Abstract
Covalent attachment of ubiquitin to target proteins, or ubiquitylation, has emerged as one of the most prevalent posttranslational modifications (PTMs), regulating nearly every cellular pathway. The diversity of functions associated with this particular PTM stems from the myriad ways in which a target protein can be modified by ubiquitin, e.g., monoubiquitin, multi-monoubiquitin and polyubiquitin linkages. In the current study, we took a systematic approach to analyze the ubiquitylation profiles of the yeast Saccharomyces cerevisiae nuclear pore complex (NPC) proteins or nucleoporins. We found the yeast NPC to be extensively modified by ubiquitin with highly variable ubiquitylation profiles, suggesting that dissection of these modifications may provide new insights into the regulation of NPC functions and reveal additional roles for nucleoporins beyond nuclear transport.
Keywords: dynein, mitosis, nuclear pore complex, Nup 159, ubiquitin
Eukaryotic cell function depends on the physical separation of membrane-enclosed compartments that enable it to concomitantly perform numerous cellular tasks in specialized microenvironments. This subcellular division is achieved by intricate intracellular membrane systems. The nuclear envelope (NE), a double lipid membrane perforated by nuclear pore complexes, segregates the nucleoplasm containing the genetic material from the cytoplasm. The NPC represents one of the largest and most complex proteinaceous assemblies in the eukaryotic cell. The yeast NPC is a ~50–60 MDa protein complex consisting in multiple copies of 30 different conserved proteins called nucleoporins or Nups whose canonical function is the selective and directional transport of proteins and ribonucleoparticles between the nucleus and the cytoplasm.1-3 Moreover, in eukaryotes with open mitosis, some nucleoporins have a well-documented role during cell division. In particular, the best-characterized example of an NPC component with a cell cycle-specific function is the vertebrate Nup107–160 complex that is targeted to kinetochores during mitosis, where it functions in spindle assembly.4-6 Although electron microscopy has provided valuable insights into the general shape of the NPC,7-9 the spatial configuration of the yeast NPC components has only recently started to be revealed thanks to a combination of proteomics, structural and computational data.1 Overall, the NPC displays an 8-fold symmetry cylindrical structure around the axis of transport and a planar pseudo-symmetry through the nuclear envelope and consists in five distinct substructures. The membrane ring anchors the NPC to the nuclear envelope, inner and outer rings form the core scaffold of the NPC, FG-repeat nucleoporins cover the inner surface of the NPC and play major functions in nuclear translocation via their ability to interact with transport receptors, and linker nucleoporins bridges FG Nups to the core scaffold.
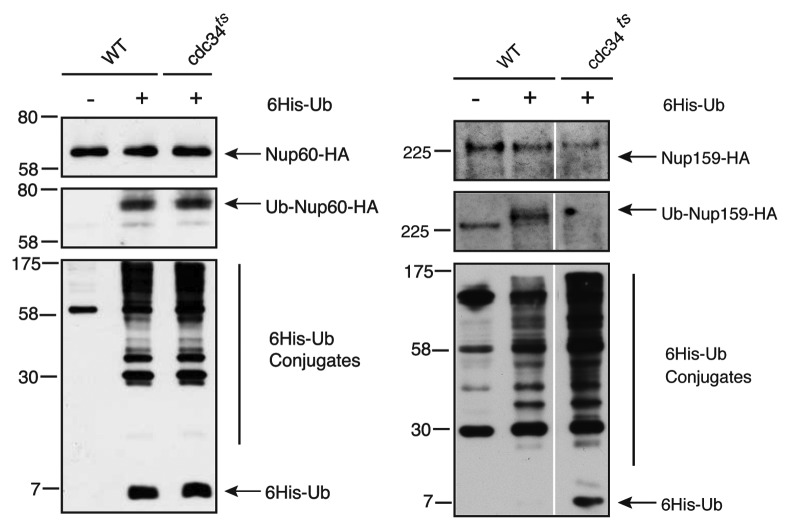
Although extensive research shed light onto the structure and roles of the NPC, the regulation of its functions by post-translational modifications remains ill defined. Since protein ubiquitylation is one of the most prevalent mechanism for regulating protein function and stability in eukaryotes, we rationalized that the extensive analysis of nucleoporins modification by ubiquitin would represent a valuable set of data to unravel either the mechanisms of regulation of known NPC functions or the discovery of new functions.10 Surprisingly, the systematic analysis of the ubiquitylation pattern of the yeast Nups revealed that more than 50% of them are conjugated to ubiquitin, mostly to monoubiquitin. No correlation was found between modification and localization of Nups within the NPC. In addition, none of the trans-membrane proteins anchoring the NPC in the nuclear envelope was modified, confirming that accessibility of Nups for the conjugation machinery is required for ubiquitylation to occur. Prevalence of monoubiquitylation suggests a non-degradative role of this PTM. This is indeed exemplified by the finding that mono-ubiquitylation of Nup159, a nucleoporin located exclusively on the cytoplasmic side of the NPC, is involved in anchoring the dynein light chain to the NPC. Preventing this ubiquitylation neither affects NPC organization nor nuclear transport pathways but alters the nuclear segregation and the spindle positioning at the onset of mitosis. Hence, this general approach allowed unraveling a yet unappreciated function for the yeast NPC. Importantly, ubiquitylation of Nups involves distinct components of the ubiquitin conjugation machinery. Cdc34, the ubiquitin-conjugating enzyme (E2) required for Nup159 mono-ubiquitylation is indeed not responsible for the mono-ubiquitylation of Nup60, a Nup exclusively localized on the nuclear side of the NPC (Fig. 1). This indicated that the NPC cannot be considered as a single target toward the ubiquitin/proteasome system but is rather the target of multiple ubiquitin-modifying enzymes.
Figure 1. Cdc34 is not the general E2 of the yeast NPC. Ni-purified 6His-ubiquitin conjugated forms of genomically HA-tagged Nup60 (left) or Nup159 (right) were extracted from cells transformed (+) or not (–) with a plasmid encoding 6His-ubiquitin under control of the CUP1 promoter. Cell lysates (upper panels) and Ni-purified material (middle panels) were examined by western blotting with an anti-HA antibody. Ubiquitin expression and efficiency of purification were controlled using an anti-6His antibody (lower panels).
In recent years, the NPC has been involved in transport-independent functions such as chromatin organization, replication-coupled DNA repair, and regulation of gene expression.11 Based on our findings, we anticipate that the detailed analysis of Nups post-translational modifications, and in particular ubiquitylation, could unravel new mechanisms of regulation and/or functions of the yeast NPC.
Acknowledgments
This study was funded by grants from the Agence Nationale pour la Recherche (grant 2010 BLAN1227-01 to C.D.) and the Ligue contre le Cancer (C.D. is “Equipe labellise´e”).
References
- 1.Alber F, Dokudovskaya S, Veenhoff LM, Zhang W, Kipper J, Devos D, et al. The molecular architecture of the nuclear pore complex. Nature. 2007;450:695–701. doi: 10.1038/nature06405. [DOI] [PubMed] [Google Scholar]
- 2.Cronshaw JM, Krutchinsky AN, Zhang W, Chait BT, Matunis MJ. Proteomic analysis of the mammalian nuclear pore complex. J Cell Biol. 2002;158:915–27. doi: 10.1083/jcb.200206106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Rout MP, Aitchison JD, Suprapto A, Hjertaas K, Zhao Y, Chait BT. The yeast nuclear pore complex: composition, architecture, and transport mechanism. J Cell Biol. 2000;148:635–51. doi: 10.1083/jcb.148.4.635. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Glavy JS, Krutchinsky AN, Cristea IM, Berke IC, Boehmer T, Blobel G, et al. Cell-cycle-dependent phosphorylation of the nuclear pore Nup107-160 subcomplex. Proc Natl Acad Sci U S A. 2007;104:3811–6. doi: 10.1073/pnas.0700058104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Zuccolo M, Alves A, Galy V, Bolhy S, Formstecher E, Racine V, et al. The human Nup107-160 nuclear pore subcomplex contributes to proper kinetochore functions. EMBO J. 2007;26:1853–64. doi: 10.1038/sj.emboj.7601642. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Mishra RK, Chakraborty P, Arnaoutov A, Fontoura BM, Dasso M. The Nup107-160 complex and gamma-TuRC regulate microtubule polymerization at kinetochores. Nat Cell Biol. 2010;12:164–9. doi: 10.1038/ncb2016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Stoffler D, Feja B, Fahrenkrog B, Walz J, Typke D, Aebi U. Cryo-electron tomography provides novel insights into nuclear pore architecture: implications for nucleocytoplasmic transport. J Mol Biol. 2003;328:119–30. doi: 10.1016/S0022-2836(03)00266-3. [DOI] [PubMed] [Google Scholar]
- 8.Yang Q, Rout MP, Akey CW. Three-dimensional architecture of the isolated yeast nuclear pore complex: functional and evolutionary implications. Mol Cell. 1998;1:223–34. doi: 10.1016/S1097-2765(00)80023-4. [DOI] [PubMed] [Google Scholar]
- 9.Beck M, Förster F, Ecke M, Plitzko JM, Melchior F, Gerisch G, et al. Nuclear pore complex structure and dynamics revealed by cryoelectron tomography. Science. 2004;306:1387–90. doi: 10.1126/science.1104808. [DOI] [PubMed] [Google Scholar]
- 10.Hayakawa A, Babour A, Sengmanivong L, Dargemont C. Ubiquitylation of the nuclear pore complex controls nuclear migration during mitosis in S. cerevisiae. J Cell Biol. 2012;196:19–27. doi: 10.1083/jcb.201108124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Strambio-De-Castillia C, Niepel M, Rout MP. The nuclear pore complex: bridging nuclear transport and gene regulation. Nat Rev Mol Cell Biol. 2010;11:490–501. doi: 10.1038/nrm2928. [DOI] [PubMed] [Google Scholar]