Abstract
Insulin-like growth factor binding proteins (IGFBP-1 to -6) bind insulin-like growth factors-I and -II (IGF-I and IGF-II) with high affinity. These binding proteins maintain IGFs in the circulation and direct them to target tissues, where they promote cell growth, proliferation, differentiation, and survival via the type 1 IGF receptor. IGFBPs also interact with many other molecules, which not only influence their modulation of IGF action but also mediate IGF-independent activities that regulate processes such as cell migration and apoptosis by modulating gene transcription. IGFBPs-1 to -6 are structurally similar proteins consisting of three distinct domains, N-terminal, linker, and C-terminal. There have been major advances in our understanding of IGFBP structure in the last decade and a half. While there is still no structure of an intact IGFBP, several structures of individual N- and C-domains have been solved. The structure of a complex of N-BP-4:IGF-I:C-BP-4 has also been solved, providing a detailed picture of the structural features of the IGF binding site and the mechanism of binding. Structural studies have also identified features important for interaction with extracellular matrix components and integrins. This review summarizes structural studies reported so far and highlights features important for binding not only IGF but also other partners. We also highlight future directions in which structural studies will add to our knowledge of the role played by the IGFBP family in normal growth and development, as well as in disease.
Keywords: insulin-like growth factor, IGF binding protein, protein structure
Introduction
The insulin-like growth factor binding protein (IGFBP) family consists of six structurally similar proteins (IGFBP-1 to -6), which bind insulin-like growth factors (IGFs) with high affinity (Firth and Baxter, 2002). The two IGF ligands (IGF-I and IGF-II) are expressed in a wide variety of tissues and act primarily via the type 1 IGF receptor (IGF-1R) to promote cell proliferation, survival, differentiation, and migration as well as metabolic processes (Denley et al., 2005a). IGF-II also interacts with one of the two splice variants of the insulin receptor (IR-A), with similar signaling outcomes to those arising from the interaction with IGF-1R (Denley et al., 2003). Bioavailability of IGF-II is also controlled by an interaction with the type 2 IGF receptor (IGF-2R), which internalizes bound IGF-II, leading to its degradation (Brown et al., 2009). Through these interactions, IGFs play a critical role in normal pre and postnatal growth and development (Rosenfeld, 2007). Dysregulation of IGFs is associated with growth-related diseases such as small stature resulting from low IGF expression or acromegaly due to IGF-I overexpression (Rosenfeld, 2007). IGFs are also associated with many other diseases including atherosclerosis (Clemmons, 2007) and Alzheimer’s disease (Piriz et al., 2011). Moreover, high circulating IGF levels are associated with an increased risk of several cancers, and IGFs promote cancer cell growth and survival (Pollak, 2008a). A significant effort is therefore being devoted to developing inhibitors of IGF action for the treatment of cancer (Pollak, 2008b).
Insulin-like growth factor binding proteins modulate IGF action by functioning as IGF carriers within the circulation. In serum, IGFs are predominantly found in a ternary complex formed when IGF bound to IGFBP-3 forms a complex with the acid labile subunit (ALS; Boisclair et al., 2001). A small proportion of IGFs can also be found in a ternary complex of IGF:IGFBP-5:ALS (Twigg and Baxter, 1998). These ternary complexes are unable to cross the vascular epithelial layer and serve as an IGF reservoir within the circulation. The other IGFBPs (IGFBP-1, -2, -4, -6) are also present in serum, with the most abundant being IGFBP-2, and are all competent to bind IGFs. These smaller IGF:IGFBP binary complexes can cross the vascular epithelial layer, and thus deliver IGFs to target tissues (Boes et al., 1992; Lewitt et al., 1994).
Locally expressed IGFBPs also modulate IGF action by inhibiting binding to the IGF-1R. Two mechanisms of IGF release have been identified, one involving extracellular matrix (ECM) binding and the other IGFBP proteolysis (Firth and Baxter, 2002). Both of these processes lower the affinity of IGFBPs for IGFs and thus increase the local concentration of bioactive IGF.
Almost all IGFBPs have been shown to have IGF-independent actions (Bach et al., 2005; Wheatcroft and Kearney, 2009). These are a consequence of their ability to enter cells and locate to the nucleus as well as their interaction with ECM components, integrins, or other binding partners. IGF-independent actions are much less well understood than IGF-dependent actions.
This review outlines the structural features of IGFBPs involved in their intermolecular interactions, and focuses on our understanding of the molecular mechanisms underlying the control of IGF action by the IGFBPs
Structural Features of the IGFBPs
Overall structure
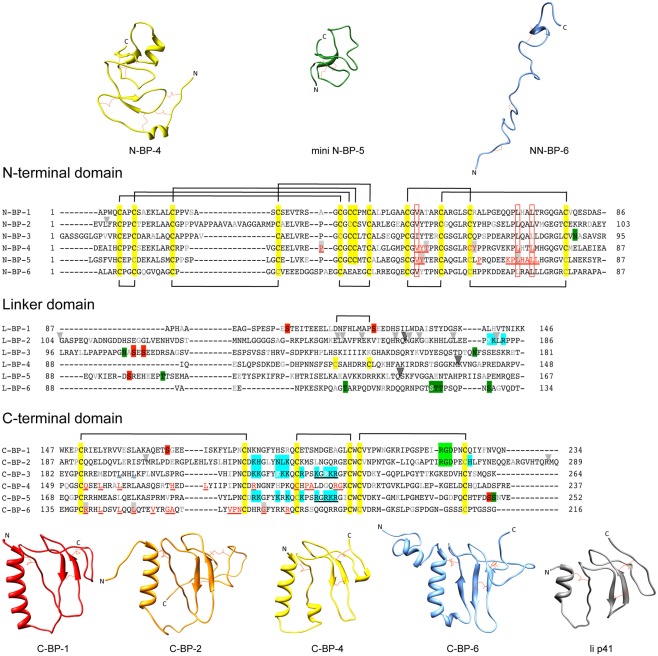
The IGFBPs range in mass from ∼24 to 50 kDa (216–289 amino acids) and share a similar overall three-domain structure, with the structured N- and C-terminal domains connected by a less structured linker domain (Figure 1). IGFBPs-1 to -5 have six conserved disulfide bonds and a characteristic GCGCC motif in the N-domain, whereas the IGFBP-6 N-domain lacks the last two cysteines of the motif and therefore has only five N-domain disulfide bonds (Neumann et al., 1998; Bach et al., 2005) resulting in a different fold from IGFBPs-1 to -5. All six IGFBP C-domains have three conserved disulfide bonds, making a total of nine disulfide bonds throughout (except for IGFBP-6, which has 8, and IGFBP-4, which has 10 due to an extra disulfide bond in the linker domain, Chelius et al., 2001).
Figure 1.
Sequences and structures of IGFBP domains. Sequence alignments of IGFBP-1 to-6 N-terminal (N-), linker (L-), and C-terminal (C-) domains, highlighting cysteine residues (yellow box) and the disulfide bonding connectivity (black lines above and below sequences). Residue numbers are shown next to the sequences and every 10th residues is gray or white. The following distinctive features are highlighted: Red underlined text highlights IGF binding residues defined in structural studies of the N-domain [IGFBP-4 (Sitar et al., 2006) and mini IGFBP-5 (Kalus et al., 1998)] and C-domain [IGFBP-4 (Sitar et al., 2006) and IGFBP-6 (Headey et al., 2004a)]. Open red boxes, key residues involved in binding conserved across all IGFBPs; Light gray arrowheads, IGFBP-2 proteolysis sites (Ho and Baxter, 1997; Rehault et al., 2001; Monget et al., 2003; Standker et al., 2003; Mark et al., 2005; Berg et al., 2007; Miyamoto et al., 2007); Dark gray arrowhead, PAPP-A cleavage sites on IGFBP-2, -4, and -5 (Conover et al., 1995; Laursen et al., 2001; Monget et al., 2003); Blue boxes, IGFBP heparin-binding domains confirmed by site-directed mutagenesis or NMR-IGFBP-2 (Russo et al., 2005; Kuang et al., 2006), IGFBP-3 (Firth et al., 1998), and IGFBP-5 (Arai et al., 1996b); Filled red boxes, phosphorylation sites (Jones et al., 1993a; Coverley et al., 2000; Gibson et al., 2001; Graham et al., 2007; Dolcini et al., 2009); Light green boxes, integrin binding sites (Jones et al., 1993c; Kuang et al., 2006), Dark green boxes; glycosylation sites (Neumann et al., 1998; Firth and Baxter, 1999; Graham et al., 2007); Black underlined text, nuclear localization sequences (Schedlich et al., 1998; Iosef et al., 2008); Blue underlined text, Leu194 and Leu197 of IGFBP-3 involved in nuclear export (Paharkova-Vatchkova and Lee, 2010). Protein sequence SwissProt accession numbers are as follows: IGFBP-1 (P08833), IGFBP-2 (P18065), IGFBP-3 (P17936), IGFBP-4 (P22692), IGFBP-5 (P24593), IGFBP-6 (P24592). N-domain and C-domain ribbon structures are from the following database files: N-BP-4 (PDB 2DSR), mini N-BP-5 (PDB 1BOE, conformer 1), NN-BP-6 (PDB 2JM2), C-BP-1 (PDB 1ZT3), C-BP-2 (PDB 2H7T), C-BP-4 (PDB 2DSR), C-BP-6 (PDB 1RMJ), and the Ii p41 thryglobulin type 1 domain (PDB 1ICF). Some of these structures are derived from IGF:IGFBP complexes (see Table 1). For those NMR PDB files containing more than one conformer the first entry was used in this figure. Molecular graphic images were produced using UCSF Chimera program from Resource for Biocomputing, Visualization, and Informatics at the University of California at San Francisco (http://www.cgl.ucsf.edu/chimera; Pettersen et al., 2004).
In the Protein Data Bank1 (Berman et al., 2000) there are currently four N-domain and three C-domain structures solved by either NMR or X-ray crystallography (Table 1). The N-domains of IGFBP-2, -4, and -5 are rigid globular structures, whereas the C-domains of IGFBPs-1, -2, -4, and -6 adopt a thyroglobulin type 1 fold and contain some more flexible regions (Figure 1). The N-terminal region of IGFBP-6 (residues 1–45, NN-BP-6) adopts a completely different extended structure from that seen in the short two-stranded β-sheet structure of the equivalent subdomain of IGFBP-2 and -4 (Chandrashekaran et al., 2007; Figure 1) as a consequence of the different disulfide connectivity of NN-BP-6 compared to the other IGFBPs in this region. No IGFBP-3 structures have been reported so far, and the structures of the IGFBP linker domains are unsolved, but are believed to be relatively flexible (Siwanowicz et al., 2005; Sitar et al., 2006). Recently, a covalently cross-linked dimer of the IGFBP-5 linker domain has been reported to show helical properties, as observed using circular dichroism (Sung et al., 2010).
Table 1.
Structures of IGFBP domains found in the Protein Data Bank (PDB) and Biological Magnetic Resonance Bank (BMRB) databases.
| N-domain | C-domain | IGF-I:N-BP | IGF-I:N-BP:C-BP | |
|---|---|---|---|---|
| IGFBP-1 | 1ZT3 (aa 166–259), 1ZT5 (aa 166–259, Fe2+; Sala et al., 2005)* | |||
| IGFBP-2 | http://rest.bmrb.wisc.edu/bmrb/NMR-STAR2/17743 (Galea et al., 2012)** | 2H7T (aa 183–289; Kuang et al., 2006)# | ||
| IGFBP-4 | 2DSP (IGF-I:N-BP-4 aa 1–92; Sitar et al., 2006)* | 2DSR (IGF-I:N-BP-4 aa 1–92: C-BP-4 aa 151–232; Sitar et al., 2006)* | ||
| IWQJ (IGF-I:N-BP-4, aa 3–82; Siwanowicz et al., 2005)* | 2DSQ (IGF-I:N-BP-4 aa 1–92: C-BP-1 aa 141–234; Sitar et al., 2006)* | |||
| IGFBP-5 | 1BOE (aa 40–92; Kalus et al., 1998)# | 1H59 (IGF-I:N-BP-5, aa 58–111; Zeslawski et al., 2001)* | ||
| IGFBP-6 | 2JM2 (aa 25–69; Chandrashekaran et al., 2007)# | 1RMJ (aa 136–216; Headey et al., 2004a)# |
*Structures determined by X-ray crystallography.
#Structures determined by NMR.
**Model based on NMR data and homology modeling using N-BP-4 coordinates (PDB 2DSP and 2DSR).
The structures of an intact IGFBP or an intact IGFBP:IGF complex have also not been solved as yet. While proteins of up to 45 kDa have been solved by NMR (Kainosho et al., 2006), application of this technique to an intact IGFBP has been hampered by the tendency of IGFBPs to aggregate under the conditions of NMR (Galea et al., 2012). Furthermore, the predicted flexibility of the linker domain has been suggested to hinder the formation of intact IGFBP crystals (Siwanowicz et al., 2005; Sitar et al., 2006). However, the crystal structure of a ternary complex of IGF-I with the isolated N- and C-domains of IGFBP-4 (N-BP-4 and C-BP-4, 2DSR) has been solved (as well as a structure of the IGF-I:N-BP-4:C-BP-1 ternary complex, 2DSQ; Siwanowicz et al., 2005; Sitar et al., 2006). The ternary structure shows that the IGFBP-4 N- and C-termini are in close contact. Similarly, NMR studies identified an interaction in solution between the isolated N- and C-domains of IGFBP-2 (Kuang et al., 2007). In the case of IGFBP-6, it has also been shown that the NN- and C-domains are located in close proximity upon IGF binding (Chandrashekaran et al., 2007). The NN-domain of IGFBP-6 is unique as it is unstructured in the absence of ligand. These structural studies have been invaluable in providing an understanding of the mechanism by which IGFBPs interact with the IGFs.
IGF binding determinants
The IGFBPs bind IGFs with nanomolar affinities. IGFBPs-1 to -5 bind both IGF-I and IGF-II with affinities that differ by only 2- to 10-fold between the two ligands (Kalus et al., 1998; Neumann and Bach, 1999; Carrick et al., 2001; Galanis et al., 2001; Siwanowicz et al., 2005), whereas IGFBP-6 has a distinct preference for IGF-II, having >100-fold higher affinity for IGF-II than IGF-I (Headey et al., 2004c). All six IGFBPs have similar IGF binding determinants located in both the N- and C-domains (Figure 1, in red underlined text), and both of these domains are required for high-affinity IGF binding (Forbes et al., 1998; Carrick et al., 2001). Isolated N- and C-domains of the IGFBPs have significantly lower affinities for the IGFs than the intact IGFBPs (Clemmons, 2001). Isolated N-domains have affinities up to 100-fold lower than intact IGFBPs; for example, N-BP-3 has a 100-fold lower affinity for IGF-I (Payet et al., 2003) than IGFBP-3 and N-BP-2 has a 10-fold lower affinity for IGF-I than IGFBP-2 (Kuang et al., 2007). Isolated C-domains have lower affinities than N- domains and intact IGFBPs, with C-BP-3 and C-BP-2, for example, having ∼1000-fold lower affinity for IGF-I than IGFBP-3 and IGFBP-2, respectively (Payet et al., 2003, 2004; Kuang et al., 2007).
An intriguing finding from NMR studies is that the N- and C-terminal domains of IGFBP-2 bind IGF cooperatively (Kuang et al., 2007). C-BP-2 binding to the IGF-I:N-BP-2 binary complex is significantly stronger than the binding of C-BP-2 to IGF-I alone, as shown by a switch from intermediate exchange to slow exchange on the NMR time scale. The IGF-I Phe49–Leu54 region and the Phe49 aromatic ring undergo a conformational change or stabilization upon binding to the N-domain. Both this effect on the ligand and the inter-domain interaction between N-BP-2 and C-BP-2 (which is also detectable in the absence of ligand), may contribute to this cooperativity in IGF binding. Furthermore, combining individual IGFBP-3 N- and C-domains in solution results in a binary complex with an affinity only ∼10-fold lower than native IGFBP-3, 10- to 100-fold greater than the affinities observed for the individual domains (Payet et al., 2003; Yan et al., 2004). A similar result was obtained with the combination of C-BP-4 and N-BP-4 with IGF-I (Siwanowicz et al., 2005), although enhanced binding was not observed with N- and C-domains of IGFBP-2 and -6 using surface Plasmon resonance techniques (Carrick et al., 2001; Headey et al., 2004c). It appears that in solution the N- and C-domains can cooperate to bind IGF with high affinity. Presumably, flexibility in the inter-domain binding sites (including loops I and II of the C-domain) is required to enable conformational changes upon ligand binding and this may be hindered upon tethering to a biosensor chip.
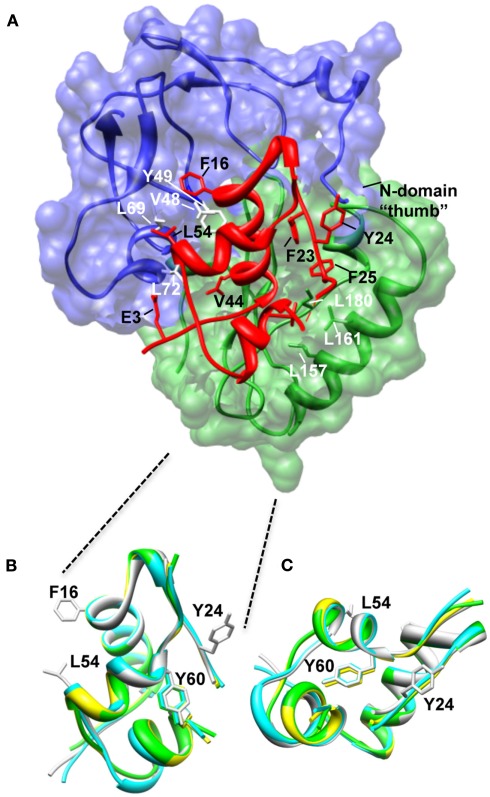
Structural studies have identified three key N-domain residues [Val49, Leu70, and Leu74 of IGFBP-5 (Zeslawski et al., 2001) or Val48, Leu69, and Leu72 of IGFBP-4 (Sitar et al., 2006)] and several other N-domain residues (residues 40–92 of mini-IGFBP-5; Kalus et al., 1998) that interact with IGF (Figure 1). These residues form a deep binding cleft into which IGFs dock (Figure 2A). Mutagenesis studies confirmed the role of IGFBP-5 residues Val49, Leu70, and Leu74 in IGF-I and IGF-II binding and sequence comparisons show that these residues are conserved throughout all six IGFBPs (Kalus et al., 1998). Equivalent residues have been mutated in IGFBPs -3,-4, -5, and -6 to produce IGFBP analogues with <1000-fold lower affinities for IGFs than the wild-type IGFBPs (Qin et al., 1998b; Imai et al., 2000; Yan et al., 2004; Fu et al., 2007). These findings correlated well with a chemical modification study that identified Tyr60 of bovine IGFBP-2 as being part of the interaction surface (Hobba et al., 1996, 1998); Tyr60 of bovine IGFBP-2 lies adjacent to Val59 (equivalent to Val48 and Tyr49 of IGFBP-4; Figure 2A).
Figure 2.
Structure of the N-BP-4:IGF-I:C-BP-4 complex (PDB 2DSR; Sitar et al., 2006). (A) The N-domain of BP-4 (blue, residues 3–82) contacts the C-domain of BP-4 (green, residues 151–232) to form a high-affinity complex with IGF-I (red). IGF-I residues Phe16 and Leu54 insert into a cleft in the N-BP-4 encompassing residues Val48, Tyr49, Leu69, and Leu72 (Kalus et al., 1998). The “thumb” region of the N-domain and residues Leu157, Leu161, and Ile180 of the IGFBP-4 C-domain are in contact with IGF-I residues Phe23, Tyr24, Phe25, and Val44, which are involved in IGF-1R binding, resulting in steric hindrance of IGF-I binding to the receptor (PDB 2DSR, Sitar et al., 2006). (B,C) Superposition of IGF-I crystal structures highlighting that the Tyr60 side chain remains in the same position whether IGF-I is bound to IGFBPs or free. Ribbon views of IGF-I in the free form (bound to detergent molecules) PDB 1GZR (green; Brzozowski et al., 2002) and PDB 1IMX (yellow; Vajdos et al., 2001), the IGF-I‚ mini-N-BP-5 binary complex (PDB 1H59, blue; Zeslawski et al., 2001), and the IGF-I‚ N-BP-4‚ C-BP-4 ternary complex (PDB 2DSR, light gray; Sitar et al., 2006) are superimposed over backbone heavy atoms of the three helices (Ala8–Cys18, Gly42–Cys48, Leu54–Cys61). The side chains of Phe16, Phe49, and Leu54 are shown. The structures are shown in two different orientations (B,C). The structures in (B) are in an orientation equivalent to that of the IGF-I structure in (A). Parts of the flexible regions (residues 1–2, 26–42, and 63–70) are excluded for clarity. Molecular graphic images were produced with the UCSF Chimera program from Resource for Biocomputing, Visualization, and Informatics at the University of California at San Francisco.
Looking at IGF-I, the core of the N-domain interaction with IGF-I involves the side chains of Phe16 and Leu54 of IGF-I that are inserted into the deep binding cleft of mini N-BP-5 (Zeslawski et al., 2001), which forms a “palm”-like structure (Sitar et al., 2006; Figure 2A). Mutagenesis and NMR studies previously mapped specific residues on IGFs involved in IGFBP binding (Headey et al., 2004b; Carrick et al., 2005; Denley et al., 2005a), and these findings agreed with the sites of interaction revealed by the mini-IGFBP-5 structure. The critical role of Glu3 in IGF-I in binding IGFBPs (Francis et al., 1992) could be attributed to hydrogen bonds between this residue (and IGF-I Glu9) and IGFBP-5 residues Tyr50 (equivalent to Tyr60 in IGFBP-2) and His71 (Zeslawski et al., 2001; Figure 2A).
Further structural studies with the intact IGFBP-4 N-domain (residues 1–92) and NN-BP-6 revealed that N-terminal residues wrap around IGF-I in a “thumb”-like manner (Siwanowicz et al., 2005; Chandrashekaran et al., 2007) and contact residues Phe23, Tyr24, Phe25 of IGF-I (Figure 2A). This hydrophobic patch of residues on IGF-I is critical for IGF-1R binding (Denley et al., 2005a), and thus binding of IGF-I to IGFBPs sterically hinders its interaction with the IGF-1R. Importantly, in addition to contributing to overall IGF binding affinity, the NN-subdomain of IGFBP-6 preferentially binds IGF-II over IGF-I, suggesting that this region is important in determining the marked preference of IGFBP-6 for IGF-II (Sitar et al., 2006; Chandrashekaran et al., 2007). The details of the mechanism underlying this IGF-II preference are yet to be elucidated.
Sites of ligand interaction with the C-domain have also been defined in structural studies. NMR techniques have been used to map the large IGF binding surface of C-BP-6 to one side of the flat thyroglobulin type I domain encompassing the helix, the first and second strands of the β-sheet, and the first and second loops (Headey et al., 2004b; residues in red underlined text in Figure 1). Site-directed mutagenesis confirmed the involvement of Asn166 in binding both IGF-I and IGF-II and identified Ser180, Ser181, Gln182 as being determinants contributing to IGFBP-6’s marked specificity for IGF-II. This region was previously identified as an IGF binding surface of IGFBP-2 by deletion mutagenesis (Forbes et al., 1998) and cross-linking studies (Horney et al., 2001). Interestingly, crystal structures of the ternary IGF-I:N-BP-4:C-BP-1 complex suggest that the IGF binding surface of C-BP-4 is located in the same region, but the overall binding surface is much smaller than the C-BP-6 surface defined by NMR (Figure 2; Sitar et al., 2006), as is often the case in such comparisons. A similar region of IGFBP-3 and -5 is involved in IGF binding, as defined by mutagenesis studies (Song et al., 2000; Yan et al., 2004). Mutation of Gly217 and Gln223 in the C terminus (Gly217Ser, Gln223Ala IGFBP-3) disrupted IGF binding by 4- to 11-fold, with a greater effect on IGF-II binding. Strikingly, IGF binding was completely abolished when Gly217 was mutated to Ser and Gln223 to Ala in the C terminus in combination with mutations of the IGFBP-3 N-domain binding site (Leu77Gln, Leu80Gln, and Leu81Gln; Yan et al., 2004).
C-BP-4 also contacts IGF residues critical for IGF-1R binding, including Phe25 (contacting Leu161 and Ile180 of C-BP-4) and Val44 (contacting Leu157 of C-BP-4; Denley et al., 2005b) and equivalent residues of C-BP-6 are involved in the interaction with IGF-II (Headey et al., 2004a). Therefore, the interaction of both the N- and C-domains of the IGFBPs with IGFs sterically hinders IGF receptor binding and thus inhibits IGF action.
While structural studies to date suggest that the major binding determinants lie in the N- and C-domains, there is some indirect evidence for a role of the linker in IGF binding. As described above, high-affinity binding is achieved by incubating individual domains with IGF. However, in the absence of a flexible linker domain, the affinity is 10-fold lower than wild-type. This suggests that the linker domain plays a role in the formation of a high-affinity IGF binding complex, perhaps by controlling inter-domain movement upon ligand binding (Kuang et al., 2007). Further evidence for a role of the linker domains comes from a chemical modification study where Tyr60 of IGF-I was protected from iodination when in complex with IGFBP-2 (Hobba et al., 1996). This suggests that either IGFBP-2 is blocking access or the IGF-I undergoes a structural change upon binding to prohibit access of the labeling reagent to this residue, as we know from structural studies that Tyr60 does not interact with either the N- or C-domains (Figure 2A). However, when comparing the IGF-I structures in the IGFBP-bound and free states we see that there is no major perturbation of the overall IGF-I structure or local structure around Tyr60 (Figure 2B). Therefore a likely explanation is that the linker domain (not present in the IGF-I:N-BP-4:C-BP-1 structure) plays a role in protecting Tyr60 from iodination, although this is yet to be proven experimentally.
Other functional motifs
Insulin-like growth factor binding proteins engage in a range of interactions with binding partners other than IGFs. This includes interactions with serum, cell-surface, and ECM proteins, as well as metal ions (reviewed in Clemmons, 2001; Firth and Baxter, 2002; Bach et al., 2005; Duan and Xu, 2005). The IGFBPs can also undergo post-translational modification (some of which are shown in Figure 1). These interactions and modifications confer on each IGFBP some of its unique characteristics and functions; here we discuss the best characterized of these interactions with respect to their effect on IGFBP structure and function.
Acid labile subunit binding site
Acid labile subunit is an ∼85-kDa glycosylated protein that binds to IGFBP-3:IGF and IGFBP-5:IGF binary complexes (Boisclair et al., 2001). ALS belongs to the leucine-rich repeats (LRR) family of proteins, whose members participate in protein–protein interactions. ALS consists of 20 LRR(s), modeled to be arranged in a donut shape, with negatively charged patches within its center proposed to form the sites of interaction with the binary complex (Janosi et al., 1999). On IGFBP-3 and IGFBP-5 the ALS site of interaction has been mapped by mutagenesis studies to the C-domain; it involves residues 215–232 in IGFBP-3 [more specifically residues 228–232 (Firth et al., 1998) and residues 201–218 in IGFBP-5; Figure 1; Twigg et al., 1998]. Interestingly, ALS does not bind either IGFBP unless they are in complex with IGF (Boisclair et al., 2001).
The main function of ALS is to increase the half-life of IGFs in the circulation. Free IGF-I has a half-life of ∼12 min, in marked contrast to >12 h when in complex with IGFBP-3 and ALS (Boisclair et al., 2001). Almost all serum IGF is found in 150-kDa complexes, with ∼90% in the ALS:IGFBP-3:IGF complex and ∼10% in the ALS:IGFBP-5:IGF form. The affinity of ALS for the IGF-I:IGFBP-3 binary complex is ∼300-fold lower than the binding constant for the binary complex, in line with the 150-kDa complex playing an important role in maintaining a readily available pool of bioavailable IGF (Holman and Baxter, 1996). Knockout mice (Ueki et al., 2000; Yakar et al., 2002) and mutations in the human ALS gene clearly demonstrate this role (Domene et al., 2011). At least 16 naturally occurring mutations in the human ALS gene have been reported so far, including missense, nonsense, frameshift, and duplication mutants (Domene et al., 2011). The lack of functional ALS results in low serum IGF-I and IGFBP-3 and leads to small mice or short stature in humans.
Extracellular matrix binding
Insulin-like growth factor binding protein-2, -3, -5, and -6 have all been shown to interact with glycosaminoglycans (GAGs; Booth et al., 1995; Fowlkes et al., 1997; Firth et al., 1998; Russo et al., 2005; Kuang et al., 2006). A region spanning residues 215–232 of IGFBP-3 is rich in basic residues. Synthetic peptides corresponding to this sequence and mutation of the Lys-Gly-Arg-Lys-Arg consensus sequence (residues 227–232) in IGFBP-3 disrupt binding to GAGs (Booth et al., 1995; Fowlkes et al., 1997). Similar experiments demonstrated the specificity of GAG binding by IGFBP-5 and -6 via their equivalent basic regions (Arai et al., 1996b; Fowlkes et al., 1997). The heparin-binding motif consensus sequence (B-B-B-X-X-B) found within the IGFBP-3, -5, and -6 C-domain basic regions (Figure 1, blue boxes) is lacking in IGFBPs-1 and -4.
A heparin-binding surface identified in IGFBP-2 by NMR encompasses Lys227, His228, Asn232, Leu233, Lys234, and His271 and overlaps the equivalent IGFBP-3 heparin-binding region (Kuang et al., 2006). These heparin-binding residues of IGFBP-2 are located in the thyroglobulin type I domain within the β-turn connecting the first and second strands, part of the third strand, and the beginning of the C-terminal tail (Kuang et al., 2006). The interaction of IGFBP-2 with heparin via this site is pH-dependent, presumably as His becomes protonated at the optimal binding pH of 6.0, thereby improving the electrostatic interaction. The pH-dependent interaction with GAGs may be relevant in situations where extracellular pH is low, as is the case in sites of wound healing or with highly metabolically active cancer cells. IGFBP-2 is unique in that it also contains a second GAG binding site within the linker domain (residues 179–184), which matches a secondary heparin-binding motif consensus sequence (B-B-X-B; Russo et al., 2005). Interestingly, an increase in GAG binding affinity by IGFBP-2 is achieved in the presence of IGF-I and IGF-II, suggesting that a structural change occurs upon IGF binding to expose the heparin-binding surfaces (Arai et al., 1996a).
The interaction with GAGs modulates IGF action. The GAG binding region is adjacent to residues important for IGF binding (Song et al., 2000; Kuang et al., 2006; Figure 1). Once bound to GAGs, the affinity of IGFBPs for IGFs is much lower, leading to an increase in the concentration of bioavailable IGF to bind to the IGF-1R (Jones et al., 1993b; Arai et al., 1994). The heparin-binding region also overlaps the ALS binding sites of IGFBPs-3 and -5 (Figure 1). As such, binding to ECM components can result in IGF release from the 150-kDa complex, thereby promoting delivery of IGFs to targets (Firth and Baxter, 2002). Interestingly, IGFBP-2 and IGFBP-6 bind a broad range of GAGs whereas IGFBP-3 and IGFBP-5 preferentially bind to heparin, heparan sulfate, dermatan sulfate and minimally to chondroitin sulfates and hyaluronic acid (Fowlkes and Serra, 1996). These differences in GAG binding specificity are likely to confer some tissue-specific actions upon the IGFBPs.
GAG binding can also alter IGFBP interactions with other ligands, thereby modulating the IGF-independent actions of IGFBPs. For example, activation of plasminogen to plasmin is promoted by the interaction of IGFBP-5 and plasminogen activator inhibitor-1 (PAI-1), leading to ECM degradation and remodeling independently of IGF-I. This process is important in mammary gland involution. However, binding of heparin to IGFBP-5 perturbs the IGFBP-5:PAI-1 interaction, thus inhibiting this IGF-independent action of IGFBP-5 (Nam et al., 1997). Further interaction with ECM proteins via IGFBP C-domains has been reported (Bach et al., 2005). For example, IGFBP-3 binds fibrin/fibrinogen, fibronectin, vitronectin (Kricker et al., 2010), and plasminogen, which then can influence both its IGF-dependent and IGF-independent actions. Also, fibronectin binds IGFBP-5 and inhibits its ability to promote IGF-dependent cell migration (Xu et al., 2004).
Nuclear localization and interaction with importins
In some circumstances IGFBPs-2, -3, -5, and -6 are able to localize to the nucleus and act in an IGF-independent manner to influence cell signaling and gene expression (Xu et al., 2004; Miyako et al., 2009; Azar et al., 2011; Lee et al., 2011). Nuclear localization sequences have been located in the C-domain of IGFBPs-3 and -5 within the basic region important for ALS and ECM binding (Schedlich et al., 1998; Figure 1, black underlined text), and within the corresponding region of IGFBP-6 (Iosef et al., 2008). In the case of IGFBP-3, nuclear localization results in binding to the retinoid-X-receptor (RXR) α transcription factor and control of its activity to promote apoptosis (Liu et al., 2000). Recently, nuclear IGFBP-6 has been shown to bind the vitamin D receptor (VDR), which inhibits the VDR function and its interaction with RXR, thereby blocking osteoblastic differentiation (Cui et al., 2011). Finally, site-directed mutagenesis identified Leu194 and Leu197 in the IGFBP-3 C-domain as playing a role in nuclear export (Paharkova-Vatchkova and Lee, 2010; Figure 1, blue underlined text). Nuclear export sequences have not been reported for other IGFBPs so far.
Integrin binding – RGD motifs
Other IGF-independent activities ascribed to IGFBPs-1 and -2 involve Arg-Gly-Asp (RGD) motifs present in their C-domains (Figure 1, light green boxes), which mediate interaction with α1β5 integrins (Jones et al., 1993c). The IGFBP-2 RGD motif is located in loop III of the C-domain (Kuang et al., 2006). The interaction decreases cell adhesion and proliferation and promotes cell migration, all processes involved in promoting cancer growth and metastasis (Schutt et al., 2004; Russo et al., 2005; Wang et al., 2006). For example, IGFBP-2/β5 integrin binding stimulates migration of glioma cells via activation of the MAP kinase JNK (Mendes et al., 2010). Similarly, interaction of IGFBP-1 with α1β5 integrin via its RGD motif promotes extravillous trophoblast migration (Gleeson et al., 2001).
Sites of proteolysis
All IGFBPs are susceptible to proteolysis by a range of proteases (Firth and Baxter, 2002; Bunn and Fowlkes, 2003), with most cleavage sites identified so far being located in the linker domain (Figure 1, arrowheads). The importance of IGFBP proteolysis was first recognized over 20 years ago when IGFBP fragments were detected in sera of pregnant women. A pregnancy-associated zinc-binding metalloproteinase, pregnancy-associated plasma protein A (PAPP-A), was identified, and since then its cleavage of IGFBP-4 on the N-terminal side of residue Lys136 has been studied in detail (Boldt and Conover, 2007; Conover, 2010; Table 2). Interestingly PAPP-A cleavage of IGFBP-4 is dependent on IGF binding, suggesting that the cleavage site becomes more accessible upon a conformational change of the linker domain (Conover et al., 1993; Gyrup and Oxvig, 2007). IGFBP-2 cleavage between Gln165 and Met166 by PAPP-A is also IGF-dependent, whereas IGFBP-5 is cleaved between Ser143 and Lys144 independently of IGF binding, suggesting that the cleavage site is already exposed in the case of IGFBP-5 (Laursen et al., 2001; Gyrup and Oxvig, 2007; Table 2). Thus, these studies highlight subtle differences in local structure within the linker domains of individual IGFBPs.
Table 2.
IGFBP-2 proteolytic cleavage sites and PAPP-A cleavage sites on IGFBPs-4, and -5 as identified by mass spectrometry and/or N-terminal sequencing.
| IGFBP | Protease | Cleavage site | Reference |
|---|---|---|---|
| IGFBP-2 | Calpain (Ca2+ activated) | His165–Arg166 | Berg et al. (2007) |
| Unknown in hemofiltrate | Tyr103–Gly104, Leu152–Ala153, Arg156–Glu157, Gln165–Met166, Thr205–Met206, Arg287–Met288 | Mark et al. (2005) | |
| Unknown in hemofiltrate | Met166–Gly167, Lys168–Gly169 | Standker et al. (2003) | |
| Unknown in milk | Leu3–Phe4, Lys168–Gly169, Lys181–Leu182 | Ho and Baxter (1997) | |
| Human kallikrein-2 | Arg164–Gln165 | Rehault et al. (2001) | |
| Matrix metalloproteinase-7 | Glu151–Leu152, Gly175–Leu176, Lys181–Leu182 | Miyamoto et al. (2007) | |
| PAPP-A | Gln165–Met166 | Monget et al. (2003) | |
| IGFBP-4 | PAPP-A | Met135–Lys136 | Conover et al. (1995) |
| IGFBP-5 | PAPP-A | Gln142–Ser143 | Laursen et al. (2001) |
Many other proteases, including pepsin, cathepsins, matrix metalloproteinases, and kallikreins, also cleave IGFBPs predominantly in the linker domain and are not dependent on IGF binding (Firth and Baxter, 2002; Bunn and Fowlkes, 2003). These observations again provide indirect evidence that the linker domain is predominantly unstructured and solvent accessible. To exemplify the complexity of IGFBP regulation by proteolysis, the range of proteases reported to cleave IGFBP-2, and their sites of cleavage, is presented in Table 2.
Proteolytic fragments of IGFBPs have lower affinity for the IGFs than the intact proteins. Proteolysis therefore provides a mechanism by which the concentration of freely bioavailable IGFs is increased, leading to subsequent activation of the IGF-1R. This mechanism has been elegantly demonstrated by in vivo studies in which, for example, PAPP-A and PAPP-A2 (a PAPP-A homologue)-knockout mice exhibit pre and postnatal growth retardation due to their lack of ability to cleave IGFBP-4 or IGFBP-5, respectively (Conover et al., 2004, 2011). The growth defect is reversed in the PAPP-A (−/−) and IGFBP-4 (−/−) double knockouts, thus demonstrating IGFBP-4 is acting as a reservoir for IGF-II and regulates IGF-II release during development (Ning et al., 2008).
Sites of post-translational modification (glycosylation and phosphorylation)
Glycosylation of all IGFBPs has been reported, although the biological significance of these modifications is not fully understood (Firth and Baxter, 2002; Graham et al., 2007). The specific sites of N-linked glycosylation have been identified in the IGFBP-3 N- and linker domains (Asn89, Asn109, and Asn172; Figure 1, dark green boxes). Glycosylated and non-glycosylated forms of IGFBP-3 have been shown to have identical affinity for the IGFs (Firth and Baxter, 1999), although lack of glycosylation increases the affinity of IGFBP-3 for the cell membrane (Firth and Baxter, 1999). IGFBP-5 is also glycosylated, predominantly at Thr103 in the linker domain and to a lesser extent at Ser249 in the C-domain. This similarly has no effect on IGF binding but increases the affinity for heparin, suggesting a role in modulating ECM binding (Graham et al., 2007). IGFBP-6 is O-glycosylated on Thr126, Ser144, Thr145, Thr146, and Ser152, and this protects IGFBP-6 from proteolytic degradation, thereby increasing its circulating half-life (Neumann et al., 1998). Therefore, glycosylation can influence both the IGF-dependent and IGF-independent actions of some IGFBPs.
Phosphorylation can also influence IGFBP actions. IGFBPs-1, -3, and -5 can be phosphorylated on sites found mainly in the linker domain (Figure 1, filled red boxes). IGFBP-1 is phosphorylated by casein kinase-2, predominantly on Ser101, with Ser119 and Ser169 being minor sites (Jones et al., 1993a). Increasing phosphorylation was shown to increase susceptibility to proteolysis by amniotic fluid proteases (Dolcini et al., 2009), but provide protection against plasmin cleavage (Gibson et al., 2001), suggesting that this modification alters the recognition motif for each protease in different ways. Phosphorylation of human IGFBP-1 also increases its affinity for IGFs, and thus the phosphorylation status is thought to influence whether IGFBP-1 inhibits or potentiates IGF action (Jones et al., 1993a). The same enzyme phosphorylates IGFBP-3 on Ser111 and Ser113, but this modification does not alter IGFBP-3’s affinity for IGF-I or IGF-II (Coverley et al., 2000). Phosphorylation of IGFBP-3 protects it from proteolytic cleavage and disrupts its cell membrane association and ability to complex with ALS (Coverley et al., 2000). IGFBP-5 is also a substrate of casein kinase-2 and is phosphorylated on Ser96 and to a lesser extent on Ser248. This modification does not affect IGF or ALS binding but disrupts heparin binding (Graham et al., 2007). Therefore, phosphorylation of each IGFBP has distinct effects on its interactions with binding partners and influences the IGFBP modulation of both IGF-dependent and IGF-independent actions.
Metal ion binding
Insulin-like growth factor binding protein-3 possesses a metal binding domain, which overlaps the heparin-binding domain in the C-domain (Singh et al., 2004). Metal binding influences its interaction with integrins and transferrin and subsequent uptake into cells, a property that has been exploited for the delivery of pro-apoptotic molecules into cancer cells (Huq et al., 2009). Zn2+ also influences binding of IGFs to cell surface associated IGFBP-3 and -5, possibly affecting IGF bioavailability (McCusker, 1998). NMR studies showed that IGFBP-6 residues Gln13, Glu23, Asp25, Ser28, Glu31, Glu35, and Glu37 interact with the lanthanide ion Yb3+ but binding to physiologically relevant metal ions like Ca2+ or Mg2+ was not detected (Chandrashekaran et al., 2007).
Implications of IGFBP Structural Studies for the Design of Disease Therapies
Significant advances have been made in determining the structures of IGFBPs and relating their specific structural motifs to function. These studies have provided the basic understanding required for development of IGFBPs as disease therapies. For example, the product Mecasermin rinfabate (iPLEX™, a complex between IGF-I and IGFBP-3 intact proteins) was originally developed for the treatment of primary IGF-I deficiency in children with short stature and patients with a growth hormone (GH) gene deletion who have developed neutralizing antibodies to GH. However, use of Mecasermin for this application is currently suspended for legal reasons (Williams et al., 2008). While IGF-I treatment alone is also effective, the aim is to increase the half-life of IGF-I by co-administration with IGFBP-3. Mecasermin rinfabate is currently in clinical trials for the treatment of other diseases associated with poor growth, including amyotrophic lateral sclerosis (Lou Gehrig’s disease) and myotonic muscular dystrophy2. Improved insulin sensitivity in patients with severe insulin resistance was achieved recently by treatment with Mecasermin rinfabate, possibly by promoting improve beta cell function (Regan et al., 2010).
Insulin-like growth factor binding proteins have also been investigated as potential cancer therapies based on their ability to inhibit IGF binding to the IGF-1R. Overexpression of IGFBP-4 and -6 in cancer xenografts inhibited tumor growth (Damon et al., 1998; Gallicchio et al., 2001). In addition, PAPP-A resistant IGFBP-4 effectively inhibited mammary tumor xenograft growth when overexpressed in the tumor cells producing the protease (Ryan et al., 2009). Therefore, there is significant potential for the use of a modified protease-resistant IGFBP for the treatment of IGF-dependent cancers (Rosenzweig and Atreya, 2010). Toward this aim we have recently demonstrated efficacy of subcutaneous administration of a protease-resistant IGFBP-2 in inhibiting breast cancer xenografts (unpublished).
The trimeric complex involving IGF-I, vitronectin, and IGFBP-3 has been developed to promote cell migration for application in tissue repair and wound healing (Vitrogro®; Kricker et al., 2003, 2010). This complex leads to activation of the phosphatidylinositide 3-Kinase/AKT and MAPK/ERK 1/2 signaling pathways via co-activation of the αv integrins and the IGF-1R (Hollier et al., 2008). It is currently in clinical trials for the treatment of diabetic ulcers. A chimera of vitronectin and IGF-I also has similar properties (Van Lonkhuyzen et al., 2007).
Further understanding of the molecular interactions with other binding partners and the relevance to IGFBP action in normal and diseased states will ultimately lead to the development of novel therapies targeting these interactions.
Summary: Questions Remaining
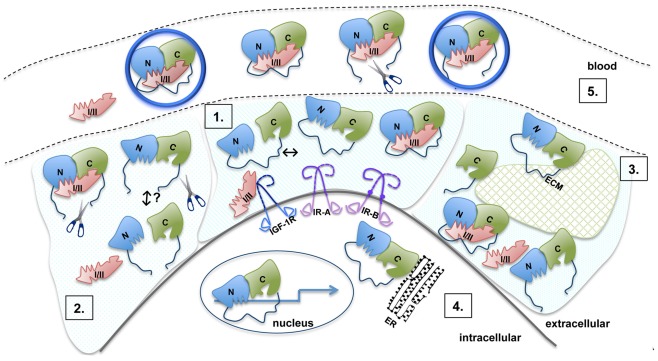
Recent significant advances in understanding the structure and function of IGFBPs provide us with a growing and detailed model of the molecular mechanisms of IGFBP action (Figure 3). Within this model, it is clear that IGFBPs act primarily as inhibitors of IGF binding to the IGF-1R (or IR-A in the case of IGF-II), as residues critical for receptor binding interact directly with the N- and C-terminal domains of the IGFBPs. Importantly, the role of the IGFBP linker domain in IGF binding remains unclear. A significant proportion of the linker domain can be removed with little effect on overall binding affinity (Qin et al., 1998a) and individual N- and C-domains can bind IGFs with only a ∼10-fold lower affinity compared to intact IGFBPs (Payet et al., 2003; Siwanowicz et al., 2005). Experience from fragment-based drug design shows that linkage of two separate binding entities results in a molecule with a binding affinity that approximates the product of the two individual affinities (Hajduk and Greer, 2007). For instance, two chemical entities with Kd values of 10−4 and 10−5 M, respectively, could be linked to produce a molecule with a Kd of 10−9 M. In the case of the IGFBPs, individual N- and C-domains have Kd values of micromolar or better, which implies that it should be possible to link them to produce an intact IGFBP with a Kd in the picomolar range (depending on the length and flexibility of the linker). That this is not the case further supports the observations of a pre-existing interaction between the individual domains even in the absence of the linker domain (Sitar et al., 2006; Kuang et al., 2007). These considerations further emphasize the need to understand the structure and dynamics of the linker domain and its proximity to the IGF ligands in order to clarify the molecular basis for its function.
Figure 3.
Model of the molecular mechanisms of IGFBP action. 1. The N- (blue) and C- (green) domains are connected by a flexible linker (dark blue). Inter-domain interaction between the N- and C-domains is detected in the absence of ligand, but this is of low affinity (Kuang et al., 2007). The interface between the N- and C-domains lies immediately adjacent to their IGF binding surfaces and both domains act cooperatively to bind IGF. Upon IGF-I or IGF-II (pink) binding, the C-domain undergoes a structural change. Residues in the N- and C-domains form the IGF binding site, with the linker playing an as yet unknown role. 2. Proteolysis of IGFBPs occurs primarily in the linker domain (indicated by scissors) resulting in an increased concentration of free IGFs. Individual N- and C-domains have low affinity for IGFs, whereas in vitro, reconstituted binary complexes of the N- and C-domains generally bind IGFs with an affinity only 10-fold lower than intact IGFBPs. It is not clear whether in vivo cleavage results in the dissociation of IGFBP into separate domains or whether cleaved IGFBP domains remain associated via the inter-domain interface. Cleavage products are susceptible to further breakdown, which possibly leads to the equilibrium being pushed toward free IGF and individual IGFBP domains. 3. Intact and cleaved IGFBPs bind extracellular matrix components (ECM, glycosaminoglycans, and proteins) via the C-domain (and the linker domain in IGFBP-2). Binding to ECM (hatched green shape) lowers the affinity for IGFs. 4. IGFBP-2, -3, -5, and -6 are found within the nucleus and can affect gene expression (arrow). IGFBP-3 can be associated with the endoplasmic reticulum (ER). 5. Binary IGFBP:IGF complexes, intact IGFBPs, and cleavage products can be found in the blood. IGFBP-3 and -5 form a trimeric 150 kDa complex with IGF and the acid labile subunit (ALS, large blue circle). Even with limited proteolysis in the L-domain, IGFBP fragments can also form a trimeric complex. Only the binary complexes are small enough to cross the vascular epithelium into the extracellular space to promote IGF-dependent biological activities.
An understanding of the role of the linker domain in IGF binding will also have implications in understanding the mechanism of IGF release upon IGFBP proteolysis. Experimental evidence in vitro and in vivo suggests that IGFBP proteolysis of the linker domain leads to increased concentrations of bioavailable IGF. A basic interpretation of these observations has been that proteolysis leads to the complex dissociating into low affinity fragments. However, it has been clearly shown that in pregnancy serum the majority of IGF remains in the large molecular weight IGFBP-3:ALS:IGF complex despite most of the IGFBP-3 being proteolyzed (Suikkari and Baxter, 1992; Yan et al., 2009). It is not clear whether proteolytic fragments also remain associated at the tissue level. Therefore the mechanism of IGF release remains to be elucidated.
Finally, a full understanding of the determinants of IGF specificity for all IGFBPs has not yet been achieved, particularly in the case of the IGF-II preference for IGFBP-6, although we do know that this involves residues in both the C- and NN-domains (Headey et al., 2004b; Chandrashekaran et al., 2007).
In contrast to the interaction with IGFs, little is known about the mechanism of interaction with other binding partners. In some cases, binding sites have been identified but no structural detail is available for complexes formed with other binding partners. Therefore much remains to be understood about the IGF-independent binding interactions.
In summary, studying the structural features of the individual domains has revealed a range of different functions of the IGFBPs. A complex picture has emerged of IGFBPs as multi-functional proteins able to interact with a broad range of proteins and GAGs. These interactions confer on the IGFBPs the ability to modulate IGF actions and/or to influence IGF-independent actions, such as ECM and cell membrane binding leading to cell migration. An understanding of these molecular interactions has allowed researchers to probe the role of IGFBPs in normal growth and development as well as in disease. Future understanding of the linker domain structure, as well as solution of the structure of an intact IGFBP alone or bound to its various ligands, will provide the basis for development of specific tools to inhibit IGF-dependent and IGF-independent actions for the treatment of diseases such as cancer.
Conflict of Interest Statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Acknowledgments
Raymond S. Norton acknowledges fellowship support from the NHMRC.
Footnotes
References
- Arai T., Busby W., Jr., Clemmons D. R. (1996a). Binding of insulin-like growth factor (IGF) I or II to IGF-binding protein-2 enables it to bind to heparin and extracellular matrix. Endocrinology 137, 4571–4575 10.1210/en.137.11.4571 [DOI] [PubMed] [Google Scholar]
- Arai T., Clarke J., Parker A., Busby W., Jr., Nam T., Clemmons D. R. (1996b). Substitution of specific amino acids in insulin-like growth factor (IGF) binding protein 5 alters heparin binding and its change in affinity for IGF-I response to heparin. J. Biol. Chem. 271, 6099–6106 10.1074/jbc.271.11.6099 [DOI] [PubMed] [Google Scholar]
- Arai T., Parker A., Busby W., Jr., Clemmons D. R. (1994). Heparin, heparan sulfate, and dermatan sulfate regulate formation of the insulin-like growth factor-I and insulin-like growth factor-binding protein complexes. J. Biol. Chem. 269, 20388–20393 [PubMed] [Google Scholar]
- Azar W. J., Azar S. H., Higgins S., Hu J. F., Hoffman A. R., Newgreen D. F., Werther G. A., Russo V. C. (2011). IGFBP-2 enhances VEGF gene promoter activity and consequent promotion of angiogenesis by neuroblastoma cells. Endocrinology 152, 3332–3342 10.1210/en.2011-1121 [DOI] [PubMed] [Google Scholar]
- Bach L. A., Headey S. J., Norton R. S. (2005). IGF-binding proteins – the pieces are falling into place. Trends Endocrinol. Metab. 16, 228–234 10.1016/j.tem.2005.05.005 [DOI] [PubMed] [Google Scholar]
- Berg U., Bang P., Carlsson-Skwirut C. (2007). Calpain proteolysis of insulin-like growth factor binding protein (IGFBP) -2 and -3, but not of IGFBP-1. Biol. Chem. 388, 859–863 10.1515/BC.2007.098 [DOI] [PubMed] [Google Scholar]
- Berman H. M., Westbrook J., Feng Z., Gilliland G., Bhat T. N., Weissig H., Shindyalov I. N., Bourne P. E. (2000). The protein data bank. Nucleic Acids Res. 28, 235–242 10.1093/nar/28.1.235 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boes M., Booth B. A., Sandra A., Dake B. L., Bergold A., Bar R. S. (1992). Insulin-like growth factor binding protein (IGFBP)4 accounts for the connective tissue distribution of endothelial cell IGFBPs perfused through the isolated heart. Endocrinology 131, 327–330 10.1210/en.131.1.327 [DOI] [PubMed] [Google Scholar]
- Boisclair Y. R., Rhoads R. P., Ueki I., Wang J., Ooi G. T. (2001). The acid-labile subunit (ALS) of the 150 kDa IGF-binding protein complex: an important but forgotten component of the circulating IGF system. J. Endocrinol. 170, 63–70 10.1677/joe.0.1700063 [DOI] [PubMed] [Google Scholar]
- Boldt H. B., Conover C. A. (2007). Pregnancy-associated plasma protein-A (PAPP-A): a local regulator of IGF bioavailability through cleavage of IGFBPs. Growth Horm. IGF Res. 17, 10–18 10.1016/j.ghir.2006.11.003 [DOI] [PubMed] [Google Scholar]
- Booth B. A., Boes M., Andress D. L., Dake B. L., Kiefer M. C., Maack C., Linhardt R. J., Bar K., Caldwell E. E., Weiler J., Bar R. S. (1995). IGFBP-3 and IGFBP-5 association with endothelial cells: role of C-terminal heparin binding domain. Growth Regul. 5, 1–17 [PubMed] [Google Scholar]
- Brown J., Jones E. Y., Forbes B. E. (2009). Keeping IGF-II under control: lessons from the IGF-II-IGF2R crystal structure. Trends Biochem. Sci. 34, 612–619 10.1016/j.tibs.2009.07.003 [DOI] [PubMed] [Google Scholar]
- Brzozowski A. M., Dodson E. J., Dodson G. G., Murshudov G. N., Verma C., Turkenburg J. P., De Bree F. M., Dauter Z. (2002). Structural origins of the functional divergence of human insulin-like growth factor-I and insulin. Biochemistry 41, 9389–9397 10.1021/bi020084j [DOI] [PubMed] [Google Scholar]
- Bunn R. C., Fowlkes J. L. (2003). Insulin-like growth factor binding protein proteolysis. Trends Endocrinol. Metab. 14, 176–181 10.1016/S1043-2760(03)00049-3 [DOI] [PubMed] [Google Scholar]
- Carrick F. E., Forbes B. E., Wallace J. C. (2001). BIAcore analysis of bovine insulin-like growth factor (IGF)-binding protein-2 identifies major IGF binding site determinants in both the amino- and carboxyl-terminal domains. J. Biol. Chem. 276, 27120–27128 10.1074/jbc.M101317200 [DOI] [PubMed] [Google Scholar]
- Carrick F. E., Hinds M. G., McNeil K. A., Wallace J. C., Forbes B. E., Norton R. S. (2005). Interaction of insulin-like growth factor (IGF)-I and -II with IGF binding protein-2: mapping the binding surfaces by nuclear magnetic resonance. J. Mol. Endocrinol. 34, 685–698 10.1677/jme.1.01756 [DOI] [PubMed] [Google Scholar]
- Chandrashekaran I. R., Yao S., Wang C. C., Bansal P. S., Alewood P. F., Forbes B. E., Wallace J. C., Bach L. A., Norton R. S. (2007). The N-terminal subdomain of insulin-like growth factor (IGF) binding protein 6. Structure and interaction with IGFs. Biochemistry 46, 3065–3074 10.1021/bi0619876 [DOI] [PubMed] [Google Scholar]
- Chelius D., Baldwin M. A., Lu X., Spencer E. M. (2001). Expression, purification and characterization of the structure and disulfide linkages of insulin-like growth factor binding protein-4. J. Endocrinol. 168, 283–296 10.1677/joe.0.1680283 [DOI] [PubMed] [Google Scholar]
- Clemmons D. R. (2001). Use of mutagenesis to probe IGF-binding protein structure/function relationships. Endocr. Rev. 22, 800–817 10.1210/er.22.6.800 [DOI] [PubMed] [Google Scholar]
- Clemmons D. R. (2007). Modifying IGF1 activity: an approach to treat endocrine disorders, atherosclerosis and cancer. Nat. Rev. Drug Discov. 6, 821–833 10.1038/nrd2359 [DOI] [PubMed] [Google Scholar]
- Conover C. A. (2010). PAPP-A: a new anti-aging target? Aging Cell 9, 942–946 10.1111/j.1474-9726.2010.00589.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- Conover C. A., Bale L. K., Overgaard M. T., Johnstone E. W., Laursen U. H., Fuchtbauer E. M., Oxvig C., Van Deursen J. (2004). Metalloproteinase pregnancy-associated plasma protein A is a critical growth regulatory factor during fetal development. Development 131, 1187–1194 10.1242/dev.00997 [DOI] [PubMed] [Google Scholar]
- Conover C. A., Boldt H. B., Bale L. K., Clifton K. B., Grell J. A., Mader J. R., Mason E. J., Powell D. R. (2011). Pregnancy-associated plasma protein-A2 (PAPP-A2): tissue expression and biological consequences of gene knockout in mice. Endocrinology 152, 2837–2844 10.1210/en.2011-0036 [DOI] [PubMed] [Google Scholar]
- Conover C. A., Durham S. K., Zapf J., Masiarz F. R., Kiefer M. C. (1995). Cleavage analysis of insulin-like growth factor (IGF)-dependent IGF-binding protein-4 proteolysis and expression of protease-resistant IGF-binding protein-4 mutants. J. Biol. Chem. 270, 4395–4400 10.1074/jbc.270.9.4395 [DOI] [PubMed] [Google Scholar]
- Conover C. A., Kiefer M. C., Zapf J. (1993). Posttranslational regulation of insulin-like growth factor binding protein-4 in normal and transformed human fibroblasts. Insulin-like growth factor dependence and biological studies. J. Clin. Invest. 91, 1129–1137 10.1172/JCI116272 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Coverley J. A., Martin J. L., Baxter R. C. (2000). The effect of phosphorylation by casein kinase 2 on the activity of insulin-like growth factor-binding protein-3. Endocrinology 141, 564–570 10.1210/en.141.2.564 [DOI] [PubMed] [Google Scholar]
- Cui J., Ma C., Qiu J., Ma X., Wang X., Chen H., Huang B. (2011). A novel interaction between insulin-like growth factor binding protein-6 and the vitamin D receptor inhibits the role of vitamin D3 in osteoblast differentiation. Mol. Cell. Endocrinol. 338, 84–92 10.1016/j.mce.2011.03.011 [DOI] [PubMed] [Google Scholar]
- Damon S. E., Maddison L., Ware J. L., Plymate S. R. (1998). Overexpression of an inhibitory insulin-like growth factor binding protein (IGFBP), IGFBP-4, delays onset of prostate tumor formation. Endocrinology 139, 3456–3464 10.1210/en.139.8.3456 [DOI] [PubMed] [Google Scholar]
- Denley A., Cosgrove L. J., Booker G. W., Wallace J. C., Forbes B. E. (2005a). Molecular interactions of the IGF system. Cytokine Growth Factor Rev. 16, 421–439 10.1016/j.cytogfr.2005.04.004 [DOI] [PubMed] [Google Scholar]
- Denley A., Wang C. C., McNeil K. A., Walenkamp M. J., Van Duyvenvoorde H., Wit J. M., Wallace J. C., Norton R. S., Karperien M., Forbes B. E. (2005b). Structural and functional characteristics of the Val44Met insulin-like growth factor I missense mutation: correlation with effects on growth and development. Mol. Endocrinol. 19, 711–721 10.1210/me.2004-0409 [DOI] [PubMed] [Google Scholar]
- Denley A., Wallace J. C., Cosgrove L. J., Forbes B. E. (2003). The insulin receptor isoform exon 11- (IR-A) in cancer and other diseases: a review. Horm. Metab. Res. 35, 778–785 10.1055/s-2004-814157 [DOI] [PubMed] [Google Scholar]
- Dolcini L., Sala A., Campagnoli M., Labo S., Valli M., Visai L., Minchiotti L., Monaco H. L., Galliano M. (2009). Identification of the amniotic fluid insulin-like growth factor binding protein-1 phosphorylation sites and propensity to proteolysis of the isoforms. FEBS J. 276, 6033–6046 10.1111/j.1742-4658.2009.07318.x [DOI] [PubMed] [Google Scholar]
- Domene H. M., Hwa V., Jasper H. G., Rosenfeld R. G. (2011). Acid-labile subunit (ALS) deficiency. Best Pract. Res. Clin. Endocrinol. Metab. 25, 101–113 10.1016/j.beem.2010.08.010 [DOI] [PubMed] [Google Scholar]
- Duan C., Xu Q. (2005). Roles of insulin-like growth factor (IGF) binding proteins in regulating IGF actions. Gen. Comp. Endocrinol. 142, 44–52 10.1016/j.ygcen.2004.12.022 [DOI] [PubMed] [Google Scholar]
- Firth S. M., Baxter R. C. (1999). Characterisation of recombinant glycosylation variants of insulin-like growth factor binding protein-3. J. Endocrinol. 160, 379–387 10.1677/joe.0.1600379 [DOI] [PubMed] [Google Scholar]
- Firth S. M., Baxter R. C. (2002). Cellular actions of the insulin-like growth factor binding proteins. Endocr. Rev. 23, 824–854 10.1210/er.2001-0033 [DOI] [PubMed] [Google Scholar]
- Firth S. M., Ganeshprasad U., Baxter R. C. (1998). Structural determinants of ligand and cell surface binding of insulin-like growth factor-binding protein-3. J. Biol. Chem. 273, 2631–2638 10.1074/jbc.273.5.2631 [DOI] [PubMed] [Google Scholar]
- Forbes B. E., Turner D., Hodge S. J., McNeil K. A., Forsberg G., Wallace J. C. (1998). Localization of an insulin-like growth factor (IGF) binding site of bovine IGF binding protein-2 using disulfide mapping and deletion mutation analysis of the C-terminal domain. J. Biol. Chem. 273, 4647–4652 10.1074/jbc.273.8.4647 [DOI] [PubMed] [Google Scholar]
- Fowlkes J. L., Serra D. M. (1996). Characterization of glycosaminoglycan-binding domains present in insulin-like growth factor-binding protein-3. J. Biol. Chem. 271, 14676–14679 10.1074/jbc.271.25.14676 [DOI] [PubMed] [Google Scholar]
- Fowlkes J. L., Thrailkill K. M., George-Nascimento C., Rosenberg C. K., Serra D. M. (1997). Heparin-binding, highly basic regions within the thyroglobulin type-1 repeat of insulin-like growth factor (IGF)-binding proteins (IGFBPs) -3, -5, and -6 inhibit IGFBP-4 degradation. Endocrinology 138, 2280–2285 10.1210/en.138.6.2280 [DOI] [PubMed] [Google Scholar]
- Francis G. L., Ross M., Ballard F. J., Milner S. J., Senn C., McNeil K. A., Wallace J. C., King R., Wells J. R. (1992). Novel recombinant fusion protein analogues of insulin-like growth factor (IGF)-I indicate the relative importance of IGF-binding protein and receptor binding for enhanced biological potency. J. Mol. Endocrinol. 8, 213–223 10.1677/jme.0.0080213 [DOI] [PubMed] [Google Scholar]
- Fu P., Thompson J. A., Bach L. A. (2007). Promotion of cancer cell migration: an insulin-like growth factor (IGF)-independent action of IGF-binding protein-6. J. Biol. Chem. 282, 22298–22306 10.1074/jbc.M608748200 [DOI] [PubMed] [Google Scholar]
- Galanis M., Firth S. M., Bond J., Nathanielsz A., Kortt A. A., Hudson P. J., Baxter R. C. (2001). Ligand-binding characteristics of recombinant amino- and carboxyl-terminal fragments of human insulin-like growth factor-binding protein-3. J. Endocrinol. 169, 123–133 10.1677/joe.0.1690123 [DOI] [PubMed] [Google Scholar]
- Galea C. A., Mobli M., McNeil K. A., Mulhern T. D., Wallace J. C., King G. F., Forbes B. E., Norton R. S. (2012). Insulin-like growth factor binding protein-2: NMR analysis and structural characterization of the N-terminal domain. Biochimie 94, 608–616 10.1016/j.biochi.2011.09.012 [DOI] [PubMed] [Google Scholar]
- Gallicchio M. A., Kneen M., Hall C., Scott A. M., Bach L. A. (2001). Overexpression of insulin-like growth factor binding protein-6 inhibits rhabdomyosarcoma growth in vivo. Int. J. Cancer 94, 645–651 10.1002/ijc.1519 [DOI] [PubMed] [Google Scholar]
- Gibson J. M., Aplin J. D., White A., Westwood M. (2001). Regulation of IGF bioavailability in pregnancy. Mol. Hum. Reprod. 7, 79–87 10.1093/molehr/7.1.79 [DOI] [PubMed] [Google Scholar]
- Gleeson L. M., Chakraborty C., Mckinnon T., Lala P. K. (2001). Insulin-like growth factor-binding protein 1 stimulates human trophoblast migration by signaling through alpha 5 beta 1 integrin via mitogen-activated protein Kinase pathway. J. Clin. Endocrinol. Metab. 86, 2484–2493 10.1210/jc.86.6.2484 [DOI] [PubMed] [Google Scholar]
- Graham M. E., Kilby D. M., Firth S. M., Robinson P. J., Baxter R. C. (2007). The in vivo phosphorylation and glycosylation of human insulin-like growth factor-binding protein-5. Mol. Cell Proteomics 6, 1392–1405 10.1074/mcp.M700027-MCP200 [DOI] [PubMed] [Google Scholar]
- Gyrup C., Oxvig C. (2007). Quantitative analysis of insulin-like growth factor-modulated proteolysis of insulin-like growth factor binding protein-4 and -5 by pregnancy-associated plasma protein-A. Biochemistry 46, 1972–1980 10.1021/bi062229i [DOI] [PubMed] [Google Scholar]
- Hajduk P. J., Greer J. (2007). A decade of fragment-based drug design: strategic advances and lessons learned. Nat. Rev. Drug Discov. 6, 211–219 10.1038/nrd2220 [DOI] [PubMed] [Google Scholar]
- Headey S. J., Keizer D. W., Yao S., Brasier G., Kantharidis P., Bach L. A., Norton R. S. (2004a). C-terminal domain of insulin-like growth factor binding protein-6: structure and interaction with insulin-like growth factor-II. Mol. Endocrinol. 18, 2740–2750 10.1210/me.2004-0248 [DOI] [PubMed] [Google Scholar]
- Headey S. J., Keizer D. W., Yao S., Wallace J. C., Bach L. A., Norton R. S. (2004b). Binding site for the C-domain of insulin-like growth factor (IGF) binding protein-6 on IGF-II; implications for inhibition of IGF actions. FEBS Lett. 568, 19–22 10.1016/j.febslet.2004.04.091 [DOI] [PubMed] [Google Scholar]
- Headey S. J., Leeding K. S., Norton R. S., Bach L. A. (2004c). Contributions of the N- and C-terminal domains of IGF binding protein-6 to IGF binding. J. Mol. Endocrinol. 33, 377–386 10.1677/jme.1.01547 [DOI] [PubMed] [Google Scholar]
- Ho P. J., Baxter R. C. (1997). Characterization of truncated insulin-like growth factor-binding protein-2 in human milk. Endocrinology 138, 3811–3818 10.1210/en.138.9.3811 [DOI] [PubMed] [Google Scholar]
- Hobba G. D., Forbes B. E., Parkinson E. J., Francis G. L., Wallace J. C. (1996). The insulin-like growth factor (IGF) binding site of bovine insulin-like growth factor binding protein-2 (bIGFBP-2) probed by iodination. J. Biol. Chem. 271, 30529–30536 10.1074/jbc.271.48.30529 [DOI] [PubMed] [Google Scholar]
- Hobba G. D., Lothgren A., Holmberg E., Forbes B. E., Francis G. L., Wallace J. C. (1998). Alanine screening mutagenesis establishes tyrosine 60 of bovine insulin-like growth factor binding protein-2 as a determinant of insulin-like growth factor binding. J. Biol. Chem. 273, 19691–19698 10.1074/jbc.273.31.19691 [DOI] [PubMed] [Google Scholar]
- Hollier B. G., Kricker J. A., Van Lonkhuyzen D. R., Leavesley D. I., Upton Z. (2008). Substrate-bound insulin-like growth factor (IGF)-I-IGF binding protein-vitronectin-stimulated breast cell migration is enhanced by coactivation of the phosphatidylinositide 3-Kinase/AKT pathway by alphav-integrins and the IGF-I receptor. Endocrinology 149, 1075–1090 10.1210/en.2007-0740 [DOI] [PubMed] [Google Scholar]
- Holman S. R., Baxter R. C. (1996). Insulin-like growth factor binding protein-3: factors affecting binary and ternary complex formation. Growth Regul. 6, 42–47 [PubMed] [Google Scholar]
- Horney M. J., Evangelista C. A., Rosenzweig S. A. (2001). Synthesis and characterization of insulin-like growth factor (IGF)-1 photoprobes selective for the IGF-binding proteins (IGFBPS). Photoaffinity labeling of the IGF-binding domain on IGFBP-2. J. Biol. Chem. 276, 2880–2889 10.1074/jbc.M007526200 [DOI] [PubMed] [Google Scholar]
- Huq A., Singh B., Meeker T., Mascarenhas D. (2009). The metal-binding domain of IGFBP-3 selectively delivers therapeutic molecules into cancer cells. Anticancer Drugs 20, 21–31 10.1097/CAD.0b013e3283144610 [DOI] [PubMed] [Google Scholar]
- Imai Y., Moralez A., Andag U., Clarke J. B., Busby W. H., Jr., Clemmons D. R. (2000). Substitutions for hydrophobic amino acids in the N-terminal domains of IGFBP-3 and -5 markedly reduce IGF-I binding and alter their biologic actions. J. Biol. Chem. 275, 18188–18194 10.1074/jbc.C000447200 [DOI] [PubMed] [Google Scholar]
- Iosef C., Gkourasas T., Jia C. Y., Li S. S., Han V. K. (2008). A functional nuclear localization signal in insulin-like growth factor binding protein-6 mediates its nuclear import. Endocrinology 149, 1214–1226 10.1210/en.2007-0959 [DOI] [PubMed] [Google Scholar]
- Janosi J. B., Firth S. M., Bond J. J., Baxter R. C., Delhanty P. J. (1999). N-Linked glycosylation and sialylation of the acid-labile subunit. Role in complex formation with insulin-like growth factor (IGF)-binding protein-3 and the IGFs. J. Biol. Chem. 274, 5292–5298 10.1074/jbc.274.33.23328 [DOI] [PubMed] [Google Scholar]
- Jones J. I., Busby W. H., Jr., Wright G., Smith C. E., Kimack N. M., Clemmons D. R. (1993a). Identification of the sites of phosphorylation in insulin-like growth factor binding protein-1. Regulation of its affinity by phosphorylation of serine 101. J. Biol. Chem. 268, 1125–1131 [PubMed] [Google Scholar]
- Jones J. I., Gockerman A., Busby W. H., Jr., Camacho-Hubner C., Clemmons D. R. (1993b). Extracellular matrix contains insulin-like growth factor binding protein-5: potentiation of the effects of IGF-I. J. Cell Biol. 121, 679–687 10.1083/jcb.121.3.679 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jones J. I., Gockerman A., Busby W. H., Jr., Wright G., Clemmons D. R. (1993c). Insulin-like growth factor binding protein 1 stimulates cell migration and binds to the alpha 5 beta 1 integrin by means of its Arg-Gly-Asp sequence. Proc. Natl. Acad. Sci. U.S.A. 90, 10553–10557 10.1073/pnas.90.14.6557 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kainosho M., Torizawa T., Iwashita Y., Terauchi T., Mei Ono A., Guntert P. (2006). Optimal isotope labelling for NMR protein structure determinations. Nature 440, 52–57 10.1038/nature04525 [DOI] [PubMed] [Google Scholar]
- Kalus W., Zweckstetter M., Renner C., Sanchez Y., Georgescu J., Grol M., Demuth D., Schumacher R., Dony C., Lang K., Holak T. A. (1998). Structure of the IGF-binding domain of the insulin-like growth factor-binding protein-5 (IGFBP-5): implications for IGF and IGF-I receptor interactions. EMBO J. 17, 6558–6572 10.1093/emboj/17.22.6558 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kricker J. A., Hyde C. E., Van Lonkhuyzen D. R., Hollier B. G., Shooter G. K., Leavesley D. I., Herington A. C., Upton Z. (2010). Mechanistic investigations into interactions between IGF-I and IGFBPs and their impact on facilitating cell migration on vitronectin. Growth Factors 28, 359–369 10.3109/08977194.2010.494603 [DOI] [PubMed] [Google Scholar]
- Kricker J. A., Towne C. L., Firth S. M., Herington A. C., Upton Z. (2003). Structural and functional evidence for the interaction of insulin-like growth factors (IGFs) and IGF binding proteins with vitronectin. Endocrinology 144, 2807–2815 10.1210/en.2002-221086 [DOI] [PubMed] [Google Scholar]
- Kuang Z., Yao S., Keizer D. W., Wang C. C., Bach L. A., Forbes B. E., Wallace J. C., Norton R. S. (2006). Structure, dynamics and heparin binding of the C-terminal domain of insulin-like growth factor-binding protein-2 (IGFBP-2). J. Mol. Biol. 364, 690–704 10.1016/j.jmb.2006.09.006 [DOI] [PubMed] [Google Scholar]
- Kuang Z., Yao S., McNeil K. A., Thompson J. A., Bach L. A., Forbes B. E., Wallace J. C., Norton R. S. (2007). Cooperativity of the N- and C-terminal domains of insulin-like growth factor (IGF) binding protein 2 in IGF binding. Biochemistry 46, 13720–13732 10.1021/bi701251d [DOI] [PubMed] [Google Scholar]
- Laursen L. S., Overgaard M. T., Soe R., Boldt H. B., Sottrup-Jensen L., Giudice L. C., Conover C. A., Oxvig C. (2001). Pregnancy-associated plasma protein-A (PAPP-A) cleaves insulin-like growth factor binding protein (IGFBP)-5 independent of IGF: implications for the mechanism of IGFBP-4 proteolysis by PAPP-A. FEBS Lett. 504, 36–40 10.1016/S0014-5793(01)02760-0 [DOI] [PubMed] [Google Scholar]
- Lee Y. C., Jogie-Brahim S., Lee D. Y., Han J., Harada A., Murphy L. J., Oh Y. (2011). Insulin-like growth factor-binding protein-3 (IGFBP-3) blocks the effects of asthma by negatively regulating NF-kappaB signaling through IGFBP-3R-mediated activation of caspases. J. Biol. Chem. 286, 17898–17909 10.1074/jbc.M110.209379 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lewitt M. S., Saunders H., Phuyal J. L., Baxter R. C. (1994). Complex formation by human insulin-like growth factor-binding protein-3 and human acid-labile subunit in growth hormone-deficient rats. Endocrinology 134, 2404–2409 10.1210/en.134.6.2404 [DOI] [PubMed] [Google Scholar]
- Liu B., Lee H. Y., Weinzimer S. A., Powell D. R., Clifford J. L., Kurie J. M., Cohen P. (2000). Direct functional interactions between insulin-like growth factor-binding protein-3 and retinoid X receptor-alpha regulate transcriptional signaling and apoptosis. J. Biol. Chem. 275, 33607–33613 10.1074/jbc.M002547200 [DOI] [PubMed] [Google Scholar]
- Mark S., Kubler B., Honing S., Oesterreicher S., John H., Braulke T., Forssmann W. G., Standker L. (2005). Diversity of human insulin-like growth factor (IGF) binding protein-2 fragments in plasma: primary structure, IGF-binding properties, and disulfide bonding pattern. Biochemistry 44, 3644–3652 10.1021/bi0478401 [DOI] [PubMed] [Google Scholar]
- McCusker R. H. (1998). Controlling insulin-like growth factor activity and the modulation of insulin-like growth factor binding protein and receptor binding. J. Dairy Sci. 81, 1790–1800 10.3168/jds.S0022-0302(98)75748-0 [DOI] [PubMed] [Google Scholar]
- Mendes K. N., Wang G. K., Fuller G. N., Zhang W. (2010). JNK mediates insulin-like growth factor binding protein 2/integrin alpha5-dependent glioma cell migration. Int. J. Oncol. 37, 143–153 [DOI] [PubMed] [Google Scholar]
- Miyako K., Cobb L. J., Francis M., Huang A., Peng B., Pintar J. E., Ariga H., Cohen P. (2009). PAPA-1 Is a nuclear binding partner of IGFBP-2 and modulates its growth-promoting actions. Mol. Endocrinol. 23, 169–175 10.1210/me.2008-0168 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miyamoto S., Nakamura M., Yano K., Ishii G., Hasebe T., Endoh Y., Sangai T., Maeda H., Shi-Chuang Z., Chiba T., Ochiai A. (2007). Matrix metalloproteinase-7 triggers the matricrine action of insulin-like growth factor-II via proteinase activity on insulin-like growth factor binding protein 2 in the extracellular matrix. Cancer Sci. 98, 685–691 10.1111/j.1349-7006.2007.00448.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- Monget P., Mazerbourg S., Delpuech T., Maurel M. C., Maniere S., Zapf J., Lalmanach G., Oxvig C., Overgaard M. T. (2003). Pregnancy-associated plasma protein-A is involved in insulin-like growth factor binding protein-2 (IGFBP-2) proteolytic degradation in bovine and porcine preovulatory follicles: identification of cleavage site and characterization of IGFBP-2 degradation. Biol. Reprod. 68, 77–86 10.1095/biolreprod.102.007609 [DOI] [PubMed] [Google Scholar]
- Nam T. J., Busby W., Jr., Clemmons D. R. (1997). Insulin-like growth factor binding protein-5 binds to plasminogen activator inhibitor-I. Endocrinology 138, 2972–2978 10.1210/en.138.7.2972 [DOI] [PubMed] [Google Scholar]
- Neumann G. M., Bach L. A. (1999). The N-terminal disulfide linkages of human insulin-like growth factor-binding protein-6 (hIGFBP-6) and hIGFBP-1 are different as determined by mass spectrometry. J. Biol. Chem. 274, 14587–14594 10.1074/jbc.274.21.14587 [DOI] [PubMed] [Google Scholar]
- Neumann G. M., Marinaro J. A., Bach L. A. (1998). Identification of O-glycosylation sites and partial characterization of carbohydrate structure and disulfide linkages of human insulin-like growth factor binding protein 6. Biochemistry 37, 6572–6585 10.1021/bi972894e [DOI] [PubMed] [Google Scholar]
- Ning Y., Schuller A. G., Conover C. A., Pintar J. E. (2008). Insulin-like growth factor (IGF) binding protein-4 is both a positive and negative regulator of IGF activity in vivo. Mol. Endocrinol. 22, 1213–1225 10.1210/me.2007-0480 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Paharkova-Vatchkova V., Lee K. W. (2010). Nuclear export and mitochondrial and endoplasmic reticulum localization of IGF-binding protein 3 regulate its apoptotic properties. Endocr. Relat. Cancer 17, 293–302 10.1677/ERC-09-0106 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Payet L. D., Firth S. M., Baxter R. C. (2004). The role of the acid-labile subunit in regulating insulin-like growth factor transport across human umbilical vein endothelial cell monolayers. J. Clin. Endocrinol. Metab. 89, 2382–2389 10.1210/jc.2003-031880 [DOI] [PubMed] [Google Scholar]
- Payet L. D., Wang X. H., Baxter R. C., Firth S. M. (2003). Amino- and carboxyl-terminal fragments of insulin-like growth factor (IGF) binding protein-3 cooperate to bind IGFs with high affinity and inhibit IGF receptor interactions. Endocrinology 144, 2797–2806 10.1210/en.2003-0102 [DOI] [PubMed] [Google Scholar]
- Pettersen E. F., Goddard T. D., Huang C. C., Couch G. S., Greenblatt D. M., Meng E. C., Ferrin T. E. (2004). UCSF Chimera – a visualization system for exploratory research and analysis. J. Comput. Chem. 25, 1605–1612 10.1002/jcc.20084 [DOI] [PubMed] [Google Scholar]
- Piriz J., Muller A., Trejo J. L., Torres-Aleman I. (2011). IGF-I and the aging mammalian brain. Exp. Gerontol. 46, 96–99 10.1016/j.exger.2010.08.022 [DOI] [PubMed] [Google Scholar]
- Pollak M. (2008a). Insulin and insulin-like growth factor signalling in neoplasia. Nat. Rev. Cancer 8, 915–928 10.1038/nrc2536 [DOI] [PubMed] [Google Scholar]
- Pollak M. (2008b). Targeting insulin and insulin-like growth factor signalling in oncology. Curr. Opin. Pharmacol. 8, 384–392 10.1016/j.coph.2008.07.004 [DOI] [PubMed] [Google Scholar]
- Qin X., Strong D. D., Baylink D. J., Mohan S. (1998a). Structure-function analysis of the human insulin-like growth factor binding protein-4. J. Biol. Chem. 273, 23509–23516 10.1074/jbc.273.36.23509 [DOI] [PubMed] [Google Scholar]
- Qin X., Strong D. D., Baylink D. J., Mohan S. (1998b). Structure-function analysis of the human insulin-like growth factor binding protein-4. J. Biol. Chem. 273, 23509–23516 10.1074/jbc.273.36.23509 [DOI] [PubMed] [Google Scholar]
- Regan F. M., Williams R. M., Mcdonald A., Umpleby A. M., Acerini C. L., O’Rahilly S., Hovorka R., Semple R. K., Dunger D. B. (2010). Treatment with recombinant human insulin-like growth factor (rhIGF)-I/rhIGF binding protein-3 complex improves metabolic control in subjects with severe insulin resistance. J. Clin. Endocrinol. Metab. 95, 2113–2122 10.1210/jc.2009-2088 [DOI] [PubMed] [Google Scholar]
- Rehault S., Monget P., Mazerbourg S., Tremblay R., Gutman N., Gauthier F., Moreau T. (2001). Insulin-like growth factor binding proteins (IGFBPs) as potential physiological substrates for human kallikreins hK2 and hK3. Eur. J. Biochem. 268, 2960–2968 10.1046/j.1432-1327.2001.02185.x [DOI] [PubMed] [Google Scholar]
- Rosenfeld R. G. (2007). IGF-I therapy in growth disorders. Eur. J. Endocrinol. 157, S57–S60 10.1530/EJE-07-0187 [DOI] [PubMed] [Google Scholar]
- Rosenzweig S. A., Atreya H. S. (2010). Defining the pathway to insulin-like growth factor system targeting in cancer. Biochem. Pharmacol. 80, 1115–1124 10.1016/j.bcp.2010.06.013 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Russo V. C., Schutt B. S., Andaloro E., Ymer S. I., Hoeflich A., Ranke M. B., Bach L. A., Werther G. A. (2005). Insulin-like growth factor binding protein-2 binding to extracellular matrix plays a critical role in neuroblastoma cell proliferation, migration, and invasion. Endocrinology 146, 4445–4455 10.1210/en.2005-0467 [DOI] [PubMed] [Google Scholar]
- Ryan A. J., Napoletano S., Fitzpatrick P. A., Currid C. A., O’Sullivan N. C., Harmey J. H. (2009). Expression of a protease-resistant insulin-like growth factor-binding protein-4 inhibits tumour growth in a murine model of breast cancer. Br. J. Cancer 101, 278–286 10.1038/sj.bjc.6605141 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sala A., Capaldi S., Campagnoli M., Faggion B., Labo S., Perduca M., Romano A., Carrizo M. E., Valli M., Visai L., Minchiotti L., Galliano M., Monaco H. L. (2005). Structure and properties of the C-terminal domain of insulin-like growth factor-binding protein-1 isolated from human amniotic fluid. J. Biol. Chem. 280, 29812–29819 10.1074/jbc.M411048200 [DOI] [PubMed] [Google Scholar]
- Schedlich L. J., Young T. F., Firth S. M., Baxter R. C. (1998). Insulin-like growth factor-binding protein (IGFBP)-3 and IGFBP-5 share a common nuclear transport pathway in T47D human breast carcinoma cells. J. Biol. Chem. 273, 18347–18352 10.1074/jbc.273.29.18347 [DOI] [PubMed] [Google Scholar]
- Schutt B. S., Langkamp M., Rauschnabel U., Ranke M. B., Elmlinger M. W. (2004). Integrin-mediated action of insulin-like growth factor binding protein-2 in tumor cells. J. Mol. Endocrinol. 32, 859–868 10.1677/jme.0.0320859 [DOI] [PubMed] [Google Scholar]
- Singh B., Charkowicz D., Mascarenhas D. (2004). Insulin-like growth factor-independent effects mediated by a C-terminal metal-binding domain of insulin-like growth factor binding protein-3. J. Biol. Chem. 279, 477–487 10.1074/jbc.M404235200 [DOI] [PubMed] [Google Scholar]
- Sitar T., Popowicz G. M., Siwanowicz I., Huber R., Holak T. A. (2006). Structural basis for the inhibition of insulin-like growth factors by insulin-like growth factor-binding proteins. Proc. Natl. Acad. Sci. U.S.A. 103, 13028–13033 10.1073/pnas.0605652103 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Siwanowicz I., Popowicz G. M., Wisniewska M., Huber R., Kuenkele K. P., Lang K., Engh R. A., Holak T. A. (2005). Structural basis for the regulation of insulin-like growth factors by IGF binding proteins. Structure 13, 155–167 10.1016/j.str.2004.11.009 [DOI] [PubMed] [Google Scholar]
- Song H., Beattie J., Campbell I. W., Allan G. J. (2000). Overlap of IGF- and heparin-binding sites in rat IGF-binding protein-5. J. Mol. Endocrinol. 24, 43–51 10.1677/jme.0.0240043 [DOI] [PubMed] [Google Scholar]
- Standker L., Kubler B., Obendorf M., Braulke T., Forssmann W. G., Mark S. (2003). In vivo processed fragments of IGF binding protein-2 copurified with bioactive IGF-II. Biochem. Biophys. Res. Commun. 304, 708–713 10.1016/S0006-291X(03)00658-2 [DOI] [PubMed] [Google Scholar]
- Suikkari A. M., Baxter R. C. (1992). Insulin-like growth factor-binding protein-3 is functionally normal in pregnancy serum. J. Clin. Endocrinol. Metab. 74, 177–183 10.1210/jc.74.1.177 [DOI] [PubMed] [Google Scholar]
- Sung M., Jeong M. S., Jang S. B. (2010). Biological characterization and structure based prediction of insulin-like growth factor binding protein-5. Biochem. Biophys. Res. Commun. 403, 230–236 10.1016/j.bbrc.2010.11.016 [DOI] [PubMed] [Google Scholar]
- Twigg S. M., Baxter R. C. (1998). Insulin-like growth factor (IGF)-binding protein 5 forms an alternative ternary complex with IGFs and the acid-labile subunit. J. Biol. Chem. 273, 6074–6079 10.1074/jbc.273.44.28791 [DOI] [PubMed] [Google Scholar]
- Twigg S. M., Kiefer M. C., Zapf J., Baxter R. C. (1998). Insulin-like growth factor-binding protein 5 complexes with the acid-labile subunit. Role of the carboxyl-terminal domain. J. Biol. Chem. 273, 28791–28798 10.1074/jbc.273.44.28791 [DOI] [PubMed] [Google Scholar]
- Ueki I., Ooi G. T., Tremblay M. L., Hurst K. R., Bach L. A., Boisclair Y. R. (2000). Inactivation of the acid labile subunit gene in mice results in mild retardation of postnatal growth despite profound disruptions in the circulating insulin-like growth factor system. Proc. Natl. Acad. Sci. U.S.A. 97, 6868–6873 10.1073/pnas.120172697 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vajdos F. F., Ultsch M., Schaffer M. L., Deshayes K. D., Liu J., Skelton N. J., De Vos A. M. (2001). Crystal structure of human insulin-like growth factor-1: detergent binding inhibits binding protein interactions. Biochemistry 40, 11022–11029 10.1021/bi0109111 [DOI] [PubMed] [Google Scholar]
- Van Lonkhuyzen D. R., Hollier B. G., Shooter G. K., Leavesley D. I., Upton Z. (2007). Chimeric vitronectin:insulin-like growth factor proteins enhance cell growth and migration through co-activation of receptors. Growth Factors 25, 295–308 10.1080/08977190701803752 [DOI] [PubMed] [Google Scholar]
- Wang G. K., Hu L., Fuller G. N., Zhang W. (2006). An interaction between insulin-like growth factor-binding protein 2 (IGFBP2) and integrin alpha 5 is essential for IGFBP2-induced cell mobility. J. Biol. Chem. 281, 14085–14091 10.1074/jbc.M511022200 [DOI] [PubMed] [Google Scholar]
- Wheatcroft S. B., Kearney M. T. (2009). IGF-dependent and IGF-independent actions of IGF-binding protein-1 and -2: implications for metabolic homeostasis. Trends Endocrinol. Metab. 20, 153–162 10.1016/j.tem.2009.01.002 [DOI] [PubMed] [Google Scholar]
- Williams R. M., Mcdonald A., O’Savage M., Dunger D. B. (2008). Mecasermin rinfabate: rhIGF-I/rhIGFBP-3 complex: iPLEX. Expert Opin. Drug Metab. Toxicol. 4, 311–324 10.1517/17425255.4.3.311 [DOI] [PubMed] [Google Scholar]
- Xu Q., Yan B., Li S., Duan C. (2004). Fibronectin binds insulin-like growth factor-binding protein 5 and abolishes Its ligand-dependent action on cell migration. J. Biol. Chem. 279, 4269–4277 10.1074/jbc.M406896200 [DOI] [PubMed] [Google Scholar]
- Yakar S., Rosen C. J., Beamer W. G., Ackert-Bicknell C. L., Wu Y., Liu J. L., Ooi G. T., Setser J., Frystyk J., Boisclair Y. R., Leroith D. (2002). Circulating levels of IGF-1 directly regulate bone growth and density. J. Clin. Invest. 110, 771–781 10.1172/JCI200215463 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yan X., Forbes B. E., McNeil K. A., Baxter R. C., Firth S. M. (2004). Role of N- and C-terminal residues of insulin-like growth factor (IGF)-binding protein-3 in regulating IGF complex formation and receptor activation. J. Biol. Chem. 279, 53232–53240 10.1074/jbc.M312810200 [DOI] [PubMed] [Google Scholar]
- Yan X., Payet L. D., Baxter R. C., Firth S. M. (2009). Activity of human pregnancy insulin-like growth factor binding protein-3: determination by reconstituting recombinant complexes. Endocrinology 150, 4968–4976 10.1210/en.2009-0090 [DOI] [PubMed] [Google Scholar]
- Zeslawski W., Beisel H. G., Kamionka M., Kalus W., Engh R. A., Huber R., Lang K., Holak T. A. (2001). The interaction of insulin-like growth factor-I with the N-terminal domain of IGFBP-5. EMBO J. 20, 3638–3644 10.1093/emboj/20.14.3638 [DOI] [PMC free article] [PubMed] [Google Scholar]