Abstract
Small ubiquitin-like modifier-1/2/3 (SUMO-1/2/3) and ubiquitin share similar structure and utilize analogous machinery for protein lysine conjugation. Although sumoylation and ubiquitylation have distinct functions, they are often tightly associated with each other to fine-tune protein fate in transducing signals to regulate a wide variety of cellular functions, including DNA damage response, cell proliferation, DNA replication, embryonic development, and cell differentiation. In this Perspective, we specifically highlight the role of sumoylation and ubiquitylation in ataxia-telangiectasia mutated (ATM) signaling in response to DNA double-strand breaks and hypothesize that ATM-induced phosphorylation is a unique node in regulating SUMO-targeted ubiquitylation in mammalian cells to combat DNA damage and to maintain genome integrity. A potential role for the coordination of three types of post-translational modification in dictating the tempo and extent of cellular response to genotoxic stress is speculated.
Keywords: ATM, phosphorylation, sumoylation, ubiquitylation, DNA damage response
Small ubiquitin-like modifier (SUMO) and ubiquitin (Ub) can be post-translationally attached to their target proteins. They share less than 20% identity of amino acid sequence; however, their structure and biochemical machinery catalyzing the modification are similar. Sumoylation and ubiquitylation are known to function distinctly in operating the fate of protein. Sumoylation is to conduct signals for nuclear translocation, transcriptional repression and protein recruitment, whereas lysine (Lys)-48-linked polyubiquitylation is a common mark for protein degradation in proteasome; Lys-63 linkage of Ub directs protein trafficking. Both SUMO and Ub participate in regulating a wide range of cellular functions, including DNA damage response (DDR), cell growth, differentiation, and development (Gill, 2004; Denuc and Marfany, 2010; Tang and Greenberg, 2010; Praefcke et al., 2011). Here, we specifically focus on the role of sumoylation and ubiquitylation in DDR.
DNA damage response is an evolved mechanism for cells to confront DNA lesions generated by endogenous or environmental agents. The cells sense DNA lesions, such as DNA double-strand breaks (DSB), mismatches, and bulky adducts to subsequently induce signaling pathways promoting cell cycle arrest, DNA repair, apoptosis, transcription, and chromatin remodeling (Jackson and Bartek, 2009). A large protein kinase family, phosphoinositide 3 kinase-like protein kinases (PIKKs) including ataxia-telangiectasia mutated (ATM), ataxia-telangiectasia, and Rad3 related (ATR), and DNA-dependent protein kinase catalytic subunit (DNA-PKcs), participates in DDR by phosphorylating a subset of target proteins to regulate cellular functions and maintain genome integrity. Defective DDR causes the accumulation of DNA lesions and eventually leads to genome instability, a hallmark of cancer. Particularly, ATM plays a primary role in transducing DSB signal to a broad spectrum of proteins involved in DNA repair, cell cycle checkpoints, and apoptosis (Shiloh, 2003). Mutations at the ATM locus thereby elicit phenotypes reflective of defects in cell cycle regulation and apoptosis. Ataxia telangiectasia (A-T) patients are extremely sensitive to ionizing radiation, such as X- and γ-rays (Lavin, 2008). A-T cells do not exhibit cell cycle arrest at regular checkpoints when stressed by irradiation (Morrison et al., 2000). Distinct mutations of the ATM gene also predispose some patients to cancer, particularly lymphoma cancers in 30% of A-T patients and leukemia (Hunter, 2007; Lavin, 2008). Therefore, studies of ATM-regulated pathways in conjunction with DDR are crucial to understanding maintenance of genome integrity. Particularly, in this Perspective, we review multiple types of post-translational modification (PTM): phosphorylation, sumoylation, ubiquitylation, and their combinatorial effects in ATM signaling pathways.
ATM-Regulated Sumoylation in DDR
Sumoylation regulates ATM-mediated DNA damage signaling in both transcription and chromatin remodeling. A unique example that well-illustrates the relationship between ATM-induced phosphorylation and sumoylation is Krüppel-associated box (KRAB)-associated protein 1 (KAP1; also known as TRIM28 and TIF1β). KAP1 is a transcriptional co-repressor primarily responding to DNA damage and regulating cellular functions such as checkpoint control and apoptosis (Lee et al., 2007; Li et al., 2007). KAP1 has six putative sumoylation sites and its sumoylation at Lys-779 and Lys-804 are required for the interaction with chromatin remodelers including heterochromatin protein 1(HP1), SET domain, bifurcated 1 (SETDB1), nucleosome remodeling deacetylase (NuRD), and histone deacetylases (HDACs) to establish a silent state of chromatin and to repress transcription (Ryan et al., 1999; Schultz et al., 2001, 2002; Sripathy et al., 2006; Ivanov et al., 2007; Lee et al., 2007). When cells are exposed to genotoxic stress, DSBs activate ATM to phosphorylate KAP1 at serine (Ser)-824 and result in a decrease of SUMO–KAP1 population, leading to the de-repression of KAP1-regulated DNA damage-responsive genes: p21, GADD45α, BAX, NOXA, and PUMA (White et al., 2006; Lee et al., 2007; Li et al., 2007). Furthermore, phosphorylation/sumoylation switch on KAP1 has been illuminated through research in protein phosphatases 1α and 1β (PP1α and PP1β). While PP1α directly interacts with KAP1 under basal condition, PP1β interacts with KAP1 only in response to genotoxic stress. Changes in the abundance of PP1α and PP1β have differential effects on phosphorylation and sumoylation of KAP1 under both unstressed and stressed conditions (Li et al., 2010). Although the underlying mechanism of phosphorylation-induced decrease of SUMO–KAP1 is still not clear, possibilities might include that ATM-induced phosphorylation activates SUMO proteases to de-sumoylate KAP1 or that ATM triggers the degradation of SUMO–KAP1.
Besides of transcriptional regulation, KAP1 also participates in chromatin configuration. Chromodomain helicase DNA binding protein 3 (CHD3; also known as Mi2α) interacts with SUMO–KAP1 through its SUMO-interacting motif (SIM). The binding of KAP1 and CHD3 configures KAP1-associated chromatin into a compact structure. Upon DNA damage, ATM-induced phosphorylation at Ser-824 of KAP1 interrupts the SUMO:SIM interaction between KAP1 and CHD3, leading to the relaxation of chromatin which is more accessible for DNA repair modules to come in to the damage sites and repair the lesions (Goodarzi et al., 2011). Although very few reports demonstrate the connection between ATM signaling and protein sumoylation, in the KAP1 case, the control of phosphorylation/sumoylation switch on KAP1 seems to play an indispensable role in transducing ATM signal. The switch on KAP1 has to be tightly regulated in order to respond to DNA damage and return to the basal state in a timely and proper way.
ATM-Regulated Ubiquitylation in DDR
Ubiquitylation serves as a critical signal for recruiting DNA repair machinery to the damage sites. Upon DNA damage, MRE11–Rad50–NBS1 (MRN) complex recognizes and processes the end of DSBs (Uziel et al., 2003). ATM is then recruited to phosphorylate H2AX at Ser-139, creating a docking site for the mediator of DNA damage checkpoint protein 1 (MDC1) to be recruited to the DNA damage foci. ATM further phosphorylates MDC1 in order to recruit ring finger protein 8 (RNF8)–HERC2–UBC13 complex to the foci to catalyze Lys-63 polyubiquitylation of H2A (Lou et al., 2006; Huen et al., 2007; Kolas et al., 2007; Bekker-Jensen et al., 2010). RNF168 is subsequently recruited to the Lys-63 polyubiquitylated H2A and amplifies Lys-63-linked poly Ub chains on H2A and H2AX around the foci (Doil et al., 2009). Meanwhile, RNF8 also catalyzes Lys-48 polyubiquitylation on the client proteins at damage sites. The Lys-48 polyubiquitylated substrates will then be turned over by recruiting p97-UFD1–NPL4 Ub-selective segregase (Meerang et al., 2011). The coordination of these signals ultimately recruits BRCA1–RAP80 complex, 53BP1, and Rad51 to facilitate DSB repair and checkpoint arrest (Kolas et al., 2007; Doil et al., 2009; Bekker-Jensen et al., 2010; Meerang et al., 2011). After repairing the damage, the removal of H2AX from the damage sites involves an acetylation-dependent ubiquitylation catalyzed by TIP60–UBC13 complex (Ikura et al., 2007).
Other than transducing signals to recruit repair proteins, ATM also represses transcription in cis to DSBs by establishing monoubiquitylation of H2A to inhibit RNA polymerase II function. Since monoubiquitylation at Lys-119 of H2A (uH2A) is associated with transcriptional repression, ATM-mediated transcriptional silencing was explored in tandem with uH2A. By inhibiting ATM, uH2A levels at DSBs are significantly decreased although the Lys-63-linked poly Ub chains and RAP80 levels around the foci are less affected. ATM therefore plays a critical role in maintenance of uH2A at DSBs through RNF8/RNF168, while the Lys-63-linked poly Ub chains serve as separate docking sites for recruitment of repair proteins such as BRCA1 complex. Furthermore, ATM-dependent uH2A stalls RNA polymerase II-mediated transcription in cis to the damage site. A deubiquitylation enzyme, USP16, negatively regulates uH2A-dependent function and rapidly restores transcription after the cessation of DNA damage (Shanbhag et al., 2010).
Ataxia-telangiectasia mutated-induced phosphorylation also exhibits crosstalk with ubiquitylation by mediating protein degradation through direct and indirect recruitment of Ub E3 ligases. In the case of the tumor suppressor p53, ATM-induced phosphorylation participates in the dual function of repressing and promoting the ubiquitylation of different effectors, leading to their degradation. p53 induces transcription of multiple genes important for cell cycle regulation, DNA repair, and apoptosis. ATM phosphorylates p53 at Ser-15, leading to transcription of the CDK2/cyclin-E inhibitor which functions at the G1-S checkpoint (Shiloh, 2003). ATM-induced phosphorylation inhibits negative regulators of p53, including MDM2 and constitutive morphogenic 1 (COP1). Both MDM2 and COP1 are Ub E3 ligases that ubiquitylate p53 to promote its proteasomal degradation. ATM indirectly regulates MDM2-mediated degradation of p53 through phosphorylation of Chk2 which then phosphorylates p53 at Ser-20 to prevent the formation of the MDM2–p53 complex (Dumaz et al., 2001). ATM also directly phosphorylates MDM2 at Ser-395 to prevent the export of the MDM2–p53 complex into the cytoplasm, thereby maintaining p53 in the nucleus (Maya et al., 2001; Chen et al., 2005). In addition, phosphorylation of COP1 by ATM induces autoubiquitylation of COP1 (Dornan et al., 2006). ATM phosphorylation thus selectively influences the repression and activation of ubiquitylation on different proteins in response to DNA damage. Taken together, ATM-induced phosphorylation in coordination with ubiquitylation plays an essential role in establishing a series of signals directing to transcriptional regulation, the completion of DSB repair and in determining the fate of key proteins involved in DDR.
Crosstalk between Sumoylation and Ubiquitylation in DDR
The convergence of sumoylation and ubiquitylation does take place under genotoxic condition. When DSBs occur, protein inhibitor of activated signal transducer and activator of transcription (PIAS) localizes to the damage sites. PIAS1 and PIAS4 function as SUMO E3 ligase to modify BRCA1, 53BP1 and possibly RNF8, RNF168 to modulate their activities in facilitating Ub signal amplification and DNA repair after genotoxic stress (Galanty et al., 2009). Sumoylation on BRCA1 increases its Ub E3 ligase activity, therefore termed as a SUMO-regulated Ub ligase (SRUbL; Morris et al., 2009). It is still not clear how PIAS activity is regulated under DNA damage condition, and whether this is dependent on ATM-induced phosphorylation. PIAS1 is phosphorylated by IκB kinase alpha (IKKα; Liu et al., 2007), whose activity is also regulated by ATM-dependent NF-κB essential modulator (NEMO) ubiquitylation (Wuerzberger-Davis et al., 2006), implying that the role of PIAS in DDR might also be incorporated into ATM signaling.
Another interesting example showing the interplay between sumoylation and ubiquitylation in the context of genotoxic stress is SUMO-targeted ubiquitin ligase (STUbL), including Slx8–Rfp, MIP1, Slx5–Slx8, and RNF4. It is a new class of ubiquitin E3 ligases targeting sumoylated proteins through their SIMs for ubiquitylation. STUbL has been reported to trigger the degradation of sumoylated proteins and is responsible for the maintenance of cell survival and genome stability (Lallemand-Breitenbach et al., 2008; Tatham et al., 2008; Cook et al., 2009; Heideker et al., 2009). For example, RNF4 (RING finger protein 4, RING: Really Interesting New Gene), the only human homolog of Slx8–Rfp, is involved in arsenic-trioxide (ATO)-induced polyubiquitylation and proteasomal degradation of promyelocytic leukemia (PML) by targeting the poly SUMO-2 chain on PML (Lallemand-Breitenbach et al., 2008; Tatham et al., 2008). This reveals the role of RNF4 in mediating the crosstalk between sumoylation and ubiquitylation and also provides a possible mechanism of ATO-induced damage response. In conclusion, crosstalk between sumoylation and ubiquitylation seems like a general scenario in DDR; however, it is still not fully understood how this crosstalk might be modulated and since ATM signaling requires sumoylation and ubiquitylation to respond to DSB, it is possible that ATM is involved in tuning SUMO–Ub crosstalk through RNF4 in DDR.
Is RNF4 Involved in ATM Signaling?
Given to the fact that RNF4 biologically functions as an important factor for maintaining genome integrity and its ability to recognize and possibly regulate more than 300 substrates involved in a wide variety of biological processes, including chromatin remodeling and DNA repair etc. Bruderer et al. (2011), one could speculate that RNF4 might participate in ATM-regulated DSB damage response.
RNF4 is extensively involved in DNA damage signaling pathways
Yeast homolog of RNF4, Slx5/Slx8 physically associates with DSBs to form damage foci, in a SUMO and SIM-dependent way (Nagai et al., 2008; Cook et al., 2009). Slx8 functions with Rad60, a DNA repair protein, and Nse2, a SUMO ligase to protect the genome from Topoisomerase-1 (Top-1)-induced DNA damage (Prudden et al., 2007; Heideker et al., 2011). Fission yeast Rfp1 and Rfp2 complement one another in regulating defects in cell cycle progression and Chk1-dependnet DNA repair; moreover, human RNF4 is able to functionally rescue this phenotype in rfp1/rfp2 double null mutant (Kosoy et al., 2007). RNF4 also functions specifically to demethylate DNA by interacting with base excision repair enzymes TDG and APE1 that target G:T mismatches in the DNA. In addition, RNF4 deficiency displays global DNA hypermethylation (Hu et al., 2010). Taken together, the biological function of RNF4 is conserved and tightly associated with DDR, especially DNA repair and chromatin remodeling in different organisms, supporting the idea that RNF4 plays a role in coordinating and transducing ATM-induced signaling in response to DNA damage.
Evidence supporting phosphorylation-induced SUMO-dependent protein degradation
Promyelocytic leukemia is found to be degraded upon ATO-treatment and the degradation is dependent on the phosphorylation-induced by ATO and the subsequent increase of sumoylation (Lallemand-Breitenbach et al., 2001; Hayakawa and Privalsky, 2004). ATO-induced sumoylation on Lys-160 is critical for recruiting RNF4 to ubiquitylate PML for proteasomal degradation (Lallemand-Breitenbach et al., 2001; Petrie and Zelent, 2008). Although there is no direct link showing phosphorylation of PML promotes SUMO-dependent degradation, either phosphorylation-defective, or sumoylation-defective PML mutant shows abolished downstream effects in response to ATO-treatment (Lallemand-Breitenbach et al., 2001; Yang et al., 2002; Hayakawa and Privalsky, 2004), implying that both modifications and their crosstalk are indispensable in leading to ATO-induced PML degradation. PML can be phosphorylated by several kinases including MAPK, CK2, and CHK2 (Yang et al., 2002; Hayakawa and Privalsky, 2004; Joe et al., 2006; Scaglioni et al., 2006). Interestingly, phosphorylation-defective PML is stabilized upon DNA damage triggered by γ-irradiation and results in decreased apoptotic activity, in an ATM/CHK2-dependent manner (Yang et al., 2002). Evidence from the PML studies provide some hints supporting that the degradation promoted by RNF4 is possibly regulated by phosphorylation; however, it is still unclear how phosphorylation induces sumoylation of PML and whether there is another mechanism that phosphorylation of PML might enhance the recognition by RNF4.
Unique structural characteristic of RNF4 possibly links phosphorylation, sumoylation and ubiquitylation
If one speculates that RNF4-mediated SUMO-targeted ubiquitylation is regulated by ATM-induced phosphorylation, what would the mode of regulation be? A unique region is found in RNF4 protein. Following the four SIMs, there is a region rich with arginine, named arginine-rich region (ARR) in RNF4 (Figure 1), denoting that this region provides positive charge to attract phosphorylated protein with negative charge. The electrostatic interaction between arginine and phosphate forms a covalent-like binding (Woods and Ferre, 2005). Thus, the ARR in RNF4 might enhance its interaction with target proteins phosphorylated by ATM. In summary, phosphorylation on RNF4 target proteins might be a mode to regulate RNF4-mediated, SUMO-targeted ubiquitylation and related biological function.
Figure 1.
Sequence alignment of SIMs and ARR of RNF4 family members from different organisms. Data here show that four SIM domains and ARR are conserved throughout evolution.
A potential mechanism of RNF4 targeting ATM substrates for proteasomal degradation
It was noted that phosphorylation and subsequent sumoylation of PML occur within 1 h after treating with ATO (Lallemand-Breitenbach et al., 2001; Hayakawa and Privalsky, 2004); however, significant degradation of PML is observed around 12–16 h after ATO-treatment (Lallemand-Breitenbach et al., 2001, 2008), indicating that the regulation of RNF4-mediated PML degradation might be in slow kinetics. This suggests that there might be other factors required for targeting RNF4-ubiquitylated PML to proteasome. An AAA-ATPase p97, its adaptors UFD1 and NPL4 are implicated in recognizing and extracting polyubiquitylated proteins to proteasome for degradation in various cellular context, including mitosis, DNA replication, and DNA damage (Richly et al., 2005; Ramadan et al., 2007; Mouysset et al., 2008; Meerang et al., 2011; Verma et al., 2011). To explain the observation of slow degradation of PML, rather than rapid turnover, in our view, RNF4 may serve as a signal transducer that senses SUMO signal and amplifies Ub signal on its substrates to recruit selective cargo proteins, such as p97–UFD1–NPL4 complex to extract the ubiquitylated substrates for proteasomal degradation.
Conclusion and Perspective
Sumoylation and ubiquitylation widely participate in ATM-regulated DDR. When cells are exposed to genotoxic stress, DSBs activate ATM to phosphorylate a subset of target proteins to transduce signals and to induce checkpoint control and DNA repair machinery. This process largely involves the cooperation of sumoylation and ubiquitylation to regulate cellular function in response to DSB. However, little is known about the detailed mechanism of SUMO–Ub crosstalk. Here, we hypothesize that RNF4 plays a central role in recognizing ATM-induced phosphorylation and sumoylation to provide an additional Ub signal to recruit Ub-selective segregase to target for proteasomal degradation (Figure 2). This provides a novel view on the crosstalk among multiple PTMs. The crosstalk of phosphorylation, sumoylation, and ubiquitylation denotes a cooperative network in protecting cells from DNA damage and maintaining genome integrity. Defects in this network may lead to genome instability and consequently tumorigenesis. Moreover, modulation of the players involved in the network may sensitize cancer cells to DNA damage-based cancer therapy and benefit the patients.
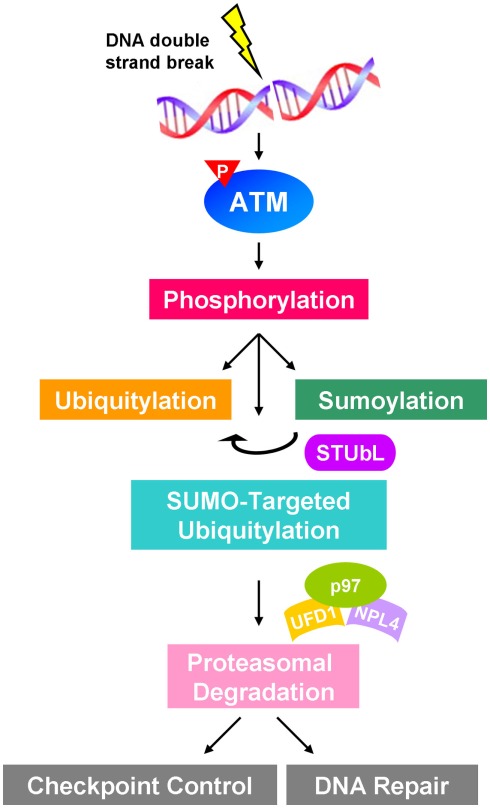
Figure 2.
Proposed schematic showing the crosstalk of phosphorylation, sumoylation, and ubiquitylation in ATM-regulated signaling in response to DNA damage. Double-strand breaks activate ATM to induce downstream signaling cascade involving sumoylation and ubiquitylation of proteins participating in DNA damage response, such as checkpoint control and DNA repair. STUbL functions as a signal transducer to coordinate SUMO and Ub signals to catalyze SUMO-targeted ubiquitylation, which may recruit p97–UFD1–NPL4 segregase complex to mediate the proteasomal degradation.
Conflict of Interest Statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Acknowledgments
We are sincerely grateful to Drs. Che-Chang Chang, Mei-Ling Kuo, and Yun Yen for their helpful suggestions and critical reading of manuscript. This work was supported in part by National Institute of Health Research Grants R01DE10742 and R01DE14183 (to David Kong Ann) and Eugene and Ruth Roberts Summer Student Academy fellowships (to Christine Shieh and Fei Cai).
References
- Bekker-Jensen S., Danielsen J. R., Fugger K., Gromova I., Nerstedt A., Lukas C., Bartek J., Lukas J., Mailand N. (2010). HERC2 coordinates ubiquitin-dependent assembly of DNA repair factors on damaged chromosomes. Nat. Cell Biol. 12, 80–86 10.1038/ncb0410-412 [DOI] [PubMed] [Google Scholar]
- Bruderer R., Tatham M. H., Plechanovova A., Matic I., Garg A. K., Hay R. T. (2011). Purification and identification of endogenous polySUMO conjugates. EMBO Rep. 12, 142–148 10.1038/embor.2010.206 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen L., Gilkes D. M., Pan Y., Lane W. S., Chen J. (2005). ATM and Chk2-dependent phosphorylation of MDMX contribute to p53 activation after DNA damage. EMBO J. 24, 3411–3422 10.1038/sj.emboj.7600666 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cook C. E., Hochstrasser M., Kerscher O. (2009). The SUMO-targeted ubiquitin ligase subunit Slx5 resides in nuclear foci and at sites of DNA breaks. Cell Cycle 8, 1080–1089 10.4161/cc.8.7.8123 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Denuc A., Marfany G. (2010). SUMO and ubiquitin paths converge. Biochem. Soc. Trans. 38, 34–39 10.1042/BST0380034 [DOI] [PubMed] [Google Scholar]
- Doil C., Mailand N., Bekker-Jensen S., Menard P., Larsen D. H., Pepperkok R., Ellenberg J., Panier S., Durocher D., Bartek J., Lukas J., Lukas C. (2009). RNF168 binds and amplifies ubiquitin conjugates on damaged chromosomes to allow accumulation of repair proteins. Cell 136, 435–446 10.1016/j.cell.2008.12.041 [DOI] [PubMed] [Google Scholar]
- Dornan D., Shimizu H., Mah A., Dudhela T., Eby M., O’rourke K., Seshagiri S., Dixit V. M. (2006). ATM Engages Autodegradation of the E3 ubiquitin ligase COP1 after DNA damage. Science 313, 1122–1126 10.1126/science.1127335 [DOI] [PubMed] [Google Scholar]
- Dumaz N., Milne D. M., Jardine L. J., Meek D. W. (2001). Critical roles for the serine 20, but not the serine 15, phosphorylation site and for the polyproline domain in regulating p53 turnover. Biochem. J. 359, 459–464 10.1042/0264-6021:3590459 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Galanty Y., Belotserkovskaya R., Coates J., Polo S., Miller K. M., Jackson S. P. (2009). Mammalian SUMO E3-ligases PIAS1 and PIAS4 promote responses to DNA double-strand breaks. Nature 462, 935–939 10.1038/nature08657 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gill G. (2004). SUMO and ubiquitin in the nucleus: different functions, similar mechanisms? Genes Dev. 18, 2046–2059 10.1101/gad.1214604 [DOI] [PubMed] [Google Scholar]
- Goodarzi A. A., Kurka T., Jeggo P. A. (2011). KAP-1 phosphorylation regulates CHD3 nucleosome remodeling during the DNA double-strand break response. Nat. Struct. Mol. Biol. 18, 831–839 10.1038/nsmb.2077 [DOI] [PubMed] [Google Scholar]
- Hayakawa F., Privalsky M. L. (2004). Phosphorylation of PML by mitogen-activated protein kinases plays a key role in arsenic trioxide-mediated apoptosis. Cancer Cell 5, 389–401 10.1016/S1535-6108(04)00082-0 [DOI] [PubMed] [Google Scholar]
- Heideker J., Perry J. J. P., Boddy M. N. (2009). Genome stability roles of SUMO-targeted ubiquitin ligases. DNA Repair (Amst.) 8, 517–524 10.1016/j.dnarep.2009.01.010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heideker J., Prudden J., Perry J. J. P., Tainer J. A., Boddy M. N. (2011). SUMO-targeted ubiquitin ligase, Rad60, and Nse2 SUMO ligase suppress spontaneous Top1 mediated DNA damage and genome instability. PLoS Genet. 7, e1001320. 10.1371/journal.pgen.1001320 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hu X. V., Rodrigues T. N. M. A., Tao H., Baker R. K., Miraglia L., Orth A. P., Lyons G. E., Schultz P. G., Wu X. (2010). Identification of RING finger protein 4 (RNF4) as a modulator of DNA demethylation through a functional genomics screen. Proc. Natl. Acad. Sci. U.S.A. 107, 15087–15092 10.1073/pnas.1001006107 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huen M. S. Y., Grant R., Manke I., Minn K., Yu X., Yaffe M. B., Chen J. (2007). RNF8 transduces the DNA-damage signal via histone ubiquitylation and checkpoint protein assembly. Cell 131, 901–914 10.1016/j.cell.2007.09.041 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hunter T. (2007). The age of crosstalk: phosphorylation, ubiquitination, and beyond. Mol. Cell 28, 730–738 10.1016/j.molcel.2007.11.019 [DOI] [PubMed] [Google Scholar]
- Ikura T., Tashiro S., Kakino A., Shima H., Jacob N., Amunugama R., Yoder K., Izumi S., Kuraoka I., Tanaka K., Kimura H., Ikura M., Nishikubo S., Ito T., Muto A., Miyagawa K., Takeda S., Fishel R., Igarashi K., Kamiya K. (2007). DNA damage-dependent acetylation and ubiquitination of H2AX enhances chromatin dynamics. Mol. Cell. Biol. 27, 7028–7040 10.1128/MCB.00579-07 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ivanov A. V., Peng H., Yurchenko V., Yap K. L., Negorev D. G., Schultz D. C., Psulkowski E., Fredericks W. J., White D. E., Maul G. G., Sadofsky M. J., Zhou M.-M., Rauscher Iii F. J. (2007). PHD domain-mediated E3 ligase activity directs intramolecular sumoylation of an adjacent bromodomain required for gene silencing. Mol. Cell 28, 823–837 10.1016/j.molcel.2007.11.012 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jackson S. P., Bartek J. (2009). The DNA-damage response in human biology and disease. Nature 461, 1071–1078 10.1038/nature08467 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Joe Y., Jeong J.-H., Yang S., Kang H., Motoyama N., Pandolfi P. P., Chung J. H., Kim M. K. (2006). ATR, PML, and CHK2 play a role in arsenic trioxide-induced apoptosis. J. Biol. Chem. 281, 28764–28771 10.1074/jbc.M604392200 [DOI] [PubMed] [Google Scholar]
- Kolas N. K., Chapman J. R., Nakada S., Ylanko J., Chahwan R., Sweeney F. D. R. D., Panier S., Mendez M., Wildenhain J., Thomson T. M., Pelletier L., Jackson S. P., Durocher D. (2007). Orchestration of the DNA-damage response by the RNF8 ubiquitin ligase. Science 318, 1637–1640 10.1126/science.1150034 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kosoy A., Calonge T. M., Outwin E. A., O’connell M. J. (2007). Fission yeast Rnf4 homologs are required for DNA repair. J. Biol. Chem. 282, 20388–20394 10.1074/jbc.M702652200 [DOI] [PubMed] [Google Scholar]
- Lallemand-Breitenbach V., Jeanne M., Benhenda S., Nasr R., Lei M., Peres L., Zhou J., Zhu J., Raught B., De Thé H. (2008). Arsenic degrades PML or PML-RAR[alpha] through a SUMO-triggered RNF4/ubiquitin-mediated pathway. Nat. Cell Biol. 10, 547–555 10.1038/ncb1717 [DOI] [PubMed] [Google Scholar]
- Lallemand-Breitenbach V., Zhu J., Puvion F., Koken M., Honoré N., Doubeikovsky A., Duprez E., Pandolfi P. P., Puvion E., Freemont P., De Thé H. (2001). Role of promyelocytic leukemia (Pml) sumolation in nuclear body formation, 11s proteasome recruitment, and as2O3-Induced Pml or Pml/retinoic acid receptor α degradation. J. Exp. Med. 193, 1361–1372 10.1084/jem.193.12.1361 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lavin M. F. (2008). Ataxia-telangiectasia: from a rare disorder to a paradigm for cell signalling and cancer. Nat. Rev. Mol. Cell Biol. 9, 759–769 10.1038/nrm2514 [DOI] [PubMed] [Google Scholar]
- Lee Y.-K., Thomas S. N., Yang A. J., Ann D. K. (2007). Doxorubicin down-regulates Kruppel-associated box domain-associated protein 1 sumoylation that relieves its transcription repression on p21WAF1/CIP1 in breast cancer MCF-7 cells. J. Biol. Chem. 282, 1595–1606 10.1074/jbc.M706372200 [DOI] [PubMed] [Google Scholar]
- Li X., Lee Y.-K., Jeng J.-C., Yen Y., Schultz D. C., Shih H.-M., Ann D. K. (2007). Role for KAP1 serine 824 phosphorylation and sumoylation/desumoylation switch in regulating KAP1-mediated transcriptional repression. J. Biol. Chem. 282, 36177–36189 10.1074/jbc.M611781200 [DOI] [PubMed] [Google Scholar]
- Li X., Lin H. H., Chen H., Xu X., Shih H.-M., Ann D. K. (2010). SUMOylation of the transcriptional co-repressor KAP1 is regulated by the serine and threonine phosphatase PP1. Sci. Signal. 3, ra32. 10.1126/scisignal.2000781 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu B., Yang Y., Chernishof V., Loo R. R. O., Jang H., Tahk S., Yang R., Mink S., Shultz D., Bellone C. J., Loo J. A., Shuai K. (2007). Proinflammatory stimuli induce IKKα-mediated phosphorylation of PIAS1 to restrict inflammation and immunity. Cell 129, 903–914 10.1016/j.cell.2007.02.047 [DOI] [PubMed] [Google Scholar]
- Lou Z., Minter-Dykhouse K., Franco S., Gostissa M., Rivera M. A., Celeste A., Manis J. P., Van Deursen J., Nussenzweig A., Paull T. T., Alt F. W., Chen J. (2006). MDC1 maintains genomic stability by participating in the amplification of ATM-dependent DNA damage signals. Mol. Cell 21, 187–200 10.1016/j.molcel.2005.11.025 [DOI] [PubMed] [Google Scholar]
- Maya R., Balass M., Kim S.-T., Shkedy D., Leal J.-F. M., Shifman O., Moas M., Buschmann T., Ronai Z. E., Shiloh Y., Kastan M. B., Katzir E., Oren M. (2001). ATM-dependent phosphorylation of Mdm2 on serine 395: role in p53 activation by DNA damage. Genes Dev. 15, 1067–1077 10.1101/gad.886901 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meerang M., Ritz D., Paliwal S., Garajova Z., Bosshard M., Mailand N., Janscak P., Hubscher U., Meyer H., Ramadan K. (2011). The ubiquitin-selective segregase VCP/p97 orchestrates the response to DNA double-strand breaks. Nat. Cell Biol. 13, 1376–1382 10.1038/ncb2367 [DOI] [PubMed] [Google Scholar]
- Morris J. R., Boutell C., Keppler M., Densham R., Weekes D., Alamshah A., Butler L., Galanty Y., Pangon L., Kiuchi T., Ng T., Solomon E. (2009). The SUMO modification pathway is involved in the BRCA1 response to genotoxic stress. Nature 462, 886–890 10.1038/nature08593 [DOI] [PubMed] [Google Scholar]
- Morrison C., Sonoda E., Takao N., Shinohara A., Yamamoto K., Takeda S. (2000). The controlling role of ATM in homologous recombination repair of DNA damage. EMBO J. 19, 463–471 10.1093/emboj/19.3.463 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mouysset J., Deichsel A., Moser S., Hoege C., Hyman A. A., Gartner A., Hoppe T. (2008). Cell cycle progression requires the CDC-48UFD-1/NPL-4 complex for efficient DNA replication. Proc. Natl. Acad. Sci. U.S.A. 105, 12879–12884 10.1073/pnas.0805944105 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nagai S., Dubrana K., Tsai-Pflugfelder M., Davidson M., Robers T., Brown G., Varela E., Hedinger F., Grasser S., Nj K. (2008). Functional targeting of DNA damage to a nuclear pore-associated SUMO-dependent ubiquitin ligase. Science 322. 10.1126/science.1162790 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Petrie K., Zelent A. (2008). Marked for death. Nat. Cell Biol. 10, 507–509 10.1038/ncb0508-507 [DOI] [PubMed] [Google Scholar]
- Praefcke G. J. K., Hofmann K., Dohmen R. J. R. (2011). SUMO playing tag with ubiquitin. Trends Biochem. Sci. 1–9 [DOI] [PubMed] [Google Scholar]
- Prudden J., Pebernard S., Raffa G., Slavin D., Perry J., Tainer J., Mcgowan C., Boddy M. N. (2007). SUMO-targeted ubiquitin ligases in genome stability. EMBO J. 26, 4089–4101 10.1038/sj.emboj.7601838 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ramadan K., Bruderer R., Spiga F. M., Popp O., Baur T., Gotta M., Meyer H. H. (2007). Cdc48/p97 promotes reformation of the nucleus by extracting the kinase Aurora B from chromatin. Nature 450, 1258–1262 10.1038/nature06388 [DOI] [PubMed] [Google Scholar]
- Richly H., Rape M., Braun S., Rumpf S., Hoege C., Jentsch S. (2005). A series of ubiquitin binding factors connects CDC48/p97 to substrate multiubiquitylation and proteasomal targeting. Cell 120, 73–84 10.1016/j.cell.2004.11.013 [DOI] [PubMed] [Google Scholar]
- Ryan R., Schultz D., Ayyanathan K., Singh P., Friedman J., Fredericks W., Rauscher F. J. (1999). KAP-1 Corepressor protein interacts and colocalizes with heterochromatic and euchromatic HP1 proteins: a potential role for Krüppel-associated box–zinc finger proteins in heterochromatin-mediated gene silencing. Mol. Cell. Biol. 19, 4366–4378 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scaglioni P. P., Yung T. M., Cai L. F., Erdjument-Bromage H., Kaufman A. J., Singh B., Teruya-Feldstein J., Tempst P., Pandolfi P. P. (2006). A CK2-dependent mechanism for degradation of the PML tumor suppressor. Cell 126, 269–283 10.1016/j.cell.2006.05.041 [DOI] [PubMed] [Google Scholar]
- Schultz D., Friedman J., Rauscher F. J. (2001). Targeting histone deacetylase complexes via KRAB-zinc finger proteins: the PHD and bromodomains of KAP-1 form a cooperative unit that recruits a novel isoform of the Mi-2α subunit of NuRD. Genes Dev. 15, 428–443 10.1101/gad.869501 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schultz D. C., Ayyanathan K., Negorev D., Maul G. G., Rauscher F. J. (2002). SETDB1: a novel KAP-1-associated histone H3, lysine 9-specific methyltransferase that contributes to HP1-mediated silencing of euchromatic genes by KRAB zinc-finger proteins. Genes Dev. 16, 919–932 10.1101/gad.973302 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shanbhag N. M., Rafalska-Metcalf I. U., Balane-Bolivar C., Janicki S. M., Greenberg R. A. (2010). ATM-dependent chromatin changes silence transcription in cis to DNA double-strand breaks. Cell 141, 970–981 10.1016/j.cell.2010.04.038 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shiloh Y. (2003). ATM and related protein kinases: safeguarding genome integrity. Nat. Rev. Cancer 3, 155–168 10.1038/nrc1011 [DOI] [PubMed] [Google Scholar]
- Sripathy S. P., Stevens J., Schultz D. C. (2006). The KAP1 corepressor functions to coordinate the assembly of de novo HP1-demarcated microenvironments of heterochromatin required for KRAB zinc finger protein-mediated transcriptional repression. Mol. Cell. Biol. 26, 8623–8638 10.1128/MCB.00487-06 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tang J.-B., Greenberg R. A. (2010). Connecting the dots interplay between ubiquitylation, and SUMOylation at DNA double-strand breaks. Genes Cancer 1, 787–796 10.1177/1947601910382774 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tatham M. H., Geoffroy M.-C., Shen L., Plechanovova A., Hattersley N., Jaffray E. G., Palvimo J. J., Hay R. T. (2008). RNF4 is a poly-SUMO-specific E3 ubiquitin ligase required for arsenic-induced PML degradation. Nat. Cell Biol. 10, 538–546 10.1038/ncb1716 [DOI] [PubMed] [Google Scholar]
- Uziel T., Lerenthal Y., Moyal L., Andegeko Y., Mittelman L., Shiloh Y. (2003). Requirement of the MRN complex for ATM activation by DNA damage. EMBO J. 22, 5612–5621 10.1093/emboj/cdg541 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Verma R., Oania R., Fang R., Smith G. T., Deshaies R. J. (2011). Cdc48/p97 Mediates UV-Dependent turnover of RNA Pol II. Mol. Cell 41, 82–92 10.1016/j.molcel.2010.12.017 [DOI] [PMC free article] [PubMed] [Google Scholar]
- White D. E., Negorev D., Peng H., Ivanov A. V., Maul G. G., Rauscher F. J., III. (2006). KAP1, a novel substrate for PIKK family members, colocalizes with numerous damage response factors at DNA lesions. Cancer Res. 66, 11594–11599 10.1158/0008-5472.CAN-06-4138 [DOI] [PubMed] [Google Scholar]
- Woods A. S., Ferre S. (2005). Amazing stability of the arginine-phosphate electrostatic interaction. J. Proteome Res. 4, 1397–1402 10.1021/pr050077s [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wuerzberger-Davis S. M., Nakamura Y., Seufzer B. J., Miyamoto S. (2006). NF-[kappa]B activation by combinations of NEMO SUMOylation and ATM activation stresses in the absence of DNA damage. Oncogene 26, 641–651 10.1038/sj.onc.1209815 [DOI] [PubMed] [Google Scholar]
- Yang S., Kuo C., Bisi J. E., Kim M. K. (2002). PML-dependent apoptosis after DNA damage is regulated by the checkpoint kinase hCds1/Chk2. Nat. Cell Biol. 4, 865–870 10.1038/ncb869 [DOI] [PubMed] [Google Scholar]