed] [Google Scholar]
- Laborda J., Sausville E. A., Hoffman T., Notario V. (1993). dlk, a putative mammalian homeotic gene differentially expressed in small cell lung carcinoma and neuroendocrine tumor cell line. J. Biol. Chem. 268, 3817–3820 [PubMed] [Google Scholar]
- Lamonerie T., Tremblay J. J., Lanctot C., Therrien M., Gauthier Y., Drouin J. (1996). Ptx1, a bicoid-related homeo box transcription factor involved in transcription of the pro-opiomelanocortin gene. Genes Dev. 10, 1284–1295 10.1101/gad.10.10.1284 [DOI] [PubMed] [Google Scholar]
- Langlais P., Dong L. Q., Ramos F. J., Hu D., Li Y., Quon M. J., Liu F. (2004). Negative regulation of insulin-stimulated mitogen-activated protein kinase signaling by Grb10. Mol. Endocrinol. 18, 350–358 10.1210/me.2003-0117 [DOI] [PubMed] [Google Scholar]
- Lania A., Mantovani G., Spada A. (2001). G protein mutations in endocrine diseases. Eur. J. Endocrinol. 145, 543–559 10.1530/eje.0.1450543 [DOI] [PubMed] [Google Scholar]
- Lash A. E., Tolstoshev C. M., Wagner L., Schuler G. D., Strausberg R. L., Riggins G. J., Altschul S. F. (2000). SAGEmap: a public gene expression resource. Genome Res. 10, 1051–1060 10.1101/gr.10.7.1051 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li S. H., Yu Z. X., Li C. L., Nguyen H. P., Zhou Y. X., Deng C., Li X. J. (2003). Lack of huntingtin-associated protein-1 causes neuronal death resembling hypothalamic degeneration in Huntington’s disease. J. Neurosci. 23, 6956–6964 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li X. J., Li S. H., Sharp A. H., Nucifora F. C., Jr., Schilling G., Lanahan A., Worley P., Snyder S. H., Ross C. A. (1995). A huntingtin-associated protein enriched in brain with implications for pathology. Nature 378, 398–402 10.1038/378724a0 [DOI] [PubMed] [Google Scholar]
- Lim M. A., Riedel H., Liu F. (2004). Grb10: more than a simple adaptor protein. Front. Biosci. 9, 387–403 10.2741/1226 [DOI] [PubMed] [Google Scholar]
- Lopez-Rios J., Gallardo M. E., Rodriguez de Cordoba S., Bovolenta P. (1999). Six9 (Optx2), a new member of the six gene family of transcription factors, is expressed at early stages of vertebrate ocular and pituitary development. Mech. Dev. 83, 155–159 10.1016/S0925-4773(99)00017-9 [DOI] [PubMed] [Google Scholar]
- Miyata S., Nakai S., Kiyohara T., Hatton G. I. (2000). Calbindin-D28k and calretinin in the rat posterior pituitary; light and electron microscopic localization and upregulation with dehydration. J. Neurocytol. 29, 5–17 10.1023/A:1007180328597 [DOI] [PubMed] [Google Scholar]
- Moutoussamy S., Renaudie F., Lago F., Kelly P. A., Finidori J. (1998). Grb10 identified as a potential regulator of growth hormone (GH) signaling by cloning of GH receptor target proteins. J. Biol. Chem. 273, 15906–15912 10.1074/jbc.273.26.15906 [DOI] [PubMed] [Google Scholar]
- Nelson N. (1998). The family of Na+/Cl- neurotransmitter transporters. J. Neurochem. 71, 1785–1803 10.1046/j.1471-4159.1998.71051785.x [DOI] [PubMed] [Google Scholar]
- Nishida Y., Yoshioka M., Ray C. A., Bolduc C., Tanaka H., St-Amand J. (2009). Regulation of pituitary gene expression by adrenalectomy. Obesity 17, 114–120 10.1038/oby.2008.484 [DOI] [PubMed] [Google Scholar]
- Nishida Y., Yoshioka M., St-Amand J. (2005a). Sexually dimorphic gene expression in the hypothalamus, pituitary gland, and cortex. Genomics 85, 679–687 10.1016/j.ygeno.2005.02.013 [DOI] [PubMed] [Google Scholar]
- Nishida Y., Yoshioka M., St-Amand J. (2005b). The top 10 most abundant transcripts are sufficient to characterize the organs functional specificity: evidences from the cortex, hypothalamus and pituitary gland. Gene 344, 133–141 10.1016/j.gene.2004.09.007 [DOI] [PubMed] [Google Scholar]
- Nishida Y., Yoshioka M., St-Amand J. (2006). Regulation of hypothalamic gene expression by glucocorticoid: implications for energy homeostasis. Physiol. Genomics 25, 96–104 10.1152/physiolgenomics.00232.2005 [DOI] [PubMed] [Google Scholar]
- Olster D. H., Blaustein J. D. (1988). Progesterone facilitation of lordosis in male and female Sprague-Dawley rats following priming with estradiol pulses. Horm. Behav. 22, 294–304 10.1016/0018-506X(88)90002-5 [DOI] [PubMed] [Google Scholar]
- Paulsen S. J., Larsen L. K., Jelsing J., Janssen U., Gerstmayer B., Vrang N. (2009). Gene expression profiling of individual hypothalamic nuclei from single animals using laser capture microdissection and microarrays. J. Neurosci. Methods 177, 87–93 10.1016/j.jneumeth.2008.09.024 [DOI] [PubMed] [Google Scholar]
- Persechini A., Moncrief N. D., Kretsinger R. H. (1989). The EF-hand family of calcium-modulated proteins. Trends Neurosci. 12, 462–467 10.1016/0166-2236(89)90097-0 [DOI] [PubMed] [Google Scholar]
- Pietila M., Alhonen L., Halmekyto M., Kanter P., Janne J., Porter C. W. (1997). Activation of polyamine catabolism profoundly alters tissue polyamine pools and affects hair growth and female fertility in transgenic mice overexpressing spermidine/spermine N1-acetyltransferase. J. Biol. Chem. 272, 18746–18751 10.1074/jbc.272.30.18746 [DOI] [PubMed] [Google Scholar]
- Qian X., Jin L., Lloyd R. V. (2000). Aberrant DNA methylation of cyclin D2 and p27 genes in rodent pituitary tumor cell lines correlates with specific gene expression. Endocr. Pathol. 11, 85–96 10.1385/EP:11:1:85 [DOI] [PubMed] [Google Scholar]
- Rockett J. C., Patrizio P., Schmid J. E., Hecht N. B., Dix D. J. (2004). Gene expression patterns associated with infertility in humans and rodent models. Mutat. Res. 549, 225–240 10.1016/j.mrfmmm.2003.11.017 [DOI] [PubMed] [Google Scholar]
- Sakurai T. (2003). Orexin: a link between energy homeostasis and adaptive behaviour. Curr. Opin. Clin. Nutr. Metab. Care 6, 353–360 10.1097/00075197-200307000-00001 [DOI] [PubMed] [Google Scholar]
- Schmitt A., Gofferje V., Weber M., Meyer J., Mossner R., Lesch K. P. (2003). The brain-specific protein MLC1 implicated in megalencephalic leukoencephalopathy with subcortical cysts is expressed in glial cells in the murine brain. Glia 44, 283–295 10.1002/glia.10304 [DOI] [PubMed] [Google Scholar]
- Schulman H. (1993). The multifunctional Ca2+/calmodulin-dependent protein kinases. Curr. Opin. Cell Biol. 5, 247–253 10.1016/0955-0674(93)90111-3 [DOI] [PubMed] [Google Scholar]
- Segal J. P., Stallings N. R., Lee C. E., Zhao L., Socci N., Viale A., Harris T. M., Soares M. B., Childs G., Elmquist J. K., Parker K. L., Friedman J. M. (2005). Use of laser-capture microdissection for the identification of marker genes for the ventromedial hypothalamic nucleus. J. Neurosci. 25, 4181–4188 10.1523/JNEUROSCI.0158-05.2005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Singhal B. S., Gorospe J. R., Naidu S. (2003). Megalencephalic leukoencephalopathy with subcortical cysts. J. Child Neurol. 18, 646–652 10.1177/08830738030180091201 [DOI] [PubMed] [Google Scholar]
- St-Amand J., Okamura K., Matsumoto K., Shimizu S., Sogawa Y. (2001). Characterization of control and immobilized skeletal muscle: an overview from genetic engineering. FASEB J. 15, 684–692 10.1096/fj.00-0150com [DOI] [PubMed] [Google Scholar]
- Takai Y., Sasaki T., Matozaki T. (2001). Small GTP-binding proteins. Physiol. Rev. 81, 153–208 [DOI] [PubMed] [Google Scholar]
- Tanaka S., Tatsumi K., Okubo K., Itoh K., Kawamoto S., Matsubara K., Amino N. (2002). Expression profile of active genes in the human pituitary gland. J. Mol. Endocrinol. 28, 33–44 10.1677/jme.0.0280033 [DOI] [PubMed] [Google Scholar]
- Tartare-Deckert S., Chavey C., Monthouel M. N., Gautier N., Van Obberghen E. (2001). The matricellular protein SPARC/osteonectin as a newly identified factor up-regulated in obesity. J. Biol. Chem. 276, 22231–22237 10.1074/jbc.M010634200 [DOI] [PubMed] [Google Scholar]
- Tung Y. C., Ma M., Piper S., Coll A., O’Rahilly S., Yeo G. S. (2008). Novel leptin-regulated genes revealed by transcriptional profiling of the hypothalamic paraventricular nucleus. J. Neurosci. 28, 12419–12426 10.1523/JNEUROSCI.3412-08.2008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Usui H., Morii K., Tanaka R., Tamura T., Washiyama K., Ichikawa T., Kumanishi T. (1997). cDNA cloning and mRNA expression analysis of the human neuronatin. High level expression in human pituitary gland and pituitary adenomas. J. Mol. Neurosci. 9, 55–60 10.1007/BF02789395 [DOI] [PubMed] [Google Scholar]
- Velculescu V. E., Zhang L., Vogelstein B., Kinzler K. W. (1995). Serial analysis of gene expression. Science 270, 484–487 10.1126/science.270.5235.484 [DOI] [PubMed] [Google Scholar]
- Warrington J. A., Nair A., Mahadevappa M., Tsyganskaya M. (2000). Comparison of human adult and fetal expression and identification of 535 housekeeping/maintenance genes. Physiol. Genomics 2, 143–147 [DOI] [PubMed] [Google Scholar]
- Weidenfeld J., Siegel R. A., Chowers I. (1980). In vitro conversion of pregnenolone to progesterone by discrete brain areas of the male rat. J. Steroid Biochem. 13, 961–963 10.1016/0022-4731(80)90171-5 [DOI] [PubMed] [Google Scholar]
- Yu R., Ren S. G., Melmed S. (2002). Proteasome inhibitors induce apoptosis in growth hormone- and prolactin-secreting rat pituitary tumor cells. J. Endocrinol. 174, 379–386 10.1677/joe.0.1740379 [DOI] [PubMed] [Google Scholar]
- Zannettino A. C., Buhring H. J., Niutta S., Watt S. M., Benton M. A., Simmons P. J. (1998). The sialomucin CD164 (MGC-24v) is an adhesive glycoprotein expressed by human hematopoietic progenitors and bone marrow stromal cells that serves as a potent negative regulator of hematopoiesis. Blood 92, 2613–2628 [PubMed] [Google Scholar]
- Zhao S., Torii S., Yokota-Hashimoto H., Takeuchi T., Izumi T. (2002). Involvement of Rab27b in the regulated secretion of pituitary hormones. Endocrinology 143, 1817–1824 10.1210/en.143.5.1817 [DOI] [PubMed] [Google Scholar]
- Zhou Y. P., Sreenan S., Pan C. Y., Currie K. P., Bindokas V. P., Horikawa Y., Lee J. P., Ostrega D., Ahmed N., Baldwin A. C., Cox N. J., Fox A. P., Miller R. J., Bell G. I., Polonsky K. S. (2003). A 48-hour exposure of pancreatic islets to calpain inhibitors impairs mitochondrial fuel metabolism and the exocytosis of insulin. Metab. Clin. Exp. 52, 528–534 10.1053/meta.2003.50091 [DOI] [PubMed] [Google Scholar]
- Zwain I. H., Yen S. S. (1999). Neurosteroidogenesis in astrocytes, oligodendrocytes, and neurons of cerebral cortex of rat brain. Endocrinology 140, 3843–3852 10.1210/en.140.2.880 [DOI] [PubMed] [Google Scholar]