Abstract
The most recently discovered PTEN tumor suppressor gene has been found to be defective in a large number of human cancers. In addition, germ-line mutations in PTEN result in the dominantly inherited disease Cowden syndrome, which is characterized by multiple hamartomas and a high proclivity for developing cancer. A series of publications over the past year now suggest a mechanism by which PTEN loss of function results in tumors. PTEN appears to negatively control the phosphoinositide 3-kinase signaling pathway for regulation of cell growth and survival by dephosphorylating the 3 position of phosphoinositides.
Cancer cells escape normal growth control mechanisms as a consequence of activating (i.e., gain-of-function) mutations and/or increased expression of one or more cellular protooncogenes and/or inactivating (i.e., loss-of function) mutations and/or decreased expression of one or more tumor suppressor genes. Most oncogene and tumor suppressor gene products are components of signal transduction pathways that control cell cycle entry or exit, promote differentiation, sense DNA damage and initiate repair mechanisms, and/or regulate cell death programs. Several oncogenes and tumor suppressor genes belong to the same signaling pathway. Perhaps the best-characterized pathway includes D-type cyclin/cdk complexes, which can be oncogenes, and two tumor suppressor genes, the p16 cyclin/cdk inhibitor and the retinoblastoma gene product (reviewed in ref. 1). Nearly all tumors have mutations in multiple oncogenes and tumor suppressor genes, indicating that cells employ multiple parallel mechanisms to regulate cell growth, differentiation, DNA damage control, and death. A rational approach to understanding cancer pathogenesis and developing novel, mechanism-based therapies requires identifying the components of these signaling pathways and determining how mutations in oncogenes and tumor suppressor genes disrupt them. Two papers appearing recently in the Proceedings, as well as papers appearing elsewhere, elucidate a novel mechanism of cell growth control mediated by the PTEN/MMAC tumor suppressor gene and involving the cellular counterparts of at least two oncogenes [the genes encoding phosphoinositide 3-kinase (PI3K) and the protein-Ser/Thr kinase (AKT)].
PTEN (phosphatase and tensin homolog deleted on chromosome ten)/MMAC (mutated in multiple advanced cancers) was identified virtually simultaneously by two groups (2, 3) as a candidate tumor suppressor gene located at 10q23; another group (4) identified the same gene in a search for new dual-specificity phosphatases (see below) and named it TEP-1 (TGF-β-regulated and epithelial cell-enriched phosphatase). Deletions in 10q22–25 occur in multiple tumor types, most prominently advanced glial tumors (glioblastoma multiforme/anaplastic astrocytoma), but also prostate, endometrial, renal and small cell lung carcinoma, melanoma, and meningioma (see references in refs. 2 and 3). Early studies indicated that 10q abnormalities are more common in advanced tumors (hence the appellation MMAC). The initial cloning studies reported PTEN/MMAC/TEP-1 (hereafter, PTEN) mutations in a large fraction of glioblastoma multiforme cell lines, xenografts, and primary tumors, as well as in smaller samples of breast and prostate cancers (2, 3). Subsequent analyses confirmed that homozygotic inactivation of PTEN occurs in a large fraction of glioblastomas (at least 30% of primary tumors and 50–60% of cell lines) but not in lower-grade (i.e., less advanced) glial tumors (5–8). PTEN mutations also are extremely common in melanoma cell lines (>50%) (9), advanced prostate cancers (10, 11), and endometrial carcinomas (30–50%) (12, 13). Although PTEN mutations are found predominantly in advanced glial and prostate tumors, mutations occur with equal frequency at all stages of endometrial cancer (12), suggesting that PTEN activation is an early event in endometrial carcinogenesis. A significant percentage (≈10%) of breast cancer cell lines have inactivated PTEN (2, 3, 14). PTEN mutations are rare in sporadic breast tumors, independent of severity (15). Also, whereas germ-line PTEN mutations lead to increased breast cancer incidence (see below), PTEN mutations are not a frequent cause of familial breast cancer (16). Occasional PTEN mutations are reported in head and neck (17) and thyroid (18) cancers, but not in other tumors associated with 10q abnormalities, including meningioma (8) and lung cancer (17); these studies raise the possibility of another tumor suppressor gene distal to PTEN on chromosome 10. Overall, PTEN is one of the most common targets of mutation in human cancer, with a mutation frequency approaching that of p53.
Germ-line mutations in PTEN cause three rare autosomal dominant inherited cancer syndromes with overlapping clinical features: Cowden disease (19–21), Lhermitte–Duclos disease (19), and Bannayan–Zonana syndrome (21, 22). These syndromes are notable for hamartomas, benign tumors in which differentiation is normal, but cells are highly disorganized. Cowden disease is characterized by hamartomas in multiple sites, including the skin, thyroid, breast, oral mucosa, and intestine. In addition, about a third of patients will have macrocephaly. Affected females have a 30–50% incidence of breast cancer, and Cowden disease patients have increased risk of thyroid carcinoma (≈10% incidence) and meningiomas (see references in ref. 19). Lhermitte–Duclos patients have multiple hamartomas, together with macrocephaly, ataxia, and seizures, caused by cerebellar glial tumors. Besides their hamartomas, Bannayan–Zonana patients exhibit macrocephaly, retardation, and unusual pigmentation of the penis (see references in ref. 21). Hamartomas from Cowden disease patients exhibit loss of heterozygosity around the PTEN locus, indicating that homozygotic loss of PTEN function probably is required for hamartoma formation (19, 23). Whether the type of mutation in PTEN contributes to the distinct features of these three hamartomatous syndromes remains unclear, but other (i.e., modifying) loci probably play the primary role in determining the spectrum of abnormalities evoked by a given mutation. Indeed, recent analyses of mutant mice (see below) suggest that genetic background can significantly affect the PTEN-deficient phenotype. However, in Cowden disease patients, the type of PTEN mutation may affect the number of affected sites and/or the presence of breast disease (21).
These genetic data strongly suggest that PTEN function is required for normal development and that loss of PTEN function contributes to carcinogenesis. Gene transfer and knockout studies have confirmed these ideas. Restoration of PTEN expression in PTEN− mutant glioblastoma multiforme cells causes growth suppression (24, 25), whereas increasing expression in glioblastoma multiforme lines that retain normal PTEN expression does not inhibit cell growth (24). Subsequent studies showed that PTEN reconstitution inhibits the growth of PTEN− prostate (26), melanoma (27), and breast cancer (28) cell lines. Interestingly, PTEN appears to suppress cell growth by distinct mechanisms in different types of tumors, producing G1 cell cycle arrest in glioblastoma cells (29), but inducing apoptosis in carcinomas (26, 28).
Three different PTEN mutations have been introduced into mice (30–32). Homozygotic mutants from all three lines exhibit early embryonic lethality, consistent with an essential role for PTEN in normal development. Heterozygotes show increased tumor incidence, consistent with its identification as a tumor suppressor gene. Although the different mutant mice share these general features, some details of the phenotype differ substantially. The timing of embryonic death ranges from day 7.5 or earlier (30, 32) to day 9.5 (31). Hyperproliferative lesions of the intestine are common to all three lines, but the spectrum of other hyperproliferative and neoplastic disorders differs dramatically. Chimeric and heterozygotic PTEN mutant mice generated by Di Cristofano et al. (30) showed a high incidence of tumor formation, with colonic, testicular, thyroid, germ-cell, and hematopoietic (acute myeloid leukemia) neoplasms observed. Suzuki et al. (31) and Podsypanina et al. (32) observed a high incidence of T cell lymphomas/leukemia; moreover, T cell lymphomagenesis in PTEN+/− mice is markedly potentiated by irradiation (31). Other tumors, including prostate, early endometrial, thyroid, liver, and germ-cell neoplasms, were also observed by these workers. Finally, Podsypanina et al. (32) noted a high incidence of lymph node hyperplasia in PTEN heterozygotes, with consequent disruption of lymphoid architecture.
Notably, none of the groups reported brain tumors, despite the frequent mutation of PTEN in glioblastoma. Conceivably, the role of PTEN in glioblastomagenesis may differ in mice and humans. A more likely explanation, consistent with the occurrence of PTEN mutations late in glioblastomagenesis, is that generation of a glioblastoma requires more mutations than the other tumors found in PTEN heterozygotes, leaving insufficient time for brain tumors to develop before these mice die from other diseases. Aside from the high incidence of intestinal hyperproliferation and polyposis, and the report of acanthotic skin lesions in one line of mice (but not the others) (30), the other features of Cowden disease and related syndromes, particularly hamartomas, are absent in PTEN+/− mice. Further study will be required to determine the reasons for the differences in phenotype between the different strains of PTEN mutant mice and the discrepancy between the effects of PTEN deficiency in mice and humans.
The PTEN cDNA sequence strongly suggested that is was a member of the protein-tyrosine phosphatase (PTP) gene superfamily (2, 3). PTPs consist of conserved catalytic domains, flanked by noncatalytic, regulatory sequences (reviewed in ref. 33). All PTP catalytic domains contain the canonical sequence HCXXGXXRS/T, known as the PTP “signature motif” (reviewed in ref. 33); the presence of this motif within any protein makes it a virtual certainty that it has PTP activity. Structural and enzymatic analyses have shown that signature motif residues constitute the catalytic center of each enzyme (reviewed in ref. 33). PTP superfamily members can be further subdivided into “classic” PTP and dual-specificity phosphatase families. The former are exquisitely selective for phosphotyrosine residues in vitro and in vivo. Dual-specificity phosphatases, as their name implies, typically dephosphorylate phosphotyrosine, phosphoserine, and/or phosphothreonine in vitro. Most known dual-specificity phosphatases (e.g., MAP kinase phosphatases, cdc25 family members) dephosphorylate phosphothreonine and phosphotyrosine in specific sequence contexts in vivo, although others may target specific tyrosine- or serine/threonine-phosphorylated cellular proteins (reviewed in refs. 33–35).
The PTEN sequence suggested that it is a dual-specificity phosphatase. Moreover, sporadic and germ-line mutations in PTEN cluster within the presumptive catalytic domain, with many mutations altering residues (e.g., the essential cysteine or arginine residues within the signature motif) required for enzymatic activity (see references cited above; reviewed in ref. 36). These results strongly suggested that PTEN catalytic activity was vital for its biological function, an idea strengthened by the inability of signature motif cysteine mutants of PTEN to restore growth suppression to PTEN-deficient tumor cell lines (24, 28, 29). Besides its catalytic domain, PTEN has a potential binding site for PDZ domain-containing proteins at its C terminus; PDZ proteins have been shown to direct the assembly of multiprotein complexes, often at membrane/cytoskeletal interfaces such as synapses (37). PTEN also contains a stretch of sequence that overlaps the catalytic domain and is similar to a domain within the cytoskeletal proteins tensin and auxilin (hence the name PTEN; see above). This led to the suggestion that PTEN might participate in regulating cytoskeletal phosphorylation events. The biological significance of this tensin similarity is unclear, however, because the catalytic domains of many PTPs contain a similar region (38). Most likely, this represents convergent evolution of tensin-like proteins and PTPs, rather than implying a specific role for PTEN (and other PTPs with tensin similarity) in cytoskeletal regulation. Indeed, in view of the recent finding that PTEN actually dephosphorylates phosphatidylinositol (PtdIns) phosphates (see below), one intriguing possibility is that this “tensin domain” actually constitutes a binding site for such lipids.
Despite the clear sequence similarity between PTEN and other dual-specificity phosphatases, it was unexpectedly difficult to detect significant catalytic activity in recombinant preparations of PTEN when conventional artificial substrates were used (4, 39, 40). This finding was surprising because PTPs are generally quite robust and remarkably efficient catalysts. The first convincing demonstration of dual-specificity phosphatase activity in PTEN was provided by Myers et al. (40). Although these workers found that PTEN could dephosphorylate some serine/threonine-phosphorylated proteins, it displayed maximal activity against poly(Glu-pTyr), which suggested that it prefers extremely acidic substrates. Many, although importantly, not all, PTEN mutants derived from patient samples were inactive in this assay. Of particular interest, however, two mutants derived from unrelated Cowden disease families retained full activity against poly(Glu-pTyr). Moreover, even measured against poly(Glu-pTyr), PTEN was markedly less active than most other PTPs. These findings suggested that PTEN might be highly selective for a particular phosphorylated protein. Alternatively, PTEN might target a nonproteinaceous phospho-substrate; a precedent for the latter had been established with the demonstration that a PTP family member serves as an RNA 5-triphosphatase (41).
Work by Tamura et al. (42) suggested that PTEN might, indeed, be highly selective for a particular tyrosine-phosphorylated target. These workers found that over-expression of PTEN inhibited, whereas PTEN antisense oligonucleotides stimulated, fibroblast migration. They also reported that PTEN associated with and could dephosphorylate focal adhesion kinase (FAK) in vivo and in vitro, leading them to suggest that PTEN regulated focal adhesion structure, cell spreading, and motility, by controlling FAK activity. However, extremely high (stoichiometric, rather than catalytic) amounts of PTEN were required to dephosphorylate FAK in these experiments. Moreover, although PTEN mutated at its catalytic cysteine residue was unable to dephosphorylate FAK or affect migration and focal adhesion structure, a Cowden disease-derived mutation (G129E) that Myers et al. (40) had shown retained activity against poly(Glu-pTyr) was active in these experiments. Because Cowden disease-derived PTEN must, by definition, be biologically inactive (or at least less active than wild-type PTEN), these results imply that PTEN must have targets other than FAK in vivo.
Recent studies suggest that the biologically relevant targets probably are not phosphoproteins at all, but rather a subset of inositol phospholipids. The key initial observation was provided by Maehama et al. (43), who showed that PTEN could dephosphorylate the 3 position of PtdIns phosphates both in vitro and in vivo. Now, a recent paper from Myers et al. (26) in the Proceedings, as well as work by Furnari et al. (29), indicates that several independent patient-derived PTEN mutants retain protein [i.e., poly(Glu-pTyr)] phosphatase activity but lose the ability to dephosphorylate PtdIns phosphates. Since the mutations analyzed by both groups occur in sporadic tumors as well as Cowden disease patients, their results provide strong genetic evidence that the lipid phosphatase activity of PTEN is required for its tumor suppressor activity and for its role(s) in normal development. Consistent with this idea, PTEN-deficient tumor cell lines (26, 44), as well as immortalized fibroblasts from PTEN−/− mice (45), have elevated concentrations of PtdIns with phosphate at the 3 position.
The finding that PTEN dephosphorylates PtdIns phosphates leads to a model for how PTEN acts as a tumor suppressor, a model linking PTEN to control of at least two known cellular protooncogenes, PI3K (46) and Akt (47). PTEN inhibits PI3K-dependent activation of AKT (also called PKB), a serine/threonine-kinase, and deletion or inactivation of PTEN results in constitutive AKT activation.
PI3K was discovered more than 10 years ago as a PtdIns kinase activity copurifying with the oncoproteins pp60v-src and polyoma middle T antigen (48, 49). Although initially thought to be an enzyme in the pathway for synthesis of phosphatidylinositol 4,5-bisphosphate (PtdIns-4,5-P2), chemical analysis of the product revealed that this enzyme phosphorylates the 3 position of the inositol ring, thus defining a new lipid signaling pathway (50).
Interest in PI3K increased with evidence that the transforming ability of several oncoproteins correlated with their ability to associate with PI3K and elevate in vivo levels of specific lipid products of this enzyme (reviewed in ref. 51). When presented with appropriate substrates in vitro, oncoprotein-associated PI3K can catalyze the production of PtdIns-3-P, PtdIns-3,4-P2, or PtdIns-3,4,5-P3 (52, 53) (Fig. 1). However, growth factor stimulation and oncoprotein transformation correlate with increases in the in vivo levels of PtdIns-3,4-P2 and PtdIns-3,4,5-P3 (52, 54). These lipids are nominally absent from quiescent cells but appear within seconds to minutes of stimulation with growth/survival factors such as platelet-derived growth factor (PDGF), nerve growth factor (NGF), or insulin-like growth factor 1 (IGF-1).
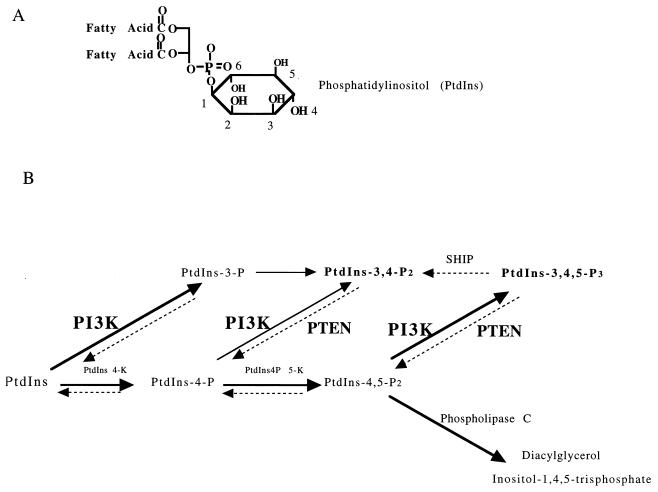
Figure 1.
Reactions catalyzed by PI3K and PTEN. (A) PtdIns contains a myo-inositol headgroup connected to diacylglycerol by a phosphodiester linkage. The numbering system of the inositol ring is indicated. (B) The class I PI3K enzymes can phosphorylate the 3 position of PtdIns, PtdIns-4-P, or PtdIns-4,5-P2 to produce PtdIns-3-P, PtdIns-3,4-P2, or PtdIns-3,4,5-P3, respectively. PtdIns-3,4-P2 can also be produced by dephosphorylating the 5 position of PtdIns-3,4,5-P3, and one enzyme that does this is an SH2-containing 5-phosphatase called SHIP. In addition, PtdIns-3,4-P2 can be produced by phosphorylating the 4 position of PtdIns-3-P [reviewed by Fruman et al. (76)]. PTEN has been shown to dephosphorylate the 3 position of both PtdIns-3,4,5-P3 (26, 43) and PtdIns-3,4-P2 (44) to reverse the reactions catalyzed by PI3K.
The acute production of PtdIns-3,4-P2 and PtdIns-3,4,5-P3 in response to cell stimulation suggested that these lipids act as membrane-embedded second messengers, analogous to diacylglycerol. It now is clear that a variety of cytosolic signaling proteins have evolved the ability to bind to one or both of these lipids as a mechanism of recruitment to the membrane (for review, see ref. 55). Several proteins bind PtdIns-3-P lipids by pleckstrin homology (PH) domains that specifically recognize the phosphoinositide headgroup. These include a PH domain-containing protein-tyrosine kinase called BTK (56, 57), an exchange factor for ARF family members called GRP-1 (58), and, of particular interest for this story, two protein-serine/threonine kinases called AKT (59, 60) and PDK1 (61, 62).
AKT was discovered as the product of a retrovirus-encoded oncogene that transforms lymphoid cells (47). The protein product has a catalytic domain with high similarity to the PKA and PKC family of protein kinases, and it was independently cloned as a PKC homologue and termed PKB (63). The first evidence that AKT is regulated via a pathway involving PI3K came from studies of PDGF-dependent AKT activation in fibroblasts (64). Further studies from several laboratories (reviewed in ref. 65) led to the model indicated in Fig. 2. In this model, the N-terminal PH domain of AKT acts as an autoinhibitory domain. This domain has a high affinity for PtdIns-3,4-P2 and somewhat lower, but significant, affinity for PtdIns-3,4,5-P3. Binding to these phosphoinositides localizes AKT to the membrane and opens up the catalytic site. A second protein-serine/threonine kinase, PDK1, also has a PH domain that binds PtdIns-3,4-P2 and PtdIns-3,4,5-P3 tightly, allowing it to colocalize with AKT and phosphorylate the activation loop of the exposed catalytic domain. Full activation of AKT requires phosphorylation at a second C-terminal site by a distinct kinase.
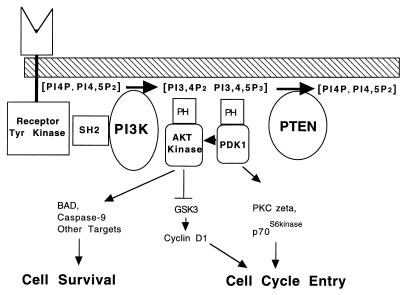
Figure 2.
Cell survival and proliferation by means of PI3K and AKT and suppression by PTEN. Growth factors and survival factors activate receptors that recruit PI3K to the membrane. Phosphorylation of the membrane lipids PtdIns-4-P and PtdIns-4,5-P2 [PI4P, PI4,5P2] by PI3K produces the second messengers PtdIns-3,4-P2 and PtdIns-3,4,5-P3 [PI3,4P2, PI3,4,5P3]. These lipids recruit the protein-serine/threonine kinases AKT and PDK1 to the membrane and induce a conformational change in AKT, exposing the activation loop. Phosphorylation of AKT at Thr-308 of the activation loop by PDK1 turns on the protein kinase activity. Phosphorylation of AKT at a C-terminal site (not shown) causes further activation. AKT phosphorylates and compromises the function of BAD and caspase-9, proteins involved in cell death pathways. AKT also phosphorylates and inhibits glycogen synthase kinase 3 (GSK3). GSK3 phosphorylates cyclin D, targeting it for proteolysis; thus, AKT may promote cyclin D accumulation. PDK1 also phosphorylates and enables activation of p70S6kinase and protein kinase C (PKC) family members. PTEN turns off the pathway by dephosphorylating the 3 position of PtdIns-3,4-P2 and PtdIns-3,4,5-P3.
Although other signaling pathways are activated downstream of PI3K, the AKT pathway has attracted much attention because of its role in cell survival. The ability to evade cell death pathways is a critical event in cancer progression, and the PI3K/AKT pathway provides such a mechanism. Activation of PI3K and AKT has been shown to provide a survival signal in response to NGF, IGF-1, PDGF, IL-3 and the extracellular matrix (reviewed in refs. 65 and 66). AKT is likely to send survival signals by phosphorylating multiple targets, including the BCL-2 family member BAD (67) and the cell death pathway enzyme caspase-9 (68). In addition to promoting cell survival, AKT may regulate cell proliferation by means of the phosphorylation of other targets. For example, AKT-catalyzed phosphorylation of another serine/threonine kinase, glycogen synthase kinase 3 (GSK3), results in GSK3 inhibition (69). Recent studies indicate that GSK3 promotes cyclin D proteolysis; thus by catalyzing GSK3 inhibition, AKT may contribute to cyclin D accumulation and cell cycle entry (70). The protein-serine/threonine kinase p70S6 kinase is also activated by the PI3K pathway (Fig. 2), and this enzyme could also contribute to cell growth by regulating translation of key mRNAs (71).
Several papers, including those by Myers et al. (26) and Wu et al. (72) in the Proceedings, now establish a link between the PI3K/AKT pathway and human cancers via defects in PTEN. Normally, AKT activity is low in the absence of growth factor stimulation. However, PTEN-deficient tumor cell lines (26, 28, 29, 44, 72), as well as immortalized fibroblasts (45) and tumors (31) derived from PTEN-deficient mice, exhibit high basal levels of AKT phosphorylation. Consistent with the anti-apoptotic actions of AKT, PTEN−/− fibroblasts are resistant to multiple pro-apoptotic stimuli. Reconstitution of wild-type PTEN expression restores normal AKT regulation and sensitivity to these stimuli (45). Moreover, PTEN action can be overcome by expression of constitutively active forms of AKT. Together, these results indicate that deregulation of the PI3K/AKT signal transduction pathway may contribute to a large fraction of human cancers. These studies in mammalian cell systems are buttressed by very recent genetic analyses of the Caenorhabditis elegans homolog of PTEN, which emphasize the evolutionary importance of this conserved pathway (73).
The prevailing paradigm of molecular oncology holds that multiple, at least partially redundant, pathways control cell growth, differentiation, and apoptosis, and, accordingly, mutations in many of these pathways are required to generate the fully neoplastic state. Available genetic evidence strongly suggests that the PTEN/PI3K/AKT pathway almost certainly constitutes a new and important regulatory pathway. Analysis of melanomas indicates that PTEN and p16 mutations coexist within the same tumors, arguing that loss of PTEN confers an additional selective advantage over loss of the retinoblastoma regulatory pathway (the main target of p16; see ref. 1). Likewise, there is no apparent correlation between epidermal growth factor receptor (EGFR) amplification and PTEN mutation in glioblastomas (5), suggesting that these two oncogenic events also lie in parallel regulatory pathways.
Although it now seems clear that the main role of PTEN is to regulate the PI3K/AKT pathway, many important issues remain unresolved. PTEN expression prevents basal activation of AKT (i.e., activation in the absence of growth factor stimulation), yet in PTEN+ cells, growth factors remain able to activate AKT via a PI3K-dependent pathway (26, 44, 45). This observation suggests that PTEN is inactivated upon growth factor stimulation or that PTEN is activated by growth factor depletion. It will be important to identify components of such a regulatory pathway(s), as they represent potential targets for oncogenic mutation as well. One attractive target for a PTEN regulator may be LKB1/STK1 (74, 75), the recently cloned gene for Peutz–Jeghers syndrome, a condition that shares a number of clinical features with Cowden disease. Alternatively, PTEN regulation could be effected by altering its intracellular localization, perhaps by means of its C-terminal PDZ- binding domain. However, C-terminal hemagglutinin epitope-tagged PTEN is able to rescue growth defects in glioblastoma cell lines, suggesting that its ability to bind PDZ-containing proteins is not absolutely required for PTEN function (24). It remains to be determined why PTEN reconstitution into prostate (26, 72) and breast (28) carcinoma cells causes apoptosis, whereas it induces G1 cell cycle arrest in glioblastoma cells (24, 26, 28, 29). Altered regulation of AKT could affect both apoptosis (via BAD and/or caspase-9 phosphorylation) and cell cycle entry (via GSK3, p70S6kinase, and possibly other pathways). However, why both pathways are not affected in any given tumor cell remains to be elucidated; most likely, other oncogenic mutations are responsible. The role of PTEN protein phosphatase activity, if any, also remains to be clarified. Although it now is clear that the lipid phosphatase activity of PTEN is required for its biological effects, these data do not rigorously establish that PTEN protein phosphatase activity is dispensable. Proof of the latter would require the generation of PTEN mutants that retain lipid phosphatase but lose protein phosphatase activity and the demonstration that such mutants retain full biological activity. In the absence of such data, it remains possible that PTEN acts as both a lipid and a protein phosphatase in vivo, and that both activities are necessary for at least some of its biological roles.
The elucidation of the PTEN/PI3K/AKT pathway stands as a potentially important advance in molecular oncology. In view of the high frequency of such mutations in human tumors, new and/or existing pharmacological agents directed against components of this pathway may have therapeutic benefit.
ABBREVIATIONS
- PI3K
phosphoinositide 3-kinase
- PTP
protein-tyrosine phosphatase
- PtdIns
phosphatidylinositol
- PH domain
pleckstrin homology domain
- PDGF
platelet-derived growth factor
- GSK3
glycogen synthase kinase 3
References
- 1.Weinberg R A. Cell. 1995;81:323–330. doi: 10.1016/0092-8674(95)90385-2. [DOI] [PubMed] [Google Scholar]
- 2.Li J, Yen C, Liaw D, Podsypanina K, Bose S, Wang S I, Puc J, Miliaresis C, Rodgers L, McCombie R, et al. Science. 1997;275:1943–1947. doi: 10.1126/science.275.5308.1943. [DOI] [PubMed] [Google Scholar]
- 3.Steck P A, Pershouse M A, Jasser S A, Yung W K, Lin H, Ligon A H, Langford L A, Baumgard M L, Hattier T, Davis T, et al. Nat Genet. 1997;15:356–362. doi: 10.1038/ng0497-356. [DOI] [PubMed] [Google Scholar]
- 4.Li D-M, Sun H. Cancer Res. 1997;57:2124–2129. [PubMed] [Google Scholar]
- 5.Liu W, James C D, Frederick L, Alderete B E, Jenkins R B. Cancer Res. 1997;57:5254–5257. [PubMed] [Google Scholar]
- 6.Rasheed B K A, Stenzel T T, McLendon R E, Parsons R, Friedman A H, Friedman H S, Bigner D D, Bigner S H. Cancer Res. 1997;57:4187–4190. [PubMed] [Google Scholar]
- 7.Wang S I, Puc J, Li J, Bruce J N, Cairns P, Sidransky D, Parsons R. Cancer Res. 1997;57:4183–4186. [PubMed] [Google Scholar]
- 8.Bostrom J, Cobbers J M J L, Wolter M, Tabatabai G, Weber R G, Lichter P, Collins V P, Reifenberger G. Cancer Res. 1998;58:29–33. [PubMed] [Google Scholar]
- 9.Guldberg P, thor Straten P, Birck A, Ahrenkiel V, Kirkin A F, Zeutehn J. Cancer Res. 1997;57:3660–3663. [PubMed] [Google Scholar]
- 10.Cairns P, Okami K, Halachmi S, Halachmi N, Esteller M, Herman J G, Jen J, Isaacs W B, Bova G S, Sidransky D. Cancer Res. 1997;57:4997–5000. [PubMed] [Google Scholar]
- 11.Suzuki H, Freije D, Nusskern D R, Okami K, Cairns P, Sidransky D, Isaacs W B, Bova G S. Cancer Res. 1998;58:204–209. [PubMed] [Google Scholar]
- 12.Risinger J I, Hayes A K, Berchuck A, Barrett J C. Cancer Res. 1997;57:4736–4738. [PubMed] [Google Scholar]
- 13.Tashiro E, Blazes M S, Wu R, Cho K R, Bose S, Wang S I, Li J, Parson R, Ellenson L H. Cancer Res. 1997;57:3935–3940. [PubMed] [Google Scholar]
- 14.Teng D H-F, Hu R, Lin H, Davis T, Iliev D, Frye C, Swedlund B, Hansen K L, Vinson V L, Gumpper K L, et al. Cancer Res. 1997;57:5221–5225. [PubMed] [Google Scholar]
- 15.Ueda K, Nishijima N, Inui H, Watatani M, Yayoi E, Okamura J, Yasutomi M, Nakamura Y, Miyoshi Y. Jpn J Cancer Res. 1998;89:17–21. doi: 10.1111/j.1349-7006.1998.tb00473.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Chen J, Lindblom P, Lindblom A. Hum Genet. 1998;102:124–125. [PubMed] [Google Scholar]
- 17.Okami K, Wu L, Riggins G, Cairns P, Goggins M, Evron E, Halachmi N, Ahrendt S A, Reed A L, Hilgers W, et al. Cancer Res. 1998;58:509–511. [PubMed] [Google Scholar]
- 18.Dahia P L M, Marsh D J, Zheng Z, Zedenius J, Komminoth P, Frisk T, Wallin G, Parsons R, Longy M, Larsson C, Eng C. Cancer Res. 1997;57:4710–4713. [PubMed] [Google Scholar]
- 19.Liaw D, Marsh D J, Li J, Dahia P L M, Wang S I, Zheng Z, Bose S, Call K M, Tsou H C, Peacocke M, et al. Nat Genet. 1997;16:64–67. doi: 10.1038/ng0597-64. [DOI] [PubMed] [Google Scholar]
- 20.Nelen M R, van Staverene W C, Peeters E A, Hassel M B, Gorlin R J, Hamm H, Lindboe C F, Fryns J P, Sijmons R H, Woods D G, et al. Hum Mol Genet. 1997;6:1383–1387. doi: 10.1093/hmg/6.8.1383. [DOI] [PubMed] [Google Scholar]
- 21.Marsh D J, Coulon V, Lunetta K L, Rocca-Serra P, Dahia P L M, Zheng Z, Liaw D, Caron S, Duboue B, Lin A Y, et al. Hum Mol Genet. 1998;7:507–515. doi: 10.1093/hmg/7.3.507. [DOI] [PubMed] [Google Scholar]
- 22.Marsh D J, Roth S, Lunetta K L, Hemminki A, Dahia P L, Sistonen P, Zheng Z, Caron S, van Orsouw N H, Bodmer W F, et al. Cancer Res. 1997;57:5017–5021. [PubMed] [Google Scholar]
- 23.Marsh D J, Dahia P L M, Coulon V, Zheng Z, Dorion-Bonnet F, Call K M, Little R, Lin A Y, Eeles R A, Goldstein A M, et al. Genes Chromosomes Cancer. 1998;21:61–69. doi: 10.1002/(sici)1098-2264(199801)21:1<61::aid-gcc8>3.0.co;2-6. [DOI] [PubMed] [Google Scholar]
- 24.Furnari F B, Lin H, Huang H-J S, Cavenee W K. Proc Natl Acad Sci USA. 1997;94:12479–12484. doi: 10.1073/pnas.94.23.12479. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Cheney I W, Johnson D E, Vaillancourt M-T, Avanzini J, Morimoto A, Demers G W, Wills K N, Shabram P W, Bolen J B, Tavtigian S V, Bookstein R. Cancer Res. 1998;58:2331–2334. [PubMed] [Google Scholar]
- 26.Myers M P, Pass I, Batty I H, Van der Kaay J, Stolarov J P, Hemmings B A, Wigler M H, Downes C P, Tonks N K. Proc Natl Acad Sci USA. 1998;95:13513–13518. doi: 10.1073/pnas.95.23.13513. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Robertson G P, Furnari F B, Miele M E, Glendening M J, Welch D R, Fountain J W, Lugo T G, Huang H-J S, Cavenee W K. Proc Natl Acad Sci USA. 1998;95:9418–9423. doi: 10.1073/pnas.95.16.9418. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Li D M, Sun H. Proc Natl Acad Sci USA. 1998;95:15406–15411. doi: 10.1073/pnas.95.26.15406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Furnari F B, Huang H-J S, Cavenee W K. Cancer Res. 1998;58:5002–5008. [PubMed] [Google Scholar]
- 30.Di Cristofano A, Pesce B, Cordon-Cardo C, Pandolfi P P. Nat Genet. 1998;19:348–355. doi: 10.1038/1235. [DOI] [PubMed] [Google Scholar]
- 31.Suzuki A, de la Pompa J L, Stambolic V, Elia A J, Sasaki T, del Barco Barrantes I, Ho A, Wakeham A, Itie A, Khoo W, et al. Curr Biol. 1998;8:1169–1178. doi: 10.1016/s0960-9822(07)00488-5. [DOI] [PubMed] [Google Scholar]
- 32.Podsypanina K, Ellenson L H, Nemes A, Gu J, Tamura M, Yamada K M, Cordon-Cardo C, Cattoretti G, Fisher P E, Parsons R. Proc Natl Acad Sci USA. 1999;96:1563–1568. doi: 10.1073/pnas.96.4.1563. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Denu J E, Stuckey J A, Saper M A, Dixon J E. Cell. 1996;87:361–364. doi: 10.1016/s0092-8674(00)81356-2. [DOI] [PubMed] [Google Scholar]
- 34.Tonks N K, Neel B G. Cell. 1996;87:365–368. doi: 10.1016/s0092-8674(00)81357-4. [DOI] [PubMed] [Google Scholar]
- 35.Neel B G, Tonks N K. Curr Opin Cell Biol. 1997;9:193–204. doi: 10.1016/s0955-0674(97)80063-4. [DOI] [PubMed] [Google Scholar]
- 36.Myers M P, Tonks N K. Am J Hum Genet. 1997;61:1234–1238. doi: 10.1086/301659. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Craven S E, Bredt D S. Cell. 1998;93:495–498. doi: 10.1016/s0092-8674(00)81179-4. [DOI] [PubMed] [Google Scholar]
- 38.Haynie D T, Ponting C P. Protein Sci. 1996;5:2643–2646. doi: 10.1002/pro.5560051227. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Li L, Ernsting B R, Wishart M J, Lohse D L, Dixon J E. J Biol Chem. 1997;272:29403–29406. doi: 10.1074/jbc.272.47.29403. [DOI] [PubMed] [Google Scholar]
- 40.Myers N P, Stolarov J P, Eng C, Li J, Wang S I, Wigler M H, Parsons R, Tonks N K. Proc Natl Acad Sci USA. 1997;94:9052–9057. doi: 10.1073/pnas.94.17.9052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Takada T, Matozaki T, Takeda H, Fukunaga K, Noguchi T, Fujioka Y, Okazaki I, Tsuda M, Yamao T, Ochi F, Kasuga M. J Biol Chem. 1998;273:9234–9242. doi: 10.1074/jbc.273.15.9234. [DOI] [PubMed] [Google Scholar]
- 42.Tamura M, Gu J, Matsumoto K, Aota S, Parsons R, Yamada K M. Science. 1998;280:1614–1617. doi: 10.1126/science.280.5369.1614. [DOI] [PubMed] [Google Scholar]
- 43.Maehama T, Dixon J E. J Biol Chem. 1998;273:13375–13378. doi: 10.1074/jbc.273.22.13375. [DOI] [PubMed] [Google Scholar]
- 44.Haas-Kogan D, Shalev N, Wong M, Mills G, Young G, Stokoe D. Curr Biol. 1998;8:1195–1198. doi: 10.1016/s0960-9822(07)00493-9. [DOI] [PubMed] [Google Scholar]
- 45.Stambolic V, Suzuki A, de la Pompa J L, Brothers G M, Mirtsos C, Sasaki T, Ruland J, Penninger J M, Siderovski D P, Mak T W. Cell. 1998;95:29–39. doi: 10.1016/s0092-8674(00)81780-8. [DOI] [PubMed] [Google Scholar]
- 46.Chang H W, Aoki M, Fruman D, Auger K R, Bellacosa A, Tsichlis P N, Cantley L C, Roberts T M, Vogt P K. Science. 1997;276:1848–1850. doi: 10.1126/science.276.5320.1848. [DOI] [PubMed] [Google Scholar]
- 47.Bellacosa A, Testa J R, Staal S P, Tsichlis P N. Science. 1991;254:274–277. doi: 10.1126/science.254.5029.274. [DOI] [PubMed] [Google Scholar]
- 48.Sugimoto Y, Whitman M, Cantley L C, Erikson R L. Proc Natl Acad Sci USA. 1984;81:2117–2121. doi: 10.1073/pnas.81.7.2117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Whitman M, Kaplan D, Schaffhausen B, Cantley L, Roberts T. Nature (London) 1985;315:239–242. doi: 10.1038/315239a0. [DOI] [PubMed] [Google Scholar]
- 50.Whitman M, Downes C P, Keeler M, Keller T, Cantley L. Nature (London) 1988;332:644–646. doi: 10.1038/332644a0. [DOI] [PubMed] [Google Scholar]
- 51.Cantley L, Auger K, Carpenter C, Duckworth B, Graziani A, Kapeller R, Soltoff S. Cell. 1991;64:281–302. doi: 10.1016/0092-8674(91)90639-g. [DOI] [PubMed] [Google Scholar]
- 52.Auger K R, Serunian L A, Soltoff S P, Libby P, Cantley L. Cell. 1988;57:167–175. doi: 10.1016/0092-8674(89)90182-7. [DOI] [PubMed] [Google Scholar]
- 53.Carpenter C L, Duckworth B C, Auger K R, Cohen B, Schaffhausen B S, Cantley L C. J Biol Chem. 1990;265:19704–19711. [PubMed] [Google Scholar]
- 54.Seruninan L A, Auger K R, Robert T, Cantley L C. J Virol. 1990;64:4718–4725. doi: 10.1128/jvi.64.10.4718-4725.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Toker A, Cantley L C. Nature (London) 1997;387:673–676. doi: 10.1038/42648. [DOI] [PubMed] [Google Scholar]
- 56.Salim K, Bottomley M J, Querfurth E, Zvelebil M J, Gout I, Scaife R, Margolis R L, Gigg R, Smith C I, Driscoll P C, et al. EMBO J. 1996;15:6241–6250. [PMC free article] [PubMed] [Google Scholar]
- 57.Rameh L E, Arvidsson A, Carraway K L, Couvillion A D, Rathbun G, Crompton A, VanRenterghem B, Czech M P, Ravichandran K S, Burakoff S J, et al. J Biol Chem. 1997;272:22059–22066. doi: 10.1074/jbc.272.35.22059. [DOI] [PubMed] [Google Scholar]
- 58.Klarlund J K, Guilherme A, Holik J J, Virbasius J V, Chawla A, Czech M P. Science. 1997;275:1927–1930. doi: 10.1126/science.275.5308.1927. [DOI] [PubMed] [Google Scholar]
- 59.Franke, T. F., Kaplan, D. R., Cantley, L. C. & Toker, A. (1997) Science 665–668. [DOI] [PubMed]
- 60.Klippel A, Kavanaugh W M, Pot D, Williams L T. Mol Cell Biol. 1997;17:338–344. doi: 10.1128/mcb.17.1.338. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Stokoe D, Stephens L R, Copeland T, Gaffney P R, Reese C B, Painter G F, Holmes A B, McCormick F, Hawkins P T. Science. 1997;277:567–570. doi: 10.1126/science.277.5325.567. [DOI] [PubMed] [Google Scholar]
- 62.Alessi D R, Deak M, Casamayor A, Caudwell F B, Morrice N, Norman D G, Gaffney P, Reese C B. Curr Biol. 1997;7:776–789. doi: 10.1016/s0960-9822(06)00336-8. [DOI] [PubMed] [Google Scholar]
- 63.Jones P F, Jakubowicz T, Pitossi F J, Maurer F, Hemmings B A. Proc Natl Acad Sci USA. 1991;88:4171–4175. doi: 10.1073/pnas.88.10.4171. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Franke T F, Yang S I, Chan T O, Datta K, Kazlauskas A, Morrison D K, Kaplan D R, Tsichlis P N. Cell. 1995;81:727–736. doi: 10.1016/0092-8674(95)90534-0. [DOI] [PubMed] [Google Scholar]
- 65.Franke T, Kaplan D, Cantley L. Cell. 1997;88:435–437. doi: 10.1016/s0092-8674(00)81883-8. [DOI] [PubMed] [Google Scholar]
- 66.Downward J. Curr Opin Cell Biol. 1998;10:262–267. doi: 10.1016/s0955-0674(98)80149-x. [DOI] [PubMed] [Google Scholar]
- 67.Datta S R, Dudek H, Tao X, Masters S, Fu H, Gotoh Y, Greenberg M E. Cell. 1997;91:231–241. doi: 10.1016/s0092-8674(00)80405-5. [DOI] [PubMed] [Google Scholar]
- 68.Cardone M H, Roy N, Stennicke H R, Salvesen G S, Franke T F, Stanbridge E, Frisch S, Reed J C. Science. 1998;282:1318–1321. doi: 10.1126/science.282.5392.1318. [DOI] [PubMed] [Google Scholar]
- 69.Cross D A, Alessi D R, Cohen P, Andejelkovich M, Hemmings B A. Nature (London) 1995;378:785–789. doi: 10.1038/378785a0. [DOI] [PubMed] [Google Scholar]
- 70.Diehl J A, Cheng M, Roussel M F, Sherr C J. Genes Dev. 1998;12:3499–3511. doi: 10.1101/gad.12.22.3499. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Chung J K, Grammer T C, Lemon K P, Kazlauskas A, Blenis J. Nature (London) 1994;370:71–75. doi: 10.1038/370071a0. [DOI] [PubMed] [Google Scholar]
- 72.Wu X, Senechal K, Neshat M S, Whang Y E, Sawyers C L. Proc Natl Acad Sci USA. 1998;95:15587–15591. doi: 10.1073/pnas.95.26.15587. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Ogg S, Ruvkun G. Mol Cell. 1988;2:887–893. doi: 10.1016/s1097-2765(00)80303-2. [DOI] [PubMed] [Google Scholar]
- 74.Jenne D E, Reimann H, Nezu J, Friedel W, Loff S, Jeschke R, Muller O, Back W, Zimmer M. Nat Genet. 1998;18:38–43. doi: 10.1038/ng0198-38. [DOI] [PubMed] [Google Scholar]
- 75.Hemminki A, Markie D, Tomlinson I, Aviezienyte E, Roth S, Loukola A, Bignell G, Warren W, Aminoff M, Hoglund P, et al. Nature (London) 1998;391:184–187. doi: 10.1038/34432. [DOI] [PubMed] [Google Scholar]
- 76.Fruman D A, Meyers R E, Cantley L C. Annu Rev Biochem. 1998;67:481–507. doi: 10.1146/annurev.biochem.67.1.481. [DOI] [PubMed] [Google Scholar]