Abstract
Purpose
Identification of new therapies in small cell lung cancer (SCLC) is urgently needed. IGF1R is a tyrosine kinase receptor implicated in the pathogenesis of several malignancies and is potentially an attractive target for anti-cancer treatment. Knowledge about IGF1R protein expression, gene copy number and the prognostic relevance of these features in SCLC is limited.
Methods
We analyzed IGF1R protein expression and gene copy number in primary tumors from 90 SCLC patients (67 males and 23 females) who underwent pulmonary resection. IGF1R expression assessed by immunohistochemistry (IHC) with H-scores from 0 to 400 was evaluable in 84 patients, and IGF1R gene copy number assessed by silver in-situ hybridization (SISH) technique in 81 patients.
Results
Median H-score for IGF1R protein expression was 88 (range 0-400) and the proportion of positive immunostaining using cut-off H-score of 10 was 74%. Increased IGF1R gene copy number (an average of 4 or more copies per cell) was found in 15 cases (18.5%), 5 of whom (6.2%) showed gene amplification. There was a significant correlation between protein expression and gene copy number (r=0.49, p<0.005). IGF1R expression and gene copy number did not associate with clinicopathological factors such as patient age, tumor size, lymph node involvement, stage and survival.
Conclusions
SCLC is characterized by frequent high IGF1R protein expression, increased gene copy number and occasional occurrence of true gene amplification. These features may have important implications for future anti-IGF1R therapeutic approaches.
Keywords: lung cancer, IGF1R, protein expression, gene copy number, prognosis
INTRODUCTION
SCLC comprises approximately 15% of new lung cancer diagnoses in the United States (1) and approximately 10-20% in Europe. While the incidence of SCLC is slowly decreasing, the cure rates have increased only slightly over the last decades and remain elusive (1). Median survival of patients presenting with limited (LD) and extensive disease (ED) is approximately 16-24 months and 6-12 months, respectively. Novel treatment approaches considering biological characteristics of this tumor are therefore urgently needed.
The insulin-like growth factor (IGF) pathway regulates several important cell functions, including cell growth, proliferation, survival and invasion. The extracellular pathway components include two ligands (IGF1 and IGF2, produced under the control of growth hormone), their binding proteins (IGFBP1-6, governing ligand bioavailability in the circulation, tissue and cell), and two cell membrane receptors (IGF1R and IGF2R) (2). IGF1R is structurally related to insulin receptor, has tyrosine kinase activity capable of downstream signaling through RAS/RAF/mitogen activated protein kinase (MAPK) pathway and phosphatidylinositol-3-kinase (PI3K)-Akt pathway, whereas IGF2R acts as a decoy receptor. Increased IGF1R activity results in up-regulation of survivin expression (3), which could be a potential mechanism of chemoresistance in SCLC.
Owing to the paucity of existing data on IGF1R abnormalities in SCLC, the aim of this study was to evaluate IGF1R protein expression and gene copy number in this tumor type in relation to clinical characteristics and survival. Since adequate primary tumor specimens of SCLC are difficult to obtain and typically small, we used previously described primary tumor specimens collected over a 20 year period from series of LD patients who underwent pulmonary resection (4).
MATERIALS AND METHODS
Patient population
Archival formalin-fixed paraffin-embedded tumor samples were obtained from 90 SCLC patients who underwent pulmonary resection between 1982 and 2002 at Medical University of Gdansk. Due to difficulties in obtaining reliable biopsy material for histopathological examination, the diagnosis of SCLC in this group was established only after the pulmonary resection. In all patients, surgery was followed by one of the standard chemotherapy regimens. Patient clinical characteristics are shown in Table 1. Median follow-up was 86 months (range, 1–211 months), with a two-year survival probability of 43% and a median survival of 17.8 months.
Table 1.
Patient Characteristics
| Characteristics | |
|---|---|
| Age | |
| Median (Range) | 57 (37 - 82) |
| ≥60, n (%) | 38 (42) |
| Gender, n (%) | |
| Female | 23 (26) |
| Male | 67 (74) |
| Pathological Stage, n (%) | |
| IA | 6 (7) |
| IB | 26 (29) |
| IIA | 5 (6) |
| IIB | 13 (14) |
| IIIA | 24 (27) |
| IIIB | 12 (13) |
| Unknown | 1 (1) |
| Surgery Type, n (%) | |
| Lobectomy | 40 (44) |
| Pneumonectomy | 46 (51) |
| Segmentectomy | 4 (4) |
| Site, n (%) | |
| Left | 43 (48) |
| Right | 47 (52) |
Tissue microarray preparation
On each paraffin fixed block a morphologically representative SCLC tumor area was identified under the microscope by a pathologist, using a stained hematoxylin and eosin (H&E) section on a glass slide as a guide. Tissue microarray was constructed using manual MTA I Beecher Instrument. Three 0.6 mm cores from different tumor areas were taken for each patient.
IGF1R protein expression
IHC evaluation, on 4 μM sections, was done using the Ventana G11 (CONFIRM™, Ventana Medical Systems, Tucson, AZ) anti-IGF1R antibody following the manufacturer’s instructions. Briefly, the staining was performed on the Ventana BenchMark XT autostainer utilizing the ultraView detection kit and the primary antibody was incubated for 16 minutes. The score was determined according to the “hybrid scoring system” (H-score) criteria, which sums the products of the five staining intensity categories (0-4) by the percentage of positive cells (0-100%). Thus, the final IHC score ranged from 0 to 400. Two certified pathologists and one reader independently scored each core.
IGF1R gene copy number
IGF1R gene copy number was evaluated using automated SISH, a chromogenic assay that allows for quantification of gene copy number concurrent to visualization of cell morphology in brightfield microscopy. The TMA containing triplicate cores per patient was probed according to the Ventana Medical Systems Inc (Tucson, AZ) protocols for the INFORM IGF1R DNA probe. The probe was labeled with dinitrophenol (DNP) and optimally formulated for use with ultraView SISH Detection Kit and accessory reagents on the Benchmark® automated slide stainer. The IGF1R DNA probe was denatured at 95°C for 12 min and hybridization was performed at 52°C for 4 hrs. After hybridization, 3 stringency washes at 72°C were performed. The IGF1R DNA probe was visualized using a rabbit anti-DNP primary antibody and the ultraView SISH Detection Kit. The detection kit contained a goat anti-rabbit secondary antibody conjugated with horseradish peroxidase (HRP) utilized as the chromogenic enzyme. The chemistry of the SISH reaction, briefly described, is driven by the sequential addition of silver A (silver acetate), silver B (hydroquinone) and silver C (H2O2). Here, the silver ions (Ag+) are reduced by hydroquinone to metallic silver atoms (Ag). This reaction is fueled by the substrate for HRP, hydrogen peroxide (silver C). The silver precipitation is deposited in the nuclei and a single copy of the IGF1R gene can be visualized as a single discrete black dot whereas a tight cluster of black dots stacked so closely together that individual signals can not be resolved are considered amplified IGF1R genes. The specimen was then counterstained with Ventana’s Hematoxilin II and Bluing reagent for interpretation by light microscopy. The number of IGF1R gene copies per nucleus was determined by two certified pathologists counting 50 nuclei, or less if the tissue microarray core was partially depleted, by two methods. In the first method, termed “focused”, the pathologists scanned the core and focused on regions that appeared to have the highest copy numbers and counted 50 non-overlapping nuclei that had the highest copy numbers. In the second method, termed “consecutive”, the pathologists scanned the slide and found a region with high copy number and counted the signals in 50 consecutive non-overlapping nuclei. Individual signals, black dots, were given a score of one and if clusters were present the numbers of signals within the cluster were estimated based on size of the cluster. The scores were analyzed to determine the mean of IGF1R gene copy number per nucleus.
Statistical analysis
SAS (version 9.2; SAS Institute Inc., Cary, NC) was used for all statistical analyses. Group comparisons were conducted using two-sided Pearson’s chi-square tests for categorical data and two-sided Student’s t-tests for continuous data. Associations between continuous measures were compared using Pearson’s product-moment correlation coefficient. Overall survival was calculated as time from surgery to last follow-up date or death and plotted with 95% confidence intervals using the Kaplan-Meier method. Differences in survival between groups were assessed using the log-rank test. Multivariate Cox proportional hazards regression analysis was performed adjusting for age, gender, tumor site, tumor stage, and tumor histology. All tests were considered statistically significant at p<0.05.
RESULTS
IGF1R protein expression
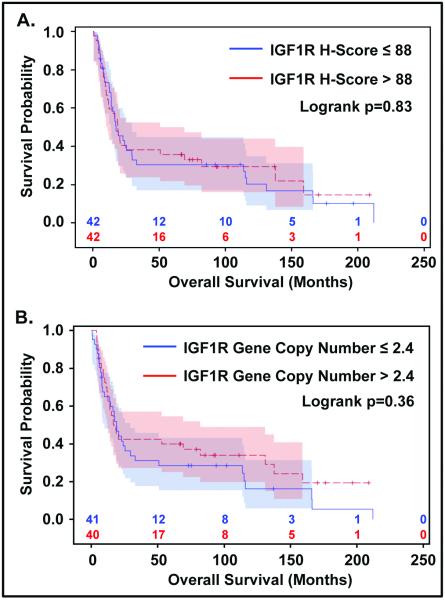
IGF1R protein expression could be evaluated in 84 samples (93%). There was a wide range (0-400) of protein expression among particular patients based on the H-scores (Figure 1). For each patient, results from three cores, or less if depleted, were averaged and compared to the core with the highest score. Figure 2 shows the distribution and frequency of scores, in addition to median and mean, based on average of cores (Figure 2A) and maximum core (Figure 2B). There is a very high correlation (r=0.97) between these two scores, resulting in excellent concordance for protein expression between all evaluators (r=0.88 DM vs MP, r=0.95 DM vs MW, r=0.86 MP vs MW) using maximum core value (data not shown). Thus, for further analyses the scores from the highest core determined by one of the pathologists (DM) were used. Median H-score for IGF1R protein expression was 88 (range 0-400) and the proportion of positive immunostaining using cut-off H-score of 10 was 74%. There was no significant correlation between IGF1R protein expression using an H-score of 88 (median) and clinicopathological factors (Table 1). No significant association was found between IGF1R protein expression and overall survival using univariate Kaplan-Meier and log-rank analysis (log rank p=0.83; Figure 4A). The same held true for the H-score considered as a continuous variable in both the univariate Cox regression (HR 0.998 [0.996-1.001], p=0.20) and multivariate Cox regression model adjusted for age, gender, tumor site, tumor histology and stage.
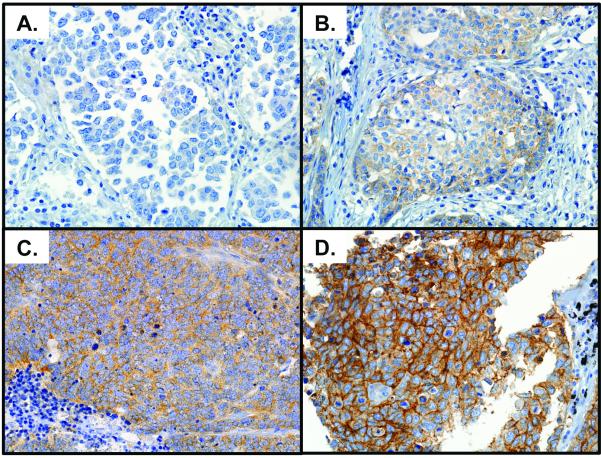
Figure 1.
Representative photographs of IGF1R protein expression visualized by immunohistochemistry. Panel A) no expression (H-score 0), Panel B) low expression (H-score 130), Panel C) moderate expression (H-score 205) and Panel D) high expression (H-score 400). Scores based on Reader DM.
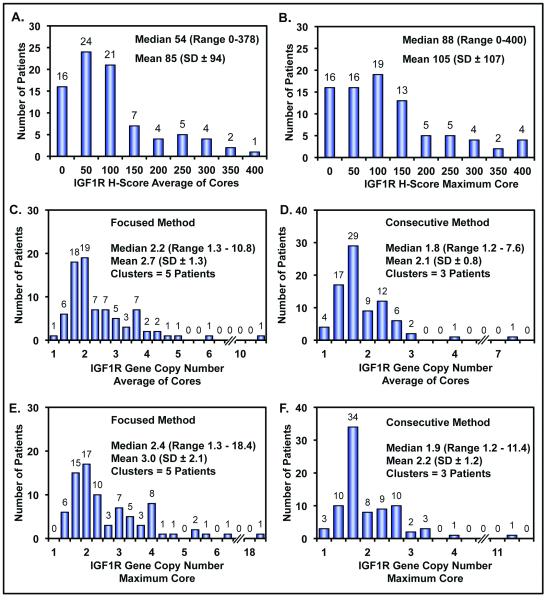
Figure 2.
Histograms showing distribution, frequency, median, range, mean and standard deviation. A and B) IGF1R protein expression from reader DM using either the average of the cores or the core with maximum H-score for each patient, respectively. C and D) IGF1R gene copy number for reader SS using the average of the cores value determined by the focused and consecutive methods. E and F) IGF1R gene copy number for reader SS using the maximum core value determined by the focused and consecutive methods.
Figure 4.
Kaplan-Meier overall survival probability curves. A) according to IGF1R protein expression and B) according to IGF1R gene copy number.
IGF1R gene copy number
IGF1R gene copy number assessed by two pathologists using both the “focused” and “consecutive” methods (see methods section) was evaluable in 81 patients (90%). The average of the three cores, or less if depleted, for each patient’s mean of IGF1R genes per nucleus per core was determined and compared to the core with the highest score from the same patient. Figure 2 shows histograms, using scores from one pathologist (SS), for average of cores with focused and consecutive methods and maximum core with both methods, C-F respectively. There was a high correlation between average of the cores and maximum core using either method (r=0.96,p<0.001, focused; r=0.97, p<0.0001, consecutive) and likewise there was a high correlation between the focused and consecutive methods using either maximum core or average of cores (r=0.98, p<0.0001, maximum core; r=0.97, p<0.0001, average of cores). Similar results were obtained by the second pathologist and therefore, for future analysis for each patient the core with highest mean IGF1R gene copy number per nucleus using the focused method assessed by one pathologist (SS) was selected. The median gene copy number was 2.4 (range 1.3-18.4). Fifteen patients (18.5%) had 4 or more copies of IGF1R per nucleus, and 5 of those had clusters (amplification). Representative pictures showing the range of IGF1R gene copy number are shown inFigure 3. High IGF1R gene copy number tended to associate with increased age (p=0.053), but otherwise there were no associations with clinical and pathological characteristics (Table 2). In the univariate analysis IGF1R gene copy number (with the median as the cut-off between high and low copy number) was not associated with overall survival (log rank p=0.36; Figure 4B). Likewise, no impact on survival was found when IGF1R gene copy number was considered as a continuous variable in the univariate Cox regression (HR 1.011 [0.898-1.139], p=0.85) or when included into multivariate Cox regression model adjusted for age, gender, tumor site, tumor histology and stage. Additionally, when considering a cut-off of 4 or more copies of IGF1R per nucleus, no association with survival was found using either Kaplan-Meier analysis (log rank p=0.32), univariate Cox regression (HR 0.723 [0.367-1.427], p=0.35), or multivariate Cox model adjusting for age, gender, tumor site, tumor histology and stage.
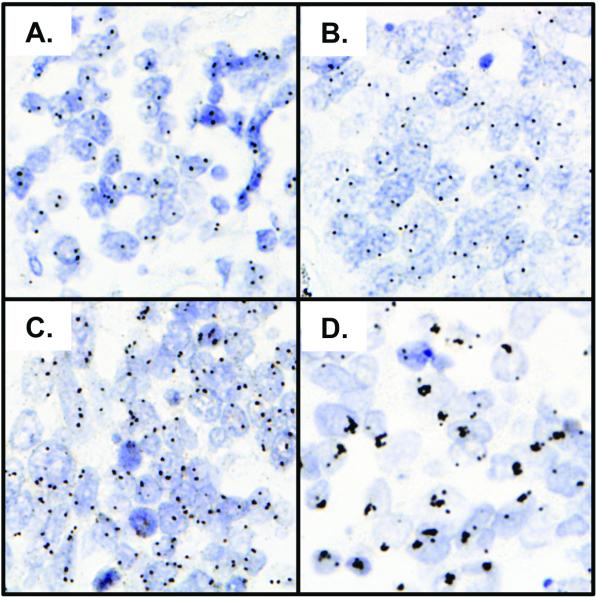
Figure 3.
Representative photographs of IGF1R gene copy number visualized by SISH. Panel A) 2.2 genes per nucleus, Panel B) 3.4 genes per nucleus, Panel C) 5.5 genes per nucleus and Panel D) 18.7 genes per nucleus. Scores based on reader SS focused reading method.
Table 2.
IGF1R Protein Expression and Gene Copy Number vs Patient Characteristics
| IGF1R Protein Expression | IGF1R Gene Copy Number | |||||
|---|---|---|---|---|---|---|
| Characteristics | IGF1R ≤88 | IGF1R >88 | p-value | IGF1R ≤2.4 | IGF1R >2.4 | p-value |
| n | 42 | 42 | 41 | 40 | ||
| Age | ||||||
| Median (Range) | 56 (40-81) | 61 (37-73) | 0.18 | 55 (40-73) | 60 (37-81) | 0.05 |
| Gender, n (%) | ||||||
| Female | 10 (24) | 12 (29) | 0.62 | 11 (27) | 9 (22) | 0.65 |
| Male | 34 (76) | 30 (71) | 30 (73) | 31 (78) | ||
| Pathological Stage, n (%) | ||||||
| IA | 2 (5) | 4(10) | 1(2) | 5(12) | ||
| IB | 11 (26) | 12(29) | 9(22) | 12(30) | ||
| IIA | 3 (7) | 2(5) | 0.76 | 3(7) | 1(2) | 0.38 |
| IIB | 4 (10) | 7(12) | 5(12) | 7(18) | ||
| IIIA | 14 (33) | 11(26) | 14(34) | 10(25) | ||
| IIIB | 8 (19) | 5(12) | 8(19) | 5(12) | ||
| Missing | 0 | 1(2) | 1(2) | 0 | ||
Combined IGF1R protein expression and gene copy number analysis
At least one core with a valid score for combined IGF1R protein expression and gene copy number analysis was obtained in 79 out of 90 patients (88%). There was a significant correlation between IGF1R protein expression and gene copy number (r=0.49, p<0.005).
DISCUSSION
The current study demonstrated that human SCLCs frequently overexpress IGF1R protein and often have increased IGF1R gene copy numbers. According to our knowledge this is the first study to report true IGF1R gene amplification in SCLC. Using the cut-off H-score of 10, IGF1R protein was present in 74% of patients, whereas the median H-score of 88 is comparable to that in non small cell lung cancer (NSCLC) (5,6). The IGF1R gene copy number was increased in 18.5% of tumors (assuming cut off point of average gene copies per nucleus ≥ 4) and IGF1R gene amplifications in 6%. In this study we used 0.6 mm core tissue microarray, which provides sufficient amount of tumor tissue for adequate protein and gene copy number assessment. However, we cannot exclude the possibility of missing some focal gene amplifications as compared with the whole tissue sections. Thus, the real frequency of amplification might be higher in larger tumor specimens.
Insulin and insulin-like growth factor have been known to be essential for SCLC growth for many years, exemplified by in vitro preclinical studies where SCLC used the HITES medium including insulin (7). The first data on IGF receptors in human cancers were published in 1989, and already at that time suggestion was made on their role in controlling tumor growth (8,9). Further studies showed that expression of IGF1 gene is present in various cancer cell lines (10) including SCLC, where expression of IGF1 mRNA was present in 11 of 12 analyzed SCLC cell lines (11). There are only a few studies addressing the influence of IGF1R signaling on SCLC cell line growth. Activation of IGF1R pathway was reported to stimulate SCLC cell line proliferation (12), whereas targeting IGF1R pathway resulted in growth inhibition and induction of apoptosis (13,14). These effects were also seen when SCLC cell lines were exposed to novel selective IGF1R tyrosine kinase inhibitor NVP-ADW742 (15). Further experiments demonstrated that NVP-ADW742 combined with imatinib sensitizes SCLC cells to chemotherapy (16).
Improved therapy for SCLC is desperately needed. Targeting IGF1R receptor is a new promising treatment strategy in oncology. Two different classes of agents are in preclinical and clinical development, including monoclonal antibodies directed against the extracellular domain of IGF1 receptor and tyrosine kinase inhibitors (TKI) targeting its intracellular domain. While studies of IGF1R TKIs are mainly in preclinical phase (17,18), those using monoclonal antibodies already showed its potential to inhibit downstream signaling of IGF1R and to suppress growth of various cell lines (19-21). Anti-IGF1R antibody CP-751,871 was tested in monotherapy (22) and in combination with chemotherapy in patients with NSCLC in a randomized phase II study, showing increased response rate as compared to chemotherapy alone, with acceptable safety profile (23).
Our results showing the presence of IGF1R protein and increased IGF1R gene copy number in a proportion of SCLCs justify further clinical studies using anti-IGF1R approaches in this tumor. Two randomized studies designed to evaluate the safety and efficacy of monoclonal antibodies against IGF1R (CP-751,871 and MK-0646) in combination with standard chemotherapy in ED SCLC will be activated shortly (www.clinicaltrials.gov accessed in March, 2010).
We did not find any association of IGF1R protein expression or IGF1R gene copy number with clinical variables and overall survival. There are no published data on prognostic impact of IGF1R gene copy number in SCLC, whereas the data on prognostic significance of IGF1R protein expression are inconsistent. In one study IGF1R protein expression was found to be associated with longer overall survival in ED SCLC, but not in LD (24), whereas another study of 124 SCLC patients failed to show prognostic relevance of this feature (25). No prognostic impact of IGF1R protein expression was found in surgical series of NSCLC patients (6,26). Increased gene copy number has prognostic value in various malignancies, as exemplified by poor prognosis related to amplification of HER2 in patients with breast cancer, or high EGFR gene copy number in NSCLC (27). The lack of such correlation in our series may partly be due to patient selection and nonstandard treatment. Surgery is not a standard therapeutic option in SCLC and most patients are diagnosed with bronchoscopy or fine needle biopsy. In consequence, tumor specimens are often tiny and insufficient for molecular testing. In our study, SCLC primary tumor samples were obtained from a unique series of patients treated with surgery, as their definite diagnosis of SCLC was established only after the resection. Thus, this group is highly selected with regard to disease stage and general status, and may not be representative for the overall population of SCLC patients. Indeed, prognosis in this group was significantly better in comparison to patients with disseminated SCLC, and more dependent on local control. Thus, our results should be interpreted regarding the specific subset of patients, and require confirmation in a typical population of SCLC patients.
In the current study we had no detailed information on smoking status. We presume that typically for this tumor virtually all patients were smokers, but whether they were current- or former smokers could not be established. Thus, we were not able to correlate IGF-1R protein expression or gene copy number to detailed smoking history. However, in our previous study in NSCLC patients, we did not demonstrate any difference in IGF-1R protein expression or gene copy number related to former- or current smoking status (6). Similarly, no correlation between IGF1R protein expression and smoking status was reported in a recent study in SCLC (24).
As demonstrated in our study, IGF1R protein expression is correlated with IGF1R gene copy number, validating our methodology of IGF1R assessments. Similar findings were recently reported in NSCLC (6). In the present study we applied two different scoring methods and came up with highly consistent results. One minor difference observed was that with the “focused” assessment we detected a slightly higher gene copy number (a median of 2.4 vs. 2.2 genes per nuclei for the cohort), and a few more patients with focal amplification (5 vs. 3), which may reflect the heterogeneity of SCLCs.
Our results demonstrating high IGF1R protein expression and increased IGF1R gene copy number in SCLC should indicate that this disease may be considered for therapies using IGF1R inhibitors. Future studies should include predictive assessment of IGF1R protein and gene copy number for response and outcome to IGF1R inhibitor therapy.
In summary, our study showed that increased IGF1R protein expression and increased gene copy number are common events in SCLC. Furthermore, some patients had tumors with true IGF1R gene amplification. The lack of prognostic implications of both protein expression and gene copy number does not preclude the potential predictive value of these markers for anti-IGF1R therapies.
Acknowledgments
This study was sponsored by a fellowship from the International Association for the Study of Lung Cancer (IASLC) (RD), the Colorado Cancer League (FRH) and from the National Cancer Institute Grant for Specialized Program for Research Excellence (SPORE, 5 P50 CA058187) (FRH).
References
- 1.Govindan R, Page N, Morgensztern D, et al. Changing epidemiology of small-cell lung cancer in the United States over the last 30 years: analysis of the surveillance, epidemiologic, and end results database. J Clin Oncol. 2006;24:4539–4544. doi: 10.1200/JCO.2005.04.4859. [DOI] [PubMed] [Google Scholar]
- 2.Camidge DR, Dziadziuszko R, Hirsch FR. The rationale and development of therapeutic insulin-like growth factor axis inhibition for lung and other cancers. Clin Lung Cancer. 2009;10:262–272. doi: 10.3816/CLC.2009.n.037. [DOI] [PubMed] [Google Scholar]
- 3.Vaira V, Lee CW, Goel HL, et al. Regulation of survivin expression by IGF-1/mTOR signaling. Oncogene. 2007;26:2678–2684. doi: 10.1038/sj.onc.1210094. [DOI] [PubMed] [Google Scholar]
- 4.Badzio A, Kurowski K, Karnicka-Mlodkowska H, et al. A retrospective comparative study of surgery followed by chemotherapy vs. non-surgical management in limited-disease small cell lung cancer. Eur J Cardiothorac Surg. 2004;26:183–188. doi: 10.1016/j.ejcts.2004.04.012. [DOI] [PubMed] [Google Scholar]
- 5.Gong Y, Yao E, Shen R, et al. High expression levels of total IGF-1R and sensitivity of NSCLC cells in vitro to an anti-IGF-1R antibody (R1507) PLoS One. 2009;4:e7273. doi: 10.1371/journal.pone.0007273. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Dziadziuszko R, Merrick DT, Witta SE, et al. Insulin-like Growth Factor Receptor 1 Gene Copy Number is Associated with Survival in Operable Non-Small Cell Lung Cancer (NSCLC): A Comparison between IGF1R FISH, Protein Expression, and mRNA Expression. J Clin Oncol. 2010;28:2174–2180. doi: 10.1200/JCO.2009.24.6611. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Nakanishi Y, Mulshine JL, Kasprzyk PG, et al. Insulin-like growth factor-I can mediate autocrine proliferation of human small cell lung cancer cell lines in vitro. J Clin Invest. 1988;82:354–359. doi: 10.1172/JCI113594. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Arteaga CL, Kitten LJ, Coronado EB, et al. Blockade of the type I somatomedin receptor inhibits growth of human breast cancer cells in athymic mice. J Clin Invest. 1989;84:1418–1423. doi: 10.1172/JCI114315. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Ankrapp DP, Bevan DR. Insulin-like growth factor-I and human lung fibroblast-derived insulin-like growth factor-I stimulate the proliferation of human lung carcinoma cells in vitro. Cancer Res. 1993;53:3399–3404. [PubMed] [Google Scholar]
- 10.Werner H, LeRoith D. The role of the insulin-like growth factor system in human cancer. Adv Cancer Res. 1996;68:183–223. doi: 10.1016/s0065-230x(08)60354-1. [DOI] [PubMed] [Google Scholar]
- 11.Quinn KA, Treston AM, Unsworth EJ, et al. Insulin-like growth factor expression in human cancer cell lines. J Biol Chem. 1996;271:11477–11483. doi: 10.1074/jbc.271.19.11477. [DOI] [PubMed] [Google Scholar]
- 12.Nakanishi Y, Mulshine JL, Kasprzyk PG, et al. Insulin-like growth factor-I can mediate autocrine proliferation of human small cell lung cancer cell lines in vitro. J Clin Invest. 1988;82:354–359. doi: 10.1172/JCI113594. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Camirand A, Pollak M. Co-targeting IGF-1R and c-kit: synergistic inhibition of proliferation and induction of apoptosis in H 209 small cell lung cancer cells. Br J Cancer. 2004;90:1825–1829. doi: 10.1038/sj.bjc.6601682. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Yeh J, Litz J, Hauck P, et al. Selective inhibition of SCLC growth by the A12 anti-IGF-1R monoclonal antibody correlates with inhibition of Akt. Lung Cancer. 2008;60:166–174. doi: 10.1016/j.lungcan.2007.09.023. [DOI] [PubMed] [Google Scholar]
- 15.Warshamana-Greene GS, Litz J, Buchdunger E, et al. The insulin-like growth factor-I (IGF-I) receptor kinase inhibitor NVP-ADW742, in combination with STI571, delineates a spectrum of dependence of small cell lung cancer on IGF-I and stem cell factor signaling. Mol Cancer Ther. 2004;3:527–535. [PubMed] [Google Scholar]
- 16.Warshamana-Greene GS, Litz J, Buchdunger E, et al. The insulin-like growth factor-I receptor kinase inhibitor, NVP-ADW742, sensitizes small cell lung cancer cell lines to the effects of chemotherapy. Clin Cancer Res. 2005;11:1563–1571. doi: 10.1158/1078-0432.CCR-04-1544. [DOI] [PubMed] [Google Scholar]
- 17.Ji QS, Mulvihill MJ, Rosenfeld-Franklin M, et al. A novel, potent, and selective insulin-like growth factor-I receptor kinase inhibitor blocks insulin-like growth factor-I receptor signaling in vitro and inhibits insulin-like growth factor-I receptor dependent tumor growth in vivo. Mol Cancer Ther. 2007;6:2158–2167. doi: 10.1158/1535-7163.MCT-07-0070. [DOI] [PubMed] [Google Scholar]
- 18.Wang Y, Ji QS, Mulvihill M, et al. Inhibition of the IGF-I receptor for treatment of cancer. Kinase inhibitors and monoclonal antibodies as alternative approaches. Recent Results Cancer Res. 2007;172:59–76. doi: 10.1007/978-3-540-31209-3_5. [DOI] [PubMed] [Google Scholar]
- 19.Maloney EK, McLaughlin JL, Dagdigian NE, et al. An anti-insulin-like growth factor I receptor antibody that is a potent inhibitor of cancer cell proliferation. Cancer Res. 2003;63:5073–5083. [PubMed] [Google Scholar]
- 20.Olmos D, Postel-Vinay S, Molife LR, et al. Safety, pharmacokinetics, and preliminary activity of the anti-IGF-1R antibody figitumumab (CP-751,871) in patients with sarcoma and Ewing’s sarcoma: a phase 1 expansion cohort study. Lancet Oncol. 2010;11:129–135. doi: 10.1016/S1470-2045(09)70354-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Tolcher AW, Sarantopoulos J, Patnaik A, et al. Phase I, pharmacokinetic, and pharmacodynamic study of AMG 479, a fully human monoclonal antibody to insulin-like growth factor receptor 1. J Clin Oncol. 2009;27:5800–5807. doi: 10.1200/JCO.2009.23.6745. [DOI] [PubMed] [Google Scholar]
- 22.Haluska P, Shaw HM, Batzel GN, et al. Phase I dose escalation study of the anti insulin-like growth factor-I receptor monoclonal antibody CP-751,871 in patients with refractory solid tumors. Clin Cancer Res. 2007;13:5834–5840. doi: 10.1158/1078-0432.CCR-07-1118. [DOI] [PubMed] [Google Scholar]
- 23.Karp DD, Paz-Ares LG, Novello S, et al. Phase II study of the anti-insulin-like growth factor type 1 receptor antibody CP-751,871 in combination with paclitaxel and carboplatin in previously untreated, locally advanced, or metastatic non-small-cell lung cancer. J Clin Oncol. 2009;27:2516–2522. doi: 10.1200/JCO.2008.19.9331. [DOI] [PubMed] [Google Scholar]
- 24.Chang MH, Lee J, Han J, et al. Prognostic role of insulin-like growth factor receptor-1 expression in small cell lung cancer. APMIS. 2009;117:861–869. doi: 10.1111/j.1600-0463.2009.02545.x. [DOI] [PubMed] [Google Scholar]
- 25.Batus M, Myint R, Coon J, et al. N-cadherin, E-cadherin, ERCC1, c-kit, and IGFR1 expression in small cell lung cancer (SCLC) and potential for new therapeutic targets. IASLC. 2009;P2:191. [Google Scholar]
- 26.Ludovini V, Bellezza G, Pistola L, et al. High coexpression of both insulin-like growth factor receptor-1 (IGFR-1) and epidermal growth factor receptor (EGFR) is associated with shorter disease-free survival in resected non-small-cell lung cancer patients. Ann Oncol. 2009;20:842–849. doi: 10.1093/annonc/mdn727. [DOI] [PubMed] [Google Scholar]
- 27.Hirsch FR, Varella-Garcia M, Bunn PA, Jr., et al. Epidermal growth factor receptor in non-small-cell lung carcinomas: correlation between gene copy number and protein expression and impact on prognosis. J Clin Oncol. 2003;21:3798–3807. doi: 10.1200/JCO.2003.11.069. [DOI] [PubMed] [Google Scholar]