Abstract
Sex pheromone production in most moths is initiated following pheromone biosynthesis activating neuropeptide receptor (PBANR) activation. PBANR was initially cloned from pheromone glands (PGs) of Helicoverpa zea and Bombyx mori. The B. mori PBANR is characterized by a relatively long C-terminus that is essential for ligand-induced internalization, whereas the H. zea PBANR has a shorter C-terminus that lacks features present in the B. mori PBANR critical for internalization. Multiple PBANRs have been reported to be concurrently expressed in the larval CNS of Heliothis virescens. In the current study, we sought to examine the prevalence of multiple PBANRs in the PGs of three moths and to ascertain their potential functional relevance. Multiple PBANR variants (As, A, B, and C) were cloned from the PGs of all species examined with PBANR-C the most highly expressed. Alternative splicing of the C-terminal coding sequence of the PBAN gene gives rise to the variants, which are distinguishable only by the length and composition of their respective C-terminal tails. Transient expression of fluorescent PBANR chimeras in insect cells revealed that PBANR-B and PBANR-C localized exclusively to the cell surface while PBANR-As and PBANR-A exhibited varying degrees of cytosolic localization. Similarly, only the PBANR-B and PBANR-C variants underwent ligand-induced internalization. Taken together, our results suggest that PBANR-C is the principal receptor molecule involved in PBAN signaling regardless of moth species. The high GC content of the C-terminal coding sequence in the B and C variants, which makes amplification using conventional polymerases difficult, likely accounts for previous “preferential” amplification of PBANR-A like receptors from other species.
Keywords: PBAN, receptor, alternative splicing, pheromone gland, receptor internalization, confocal microscopy, GC-rich sequence
Introduction
In most moth species, a 33–34 aa neuropeptide termed pheromone biosynthesis activating neuropeptide (PBAN) regulates sex pheromone production and release. First isolated from Helicoverpa zea (Raina et al., 1989) and Bombyx mori (Kitamura et al., 1989), PBAN has subsequently been identified in a variety of species. PBAN is a member of the pyrokinin/PBAN family of peptides that are characterized by a FXPRLamide C-terminal pentapeptide motif. This motif is essential for biological activity, which, in addition to regulation of moth sex pheromone biosynthesis, includes melanization in lepidopteran larvae, induction of embryonic diapause in B. mori, and ecdysone biosynthesis in prothoracic glands of B. mori (Rafaeli, 2009). The role of PBAN in sex pheromone biosynthesis is governed by species-specifically defined photoperiods in which PBAN is released from the subesophageal ganglion into the hemolymph. Circulating PBAN acts directly on the modified epidermal cells of the eighth and ninth abdominal segment that comprise the pheromone gland (PG) via its cognate G protein-coupled receptor (GPCR). The pheromone biosynthesis activating neuropeptide receptor (PBANR) consequently plays a pivotal role in turning the extracellular PBAN signal into the biological response of sex pheromone production. Indeed, in vivo dsRNA-mediated knockdown of PBANR reduced sex pheromone production in the silkmoth (B. mori; Ohnishi et al., 2006) and negatively affected mating frequency in the diamondback moth (Plutella xylostella; Lee et al., 2011).
Pheromone biosynthesis activating neuropeptide receptor was initially cloned from PGs of the corn earworm, H. zea (Choi et al., 2003) and B. mori (Hull et al., 2004). While the two PBANRs share significant sequence similarity (82%), the B. mori PBANR (BommoPBANR) is structurally differentiated by a 67-aa C-terminal extension that is absent in the H. zea PBANR (HelzePBANR). BommoPBANR, like most GPCRs, undergoes ligand-induced internalization, a common endocytotic regulatory mechanism involved in GPCR desensitization (Moore et al., 2007; Marchese et al., 2008). Truncation of the BommoPBANR C-terminal extension prevented this internalization event (Hull et al., 2004). Further studies revealed that a 10 residue segment (Arg358–Gln367) of the BommoPBANR C-terminal extension is essential for internalization and that endocytosis was phosphorylation dependent, proceeded via clathrin-coated pits, and involved a YXXΦ motif (Hull et al., 2005).
Pheromone biosynthesis activating neuropeptide receptors that have C-terminal sequences more similar to HelzePBANR than to BommoPBANR have since been cloned from other moth species (Rafaeli et al., 2007; Zheng et al., 2007; Cheng et al., 2010; Lee et al., 2011). However, multiple PBANR subtypes have recently been identified in the tobacco budworm Heliothis virescens and the tobacco hornworm Manduca sexta (Kim et al., 2008). Similar to HelzePBANR, H. virescens PBANR (HelviPBANR)-A has a relatively short C-terminus, while HelviPBANR-C has an extended C-terminus that is ~78% identical to BommoPBANR and which contains the YXXΦ motif. Surprisingly, even though H. virescens and H. zea are closely related species, the HelviPBANR-C variant rather than HelviPBANR-A (the HelzePBANR ortholog) was preferentially amplified from H. virescens PGs (Kim et al., 2008). Sequence analyses suggest that the HelviPBANR subtypes arise from alternative splicing, a common transcriptional regulation event in GPCR genes (Minneman, 2001; Markovic and Challiss, 2009). Because alternative splicing can generate protein isoforms that are structurally, functionally, and/or spatially distinct, the presence of multiple PBANR variants raises questions regarding the functional role and relevance of the individual variants in regulating sex pheromone biosynthesis. To address these questions, we sought to examine and characterize PBANR variants from the PGs of multiple moth species.
Materials and Methods
Insects
Insects were maintained in a rearing chamber at 25°C under a 16L (light): 8D (dark) regime. Larvae of the inbred p50 strain of B. mori were reared on an artificial diet as described previously (Fónagy et al., 1992). Pupal age was determined based on morphological characteristics as described (Matsumoto et al., 2002). Larvae of Pseudaletia separata and Helicoverpa armigera were reared on an artificial diet (Insecta-LFS; Nihon Nosan Kogyo Ltd., Yokohama, Japan) under the same conditions as described (Fónagy et al., 2011). Pupae were sexed such that the newly emerged females were collected and kept separately in boxes and provided with a special sucrose solution energy drink (Pocari sweat®; Otsuka Pharmaceutical Co., Ltd, Tokyo, Japan). The newly emerged females were designated as day 0. H. zea pupae were purchased from Benzon Research (Carlisle, PA, USA) and maintained in a rearing chamber at 25°C under a 16L (light): 8D (dark) regime until adult emergence.
Degenerate PCR and RACE-based cloning of PBANR variants
Total RNA was isolated from PGs of B. mori (p50), P. separata, H. armigera, and H. zea with first strand cDNAs synthesized using a SMARTer™ RACE cDNA Amplification kit (Clontech, Palo Alto, CA, USA) according to the manufacturer’s instructions. Fragments of Pseudaletia separata PBANR (PsesePBANR) and Helicoverpa armigera PBANR (HelarPBANR) transcripts were amplified with Advantage 2 polymerase (Clontech, Palo Alto, CA, USA) using multiple combinations of degenerate oligonucleotide primers described previously (Hull et al., 2004). PCR products of the expected sizes were sub-cloned using a TOPO TA cloning kit (Invitrogen Co., Ltd., Tokyo, Japan) and sequenced. RACE was performed using cDNAs generated above with Advantage 2 polymerase in conjunction with adaptor specific primers (UPM and NUP) and gene-specific primers (Table 1; P1–P7). Thermacycler conditions consisted of 94°C for 2 min, followed by 35 cycles at 94°C for 30 s, 65°C for 30 s, and 68°C for 3 min, and a final extension at 68°C for 7 min. PCR products were sub-cloned as before and sequenced.
Table 1.
Species-specific PCR primers used to amplify the PBANR variants.
| Primer | Name | Sequence (5′-3′) |
|---|---|---|
| P1 | BommoPBANR3′F1 | CAGGAACGCTTTCAAGGTAAGATTAAACTAG |
| P2 | PsesePBANR3′F1 | CACACCATGTCGAAGCTGTCAAGAG |
| P3 | PsesePBANR3′F2 | GCATCTCCAACTCCAGTCTTCGCGAG |
| P4 | PsesePBANR5′R | CAACTTAATGCCTATCAACGCGTACAAC |
| P5 | HelarPBANR3′F1 | GCGATGCAGTTCGGTATAGTGTCGTAT |
| P6 | HelarPBANR3′F2 | GCATCTCCAACTCCAGTCTTCGCGAG |
| P7 | HelarPBANR5′R | GCATCTGACCAGGTCTTTCATTGCT |
| P8 | BommoPBANR | ATGATGGCAGATGAAACCGTCAAC |
| P9 | BommoPBANR-As-A | CCTTTAAGAGTTTCGTACTAGTTTAATCTTACC |
| P10 | BommoPBANR-B-C | TCCTAATGAAACCCACAACAGCTGAATC |
| P11 | PsesePBANR | ATGACCTTACCAGCGCCTCCGAGCATC |
| P12 | PsesePBANR-As-A | CAGTCAGCCGGCTGCCGGCCTGAAT |
| P13 | HelarPBANR | ATGACATTGTCAGCGCCCCCGAGCATCG |
| P14 | HelarPBANR-As-A | TTAATCATAGACTCTTACCTTAAAGGCGTTC |
| P15 | PsesePBANR-B-C | TCAGGTGAGTCCGCCGATGTTACAGTTC |
| P16 | Bommo-AF | CGTATTTTGCATCCCGTTAAGAAGCTG |
| P17 | Bommo-AR | CCTTTAAGAGTTTCGTACTAGTTTAATCTTACC |
| P18 | Bommo-BCF | TCGTTTTTCATCTGTTGGGCTCCATTT |
| P19 | Bommo-BR | ATCGCGATTTTGGTAGCACTGCG |
| P20 | Bommo-CR | CGCGATTTTGGTAGCACTCACCT |
| P21 | Psese-AF | TGATAGGCATTAAGTTGCGGACCTCTC |
| P22 | Psese-AR | GTTCCGCGAGGTAACAATACAAGTAG |
| P23 | Psese-BCF | TGCAGTATAGGAACGGAGCATCACA |
| P24 | Psese-BR | CGCGCCCGTTGTAGCACTGCG |
| P25 | Psese-CR | GCCCGTTGTAGCACTCACCTG |
| P26 | Helar-AF | CAGTGTTGTACGCGTTGATAGGCATT |
| P27 | Helar-AR | GTCGTGAGATGTAAGACAACAAGGAG |
| P28 | Helar-BCF | AGTCATCAGAATGCTCGTTGCAGTG |
| P29 | Helar-BR | CGCGCCCGTTGTAGCACTGCG |
| P30 | Helar-CR | GCCCGTTGTAGCACTCACCTG |
Specific amplification of Bommo, Psese, Helar, and Helze PBANR variants
To amplify the BommoPBANR variants, total RNA was isolated from 10 B. mori (p50) PGs immediately after adult eclosion with first strand cDNAs synthesized as before. BommoPBANR-As and BommoPBANR-A were amplified using Advantage 2 polymerase with a general BommoPBANR sense primer (Table 1; P8) and a short variant antisense primer (Table 1; P9), while BommoPBANR-B and BommoPBANR-C were amplified using the same sense primer and a long variant antisense primer (Table 1; P10). Thermacycler conditions consisted of 94°C for 2 min, followed by 35 cycles at 94°C for 30 s, 62°C for 30 s, and 68°C for 3 min with a final extension at 68°C for 7 min. PCR products were sub-cloned as before and sequenced. To amplify the PsesePBANR, HelarPBANR, and HelzePBANR variants, total RNA was isolated from 120 P. separate, 850 H. armigera, and 30 H. zea PGs dissected 1–3 days after adult eclosion. First strand cDNA was synthesized as before. PsesePBANR-As and PsesePBANR-A transcripts were amplified using Advantage 2 polymerase with a general PsesePBANR sense primer (Table 1; P11) and a short variant antisense primer (Table 1; P12). Thermacycler conditions consisted of 94°C for 2 min, followed by 35 cycles at 94°C for 30 s, 65°C for 30 s, and 68°C for 2 min with a final extension at 68°C for 7 min. HelarPBANR-As and HelarPBANR-A were amplified using a general HelarPBANR sense primer (Table 1; P13) and a short variant antisense primer (Table 1; P14). The resulting PCR products were sub-cloned and sequenced. Full-length PsesePBANR-B and PsesePBANR-C transcripts were amplified using a GC-optimized polymerase, KOD-FX (Toyobo, Osaka, Japan), with the previous PsesePBANR sense primer and a long variant-specific antisense primer (Table 1; P15) with thermacycler conditions consisting of 94°C for 2 min, followed by 32 cycles at 98°C for 10 s, 65°C for 30 s, and 68°C for 2 min with a final extension at 68°C for 7 min. HelarPBANR-B and HelarPBANR-C transcripts were similarly amplified using the HelarPBANR sense primer and the long variant-specific PsesePBANR antisense primer (Table 1; P15). Transcripts for the HelzePBANR variants were amplified using PCR conditions identical to those of the HelarPBANR variants. All resulting PCR products were sub-cloned and sequenced. The sequences reported in this paper have been deposited in the GenBank database (B. mori accession nos. JN228346–228349; H. armigera accession nos. JN228350–228353; P. separata accession nos. JN228354–JN228357; H. zea accession nos. JN206677 and JQ255024).
Primer check PCR using gene-specific primers
Primer check PCR was performed using 1 ng plasmid DNA containing the full-length sequence of each PBANR variant (PBANR-As, -A, -B, or -C) as template with primer sets designed for specific amplification of each PBANR sequence (Table 1; P16–P30). Primer check PCR using a general polymerase, Ex Taq DNA polymerase (Takara Bio Inc., Otsu, Japan), was performed using thermacycler conditions consisting of 94°C for 2 min, followed by 30 cycles at 94°C for 30 s and 68°C for 1 min. Primer check PCR using KOD-FX was performed using thermacycler conditions consisting of 94°C for 2 min, followed by 30 cycles at 98°C for 10 s and 68°C for 1 min. PCR products were analyzed on a 2% agarose gel and visualized with ethidium bromide.
RT-PCR tissue expression analysis
Total RNAs isolated using TRIzol reagent (Invitrogen) were prepared from various newly emerged adult B. mori female tissues or adult female P. separata and H. armigera tissues 3 days after adult eclosion. cDNAs were generated using a Onestep RT-PCR kit (Qiagen, Tokyo, Japan) with RT-PCR performed using HotStarTaq DNA polymerase (Qiagen) and primer sets designed for specific amplification of each PBANR sequence (Table 1; P16–P30). Thermacycler conditions consisted of reverse transcription for 30 min at 50°C, then 95°C for 15 min followed by 20–30 cycles at 94°C for 30 s, 62°C for 30 s, and 72°C for 1 min. PCR products were analyzed on a 2% agarose gel and visualized with ethidium bromide. A separate cDNA synthesis was carried out in parallel using 500 ng total RNA with an RNA PCR kit (Takara Bio Inc.,) according to the manufacturer’s instructions. RT-PCR with Ex Taq DNA polymerase was performed using thermacycler conditions consisting of 94°C for 2 min, followed by 26–32 cycles at 94°C for 30 s and 68°C for 1 min. RT-PCR using KOD-FX DNA polymerase was performed with thermacycler conditions consisting of 94°C for 2 min, followed by 26–32 cycles at 98°C for 10 s and 68°C for 1 min. PCR products were analyzed on a 2% agarose gel and visualized with ethidium bromide.
Sequence analysis
The guanine–cytosine (GC) content of the full-length cDNA sequences for the PBANR-As, -A, -B, and -C variants of B. mori, P. separata, and H. armigera was determined following alignment of the putative translation initiation sites. The average GC content was calculated using 100 base windows over a range of 700 bases. GC content distribution diagrams were generated using GENETYX-MAC Version 12.0.0 (Genetyx, Tokyo, Japan).
Construction of expression plasmids
Expression plasmids encoding the respective BommoPBANRs, PsesePBANRs, and HelarPBANRs fused at their respective C termini to enhanced green fluorescent protein (EGFP) were generated using a pIB/V5-His-TOPO TA Expression kit (Invitrogen). Chimeric genes were constructed via overlap extension PCR using plasmid DNAs with KOD-plus (Toyobo) and KOD-FX. Gene-specific primers described above were used with species-specific chimeric primers (Table 2) and an EGFP antisense primer. The resulting products were sub-cloned into the pIB/V5-His expression vector and sequenced to confirm presence and orientation of the insert.
Table 2.
Primer sets used for generating fluorescent PBANR chimeras.
| Name | Orientation | Sequence (5′-3′) |
|---|---|---|
| BommoPBANR-As–EGFP | Sense | CTGATACATTTTATCTGGTAATGGTGAGCAAGGGC |
| BommoPBANR-As–EGFP | Antisense | GCCCTTGCTCACCATTACCAGATAAAATGTATCAG |
| BommoPBANR-A–EGFP | Sense | TTTCAAGGTAAGATTAAACATGGTGAGCAAGGGC |
| BommoPBANR-A–EGFP | Antisense | GCCCTTGCTCACCATGTTTAATCTTACCTTGAAA |
| BommoPBANR-B–EGFP | Sense | TAATATAGAAGGACTTACCATGGTGAGCAAGGGC |
| BommoPBANR-B–EGFP | Antisense | GCCCTTGCTCACCATGGTAAGTCCTTCTATATTA |
| BommoPBANR-C–EGFP | Sense | ATCGCGATCTCTCCAATGGTGAGCAAGGGC |
| BommoPBANR-C–EGFP | Antisense | GCCCTTGCTCACCATTGGAGAGATCGCGAT |
| PsesePBANR-As–EGFP | Sense | CTTTTACCTGGTAAACTTGATGGTGAGCAAGGGC |
| PsesePBANR-As–EGFP | Antisense | GCCCTTGCTCACCATCAAGTTTACCAGGTAAAAG |
| PsesePBANR-A–EGFP | Sense | CGGCTTTGTTATTAATACTTATGGTGAGCAAGGGC |
| PsesePBANR-A–EGFP | Antisense | GCCCTTGCTCACCATAAGTATTAATAACAAAGCCG |
| PsesePBANR-B–EGFP | Sense | ATCGGCGGACTCACCATGGTGAGCAAGGGC |
| PsesePBANR-B–EGFP | Antisense | GCCCTTGCTCACCATGGTGAGTCCGCCGAT |
| PsesePBANR-C–EGFP | Sense | CCTACATGTACCACGATGGTGAGCAAGGGC |
| PsesePBANR-C–EGFP | Antisense | GCCCTTGCTCACCATCGTGGTACATGTAGG |
| HelarPBANR-As–EGFP | Sense | GGAATCTATAGATACATGGTGAGCAAGGGC |
| HelarPBANR-As–EGFP | Antisense | GCCCTTGCTCACCATGTATCTATAGATTCC |
| HelarPBANR-A–EGFP | Sense | GTAAGAGTCTATGATATGGTGAGCAAGGGC |
| HelarPBANR-A–EGFP | Antisense | GCCCTTGCTCACCATATCATAGACTCTTAC |
| HelarPBANR-B–EGFP | Sense | ATCGGCGGACTCACCATGGTGAGCAAGGGC |
| HelarPBANR-B–EGFP | Antisense | GCCCTTGCTCACCATGGTGAGTCCGCCGAT |
| HelarPBANR-C–EGFP | Sense | CCTACATGTACCACGATGGTGAGCAAGGGC |
| HelarPBANR-C–EGFP | Antisense | GCCCTTGCTCACCATCGTGGTACATGTAGG |
| EGFP | Antisense | TTACTTGTACAGCTCGTCCAT |
The EGFP portion of the chimeric oligonucleotides is underlined.
Preparation of a fluorescent PBAN analog
A fluorescent PBAN analog was prepared from a synthetic peptide corresponding to the terminal 10 residues of B. mori PBAN (i.e., SRTRYFSPRLamide) with Lys substitution of Arg2. The Lys residue was labeled with Rhodamine Red succinimidyl ester (Molecular Probes, Eugene, OR, USA) following overnight incubation in 0.1 M sodium bicarbonate (pH 8.2). The conjugated peptide, designated RR-C10PBANR2K, was purified by reversed-phase high-performance liquid chromatography on a Senshu Pak PEGASIL ODS column (10 mm × 150 mm; Senshu Scientific Co., Ltd., Tokyo, Japan) with absorbance monitored at 225 nm. RR-C10PBANR2K was stored at 4°C until needed.
Fluorescent confocal microscopy imaging
A monolayer of adherent Sf9 insect cells (Smith et al., 1985) was transfected with 1 μg plasmid DNA encoding the respective fluorescent PBANR chimeras and 8 μl Cellfectin II (Invitrogen) according to the manufacturer’s instructions. Ligand-induced internalization of the fluorescent PBANR chimeras was performed as described previously using a Leica TCS NT confocal system (Hull et al., 2004) but with 50 nM RR-C10PBANR2K. Fluorescence images were obtained. Images were processed using Photoshop 6.0 (Adobe Systems Inc., San Jose, CA, USA).
Northern blot analysis
Northern blots were performed using denatured total RNAs (1 μg of each) prepared from various B. mori, P. separata, and H. armigera tissues electrophoresed on a 1.0% gel in 2 M formaldehyde/1X MOPS buffer and then transferred to a nylon membrane (Hybond N+, Amersham Biosciences, Piscataway, NJ, USA) by capillary blotting. cDNA probes (BommoPBANR-A nt 1–1265, BommoPBANR-C nt 1591–2616, PsesePBANR-A nt 254–1378, PsesePBANR-C nt 1586–2608, HelarPBANR-A nt 242–1282 and HelarPBANR-C nt 1574–2597) were labeled with DIG (digoxigenin-11-UTP) using a DIG northern starter kit (Roche Applied Science, Indianapolis, IN, USA). Probe hybridization was performed at 68°C for 18 h at which point the blot was washed twice in an initial solution of 0.1% SDS/2 × SSC for 5 min at 22°C and then transferred to 0.1% SDS/0.1 × SSC for two 15 min washes at 68°C. Signals were detected using a LAS-3000 image analyzer (Fujifilm, Tokyo, Japan).
Results
Identification of multiple BommoPBANR variants
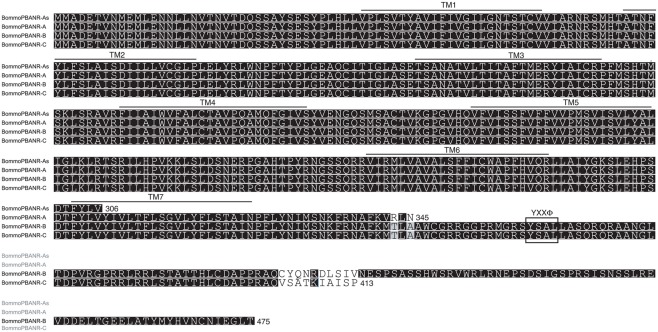
To determine if multiple PBANR variants are expressed in the B. mori PG, we PCR-screened our B. mori PG cDNA library (Yoshiga et al., 2000) using primers designed to BommoPBANR (AB181298). Sequencing multiple cDNA clones resulted in the identification of transcripts encoding the 413-aa BommoPBANR reported previously (Hull et al., 2004) and a new 475-aa protein (Figure 1). Despite large differences in the amino acid sequences of the respective C-terminal tails, the nucleotide sequence of the clones differed by five nucleotides. The larger cDNA was only present in ~10% of the clones sequenced, suggesting that it was poorly represented in our cDNA library and thus that its PG transcript levels are lower than the 413-aa variant. Because the 475-aa variant exhibited significant homology (82%) to HelviPBANR-B, we sought to present a uniform nomenclature system for the PBANR variants. Consequently, we designated the 475-aa variant as BommoPBANR-B and the 413-aa variant, which shares 81% sequence homology with HelviPBANR-C, as BommoPBANR-C.
Figure 1.
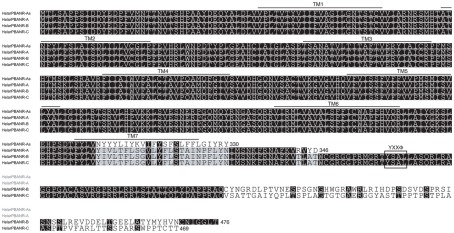
Multiple sequence alignment of B. mori PBANR variants. The seven predicted TM domains are over-lined. Location of the YXXΦ endosomal sorting motif is outlined. Alignment was performed using MAFFT v6.814b (Katoh et al., 2002) with certain gaps removed. Species abbreviations are per National Center for Biotechnology Information/Swiss Prot: Bommo, B. mori.
PCR primers designed from B. mori genomic data based on sequence similarity with HelviPBANR-A lead to the identification of cDNA clones encoding 345-aa and 306-aa proteins. Because the 345-aa variant is similar in size to HelviPBANR-A, we designated it BommoPBANR-A. The 306-aa variant, which has an incomplete seventh transmembrane domain (TM7), was designated BommoPBANR-As. The four BommoPBANR variants identified in these experiments are differentiated only by the size and sequence of their respective C-terminal tails (Figure 1). The sequence data for all four variants have been deposited with GenBank (JN228346–228349).
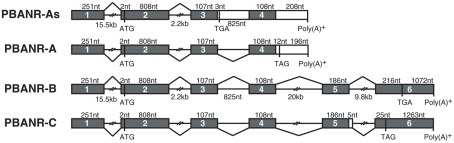
Genomic structure and alternative splicing of the BommoPBANR gene
Using the Silkworm Genome Research Program (http://sgp.dna.affrc.go.jp/KAIKObase/), we localized the BommoPBANR gene to scaffold Bm_scaf84 of chromosome 12 and determined that it is composed of six exons and five introns covering >50 kb of sequence (Figure 2). The majority of the BommoPBANR coding sequence (N-terminus through TM7) is carried on exons 2–4 while exons 5–6 encode the C-terminal tail and exon 1 the 5′ untranslated region. BommoPBANR-As and A arise from alternative splice events that retain introns 3 and 4 respectively. The inclusion of intron 3 introduces a premature stop codon (TGA) in BommoPBANR-As that truncates TM7 at residue 306, whereas the inclusion of intron 4 in BommoPBANR-A results in a premature stop codon (TAG) that truncates the protein at residue 345. BommoPBANR-C arises from an insertion of 5 nt at the 3′ end of exon 5 that shifts the reading frame of the last 10-aa (residues 404–413) and introduces a stop codon (TAG) that generates a C-terminal tail truncated 62-aa compared to BommoPBANR-B (Figure 1).
Figure 2.
Schematic diagram of the B. mori PBANR genomic structure and alternative splice sites. The four BommoPBANR variants (As, A, B, and C) are depicted. The filled boxes represent exons. Initiation (ATG) and stop sites (TGA or TAG) are indicated by their respective codons.
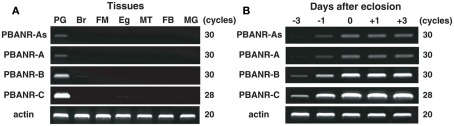
Expression analysis of BommoPBANR variants
RT-PCR analyses showed that all four PBANR variants are preferentially expressed within the PG (Figure 3A). Amplification of BommoPBANR-As and -A was barely detectable at cycle 30 while BommoPBANR-B generated a robust, but non-saturating, signal (Figure 3A). In contrast, BommoPBANR-C was saturating by PCR cycle 28, indicating that it is the most abundant PBANR transcript expressed in the PG. Amplimers corresponding to the PBANR-B and C variants were also amplified in other tissues, albeit at significantly lower levels (Figure 3A). The amplification of lower abundance PBANR transcripts in non-PG tissues is not without precedence (Rafaeli et al., 2007; Watanabe et al., 2007) and may either reflect a novel regulatory role for the receptor in those tissues or may be an indication of leaky transcription and thus are of questionable physiological relevance. Regardless, our results, and those of others, clearly demonstrate that PBANR expression in adult moths is predominantly PG-directed, which is congruous with its role in mediating sex pheromone production.
Figure 3.
Expression profile of BommoPBANR transcripts in adult female tissues. (A) Expression in adult tissues. Tissues were dissected from newly emerged (day 0) p50 female moths. PG, pheromone gland; Br, brain; FM, flight muscle; Eg, unfertilized egg; MT, Malpighian tubule; FB, fat body; MG, midgut. (B) Expression in the PG at different developmental stages. PGs were dissected from p50 female pupae at 3 days (−3) and 1 day (−1) before eclosion and from p50 adults at 0 day (0), 1 day (+1), and 3 days (+3) after eclosion. cDNAs were normalized to actin expression levels. The PCR cycle number is indicated to the right of the gels.
Consistent with our previous report, which described the developmental profile of the BommoPBANR-C transcript (Hull et al., 2004), all four BommoPBANR variants are up-regulated on the day preceding adult emergence (Figure 3B). Similar up-regulation has been observed for other genes crucial to B. mori pheromonogenesis, i.e., pgFAR, Bmpgdesat1, pgACBP, BmFATP, and BmLsd1 (Matsumoto et al., 2001; Moto et al., 2003, 2004; Ohnishi et al., 2009, 2011).
Identification of PBANR variants in Noctuid species
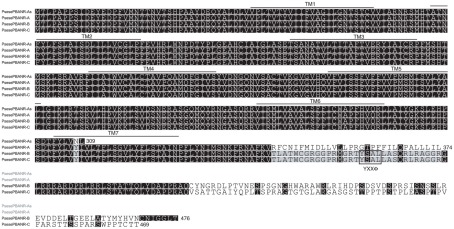
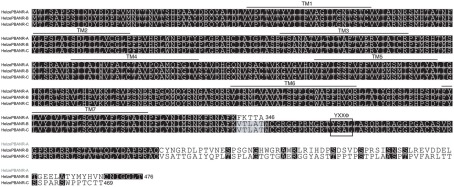
We next investigated the pervasiveness of PBANR splice variants in three other moths. In the Oriental armyworm, P. separata, we identified clones encoding 374-aa and 309-aa proteins with 98 and 82% identity to HelviPBANR-A and BommoPBANR-As respectively, which we designated PsesePBANR-A and PsesePBANR-As (Figure 4). Initial attempts to identify PsesePBANR-B and PsesePBANR-C variants via 3′-RACE using conventional polymerases were unsuccessful. Because conventional polymerases often fail to amplify GC-rich targets, we switched to a commercially available GC-optimized polymerase to identify the 469-aa PsesePBANR-C and the 476-aa PsesePBANR-B (Figure 4). As before, sequencing of multiple clones suggested that PsesePBANR-C was the predominantly expressed variant. Applying the same cloning strategy to females of the cotton bollworm, H. armigera, we isolated four PBANR variants from a PG-specific cDNA library: a 330-aa protein designated HelarPBANR-As, a 346-aa protein designated HelarPBANR-A, a 476-aa protein designated HelarPBANR-B, and a 469-aa protein designated HelarPBANR-C (Figure 5). HelarPBANR-A is 99% identical to the deposited HelarPBANR (AY792036) with nucleotide sequence variations largely restricted to the third codon position; the lone exception was Leu substitution of Trp118 (Figure 5). The sequence data for the H. armigera and P. separata PBANR variants have been deposited with GenBank (H. armigera – JN228350–228353 and P. separata – JN228354–JN228357).
Figure 4.
Multiple sequence alignment of the P. separata PBANR variants. The seven predicted TM domains are over-lined. Location of the YXXΦ endosomal sorting motif is outlined. Alignment was performed using MAFFT v6.814b (Katoh et al., 2002) with certain gaps removed. Species abbreviations are per National Center for Biotechnology Information/Swiss Prot: Psese, Pseudaleti separata.
Figure 5.
Multiple sequence alignment of the H. armigera PBANR variants. The seven predicted TM domains are over-lined. Location of the YXXΦ endosomal sorting motif is outlined. Alignment was performed using MAFFT v6.814b (Katoh et al., 2002) with certain gaps removed. Species abbreviations are per National Center for Biotechnology Information/Swiss Prot: Helar, Helicoverpa armigera.
Because the only PBANR identified in H. zea to date corresponds to the PBANR-A variant, we sought to determine if the longer PBANR-B and PBANR-C variants are also expressed in the PG of this species. Using RT-PCR methods as above, we identified H. zea homologs of both variants (Figure 6). HelzePBANR-B (JQ255024) is a 476-aa protein with 99% sequence identity to HelviPBANR-B while HelzePBANR-C (JN206677) is a 469-aa protein with 98% sequence identity to HelviPBANR-C. Based on our results and those of Kim et al. (2008), the concurrent expression of multiple PBANR variants that arise from alternative splicing of the 3′ coding region appears to be extremely prevalent, if not absolute, across moth species.
Figure 6.
Multiple sequence alignment of the H. zea PBANR variants. The seven predicted TM domains are over-lined. Location of the YXXΦ endosomal sorting motif is outlined. Alignment was performed using MAFFT v6.814b (Katoh et al., 2002) with certain gaps removed. Species abbreviations are per National Center for Biotechnology Information/Swiss Prot: Helze, Helicoverpa zea.
RT-PCR expression analyses of PBANR variants
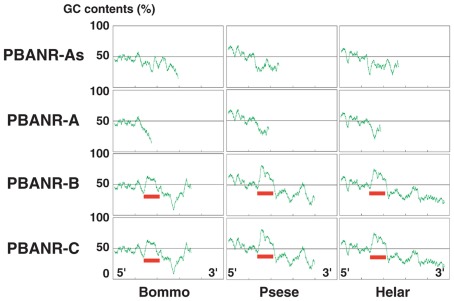
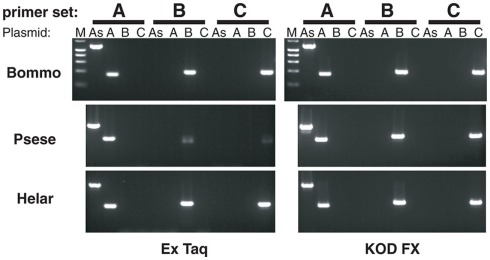
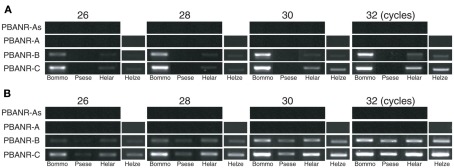
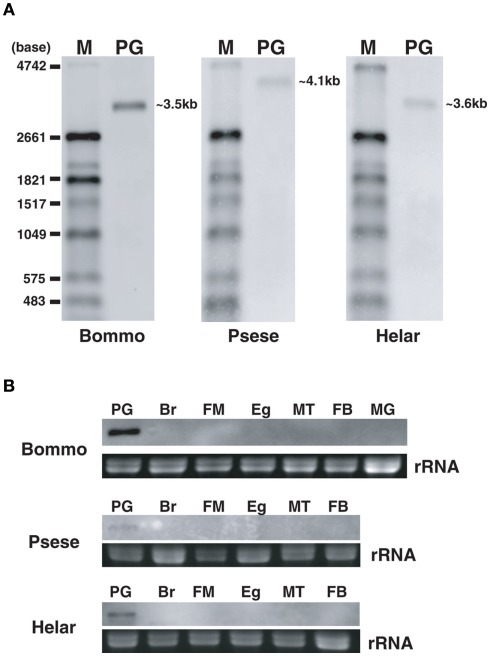
Sequence analyses revealed that, regardless of moth species, the ~300-nt coding sequence unique to the PBANR-B and -C variants (Figure 7, red bars) is GC-rich. In P. separata and H. armigera, the GC content of this region ranges from 65–80% while that of the B. mori gene is 55–60%. The C-terminal coding sequence of the PBANR-As and -A variants, in contrast, is more AT-rich (Figure 7). In preliminary experiments, we confirmed that species-specific primer sets capable of amplifying both PBANR-A and -As (Figure 8, primer set A) generated distinct amplimers of the expected sizes for each transcript from each species assayed. We also found that while the PBANR-B and -C variants were poorly amplified from P. separata with a conventional polymerase, both were robustly amplified with a commercially available GC-optimized polymerase (Figure 8). Based on these results, we performed RT-PCR using PG cDNAs with both the GC-optimized polymerase as well as the conventional DNA polymerase (Figure 9). In all moth species, we observed higher expression levels for PBANR-C than either of the shorter variants. To further confirm the expression levels of PBANR isoforms in the three moth species, we performed Northern blots using total RNA prepared from PGs during pheromonogenesis. Using probes that recognize the “short” (PBANR-As and A) and “long” (PBANR-B and C) variants, we detected a single band corresponding to the “long” variant transcript of 3.4–4.1 kb in the PG of each species (Figure 10A). No bands corresponding to the “short” variant transcript were detected. Furthermore, Northern blots using total RNAs prepared from various female tissues during this same time period confirmed PG-specific expression of the long variants as single bands were only detected in PGs (Figure 10B).
Figure 7.
Guanine–cytosine content distribution diagrams of PBANR variants. The distribution of GC content in full-length cDNA sequences for the PBANR variants of B. mori, P. separata, and H. armigera was calculated using 100 base windows over a range of 700 bases. Red bars indicate the ~300-nt coding sequence unique to the PBANR-B and -C variants.
Figure 8.
Amplification of PBANR variants using species- and variant-specific primers. Amplification efficiencies of the primers were determined using plasmids containing the full-length sequence of each PBANR variant (PBANR-As -A, -B, or -C). Specific primer sets used are listed in Table 1. Amplification was performed for 30 cycles using either a conventional polymerase (Ex Taq) or a GC-optimized polymerase (KOD-FX). Species abbreviations are per National Center for Biotechnology Information/Swiss Prot: Bommo, B. mori; Psese, P. separata, and Helar, H. armigera.
Figure 9.
RT-PCR analysis of PG-derived PBANR variants. RT-PCR was performed using B. mori, P. separata, H. armigera, and H. zea PG cDNAs with either (A) a conventional DNA polymerase (Ex Taq) or (B) a GC-optimized DNA polymerase (KOD-FX). Amplifications were performed using 26–32 PCR cycles as indicated.
Figure 10.
Northern blot analyses of PBANR variants in adult female tissues. (A) PG. (B) Adult tissues. Using two probes (BommoPBANR-A: 1–1265, BommoPBANR-C: 1591–2616, PsesePBANR-A: 254–1378, PsesePBANR-C: 1586–2608, HelarPBANR-A: 242–1282 and HelarPBANR-C: 1574–2597) that recognize either the “short” PBANR variants or the “long” variants. A single 3.4–4.1 kb band corresponding to the “long” (i. e., PBANR-B and -C) transcripts is detectable only in the PG for each species. PG, pheromone gland; Br, brain; FM, flight muscle; Eg, unfertilized egg; MT, malpighian tubule; FB, fat body; MG, midgut.
Cell surface localization and functional activation of the PBANR variants
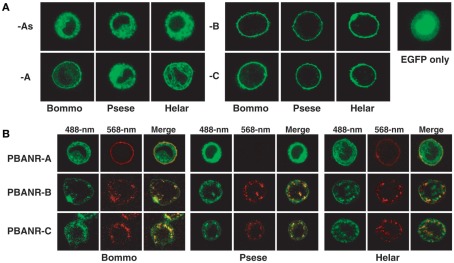
To confirm the functionality of the PBANR variants, cultured Sf9 insect cells were transfected with plasmid DNAs encoding fluorescent chimeras of the B. mori, P. separata, and H. armigera PBANR variants in which EGFP was fused in frame to the receptor C-terminus. Cell surface localization of each variant was confirmed using laser confocal microscopy (Figure 11). Regardless of moth species, the PBANR-B and -C variants localized exclusively to the plasma membrane (Figure 11A). The BommoPBANR-A and HelarPBANR-A variants exhibited varying degrees of intracellular localization with some cell surface localization (Figure 11A). The PsesePBANR-A variant, in contrast, was completely intracellular (Figure 11A), as were all of the PBANR-As variants (Figure 11A), suggesting that an intact TM7 is necessary for cell surface trafficking and localization.
Figure 11.
Confocal imaging of fluorescent chimeras of the PBANR variants transiently expressed in Sf9 cells. (A) Localization of EGFP-tagged PBANR variants. For control purposes, Sf9 cells were transfected with an expression plasmid containing EGFP. (B) Ligand-induced internalization of EGFP-tagged PBANR variants. Internalization was triggered with 50 nM RR-C10PBANR2K. Co-localization is indicated by yellow in the merged images.
Ligand-induced receptor internalization is a key element in regulating the strength and duration of receptor-mediated cell signaling. We have previously shown that PBAN induces BommoPBANR-C (formerly referred to as BomPBANR) internalization (Hull et al., 2004, 2005, 2011). We consequently sought to determine if the other PBANR variants are regulated in a similar fashion. Cells transiently expressing the fluorescent PBANR variant chimeras were incubated with a Rhodamine Red-labeled PBAN derivative (RR-C10PBANR2K). RR-C10PBANR2K induced internalization of the PBANR-C and -B variants from each species as evidenced by the intracellular accumulation of red fluorescent vesicles that co-localized with the EGFP chimera-derived green fluorescent signals (Figure 11B). In contrast, despite clear cell surface binding, RR-C10PBANR2K failed to induce internalization of BommoPBANR-A and HelarPBANR-A (Figure 11B). In addition, consistent with a lack of cell surface localization, no RR-C10PBANR2K binding was observed in cells expressing PsesePBANR-A (Figure 11B).
Discussion
Following ligand binding, receptor trafficking is crucial for the temporal and spatial control of GPCR signaling. The magnitude and duration of GPCR signaling is tightly regulated by mechanisms that terminate initial signaling and prime the cells to respond to new ligand exposure. Consequently, endocytosis of GPCRs from the cell surface allows for the fine-tuning of signal magnitude and duration by playing a crucial role in signal desensitization, re-sensitization, and down-regulation (Moore et al., 2007; Marchese et al., 2008). Initially, our understanding of the molecular mechanisms underlying PBANR signaling was restricted to the PBANRs identified in H. zea and B. mori (Choi et al., 2003; Hull et al., 2004). These receptors, however, are structurally differentiated by a 67-aa C-terminal extension in BommoPBANR that is essential for receptor internalization (Hull et al., 2004). Given the significance of GPCR endocytotic trafficking in signal termination, it seems clear that the C-terminal extension present in BommoPBANR plays an essential role in regulating the duration of the PBAN signal. This conclusion, however, raises questions regarding PBANR signaling in general given that HelzePBANR lacks this domain. The functional significance of the two receptor subtypes was initially ascribed to differences in signal transduction cascades activated (H. zea: cAMP dependent vs. B. mori: cAMP independent). However, it was recently reported that multiple PBANRs, two of which have extended C-terminal tails and YXXΦ endosomal sorting motifs similar to BommoPBANR, are concurrently expressed in the H. virescens larval CNS (Kim et al., 2008). The presence of multiple PBANR transcripts that are concomitantly expressed and that differ only in the length of their C-terminal loop suggests that our understanding of PBAN signaling may be more incomplete than previously thought and raises questions regarding their physiological role.
In the current study, we sought to assess the prevalence of PBANR variants in the PGs of various moth species, and found that multiple variants were expressed in the PGs of every moth examined (Figures 1, 4–6). Genomic structure analysis of the B. mori gene showed that the PBANR variants arise from alternative splicing at the 3′-end of the receptor gene on chromosome 12 (Figure 2). Taken together, our findings and those of Kim et al. (2008) provide clear evidence that the PBANR gene in moths undergoes a number of alternative splice events involving the 3′-portion of the gene and suggest that expression of multiple PBANR variants in moth PGs is pervasive.
To determine the potential functionality of the PBANR variants, we expressed EGFP-tagged PBANR variants in Sf9 cells and examined their localization and ability to undergo ligand-induced internalization (Figure 11). The PBANR-As from all the species studied failed to localize to the plasma membrane, a functional deficiency likely attributable to the incomplete TM7 (Markovic and Challiss, 2009). Even though the PBANR-A variant of H. armigera and B. mori partially localized to the plasma membrane and specifically bound RR-C10PBANR2K, there was no indication of ligand-induced internalization (Figure 11B). In contrast, all of the PBANR-B and -C variants localized to the plasma membrane and exhibited ligand-induced internalization (Figure 11B). Because internalization is dependent on an active signaling cascade (Hull et al., 2005, 2011), we concluded that the PBANR-A variant is either functionally deficient or has distinct intracellular signaling properties. The lack of an internalization response in cells expressing the PBANR-A variants, however, was not completely unexpected as C-terminal truncations lacking the YXXΦ motif, which is present in the PBANR-B and -C variants but not the PBANR-A variants, exhibited drastically different internalization kinetics (Hull et al., 2005). Interestingly, while capable of mobilizing extracellular Ca2+ in response to PBAN, the initial HelzePBANR variant identified by Choi et al. (2003) also lacks this motif. This suggests that functional differences between the H. zea receptor and the B. mori receptor likely extend beyond the C-terminus. Notable sequence variations between BommoPBANR-A and HelzePBANR-A include the last four to five residues (BommoPBANR-A: VRLN vs. HelzePBANR: FKTTA) of the respective C-terminal tails and a four residue span in the third intracellular loop (BommoPBANR-A: AHTP vs. HelzePBARN: QMQ). What role, if any, these residues may play in PBANR signal transduction remains to be determined.
In the course of our efforts to clone transcripts encoding PBANR variants, we found that conventional 3′-RACE PCR failed to amplify cDNAs for the PBANR-B and -C variants. In addition, RT-PCR using gene-specific primers with conventional polymerases likewise failed to amplify the cDNAs encoding PsesePBANR-B and -C. In contrast, these transcripts were successfully amplified with a GC-optimized polymerase (Figure 9). Regardless of moth species, the coding sequence unique to the PBANR-B and -C variants is GC abundant, while the C-terminal coding sequence of the PBANR-As and -A variants is AT-rich (Figure 7). We speculate that this may be the reason why transcripts encoding PBANR-A have been so frequently amplified (Rafaeli et al., 2007; Zheng et al., 2007; Cheng et al., 2010; Lee et al., 2011) despite the higher expression levels of PBANR-B and -C (see Figure 9). As a consequence, we believe that conclusions drawn regarding the in vivo functional role of the PBANR-A variant should be reconsidered within the context of coincident PBANR-B and -C expression.
While evidence for the four PBANR splice variants is currently limited to the transcript level, their conservation across multiple species, in conjunction with similar observations of conserved splicing patterns in mammalian receptors (Markovic and Grammatopoulos, 2009), suggests that the variants are likely physiological relevant. Concomitant expression of the multiple variants within a single cell type could represent a fine-tuning mechanism for cellular responsiveness to the extracellular signal. In one scenario, a non-responsive receptor (e.g., BommoPBANR-A) expressed at the cell surface might function as a decoy receptor that competes with the wildtype receptor (e.g., BommoPBANR-C) for ligand binding. The net result would be less bioactive peptide available to trigger the cellular response thus decreasing overall sensitivity. Alternatively, heterodimerization of the short receptors (e.g., BommoPBANR-A and/or -As) with the longer wildtype receptors (e.g., BommoPBANR-C or-B) could trap the active receptors within the secretory pathway, thereby decreasing the pool of available receptor for ligand binding and effectively decreasing overall cellular sensitivity. Truncated variants of the calcitonin receptor, corticotropin releasing factor receptor type 1, and, more recently, growth hormone secretagogue receptor type 1a have all been shown to exhibit dominant negative effects on signaling when co-expressed with wildtype variants (Seck et al., 2005; Zmijewski and Slominski, 2009; Chow et al., 2012). Receptor variants have also been shown to be functionally distinct with respect to spatial and temporal expression, ligand binding, regulation, and downstream effector pathways (Markovic and Challiss, 2009). A naturally occurring variant of the neurokinin 1 receptor that essentially lacks a C-terminus has been shown to mediate intracellular signaling mechanisms distinct from those initiated by the full-length receptor including: an inability to mobilize extracellular Ca2+, decreased protein kinase δ phosphorylation, slowed activation of the extracellular signal-regulated kinase (ERK) pathway, and decreased desensitization and internalization (DeFea et al., 2000; Lai et al., 2008). A similar differentiation in signaling pathways activated downstream of ligand binding has been observed for the PBANR variants. When expressed in mammalian CHO-WTA11 cells, only the HelviPBANR-C variant generated a robust Ca2+ signal in response to PBAN, little to no Ca2+ mobilization was observed in cells expressing the HelviPBANR-A or -B variants (Kim et al., 2008). Furthermore, given the pleiotropic nature of PBAN and PBAN-like peptides (Rafaeli, 2009), it is likely that PBANR variants expressed in other tissues/developmental stages may govern tissue-dependent physiological functions. Indeed, the PBANR-A variant in Spodoptera littoralis was cloned from fifth instar larvae as part of an effort to identify the receptor responsible for cuticular pigmentation, a physiological effect mediated by the PBAN family of peptides (Zheng et al., 2007).
In conclusion, our results show that PBANR-C is the predominant variant expressed in the PG during pheromonogenesis regardless of moth species, and, given the functional significance of the C-terminus (Hull et al., 2004, 2005, 2011), strongly suggest that this variant is the principal receptor molecule involved in PBAN signaling. This conclusion is supported by Northern blot analyses (Figure 10) as well as specific photoaffinity-based binding of biotin-labeled PBAN to a membrane-bound protein of ~50 kDa in H. armigera PGs (Rafaeli et al., 2003), which is more consistent with the molecular weight of HelarPBANR-C (51.1 kDa) than HelarPBANR-A (38.7 kDa). Furthermore, preferential amplification of the PBANR-C variant was also demonstrated in H. virescens PGs (Kim et al., 2008). Lastly, in contrast to PBANR-C, the shorter PBANR-A variant localizes poorly at the cell surface and fails to undergo typical ligand-induced internalization (Figure 11), which suggests that desensitization of this variant either does not occur, or at the very least that its regulation is mediated by a distinct intracellular pathway.
Conflict of Interest Statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Acknowledgments
This work was supported by the Lipid Dynamics Research Program from RIKEN and the Targeted Proteins Research Program (TPRP) from the Ministry of Education, Culture, Sports, Science, and Technology of Japan.
References
- Cheng Y., Luo L., Jiang X., Zhang L., Niu C. (2010). Expression of pheromone biosynthesis activating neuropeptide and its receptor (PBANR) mRNA in adult female Spodoptera exigua (Lepidoptera: Noctuidae). Arch. Insect Biochem. Physiol. 75, 13–27 10.1002/arch.20379 [DOI] [PubMed] [Google Scholar]
- Choi M. Y., Fuerst E. J., Rafaeli A., Jurenka R. (2003). Identification of a G protein-coupled receptor for pheromone biosynthesis activating neuropeptide from pheromone glands of the moth Helicoverpa zea. Proc. Natl. Acad. Sci. U.S.A. 100, 9721–9726 10.1073/pnas.1632485100 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chow K. B. S., Sun J., Chu K. M., Cheung W. T., Cheng C. H. K., Wise H. (2012). The truncated ghrelin receptor polypeptide (GHS-R1b) is localized in the endoplasmic reticulum where it forms heterodimers with ghrelin receptors (GHS-R1a) to attenuate their cell surface expression. Mol. Cell. Endocrinol. 348, 247–254 10.1016/j.mce.2011.08.034 [DOI] [PubMed] [Google Scholar]
- DeFea K. A., Vaughn Z. D., O’Bryan E. M., Nishijima D., Déry O., Bunnett N. W. (2000). The proliferative and antiapoptotic effects of substance P are facilitated by formation of a β-arrestin-dependent scaffolding complex. Proc. Natl. Acad. Sci. U.S.A. 97, 11086–11091 10.1073/pnas.190276697 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fónagy A., Matsumoto S., Uchiumi K., Orikasa C., Mitsui T. (1992). Action of pheromone biosynthesis activating neuropeptide on pheromone glands of Bombyx mori and Spodoptera litura. J. Pest Sci. 17, 47–54 10.1584/jpestics.17.47 [DOI] [Google Scholar]
- Fónagy A., Moto K., Ohnishi A., Kurihara M., Kis J., Matsumoto S. (2011). Studies of sex pheromone production under neuroendocrine control by analytical and morphological means in the oriental armyworm, Pseudaletia separata, Walker (Lepidoptera: Noctuidae). Gen. Comp. Endocrinol. 172, 62–76 10.1016/j.ygcen.2011.02.018 [DOI] [PubMed] [Google Scholar]
- Hull J. J., Lee J. M., Matsumoto S. (2011). Identification of specific sites in the third intracellular loop and carboxyl terminus of the Bombyx mori pheromone biosynthesis activating neuropeptide receptor crucial for ligand-induced internalization. Insect Mol. Biol. 6, 801–811 10.1111/j.1365-2583.2011.01110.x [DOI] [PubMed] [Google Scholar]
- Hull J. J., Ohnishi A., Matsumoto S. (2005). Regulatory mechanisms underlying pheromone biosynthesis activating neuropeptide (PBAN)-induced internalization of the Bombyx mori PBAN receptor. Biochem. Biophys. Res. Commun. 334, 69–78 10.1016/j.bbrc.2005.06.050 [DOI] [PubMed] [Google Scholar]
- Hull J. J., Ohnishi A., Moto K., Kawasaki Y., Kurata R., Suzuki M. G., Matsumoto S. (2004). Cloning and characterization of the pheromone biosynthesis activating neuropeptide receptor from the silkmoth, Bombyx mori: significance of the carboxyl terminus in receptor internalization. J. Biol. Chem. 279, 51500–51507 10.1074/jbc.M408142200 [DOI] [PubMed] [Google Scholar]
- Katoh K., Misawa K., Kuma K., Miyata T. (2002). MAFFT: a novel method for rapid multiple sequence alignment based on fast Fourier transform. Nucleic Acids Res. 30, 3059–3066 10.1093/nar/gkf436 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim Y. J., Nachman R. J., Aimanova K., Gill S., Adams M. E. (2008). The pheromone biosynthesis activating neuropeptide (PBAN) receptor of Heliothis virescens: identification, functional expression, and structure-activity relationships of ligand analogs. Peptides 29, 268–275 10.1016/j.peptides.2008.01.002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kitamura A., Nagasawa H., Kataoka H., Inoue T., Matsumoto S., Ando T., Suzuki A. (1989). Amino acid sequence of pheromone-biosynthesis-activating neuropeptide (PBAN) of the silkworm, Bombyx mori. Biochem. Biophys. Res. Commun. 163, 520–526 10.1016/0006-291X(89)92168-2 [DOI] [PubMed] [Google Scholar]
- Lai J.-P., Lai S., Tuluc F., Tansky M. F., Kilpatrick L. E., Leeman S. E., Douglas S. D. (2008). Differences in the length of the carboxyl terminus mediate functional properties of neurokinin-1 receptor. Proc. Natl. Acad. Sci. U.S.A. 105, 12605–12610 10.1073/pnas.0806632105 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee D. W., Shrestha S., Kim A. Y., Park S. J., Yang C. Y., Kim Y., Koh Y. H. (2011). RNA interference of pheromone biosynthesis-activating neuropeptide receptor suppresses mating behavior by inhibiting sex pheromone production in Plutella xylostella (L.). Insect Biochem. Mol. Biol. 41, 236–243 10.1016/j.ibmb.2011.01.001 [DOI] [PubMed] [Google Scholar]
- Marchese A., Paing M. M., Temple B. R., Trejo J. (2008). G protein-coupled receptor sorting to endosomes and lysosomes. Annu. Rev. Pharmacol. Toxicol. 48, 601–629 10.1146/annurev.pharmtox.48.113006.094646 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Markovic D., Challiss R. A. (2009). Alternative splicing of G protein-coupled receptors: physiology and pathophysiology. Cell. Mol. Life Sci. 66, 3337–3352 10.1007/s00018-009-0093-4 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Markovic D., Grammatopoulos D. K. (2009). Focus on the splicing of secretin GPCRs transmembrane-domain 7. Trends Biochem. Sci. 34, 443–452 10.1016/j.tibs.2009.06.002 [DOI] [PubMed] [Google Scholar]
- Matsumoto S., Fónagy A., Yamamoto M., Wang F., Yokoyama N., Esumi Y., Suzuki Y. (2002). Chemical characterization of cytoplasmic lipid droplets in the pheromone-producing cells of the silkmoth, Bombyx mori. Insect Biochem. Mol. Biol. 32, 1447–1455 10.1016/S0965-1748(02)00065-6 [DOI] [PubMed] [Google Scholar]
- Matsumoto S., Yoshiga T., Yokoyama N., Iwanaga M., Koshiba S., Kigawa T., Hirota H., Yokoyama S., Okano K., Mita K., Shimada T., Tatsuki S. (2001). Characterization of acyl-CoA-binding protein (ACBP) in the pheromone gland of the silkworm, Bombyx mori. Insect Biochem. Mol. Biol. 31, 603–609 10.1016/S0965-1748(00)00165-X [DOI] [PubMed] [Google Scholar]
- Minneman K. P. (2001). Splice variants of G protein-coupled receptors. Mol. Interv. 1, 108–116 [PubMed] [Google Scholar]
- Moore C. A., Milano S. K., Benovic J. L. (2007). Regulation of receptor trafficking by GRKs and arrestins. Annu. Rev. Physiol. 69, 451–482 10.1146/annurev.physiol.69.022405.154712 [DOI] [PubMed] [Google Scholar]
- Moto K., Suzuki M. G., Hull J. J., Kurata R., Takahashi S., Yamamoto M., Okano K., Imai K., Ando T., Matsumoto S. (2004). Involvement of a bifunctional fatty-acyl desaturase in the biosynthesis of the silkmoth, Bombyx mori, sex pheromone. Proc. Natl. Acad. Sci. U.S.A. 101, 8631–8636 10.1073/pnas.0402056101 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moto K., Yoshiga T., Yamamato M., Takahashi S., Okano K., Ando T., Nakata T., Matsumoto S. (2003). Pheromone gland-specific fatty-acyl reductase of the silkmoth, Bombyx mori. Proc. Natl. Acad. Sci. U.S.A. 100, 9156–9161 10.1073/pnas.1531993100 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ohnishi A., Hashimoto K., Imai K., Matsumoto S. (2009). Functional characterization of the Bombyx mori fatty acid transport protein (BmFATP) within the silkmoth pheromone gland. J. Biol. Chem. 284, 5128–5136 10.1074/jbc.M806072200 [DOI] [PubMed] [Google Scholar]
- Ohnishi A., Hull J. J., Kaji M., Hashimoto K., Lee J. M., Tsuneizumi K., Suzuki T., Dohmae N., Matsumoto S. (2011). Hormone signaling linked to silkmoth sex pheromone biosynthesis involves Ca2+/Calmodulin-dependent protein kinase II-mediated phosphorylation of the insect PAT family protein Bombyx mori lipid storage droplet protein-1 (BmLsd1). J. Biol. Chem. 286, 24101–24112 10.1074/jbc.M111.250555 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ohnishi A., Hull J. J., Matsumoto S. (2006). Targeted disruption of genes in the Bombyx mori sex pheromone biosynthetic pathway. Proc. Natl. Acad. Sci. U.S.A. 103, 4398–4403 10.1073/pnas.0511270103 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rafaeli A. (2009). Pheromone biosynthesis activating neuropeptide (PBAN): regulatory role and mode of action. Gen. Comp. Endocrinol. 162, 69–78 10.1016/j.ygcen.2008.04.004 [DOI] [PubMed] [Google Scholar]
- Rafaeli A., Bober R., Becker L., Choi M. Y., Fuerst E. J., Jurenka R. (2007). Spatial distribution and differential expression of the PBAN receptor in tissues of adult Helicoverpa spp. (Lepidoptera: Noctuidae). Insect Mol. Biol. 16, 287–293 10.1111/j.1365-2583.2007.00725.x [DOI] [PubMed] [Google Scholar]
- Rafaeli A., Zakharova T., Lapsker Z., Jurenka R. A. (2003). The identification of an age- and female-specific putative PBAN membrane-receptor protein in pheromone glands of Helicoverpa armigera: possible up-regulation by juvenile hormone. Insect Biochem. Mol. Biol. 33, 371–380 10.1016/S0965-1748(02)00264-3 [DOI] [PubMed] [Google Scholar]
- Raina A. K., Jaffe H., Kempe T. G., Keim P., Bacher R. W., Fales H. M., Riley C. T., Klun J. A., Ridgway R. L., Hayes D. K. (1989). Identification of a neuropeptide hormone that regulates sex pheromone production in female moths. Science 244, 796–798 10.1126/science.244.4906.796 [DOI] [PubMed] [Google Scholar]
- Seck T., Pellegrini M., Florea A. M., Grignoux V., Baron R., Mierke D. F., Horne W. C. (2005). The δe13 isoform of the calcitonin receptor forms a six-transmembrane domain receptor with dominant-negative effects on receptor surface expression and signaling. Mol. Endocrinol. 19, 2132–2144 10.1210/me.2004-0472 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith G. E., Ju G., Ericson B. L., Moschera J., Lahm H. W., Chizzonite R., Summers M. D. (1985). Modification and secretion of human interleukin 2 produced in insect cells by a baculovirus expression vector. Proc. Natl. Acad. Sci. U.S.A. 82, 8404–8408 10.1073/pnas.82.5.1555 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Watanabe K., Hull J. J., Niimi T., Imai K., Matsumoto S., Yaginuma T., Kataoka H. (2007). FXPRL-amide peptides induce ecdysteroidogenesis through a G-protein coupled receptor expressed in the prothoracic gland of Bombyx mori. Mol. Cell. Endocrinol. 273, 51–58 10.1016/j.mce.2007.05.008 [DOI] [PubMed] [Google Scholar]
- Yoshiga T., Okano K., Mita K., Shimada T., Matsumoto S. (2000). cDNA cloning of acyl-CoA desaturase homologs in the silkworm, Bombyx mori. Gene 246, 339–345 10.1016/S0378-1119(00)00047-0 [DOI] [PubMed] [Google Scholar]
- Zheng L., Lytle C., Njauw C. N., Altstein M., Martins-Green M. (2007). Cloning and characterization of the pheromone biosynthesis activating neuropeptide receptor gene in Spodoptera littoralis larvae. Gene 393, 20–30 10.1016/j.gene.2006.12.025 [DOI] [PubMed] [Google Scholar]
- Zmijewski M. A., Slominski A. T. (2009). CRF1 receptor splicing in epidermal keratinocytes: potential biological role and environmental regulations. J. Cell. Physiol. 218, 593–602 10.1002/jcp.21632 [DOI] [PMC free article] [PubMed] [Google Scholar]