Abstract
Exosomes are secreted membrane vesicles that have been proposed as an effective means to detect a variety of disease states, including cancer. The properties of exosomes, including stability in biological fluids, allow for their efficient isolation and make them an ideal vehicle for studies on early disease detection and evaluation. Much data has been collected over recent years regarding the messenger RNA, microRNA, and protein contents of exosomes. In addition, many studies have described the functional role that exosomes play in disease initiation and progression. Tumor cells have been shown to secrete exosomes, often in increased amounts compared to normal cells, and these exosomes can carry the genomic and proteomic signatures characteristic of the tumor cells from which they were derived. While these unique signatures make exosomes ideal for cancer detection, exosomes derived from cancer cells have also been shown to play a functional role in cancer progression. Here, we review the unique genomic and proteomic contents of exosomes originating from cancer cells as well as their functional effects to promote tumor progression.
Keywords: exosomes, microvesicles, cancer, detection
Background
Exosomes are small membrane vesicles ranging from 40 to 100 nm in size that are formed within multivesicular bodies (MVB) and secreted upon fusion of the MVB with the plasma membrane (Denzer et al., 2000). Exosomes were first described in 1987 when Johnstone et al. (1987) noted that vesicles shed from cultured monolayer cells retained enzymatic activity reminiscent of the parent cells. It was later determined that exosomes are remnants of the endocytic pathway following endosomal sorting of components that are not degraded by lysosomes. These vesicles are then released from the cell in the form of exosomes. Thus, exosomes were first thought of as a way of disposing unnecessary membrane proteins, especially during reticulocyte maturation (Johnstone et al., 1989, 1991; Johnstone, 1992). It is now known that this is not entirely the case and exosomes can have significant biological roles (Thery et al., 2002; Johnstone, 2006; Thery, 2011; Yang and Robbins, 2011). It is worth noting that some confusion exists regarding the terms: “exosome” and “microvesicle.” Most researchers discriminate between the two mostly on the basis of size, with exosomes falling in the range of 40–100 nm and microvesicles in the range of 100–200 nm. Some of the studies cited in this review will use these terms with more stringent requirements while others use the terms interchangeably. Still others use only the term “microvesicle” as a general term to include both types of secreted vesicles. Since exosome/microvesicle research is still a fledgling field, these definitions may change over time to become more stringent or may include additional parameters to define each type of vesicle.
Exosome membranes are enriched in cholesterol, sphingomyelin, and ceramide (Simpson et al., 2008) as well as lipid raft associated proteins (de Gassart et al., 2003). These components allow exosomes to be highly stable and they can thus be collected from numerous bodily fluids (Keller et al., 2011) including blood (Li et al., 2008), urine (Gonzales et al., 2010), breast milk (Zhou et al., 2012), ascites (Navabi et al., 2005), and saliva (Ogawa et al., 2008). Once these fluids have been collected, exosomes can be easily isolated using differential ultracentrifugation in order to first remove large cells and debris and then pellet the exosomes. The ubiquitous nature of exosomes in bodily fluids makes them ideal for use in diagnostic biomarker studies. In addition, studies have shown that exosomes can also have a functional role in human biology (Figure 1).
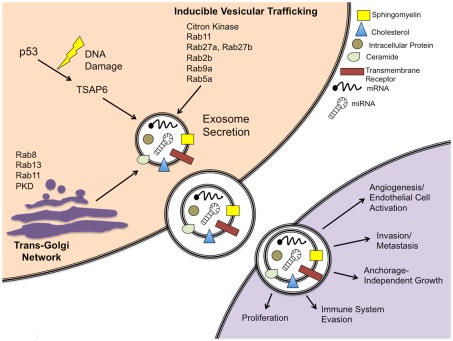
Figure 1.
Schematic of exosome secretion in a cancer cell model. Exosomes are secreted via a constitutive pathway involving the Trans-Golgi Network and/or inducible pathways, which can be activated by a p53-mediated response to DNA damage or Rab GTPases. Characteristic protein and RNA components are shown for a canonical exosome. Exosomes released from cancer cells can be taken up by neighboring cells and are capable of inducing pathways involved in cancer initiation and progression.
Exosomes are secreted by two different mechanisms, constitutive release via the Trans-Golgi Network and inducible release (Record et al., 2011). It is often noted that exosome concentrations are increased in cancer patients compared to normal controls (Taylor and Gercel-Taylor, 2008; Logozzi et al., 2009), though the reason for this is not fully understood. One study noted that the tumor suppressor protein p53 induces activation of tumor suppressor activated pathway-6 (TSAP6) following DNA damage (Yu et al., 2006). This same study found that over-expression of TSAP6 upregulates exosome production, even in the absence of a functional p53 protein. It is possible that cancer cells release prodigious amounts of exosomes due to high TSAP6, though this link has not yet been made. Another possibility is general upregulation of exosome secretion due to Rab GTPases. The Rab family of proteins are often mutated (constitutively active) or over-expressed in cancer cells. It is known that several members of the Rab GTPase family can affect exosome secretion via the Trans-Golgi Network (Ponnambalam and Baldwin, 2003) or by inducible vesicular trafficking (Loomis et al., 2006; Ostrowski et al., 2010). These factors and their role in exosome secretion are depicted in Figure 1.
Canonical exosomes contain lipids, proteins, microRNA (miRNA), and messenger RNA (mRNA). These components, after being secreted by a source cell, can be taken up by target cells wherein the proteins may affect cellular signaling and the miRNA and mRNA could modulate gene transcription and translation. Early functional studies focused on the role of exosomes in reticulocyte maturation and immune cell function. Since that time, it has been suggested that exosomes also play an important role in certain diseases such as Alzheimer’s disease (Rajendran et al., 2006), where beta-amyloid peptides are secreted from cells via exosomes. It is believed that these exosomes contribute to the pathogenesis of the disease by spreading Alzheimer’s-related proteins to surrounding cells. Additionally, the role of exosomes in immune system functions is also well documented (Li et al., 2006; Bobrie et al., 2011; Chaput and Thery, 2011).
Rather than being a random sampling of intracellular components, exosomes contain very specific types of proteins. Since exosomes form by budding of the plasma membrane, these proteins are generally cell surface receptors and components of the endocytic pathway. However, they are also known to contain select cytosolic proteins, such as heat shock proteins (Mathew et al., 1995; Clayton and Tabi, 2005). Many of the proteins, miRNA, and mRNA found in exosomes have a role in cancer development and progression (Table 1). In terms of the function of cancer cell-derived exosomes, there is conflicting evidence in the literature as studies have indicated that exosomes derived from cancer cells can have both anti-tumorigenic and pro-tumorigenic properties (Yang and Robbins, 2011). Here, we will review the genomic and proteomic data from cancer cell-derived exosomes. Furthermore, we examine how these exosomes could affect cancer progression and how they might be utilized for cancer diagnosis and treatment.
Table 1.
Summary of targets and markers isolated from cancer cell-derived exosomes.
| Marker | Type | Exosome source | Effect | Reference |
|---|---|---|---|---|
| Amphiregulin | Protein | Breast, colorectal cancer | Invasion | Higginbotham et al. (2011) |
| CD44v6 | Protein | Rat pancreatic adenocarcinoma | Pre-metastatic niche formation | Jung et al. (2009) |
| Del-1 | Protein | Mesothelioma | Angiogenesis* | Hegmans et al. (2004), Ho et al. (2004) |
| EGFR | Protein | Pancreatic cancer, brain cancer | Proliferation*, signal transduction* | Adamczyk et al. (2011), Graner et al. (2009) |
| Hsp90α | Protein | Invasive carcinomas | Migration, invasion | McCready et al. (2010) |
| LMP1 | Protein | Nasopharyngeal carcinoma | Signal transduction | Meckes et al. (2010) |
| LRG1 | Protein | Non-small cell lung | Signal transduction*, cell adhesion* | Li et al. (2011) |
| MUC1 | Protein | Breast cancer | Growth*, inhibition of Apoptosis*, invasion* | Staubach et al. (2009) |
| TSPAN8 | Protein | Rat pancreatic adenocarcinoma | Endothelial cell activation, angiogenesis | Nazarenko et al. (2010), Gesierich et al. (2006) |
| Let-7 | miRNA | Lung cancer, metastatic gastric cancer | Downregulate Ras* | Takamizawa et al. (2004), Ohshima et al. (2010) |
| miR-21 | miRNA | Ovarian cancer, glioblastoma, breast, pancreatic cancer | Downregulate PDCD4* | Taylor and Gercel-Taylor (2008), Skog et al. (2008) |
| CDK8 | mRNA | Colorectal cancer | Cell division* | Hong et al. (2009) |
| EGFRvIII | mRNA | Glioblastoma | Proliferation*, signal transduction* | Skog et al. (2008), Graner et al. (2009) |
| RAD21 | mRNA | Colorectal cancer | Mitosis*, DNA repair* | Hong et al. (2009) |
*Effect of marker is implied based on other published research.
Function of Tumor-Derived Exosomes
Although exosomes derived from tumor cells have been shown to have anti-tumorigenic properties such as inducing tumor cell apoptosis (Ristorcelli et al., 2008) and enhancing anti-tumor immunity (Zhang et al., 2010), there is increased evidence that these exosomes could play a more important role in tumor progression (Yang and Robbins, 2011). It is not often that a cancer patient dies from a lone primary tumor; it is generally the dissemination of tumor cells throughout the body and their unchecked proliferation that interferes with organ function and causes the patient to succumb to the disease. In order for this to happen, cells derived from the primary tumor must accumulate mutations in numerous genes that affect cellular functions, such as growth, apoptosis, migration, and angiogenesis (Hanahan and Weinberg, 2000). Because exosomes carry genomic and proteomic materials known to mediate these hallmarks of cancer, it is hypothesized that exosomes secreted by tumor cells have a role in the growth and dissemination of tumor cells. Indeed, many studies have demonstrated such potential in tumor-derived exosomes.
One of the hallmark properties of tumorigenicity is the ability of cells to form colonies on soft agar, which is indicative of anchorage-independent growth (AIG). Ochieng et al. (2009) demonstrated that exosomes isolated from fetal bovine serum (a common supplement in cell culture media) were capable of inducing AIG in breast cancer cells. When cells were grown using exosome-enhanced media (EEM), they formed numerous large colonies on soft agar, whereas cells grown using exosome-free media (EFM) showed little or no colony growth. Furthermore, cells grown on matrigel in combination with the EEM formed three-dimensional stellate colonies whereas cells grown with EFM on matrigel grew mainly as a monolayer. Studies on the effects of exosomes on cancer cell growth were reinforced by Qu et al. (2009) who determined that exosomes derived from a gastric cancer cell line could promote proliferation, due in part to activation of PI3K/AKT in the target cells.
In order for a tumor to successfully metastasize, cells must first extravasate from the primary tumor, intravasate into the blood or lymph, and establish a new tumor in an environment conducive to tumor cell growth. McCready et al. (2010) describes how invasive cancer cells secrete exosomes containing Hsp90α, which the study implicates in activating plasmin. The authors demonstrated that exosomes isolated from an invasive cell line could enhance cell migration but the effect was abrogated if an anti-Hsp90 antibody was added in addition to the exosomes. Many proteomic changes occur when tumor cells are induced to migrate. This is usually the result of hypoxia, where a tumor outgrows its blood supply and must induce migration and/or angiogenesis in order for the cancer cells to survive. Park et al. (2010) characterized the proteomic changes that occurred when cells were grown in hypoxic conditions and noted that 54% of the differentially regulated proteins were co-localized with the exosome fraction of the conditioned media. This suggests that cells undergoing hypoxic stress change the composition of their exosomes, potentially to facilitate migration and angiogenesis. Once tumor cells have extravasated from the primary tumor, additional conditions must be met to establish new tumors. Jung et al. (2009) noted that exosomes contributed to pre-metastatic niche formation in a rat model of pancreatic adenocarcinoma. The study noted that exosomes derived from weakly metastatic rat ASML CD44v4–v7 knockdown cells in combination with the soluble fraction of counterpart wild-type cells caused expression changes in numerous proteases, growth factors, and adhesion molecules. This suggests that exosomes can act over long distances to create a metastatic environment for tumor cells.
One of the hallmarks of angiogenesis is endothelial cell activation, wherein chronic inflammation precedes angiogenic sprout formation (Rajashekhar et al., 2006). It has been shown that exosomes enriched in TSPAN8 protein are capable of activating endothelial cells and inducing expression of many genes involved in angiogenesis (Nazarenko et al., 2010). Furthermore, the systemic role of TSPAN8-enriched exosomes in angiogenesis has also been described. Gesierich et al. (2006) noted that rats over-expressing D6.1A (the rat homolog of TSPAN8) suffered from lethally disseminated intravascular coagulation. When exosomes were isolated from isogenic cell lines, it was noted that exosomes from D6.1A-over-expressing cells induced endothelial cell branching in vitro whereas the counterpart wild-type cells did not. These effects were abrogated by the addition of an antibody against D6.1A, but remained the same when antibodies against other exosome-associated proteins were used. Importantly, the authors noted high expression of D6.1A in sprouting capillaries even if the primary tumor did not express the protein, suggesting D6.1A/TSPAN8 might be a therapeutic target for inhibition of angiogenesis.
Proteomic Analysis of Cancer Exosomes
In order to investigate the differences between tumor-derived exosomes and normal cell-exosomes, researchers often characterize the proteome of tumor exosomes. Exosomes are representative of the cell from which they were derived, so it is reasonable to assume that proteome signatures would exhibit unique qualities dependent on the cellular origin. Indeed, one of the first exosomal proteomes to be characterized was from mesothelioma cells. This study identified 38 distinct proteins, many of which were previously shown to be expressed in exosomes (Hegmans et al., 2004). However, it is interesting to note that the authors also identified developmental endothelial locus-1 (DEL-1), a protein not previously identified in exosomes but suggested to play a role in angiogenesis (Ho et al., 2004).
Another study investigated the exosomal proteome of a bladder cancer cell line. The use of exosomes in bladder cancer studies is particularly interesting since exosomes can be isolated from urine. The concentration of tumor-derived exosomes would theoretically be greater in a urine sample from a bladder cancer patient (in comparison to exosomes isolated from sera of the same patient), thus providing a potentially powerful diagnostic tool. The study by Welton et al. (2010) identified 353 proteins contained within HT1376-derived exosomes, 72 of which were not previously associated with exosomes. It will be interesting to see if any of these proteins are also found in urinary exosomes of bladder cancer patients. To this point, a Urinary Exosome Protein Database has been compiled based on two separate studies by the same group1. However, in each case, fewer than 10 patients were enrolled in the study and all patients were healthy at the time of the study (Pisitkun et al., 2004; Gonzales et al., 2009). This database currently contains 1160 identified proteins and will undoubtedly become essential in the identification of urinary biomarkers of cancer patients.
A more extensive proteomic study on urinary exosomes in cancer patients has been completed for non-small cell lung carcinoma (NSCLC; Li et al., 2011). In the study, urine was collected from 10 healthy patients and 8 NSCLC patients. The proteomic differences of the isolated exosomes were first characterized by 1D SDS-PAGE. The differentially expressed proteins were then sequenced using mass spectrometry. The authors report 18 proteins identified in urinary exosome preparations, 11 of which were found only in NSCLC exosomes, four only in normal exosomes, and three in both. Of note, LRG1, a known serum cancer biomarker (Kakisaka et al., 2007), was identified as being highly expressed in the NSCLC exosomes. The authors suggest that LRG1 might be useful in diagnosing NSCLC through the use of a simple urine test.
Oncogenic Proteins in Exosomes
The list of proteins found in exosomes is continuously expanding on ExoCarta, a database of exosomal proteins and RNA and lipids2 (Mathivanan and Simpson, 2009). Table 2 shows several of the top proteins found in exosomes as listed on ExoCarta. Oncogenes that are associated with various types of cancer can often be found in the exosomes secreted by tumor cells. Full-length EGFR has been identified in exosomes isolated from pancreatic cell lines (Adamczyk et al., 2011). Indeed, other studies have also identified EGFR in exosomes originating from brain tumors (Graner et al., 2009) and EGFR ligands, such as amphiregulin and TGFα, have been identified in exosomes isolated from breast and colorectal cell lines (Higginbotham et al., 2011). Another cell surface protein, MUC1, is commonly over-expressed in breast cancer and has been isolated in breast cancer exosomes (Staubach et al., 2009). However, in both cases it is not clear whether these oncogenes retain a functional role and affect surrounding cells or if they are simply packaged within exosomes because they are prominently expressed in the plasma membrane.
Table 2.
Proteins identified in exosomes as indicated in the ExoCarta Database.
| Number | Gene symbol | Gene name | Number of times identified |
|---|---|---|---|
| 1 | CD9 | CD9 molecule | 46 |
| 2 | HSPA8 | Heat shock 70 kDa protein 8 | 45 |
| 3 | CD63 | CD63 molecule | 41 |
| 4 | GAPDH | Glyceraldehyde-3-phosphate dehydrogenase | 38 |
| 5 | CD81 | CD81 molecule | 35 |
| 6 | SDCBP | Syndecan binding protein (syntenin) | 32 |
| 7 | PDCD6IP | Programmed cell death six interacting protein | 32 |
| 8 | ENO1 | Enolase 1, (alpha) | 32 |
| 9 | ANXA2 | Annexin A2 | 32 |
| 10 | ACTB | Actin, beta | 32 |
| 11 | YWHAZ | Tyrosine 3-monooxygenase/tryptophan 5-monooxygenase activation protein, zeta polypeptide | 29 |
| 12 | HSP90AA1 | Heat shock protein 90 kDa alpha (cytosolic), class A member 1 | 29 |
| 13 | ANXA5 | Annexin A5 | 29 |
| 14 | EEF1A1 | eukaryotic translation elongation factor 1 alpha 1 | 28 |
| 15 | YWHAE | Tyrosine 3-monooxygenase/tryptophan 5-monooxygenase activation protein, epsilon polypeptide | 26 |
| 16 | PPIA | Peptidylprolyl isomerase A (cyclophilin A) | 26 |
| 17 | MSN | Moesin | 26 |
| 18 | CFL1 | Cofilin 1 (non-muscle) | 26 |
| 19 | ALDOA | Aldolase A, fructose-bisphosphate | 26 |
| 20 | PGK1 | Phosphoglycerate kinase 1 | 25 |
Other studies have demonstrated not only the presence of oncogenic proteins in exosomes, but also their ability to influence cells that would receive these exosomes. Latent membrane protein 1 (LMP1) is produced in cells infected with Epstein–Barr virus (EBV), which is known to induce transformation in B-cell malignancies. The LMP1 protein activates multiple signaling pathways involved in apoptosis, cell cycle progression, proliferation, and migration. Cells infected with EBV produce exosomes that contain LMP1. Meckes et al. (2010) noted that the LMP1 contained within these exosomes was capable of activating the ERK and PI3K/AKT pathways in recipient cells. This study utilized isogenic cell lines, both infected with EBV but only one expressing detectable levels of LMP1. When HUVECs were treated with increasing amounts of exosomes from the LMP1 over-expressing line, the amount of LMP1 protein in the recipient cells increased in a dose-dependent manner. Exosomes isolated from the cells without LMP1 caused an increase in phospho-ERK in recipient HUVECs, but this increase was greater when cells were treated with exosomes from cells that over-expressed LMP1. Therefore, exosome-derived LMP1 can contribute to ERK and PI3K/AKT activation in recipient cells, but it is clearly not the only effector contained therein.
miRNA in Cancer Cell-Derived Exosomes
microRNA are a class of small, non-coding RNA molecules that are involved in post-transcriptional modification of gene expression. First identified in C. elegans (Lagos-Quintana et al., 2001), miRNA have been shown to possess oncogenic or tumor suppressive properties based on how each miRNA affects gene expression (Ahmed, 2007). In the bloodstream, miRNA have been found to be associated with both protein complexes (Arroyo et al., 2011) as well as microvesicles such as exosomes (Valadi et al., 2007; Kosaka et al., 2010). These miRNA have also been referred to as “exosomal shuttle RNA” (esRNA; Valadi et al., 2007). Exosomes offer protection from degradation by RNase enzymes (Mitchell et al., 2008), thereby permitting miRNA manufactured in one cell to be taken up by a neighboring cell with presumed subsequent effects on gene expression in the target cell (Valadi et al., 2007; Kosaka et al., 2010). Researchers have noted that certain cancer types, including ovarian (Taylor and Gercel-Taylor, 2008), glioblastoma (Skog et al., 2008), lung (Rosell et al., 2009), and breast (Corcoran et al., 2011) have distinct exosomal miRNA profiles.
The miRNA Let-7, which was one of the first miRNA to be characterized, was one of several miRNA identified in exosomes isolated from mouse mast cells (Valadi et al., 2007). Let-7 keeps cell proliferation in check by negatively regulating Ras GTPases (Johnson et al., 2005). Activating Ras mutations are common in cancer, most often associated with cancers of the pancreas, colon, lung, and also acute myeloid leukemia (Saxena et al., 2008). A study by Takamizawa et al. (2004) demonstrated that lower expression of Let-7 in lung cancer patients correlated with poor survival following surgical resection. Interestingly, Let-7 miRNA has been identified in exosomes isolated from a metastatic gastric cancer cell line (Ohshima et al., 2010). The authors of the study suggest that these cells selectively secrete let-7 miRNA in order to maintain tumorigenic and metastatic properties.
In contrast to the tumor suppressive activities of Let-7, miRNA have also been shown to act as oncogenes. For example, miR-21 has been shown to be present in peripheral blood exosomes (PBXs) isolated from patients with ovarian cancer (Taylor and Gercel-Taylor, 2008). This particular miRNA was also present in PBXs isolated from patients with benign ovarian disease, albeit at ∼30% of the levels of patients with stage I, II, or III ovarian cancer. It is also present at high levels in exosomes isolated from patients with glioblastoma, though levels in healthy patients were not determined by this particular study (Skog et al., 2008). miR-21 abrogates expression of the antiapoptotic gene PDCD4, thereby contributing to cell survival (Asangani et al., 2008). In addition to being a potential marker for ovarian cancer, miR-21 is over-expressed in glioblastoma (Chan et al., 2005), breast cancer (Iorio et al., 2005), and pancreatic cancer (Roldo et al., 2006; Lee et al., 2007). It has also been shown to mediate drug resistance to trastuzumab in breast cancer (Gong et al., 2011) and temozolomide in glioblastoma cells (Shi et al., 2010).
While these data regarding unique expression of miRNA in tumor-derived exosomes is promising, these studies did not demonstrate effects following uptake of these exosomes directly related to the expression of a particular miRNA. Therefore, it is not known whether these miRNA serve a functional role within tumor exosomes. There is associative evidence that would suggest they might mediate gene expression in target cells, but these studies are based on in vitro models where the miRNA levels might differ greatly from those found in exosomes.
Cancer Cell-Derived Exosomal mRNA
Exosomes are also known to contain mRNA although these species of RNA appear to be found at lower abundance compared to the small RNA components (Zomer et al., 2010). Studies have shown that these exosomal mRNA are functional and can be taken up and translated by recipient cells (Ratajczak et al., 2006; Valadi et al., 2007). Hong et al. (2009) profiled the mRNA of exosomes derived from the colorectal cancer cell line SW480. The study identified over 11,000 distinct mRNA molecules within the exosome samples. Of these, ∼2% were over-expressed at least twofold in exosomes (compared to levels in the parental cell line). The authors note that many of these mRNA are involved in cellular processes such as cell division, cell cycle, and chromosome segregation. In fact, ∼11% of the over-expressed mRNA are involved in regulating the cell cycle, specifically M-phase. When exosomes from SW480 cells were applied to HUVECs, the number of cells staining positive for a mitotic marker greatly increased. However, it is unclear if the cell cycle mRNA contained within the applied exosomes was the sole contributor to this resulting increase in mitosis.
Similar mRNA profiling was completed for exosomes derived from clinical glioblastoma samples (Skog et al., 2008). The authors noted 4,700 distinct mRNA were detected only in exosome arrays (when compared to the array of the corresponding cells from which the exosomes were derived), suggesting a selective enrichment process in the packaging of mRNA within exosomes. Approximately 8% of the exosomal mRNA were upregulated more than fivefold when compared to the corresponding cells. Many of these mRNA are known to have roles in angiogenesis, cell proliferation, immune response, migration, and/or histone modification. Interestingly, mRNA for a mutant version of EGFR (EGFRvIII) was identified in exosomes isolated from glioblastoma patient sera in 7 of the 25 patients. While this is approximately half the number of actual patients whose tumors were positive for the EGFRvIII transcript (28% positive in exosomes compared to 47% positive in corresponding tumors), the finding that mRNA encoding a mutant protein is present in circulating exosomes underscores the idea that serum exosomes may provide a powerful method for non-invasive detection of certain tumors.
Exosomes as a Potential Diagnostic
The proteomic and genomic biomarkers in tumor-derived exosomes coupled with established methodologies for isolating exosomes from body fluids has generated much promise in regards to using exosomes as a diagnostic tool. In theory, a patient would have their blood drawn using traditional methods and analysts would isolate the circulating exosomes and screen for biomarkers of disease. However, this raises a few questions. Would lab tests be able to detect the relatively small percentage of tumor exosomes in a patient’s serum? It is generally noted that tumor cells secrete more exosomes than normal counterparts (Taylor et al., 2011), so it is likely that tumor exosomes will be detectable. Another question we must address is whether exosome screening can offer any improvement over current serum biomarker tests.
The use of serum biomarkers is already established in ovarian cancer, where high levels of CA125 are often indicative of a tumor. However, the use of this marker is limited, as a significant portion of ovarian cancers do not express the protein. A study by Li et al. (2009) sought to determine if diagnostic rates of ovarian cancer could be improved using exosomes isolated from patient plasma. They found that 51% of ovarian cancer patients (32 out of 63) had circulating exosomes that expressed the protein claudin-4, which is often elevated in ovarian tumor cells. This is notably lower than the sensitivity of a CA125 test on the same patient samples, which demonstrated 71% of samples as correctly diagnosed. Both screening methods returned a false-positive diagnosis only 2% of the time (1 out of 50 normal patient samples).
Diagnosing malignant melanoma may also be aided by the use of exosome screening. Logozzi et al. (2009) demonstrated that exosomes isolated from 69% of melanoma patients enrolled in a small study contained caveolin-1. In contrast, only 7% of these patients would have been correctly diagnosed using serum LDH levels, the current-best prognostic serum factor for melanoma patients. This suggests that diagnostic rates could be improved through the use of a simple blood test. However, it is important that researchers determine which biomarker(s) are most often expressed in a given cancer type in order to prevent misdiagnosis.
One major serum-based screening method that is currently under heated debate is the PSA test for prostate cancer screening. This type of screen is plagued by high false-positives; when biopsy is performed due to elevated PSA, only 25–33% of patients actually have prostate cancer (Smith et al., 1997). One study on prostate cancer biomarkers identified high levels of extracellular delta-catenin in prostate cancer cells (Lu et al., 2009). Despite being an intracellular protein, it can be isolated in urine samples. The authors provide evidence to suggest that this is due to secretion of prostate cancer exosomes/prostasomes, which contain delta-catenin. Urinalysis for delta-catenin levels was completed on a small cohort of patients and the authors noted 87.5% sensitivity and 83.3% specificity. PSA testing on the same cohort was highly variable. In addition, a study by Tavoosidana et al. (2011) noted that the level of circulating prostasomes (microvesicles secreted by prostate acinar cells) were higher in prostate cancer patients compared to normal controls. Interestingly, higher plasma prostasome levels correlated with higher Gleason scores whereas PSA levels did not correlate with Gleason scores in the same cohort. These studies provide evidence that microvesicle-based screening could offer marked improvements in diagnosing cancer early and through non-invasive methods.
Several companies have begun developing exosome-based diagnostic tests. Exosome Diagnostics, Inc. (New York, NY, USA) has created a biofluid-based exosome test for prostate cancer, which is currently undergoing clinical validation. Another company utilizing exosomes for diagnostic purposes is Caris Life Sciences (Irving, TX, USA). Caris Life Sciences has developed a technology called Carisome™, where circulatory microvesicles are characterized to devise a biosignature, which will assist in determining diagnosis, prognosis, and predicting patient response. In contrast to these companies that utilize exosomes for diagnostics, Aethlon Medical, Inc. (San Diego, CA, USA) has developed a product called HER2osome™, which depletes circulatory HER2 protein and tumor-related exosomes in breast cancer patients. This type of therapy is proposed to enhance the benefits of traditional chemotherapies without adding drug toxicity or interaction risks.
Exosomes as Cancer Therapy
Because exosomes are capable of eliciting a potent immune response, researchers have sought to determine if exosomes can be utilized as a form of cancer immunotherapy (reviewed in (Tan et al., 2010). Many of these studies have demonstrated the application of dendritic cell-derived exosomes (DEX) in immune responses against tumors in vivo. It is known that DEX can be taken up by other dendritic cells which would likely perpetuate and amplify the immune response. In some cases, the application of DEX alone is not sufficient to elicit an immune response against an established tumor. Tumors are known to induce expansion of regulatory T (Treg) cells because of the high prevalence of self-antigens. These Treg cells are partly responsible for the tumor’s ability to evade the immune system. However, pretreatment with cyclophosphamide causes an elimination of the Treg cells and has been shown to synergize with DEX, leading to a potent secondary CD8+ T cell response and resulting in significant tumor shrinkage (Taieb et al., 2006).
Advances in exosome-based anti-cancer therapy have generally revolved around the application of DEX from dendritic cells that have been pulsed with tumor antigen. But is it possible for exosomes derived from tumor cells to elicit a similar immune response? Some studies have shown that this is possible, but the tumor cells must be engineered to express certain effector proteins (Zhang et al., 2010; Lee et al., 2011). It is unclear whether this type of therapy is feasible, since application in a clinical setting would have to involve removal of patient tumor cells, induction of ectopic expression, and harvesting of exosomes. Chen et al. (2006) demonstrated that it is possible for unmodified tumor-derived exosomes to elicit an immune response, but the effect is more potent if the tumor cells are heat shocked before harvesting the exosomes. This was true in both a therapeutic and prophylaxis model. Another study has also demonstrated the potential of tumor-derived exosomes as a prophylactic treatment (Bu et al., 2006), suggesting that exosomes could be used as a true cancer vaccine.
Conclusion
Tumor-derived exosomes can play an important role in disease progression and because exosomes are representative of parent cells, tumor-derived exosomes can provide much information about the tumor cells of origin. Studying tumor-derived exosomes will be imperative if non-invasive, early detection methods are to be developed. Though current detection methods are plagued with inaccuracies, it is likely that exosome-based detection will not be foolproof either. Cancer cells are notorious for developing mutations and changing protein expression levels in order to increase cell survival and circumvent traditional treatments. Therefore, it is important that exosome markers used in a cancer detection screen be expressed almost exclusively in tumor-derived exosomes.
Databases such as ExoCarta (Mathivanan and Simpson, 2009; Mathivanan et al., 2012) and the Urinary Exosome Protein Database have facilitated the sharing of data regarding the genomics and proteomics of exosomes. As we continue to amass this data, it will become important to determine which factors are most relevant to exosome functions. In most of the cases presented here, the authors have noted that large-scale effects, such as angiogenesis or cell growth, can be consequences of adding tumor-derived exosomes to normal cells. However, these effects are rarely due to only one protein or RNA molecule. Future exosome studies should focus on identification of the “non-reducible units” that are required for these functions. That is, which different proteins/RNA are required to mediate these effects and are these effects completely abrogated if the protein/RNA is not present? Another area of interest to the field and an area of interest to our group is the identification of exosomal markers that could be used to isolate exosomes derived by specific source cells for analysis.
The number of peer-reviewed publications focused on exosomes has increased exponentially within the past 15 years. Much excitement and promise has been generated around the use of exosomes in disease detection. Exosomes express only a small subset of the proteins and RNA expressed in a typical cell, which should permit an easier understanding of how they function and interact. These factors will hopefully expedite the clinical utilization of this data in order to better diagnose and treat cancer patients, thereby lowering mortality rates associated with such a devastating disease.
Conflict of Interest Statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Acknowledgments
The authors wish to acknowledge Kendall Jensen, Ben Rakela, and Daniel D. Von Hoff for their intellectual contributions to this work. We also thank Shilpi Arora for her critical reading of the manuscript.
Footnotes
References
- Adamczyk K. A., Klein-Scory S., Tehrani M. M., Warnken U., Schmiegel W., Schnolzer M., Schwarte-Waldhoff I. (2011). Characterization of soluble and exosomal forms of the EGFR released from pancreatic cancer cells. Life Sci. 89, 304–312 10.1016/j.lfs.2011.06.020 [DOI] [PubMed] [Google Scholar]
- Ahmed F. E. (2007). Role of miRNA in carcinogenesis and biomarker selection: a methodological view. Expert Rev. Mol. Diagn. 7, 569–603 10.1586/14737159.7.5.569 [DOI] [PubMed] [Google Scholar]
- Arroyo J. D., Chevillet J. R., Kroh E. M., Ruf I. K., Pritchard C. C., Gibson D. F., Mitchell P. S., Bennett C. F., Pogosova-Agadjanyan E. L., Stirewalt D. L., Tait J. F., Tewari M. (2011). Argonaute2 complexes carry a population of circulating microRNAs independent of vesicles in human plasma. Proc. Natl. Acad. Sci. U.S.A. 108, 5003–5008 10.1073/pnas.1019055108 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Asangani I. A., Rasheed S. A., Nikolova D. A., Leupold J. H., Colburn N. H., Post S., Allgayer H. (2008). MicroRNA-21 (miR-21) post-transcriptionally downregulates tumor suppressor Pdcd4 and stimulates invasion, intravasation and metastasis in colorectal cancer. Oncogene 27, 2128–2136 10.1038/sj.onc.1210856 [DOI] [PubMed] [Google Scholar]
- Bobrie A., Colombo M., Raposo G., Thery C. (2011). Exosome secretion: molecular mechanisms and roles in immune responses. Traffic 12, 1659–1668 10.1111/j.1600-0854.2011.01225.x [DOI] [PubMed] [Google Scholar]
- Bu N., Li Q. L., Feng Q., Sun B. Z. (2006). Immune protection effect of exosomes against attack of L1210 tumor cells. Leuk. Lymphoma 47, 913–918 10.1080/10428190500376191 [DOI] [PubMed] [Google Scholar]
- Chan J. A., Krichevsky A. M., Kosik K. S. (2005). MicroRNA-21 is an antiapoptotic factor in human glioblastoma cells. Cancer Res. 65, 6029–6033 10.1158/0008-5472.CAN-05-0137 [DOI] [PubMed] [Google Scholar]
- Chaput N., Thery C. (2011). Exosomes: immune properties and potential clinical implementations. Semin. Immunopathol. 33, 419–440 10.1007/s00281-010-0233-9 [DOI] [PubMed] [Google Scholar]
- Chen W., Wang J., Shao C., Liu S., Yu Y., Wang Q., Cao X. (2006). Efficient induction of antitumor T cell immunity by exosomes derived from heat-shocked lymphoma cells. Eur. J. Immunol. 36, 1598–1607 10.1002/eji.200635873 [DOI] [PubMed] [Google Scholar]
- Clayton A., Tabi Z. (2005). Exosomes and the MICA-NKG2D system in cancer. Blood Cells Mol. Dis. 34, 206–213 10.1016/j.bcmd.2005.03.003 [DOI] [PubMed] [Google Scholar]
- Corcoran C., Friel A. M., Duffy M. J., Crown J., O’Driscoll L. (2011). Intracellular and extracellular microRNAs in breast cancer. Clin. Chem. 57, 18–32 10.1373/clinchem.2010.150730 [DOI] [PubMed] [Google Scholar]
- de Gassart A., Geminard C., Fevrier B., Raposo G., Vidal M. (2003). Lipid raft-associated protein sorting in exosomes. Blood 102, 4336–4344 10.1182/blood-2003-03-0871 [DOI] [PubMed] [Google Scholar]
- Denzer K., Kleijmeer M. J., Heijnen H. F., Stoorvogel W., Geuze H. J. (2000). Exosome: from internal vesicle of the multivesicular body to intercellular signaling device. J. Cell Sci. 113(Pt 19), 3365–3374 [DOI] [PubMed] [Google Scholar]
- Gesierich S., Berezovskiy I., Ryschich E., Zoller M. (2006). Systemic induction of the angiogenesis switch by the tetraspanin D6.1A/CO-029. Cancer Res. 66, 7083–7094 10.1158/0008-5472.CAN-06-0391 [DOI] [PubMed] [Google Scholar]
- Gong C., Yao Y., Wang Y., Liu B., Wu W., Chen J., Su F., Yao H., Song E. (2011). Up-regulation of miR-21 mediates resistance to trastuzumab therapy for breast cancer. J. Biol. Chem. 286, 19127–19137 10.1074/jbc.M110.216887 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gonzales P. A., Pisitkun T., Hoffert J. D., Tchapyjnikov D., Star R. A., Kleta R., Wang N. S., Knepper M. A. (2009). Large-scale proteomics and phosphoproteomics of urinary exosomes. J. Am. Soc. Nephrol. 20, 363–379 10.1681/ASN.2008040406 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gonzales P. A., Zhou H., Pisitkun T., Wang N. S., Star R. A., Knepper M. A., Yuen P. S. (2010). Isolation and purification of exosomes in urine. Methods Mol. Biol. 641, 89–99 10.1007/978-1-60761-711-2_6 [DOI] [PubMed] [Google Scholar]
- Graner M. W., Alzate O., Dechkovskaia A. M., Keene J. D., Sampson J. H., Mitchell D. A., Bigner D. D. (2009). Proteomic and immunologic analyses of brain tumor exosomes. FASEB J. 23, 1541–1557 10.1096/fj.08-122184 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hanahan D., Weinberg R. A. (2000). The hallmarks of cancer. Cell 100, 57–70 10.1016/S0092-8674(00)81683-9 [DOI] [PubMed] [Google Scholar]
- Hegmans J. P., Bard M. P., Hemmes A., Luider T. M., Kleijmeer M. J., Prins J. B., Zitvogel L., Burgers S. A., Hoogsteden H. C., Lambrecht B. N. (2004). Proteomic analysis of exosomes secreted by human mesothelioma cells. Am. J. Pathol. 164, 1807–1815 10.1016/S0002-9440(10)63739-X [DOI] [PMC free article] [PubMed] [Google Scholar]
- Higginbotham J. N., Demory Beckler M., Gephart J. D., Franklin J. L., Bogatcheva G., Kremers G. J., Piston D. W., Ayers G. D., Mcconnell R. E., Tyska M. J., Coffey R. J. (2011). Amphiregulin exosomes increase cancer cell invasion. Curr. Biol. 21, 779–786 10.1016/j.cub.2011.03.043 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ho H. K., Jang J. J., Kaji S., Spektor G., Fong A., Yang P., Hu B. S., Schatzman R., Quertermous T., Cooke J. P. (2004). Developmental endothelial locus-1 (Del-1), a novel angiogenic protein: its role in ischemia. Circulation 109, 1314–1319 10.1161/01.CIR.0000118465.36018.2D [DOI] [PubMed] [Google Scholar]
- Hong B. S., Cho J. H., Kim H., Choi E. J., Rho S., Kim J., Kim J. H., Choi D. S., Kim Y. K., Hwang D., Gho Y. S. (2009). Colorectal cancer cell-derived microvesicles are enriched in cell cycle-related mRNAs that promote proliferation of endothelial cells. BMC Genomics 10, 556. 10.1186/1471-2164-10-556 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Iorio M. V., Ferracin M., Liu C. G., Veronese A., Spizzo R., Sabbioni S., Magri E., Pedriali M., Fabbri M., Campiglio M., Menard S., Palazzo J. P., Rosenberg A., Musiani P., Volinia S., Nenci I., Calin G. A., Querzoli P., Negrini M., Croce C. M. (2005). MicroRNA gene expression deregulation in human breast cancer. Cancer Res. 65, 7065–7070 10.1158/0008-5472.CAN-05-1783 [DOI] [PubMed] [Google Scholar]
- Johnson S. M., Grosshans H., Shingara J., Byrom M., Jarvis R., Cheng A., Labourier E., Reinert K. L., Brown D., Slack F. J. (2005). RAS is regulated by the let-7 microRNA family. Cell 120, 635–647 10.1016/j.cell.2005.01.014 [DOI] [PubMed] [Google Scholar]
- Johnstone R. M. (1992). The Jeanne Manery-Fisher Memorial Lecture 1991. Maturation of reticulocytes: formation of exosomes as a mechanism for shedding membrane proteins. Biochem. Cell Biol. 70, 179–190 10.1139/o92-028 [DOI] [PubMed] [Google Scholar]
- Johnstone R. M. (2006). Exosomes biological significance: a concise review. Blood Cells Mol. Dis. 36, 315–321 10.1016/j.bcmd.2005.12.001 [DOI] [PubMed] [Google Scholar]
- Johnstone R. M., Adam M., Hammond J. R., Orr L., Turbide C. (1987). Vesicle formation during reticulocyte maturation. Association of plasma membrane activities with released vesicles (exosomes). J. Biol. Chem. 262, 9412–9420 [PubMed] [Google Scholar]
- Johnstone R. M., Bianchini A., Teng K. (1989). Reticulocyte maturation and exosome release: transferrin receptor containing exosomes shows multiple plasma membrane functions. Blood 74, 1844–1851 [PubMed] [Google Scholar]
- Johnstone R. M., Mathew A., Mason A. B., Teng K. (1991). Exosome formation during maturation of mammalian and avian reticulocytes: evidence that exosome release is a major route for externalization of obsolete membrane proteins. J. Cell. Physiol. 147, 27–36 10.1002/jcp.1041470105 [DOI] [PubMed] [Google Scholar]
- Jung T., Castellana D., Klingbeil P., Cuesta Hernandez I., Vitacolonna M., Orlicky D. J., Roffler S. R., Brodt P., Zoller M. (2009). CD44v6 dependence of premetastatic niche preparation by exosomes. Neoplasia 11, 1093–1105 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kakisaka T., Kondo T., Okano T., Fujii K., Honda K., Endo M., Tsuchida A., Aoki T., Itoi T., Moriyasu F., Yamada T., Kato H., Nishimura T., Todo S., Hirohashi S. (2007). Plasma proteomics of pancreatic cancer patients by multi-dimensional liquid chromatography and two-dimensional difference gel electrophoresis (2D-DIGE): up-regulation of leucine-rich alpha-2-glycoprotein in pancreatic cancer. J. Chromatogr. B Analyt. Technol. Biomed. Life Sci. 852, 257–267 10.1016/j.jchromb.2007.01.029 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Keller S., Ridinger J., Rupp A. K., Janssen J. W., Altevogt P. (2011). Body fluid derived exosomes as a novel template for clinical diagnostics. J. Transl. Med. 9, 86. 10.1186/1479-5876-9-86 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kosaka N., Iguchi H., Yoshioka Y., Takeshita F., Matsuki Y., Ochiya T. (2010). Secretory mechanisms and intercellular transfer of microRNAs in living cells. J. Biol. Chem. 285, 17442–17452 10.1074/jbc.M110.107821 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lagos-Quintana M., Rauhut R., Lendeckel W., Tuschl T. (2001). Identification of novel genes coding for small expressed RNAs. Science 294, 853–858 10.1126/science.1064921 [DOI] [PubMed] [Google Scholar]
- Lee E. J., Gusev Y., Jiang J., Nuovo G. J., Lerner M. R., Frankel W. L., Morgan D. L., Postier R. G., Brackett D. J., Schmittgen T. D. (2007). Expression profiling identifies microRNA signature in pancreatic cancer. Int. J. Cancer 120, 1046–1054 10.1002/ijc.22486 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee Y. S., Kim S. H., Cho J. A., Kim C. W. (2011). Introduction of the CIITA gene into tumor cells produces exosomes with enhanced anti-tumor effects. Exp. Mol. Med. 43, 281–290 10.3858/emm.2011.43.1.001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li J., Sherman-Baust C. A., Tsai-Turton M., Bristow R. E., Roden R. B., Morin P. J. (2009). Claudin-containing exosomes in the peripheral circulation of women with ovarian cancer. BMC Cancer 9, 244. 10.1186/1471-2407-9-244 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li Q. L., Bu N., Yu Y. C., Hua W., Xin X. Y. (2008). Exvivo experiments of human ovarian cancer ascites-derived exosomes presented by dendritic cells derived from umbilical cord blood for immunotherapy treatment. Clin. Med. Oncol. 2, 461–467 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li X. B., Zhang Z. R., Schluesener H. J., Xu S. Q. (2006). Role of exosomes in immune regulation. J. Cell. Mol. Med. 10, 364–375 10.1111/j.1582-4934.2006.tb00405.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li Y., Zhang Y., Qiu F., Qiu Z. (2011). Proteomic identification of exosomal LRG1: a potential urinary biomarker for detecting NSCLC. Electrophoresis 32, 1976–1983 10.1002/elps.201000392 [DOI] [PubMed] [Google Scholar]
- Logozzi M., De Milito A., Lugini L., Borghi M., Calabro L., Spada M., Perdicchio M., Marino M. L., Federici C., Iessi E., Brambilla D., Venturi G., Lozupone F., Santinami M., Huber V., Maio M., Rivoltini L., Fais S. (2009). High levels of exosomes expressing CD63 and caveolin-1 in plasma of melanoma patients. PLoS ONE 4, e5219. 10.1371/journal.pone.0005219 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Loomis R. J., Holmes D. A., Elms A., Solski P. A., Der C. J., Su L. (2006). Citron kinase, a RhoA effector, enhances HIV-1 virion production by modulating exocytosis. Traffic 7, 1643–1653 10.1111/j.1600-0854.2006.00503.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lu Q., Zhang J., Allison R., Gay H., Yang W. X., Bhowmick N. A., Frelix G., Shappell S., Chen Y. H. (2009). Identification of extracellular delta-catenin accumulation for prostate cancer detection. Prostate 69, 411–418 10.1002/pros.20902 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mathew A., Bell A., Johnstone R. M. (1995). Hsp-70 is closely associated with the transferrin receptor in exosomes from maturing reticulocytes. Biochem. J. 308(Pt 3), 823–830 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mathivanan S., Fahner C. J., Reid G. E., Simpson R. J. (2012). ExoCarta 2012: database of exosomal proteins, RNA and lipids. Nucleic Acids Res. 40, D1241–D1244 10.1093/nar/gkr828 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mathivanan S., Simpson R. J. (2009). ExoCarta: a compendium of exosomal proteins and RNA. Proteomics 9, 4997–5000 10.1002/pmic.200900351 [DOI] [PubMed] [Google Scholar]
- McCready J., Sims J. D., Chan D., Jay D. G. (2010). Secretion of extracellular hsp90alpha via exosomes increases cancer cell motility: a role for plasminogen activation. BMC Cancer 10, 294. 10.1186/1471-2407-10-294 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meckes D. G., Jr., Shair K. H., Marquitz A. R., Kung C. P., Edwards R. H., Raab-Traub N. (2010). Human tumor virus utilizes exosomes for intercellular communication. Proc. Natl. Acad. Sci. U.S.A. 107, 20370–20375 10.1073/pnas.1014194107 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mitchell J. P., Court J., Mason M. D., Tabi Z., Clayton A. (2008). Increased exosome production from tumour cell cultures using the Integra CELLine Culture System. J. Immunol. Methods 335, 98–105 10.1016/j.jim.2008.03.001 [DOI] [PubMed] [Google Scholar]
- Navabi H., Croston D., Hobot J., Clayton A., Zitvogel L., Jasani B., Bailey-Wood R., Wilson K., Tabi Z., Mason M. D., Adams M. (2005). Preparation of human ovarian cancer ascites-derived exosomes for a clinical trial. Blood Cells Mol. Dis. 35, 149–152 10.1016/j.bcmd.2005.06.008 [DOI] [PubMed] [Google Scholar]
- Nazarenko I., Rana S., Baumann A., Mcalear J., Hellwig A., Trendelenburg M., Lochnit G., Preissner K. T., Zoller M. (2010). Cell surface tetraspanin Tspan8 contributes to molecular pathways of exosome-induced endothelial cell activation. Cancer Res. 70, 1668–1678 10.1158/1538-7445.AM10-1668 [DOI] [PubMed] [Google Scholar]
- Ochieng J., Pratap S., Khatua A. K., Sakwe A. M. (2009). Anchorage-independent growth of breast carcinoma cells is mediated by serum exosomes. Exp. Cell Res. 315, 1875–1888 10.1016/j.yexcr.2009.03.010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ogawa Y., Kanai-Azuma M., Akimoto Y., Kawakami H., Yanoshita R. (2008). Exosome-like vesicles with dipeptidyl peptidase IV in human saliva. Biol. Pharm. Bull. 31, 1059–1062 10.1248/bpb.31.1059 [DOI] [PubMed] [Google Scholar]
- Ohshima K., Inoue K., Fujiwara A., Hatakeyama K., Kanto K., Watanabe Y., Muramatsu K., Fukuda Y., Ogura S., Yamaguchi K., Mochizuki T. (2010). Let-7 microRNA family is selectively secreted into the extracellular environment via exosomes in a metastatic gastric cancer cell line. PLoS ONE 5, e13247. 10.1371/journal.pone.0013247 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ostrowski M., Carmo N. B., Krumeich S., Fanget I., Raposo G., Savina A., Moita C. F., Schauer K., Hume A. N., Freitas R. P., Goud B., Benaroch P., Hacohen N., Fukuda M., Desnos C., Seabra M. C., Darchen F., Amigorena S., Moita L. F., Thery C. (2010). Rab27a and Rab27b control different steps of the exosome secretion pathway. Nat. Cell Biol. 12, 19–30; sup pp. 11–13. 10.1038/ncb2000 [DOI] [PubMed] [Google Scholar]
- Park J. E., Tan H. S., Datta A., Lai R. C., Zhang H., Meng W., Lim S. K., Sze S. K. (2010). Hypoxic tumor cell modulates its microenvironment to enhance angiogenic and metastatic potential by secretion of proteins and exosomes. Mol. Cell. Proteomics 9, 1085–1099 10.1074/mcp.M900381-MCP200 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pisitkun T., Shen R. F., Knepper M. A. (2004). Identification and proteomic profiling of exosomes in human urine. Proc. Natl. Acad. Sci. U.S.A. 101, 13368–13373 10.1073/pnas.0403453101 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ponnambalam S., Baldwin S. A. (2003). Constitutive protein secretion from the trans-Golgi network to the plasma membrane. Mol. Membr. Biol. 20, 129–139 10.1080/096878031000104935 [DOI] [PubMed] [Google Scholar]
- Qu J. L., Qu X. J., Zhao M. F., Teng Y. E., Zhang Y., Hou K. Z., Jiang Y. H., Yang X. H., Liu Y. P. (2009). The role of cbl family of ubiquitin ligases in gastric cancer exosome-induced apoptosis of Jurkat T cells. Acta Oncol. (Madr.) 48, 1173–1180 10.3109/02841860903032817 [DOI] [PubMed] [Google Scholar]
- Rajashekhar G., Willuweit A., Patterson C. E., Sun P., Hilbig A., Breier G., Helisch A., Clauss M. (2006). Continuous endothelial cell activation increases angiogenesis: evidence for the direct role of endothelium linking angiogenesis and inflammation. J. Vasc. Res. 43, 193–204 10.1159/000090949 [DOI] [PubMed] [Google Scholar]
- Rajendran L., Honsho M., Zahn T. R., Keller P., Geiger K. D., Verkade P., Simons K. (2006). Alzheimer’s disease beta-amyloid peptides are released in association with exosomes. Proc. Natl. Acad. Sci. U.S.A. 103, 11172–11177 10.1073/pnas.0603838103 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ratajczak J., Miekus K., Kucia M., Zhang J., Reca R., Dvorak P., Ratajczak M. Z. (2006). Embryonic stem cell-derived microvesicles reprogram hematopoietic progenitors: evidence for horizontal transfer of mRNA and protein delivery. Leukemia 20, 847–856 10.1038/sj.leu.2404296 [DOI] [PubMed] [Google Scholar]
- Record M., Subra C., Silvente-Poirot S., Poirot M. (2011). Exosomes as intercellular signalosomes and pharmacological effectors. Biochem. Pharmacol. 81, 1171–1182 10.1016/j.bcp.2011.02.011 [DOI] [PubMed] [Google Scholar]
- Ristorcelli E., Beraud E., Verrando P., Villard C., Lafitte D., Sbarra V., Lombardo D., Verine A. (2008). Human tumor nanoparticles induce apoptosis of pancreatic cancer cells. FASEB J. 22, 3358–3369 10.1096/fj.07-102855 [DOI] [PubMed] [Google Scholar]
- Roldo C., Missiaglia E., Hagan J. P., Falconi M., Capelli P., Bersani S., Calin G. A., Volinia S., Liu C. G., Scarpa A., Croce C. M. (2006). MicroRNA expression abnormalities in pancreatic endocrine and acinar tumors are associated with distinctive pathologic features and clinical behavior. J. Clin. Oncol. 24, 4677–4684 10.1200/JCO.2005.05.5194 [DOI] [PubMed] [Google Scholar]
- Rosell R., Wei J., Taron M. (2009). Circulating microRNA signatures of tumor-derived exosomes for early diagnosis of non-small-cell lung cancer. Clin Lung Cancer 10, 8–9 10.3816/CLC.2009.n.057 [DOI] [PubMed] [Google Scholar]
- Saxena N., Lahiri S. S., Hambarde S., Tripathi R. P. (2008). RAS: target for cancer therapy. Cancer Invest. 26, 948–955 10.1080/07357900802087275 [DOI] [PubMed] [Google Scholar]
- Shi L., Chen J., Yang J., Pan T., Zhang S., Wang Z. (2010). MiR-21 protected human glioblastoma U87MG cells from chemotherapeutic drug temozolomide induced apoptosis by decreasing Bax/Bcl-2 ratio and caspase-3 activity. Brain Res. 1352, 255–264 10.1016/j.brainres.2010.07.009 [DOI] [PubMed] [Google Scholar]
- Simpson R. J., Jensen S. S., Lim J. W. (2008). Proteomic profiling of exosomes: current perspectives. Proteomics 8, 4083–4099 10.1002/pmic.200800109 [DOI] [PubMed] [Google Scholar]
- Skog J., Wurdinger T., Van Rijn S., Meijer D. H., Gainche L., Sena-Esteves M., Curry W. T., Jr., Carter B. S., Krichevsky A. M., Breakefield X. O. (2008). Glioblastoma microvesicles transport RNA and proteins that promote tumour growth and provide diagnostic biomarkers. Nat. Cell Biol. 10, 1470–1476 10.1038/ncb1800 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith D. S., Humphrey P. A., Catalona W. J. (1997). The early detection of prostate carcinoma with prostate specific antigen: the Washington University experience. Cancer 80, 1852–1856 [DOI] [PubMed] [Google Scholar]
- Staubach S., Razawi H., Hanisch F. G. (2009). Proteomics of MUC1-containing lipid rafts from plasma membranes and exosomes of human breast carcinoma cells MCF-7. Proteomics 9, 2820–2835 10.1002/pmic.200800793 [DOI] [PubMed] [Google Scholar]
- Taieb J., Chaput N., Schartz N., Roux S., Novault S., Menard C., Ghiringhelli F., Terme M., Carpentier A. F., Darrasse-Jeze G., Lemonnier F., Zitvogel L. (2006). Chemoimmunotherapy of tumors: cyclophosphamide synergizes with exosome based vaccines. J. Immunol. 176, 2722–2729 [DOI] [PubMed] [Google Scholar]
- Takamizawa J., Konishi H., Yanagisawa K., Tomida S., Osada H., Endoh H., Harano T., Yatabe Y., Nagino M., Nimura Y., Mitsudomi T., Takahashi T. (2004). Reduced expression of the let-7 microRNAs in human lung cancers in association with shortened postoperative survival. Cancer Res. 64, 3753–3756 10.1158/0008-5472.CAN-04-0637 [DOI] [PubMed] [Google Scholar]
- Tan A., De La Pena H., Seifalian A. M. (2010). The application of exosomes as a nanoscale cancer vaccine. Int. J. Nanomedicine 5, 889–900 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tavoosidana G., Ronquist G., Darmanis S., Yan J., Carlsson L., Wu D., Conze T., Ek P., Semjonow A., Eltze E., Larsson A., Landegren U. D., Kamali-Moghaddam M. (2011). Multiple recognition assay reveals prostasomes as promising plasma biomarkers for prostate cancer. Proc. Natl. Acad. Sci. U.S.A. 108, 8809–8814 10.1073/pnas.1019330108 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Taylor D. D., Gercel-Taylor C. (2008). MicroRNA signatures of tumor-derived exosomes as diagnostic biomarkers of ovarian cancer. Gynecol. Oncol. 110, 13–21 10.1016/j.ygyno.2008.04.033 [DOI] [PubMed] [Google Scholar]
- Taylor D. D., Zacharias W., Gercel-Taylor C. (2011). Exosome isolation for proteomic analyses and RNA profiling. Methods Mol. Biol. 728, 235–246 10.1007/978-1-61779-068-3_15 [DOI] [PubMed] [Google Scholar]
- Thery C. (2011). Exosomes: secreted vesicles and intercellular communications. F1000 Biol. Rep. 3, 15. 10.3410/B3-15 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thery C., Zitvogel L., Amigorena S. (2002). Exosomes: composition, biogenesis and function. Nat. Rev. Immunol. 2, 569–579 [DOI] [PubMed] [Google Scholar]
- Valadi H., Ekstrom K., Bossios A., Sjostrand M., Lee J. J., Lotvall J. O. (2007). Exosome-mediated transfer of mRNAs and microRNAs is a novel mechanism of genetic exchange between cells. Nat. Cell Biol. 9, 654–659 10.1038/ncb1596 [DOI] [PubMed] [Google Scholar]
- Welton J. L., Khanna S., Giles P. J., Brennan P., Brewis I. A., Staffurth J., Mason M. D., Clayton A. (2010). Proteomics analysis of bladder cancer exosomes. Mol. Cell. Proteomics 9, 1324–1338 10.1074/mcp.M000063-MCP201 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang C., Robbins P. D. (2011). The roles of tumor-derived exosomes in cancer pathogenesis. Clin. Dev. Immunol. 2011, 842849. 10.1155/2011/842849 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu X., Harris S. L., Levine A. J. (2006). The regulation of exosome secretion: a novel function of the p53 protein. Cancer Res. 66, 4795–4801 10.1158/0008-5472.CAN-05-3390 [DOI] [PubMed] [Google Scholar]
- Zhang Y., Luo C. L., He B. C., Zhang J. M., Cheng G., Wu X. H. (2010). Exosomes derived from IL-12-anchored renal cancer cells increase induction of specific antitumor response in vitro: a novel vaccine for renal cell carcinoma. Int. J. Oncol. 36, 133–140 [PubMed] [Google Scholar]
- Zhou Q., Li M., Wang X., Li Q., Wang T., Zhu Q., Zhou X., Gao X., Li X. (2012). Immune-related microRNAs are abundant in breast milk exosomes. Int. J. Biol. Sci. 8, 118–123 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zomer A., Vendrig T., Hopmans E. S., Van Eijndhoven M., Middeldorp J. M., Pegtel D. M. (2010). Exosomes: fit to deliver small RNA. Commun. Integr. Biol. 3, 447–450 10.4161/cib.3.5.12339 [DOI] [PMC free article] [PubMed] [Google Scholar]