Abstract
Hundreds of million people worldwide have been infected with severe acute respiratory syndrome (SARS), and the rate of global death from SARS has remarkably increased. Hence, the development of efficient drug treatments for the biological effects of SARS is highly needed. We have previously shown that quantum dots (QDs)-conjugated RNA oligonucleotide is sensitive to the specific recognition of the SARS-associated coronavirus (SARS-CoV) nucleocapsid (N) protein. In this study, we found that a designed biochip could analyze inhibitors of the SARS-CoV N protein using nanoparticle-based RNA oligonucleotide. Among the polyphenolic compounds examined, (−)-catechin gallate and (−)-gallocatechin gallate demonstrated a remarkable inhibition activity on SARS-CoV N protein. (−)-catechin gallate and (−)-gallocatechin gallate attenuated the binding affinity in a concentrated manner as evidenced by QDs-conjugated RNA oligonucleotide on a designed biochip. At a concentration of 0.05 μg mL−1, (−)-catechin gallate and (−)-gallocatechin gallate showed more than 40% inhibition activity on a nanoparticle-based RNA oligonucleotide biochip system.
Keywords: SARS, RNA oligonucleotide, quantum dots, inhibitor, screening
Introduction
Severe acute respiratory syndrome (SARS) is an infectious disease that began in Guandong, China in November 2002. It has caused serious infections in many nations, such as Asia, Europe and Canada.1–6 According to the World Health Organization (WHO), the mortality of patients afflicted with SARS is 15% on average and 50% or higher in patients aged 65 years and over.7 SARS-associated coronavirus (SARS-CoV) is an enveloped and positively single-stranded RNA virus with a typical genome size of 29.7 kb. It encodes RNA-directed RNA polymerase and structural proteins, including the spike (S), envelope (E), membrane (M), and nucleocapsid (N) proteins.8–11 The N protein is a 422 amino acids alkaline protein with a short lysine-rich region suggested as the nuclear localization signal. It plays an important role in the process of virus particle assembly by enveloping the entire genomic RNA.12 The SARS-CoV N protein is a major pathological determinant in the host and might cause host cell apoptosis, upregulate the proinflammatory cytokine production, and block innate immune responses. Moreover, the N protein of the SARS-CoV is an important antigen for both the early diagnosis of SARS and the detection of diseases.13 Since SARS broke out in 2003, researchers have made great efforts to develop fast and accurate analytical methods for its early diagnosis.14–18 In addition, due to its essential role in SARS replication, the SARS-CoV N protein is mainly regarded as a prime target for anti-SARS therapy. For this reason, the SARS-CoV N protein is an attractive and crucial target for anti-SARS therapeutic drug discovery.
Polyphenolic compounds are phytochemicals found in numerous plants and fruits.19–21 They have been reported to act as antioxidants, free radical scavengers, metal chelators, and to be antiallergic, anticancer, antioxidant, anti-inflammatory, antifungal, and antiviral and antibacterial agents. In general, these polyphenolic compounds are known to have medicinal and chemopreventive activity in human health.22–25 In particular, (−)-catechin gallate and (−)-gallocatechin gallate are known as a type of catechin. (−)-Catechin gallate and (−)-gallocatechin gallate are the most abundant catechins, particularly in tea and other plants, and they are a potent antioxidants26 with possible therapeutic properties for many disorders, including cancer.27,28 Researchers reported the benefit of catechin gallate from green tea in the treatment of human immunodeficiency virus (HIV) infection, where (−)-catechin gallate has been shown to reduce plaques related to acquired immunodeficiency syndrome-related dementia.29,30 There have also been reports showing that catechin gallate can be beneficial in treating brain,31 prostate,32 and other types of cancers.33
In this study, we report a novel approach for the inhibitor screening of SARS-CoV N protein using a quantum dots (QDs)-conjugated oligonucleotide system with wide applicability for facile and sensitive imaging analysis on a biochip. We elucidated the inhibitor on SARS-CoV N protein identified through a high-throughput screening strategy using an optical nanoparticle-based RNA oligonucleotide. To the best of our knowledge, this is the first report on the inhibition effects of (−)-catechin gallate and (−)-gallocatechin gallate on SARS-CoV N protein using an optical nanoparticle-based RNA oligonucleotide platform.
Materials and methods
Chemicals
EDC (N-(3-dimethylaminopropyl)-N′-ethylcarbodiimide hydrochloride), bovine serum albumin (BSA), ampicillin, kanamycin, (−)-catechin gallate, (−)-gallocatechin gallate, and other molecules were purchased from Sigma-Aldrich Chemical Co (St Louis, MO). Quantum dots (QD605) were purchased from Invitrogen (Carlsbad, CA). Prolinker™-terminated glass slides from Proteogen (Seoul, Republic of Korea) were used. All other chemicals were of the highest grade.
Preparation of RNA oligonucleotide
The production of designed RNA oligonucleotide and primer with T7 promoter sequence were synthesized by Bioneer Co Ltd (Seoul, Republic of Korea) and amplified by polymerase chain reaction (PCR). Template RNA was prepared by in vitro transcription using T7 RNA polymerase (Promega, Madison, WI). All RNA transcript products were separated by 8 M Urea 6% polyacrylamide gel electrophoresis (PAGE) after phenol extraction and ethanol precipitation procedures. The produced RNA oligonucleotide was solved in 0.1% diethylpyrocarbonate (DEPC) solution and stored at −70°C for further experiments.
Subcloning, expression, and purification of SARS-CoV N protein
The gene was amplified by a PCR with the primer set in the direction of sense at 5′-agtggatccatgtctgataatggacccca-3′ and in the direction of antisense at 5′-gccgtcgacttatgcctgagttgaatcagc-3′, containing restriction enzyme sites of BamHI/SalI. PCR was run with the following conditions on a thermal cycler: denaturation at 94°C for 1 minute; annealing at 60°C for 30 seconds; and an extension step at 72°C for 2.5 minutes. The sequence was repeated 35 times and followed by a 7 minute final extension step at 72°C. The PCR product was digested with BamHI/SalI, and then ligated into BamHI/SalI digested expression vector pET 24a+ (Novagen, Madison, WI), and transformed into Escherichia coli DH5α (Stratagene, La Jolla, CA). The colony with insert gene was transformed into E. coli BL21 (DE3) (Stratagene). It was then plated on Luria–Bertani (LB) agar containing 50 μg mL−1 kanamycin. GroES/EL expressing plasmid from E. coli and SARS-CoV N-expressing plasmid, which possessed ampicillin- and kanamycin-resistant markers, were cotransformed into E. coli BL21 (DE3) according to biotransformation procedures. The transformant was grown in a 250 mL flask containing 50 mL LB medium supplemented by 50 μg mL−1 of kanamycin and ampicillin at 37°C until the cell concentration reached OD600 nm of 0.6 and isopropyl-thio-β-D-galactopyranoside (IPTG) of a final concentration of 0.1 mM. It then was left to grow overnight at 25°C with shaking. The cells were harvested by centrifugation at 4000 rpm for 30 minutes at 4°C and resuspended in 100 mM potassium phosphate-buffered saline (pH 7.5) containing 1 mM phenylmethylsulfonyl fluoride (PMSF). The cells were lysed by Sonicator® (F60 Sonic Dismembrator; Fisher Scientific, Fair Lawn, NJ). The cell debris was removed by centrifugation at 13,000 rpm for 30 minutes. The supernatant was collected and the recombinant SARS-CoV N protein was purified with Ni-nitrilotriacetic acid (Ni-NTA) affinity chromatography column (Qiagen, Germany). The supernatant was equilibrated with buffer A (10 mM Tri-HCl, 500 mM NaCl, 50 mM imidazole, 1 mM PMSF, pH 8.0). The bound protein was eluted with buffer B (10 mM Tris-HCl, 500 mM NaCl, 250 mM imidazole, 1 mM PMSF, pH 8.0) at 4°C. The purity of the purified protein was estimated by sodium dodecyl sulfate (SDS)-PAGE in the eluted fractions using 12% polyacrylamide running gels.34 The purity of the enzyme was estimated by SDS-PAGE. The protein concentration was determined as described by the Bradford method.35 The purified sample was supplemented with 50% glycerol and stored at −20°C until use.
Conjugation of QDs and RNA oligonucleotide
The amine group of RNA oligonucleotide was first covalently conjugated onto the surface of the carboxyl terminated QD605 (10 pM). That is, 10 pM of QD605 were conjugated with 400 pM of RNA oligonucleotide with the coupling reagent EDC (N-(3-dimethylaminopropyl)-N-ethyl-carbodiimide hydrochloride, 40 nM), which was used to activate an amide bond formation to produce QDs-conjugated RNA oligonucleotide (QDs-based SARS-CoV N RNA oligonucleotide) at a QDs:RNA oligonucleotide molar ratio of 1:40 for 1 hour at room temperature. QDs-RNA oligonucleotide conjugate was then collected using centrifugal filtration at 15 000 rpm for 30 minutes followed by several washing steps with a Tris buffer (50 mM Tris-HCl [pH 7.4], 5 mM KCl, 100 mM NaCl, 1 mM MgCl2, and 0.1% NaN3). After centrifugal filtration and washing, the pellet of QDs-RNA oligonucleotide was dispersed by brief sonication (22 kHz, amplitude 12 μm, and sonication time 120 seconds) using a sonic dismembrator model F60 (Fisher Scientific).
Fluorescent assay in a confocal laser-scanning microscope
The recombinant SARS-CoV N protein was directly immobilized onto the functional ProLinker™-terminated surface. For the binding of the specific RNA oligonucleotide, the conjugated QDs-conjugated RNA oligonucleotide was facilitated by spotting on an immobilized SARS-CoV N protein chip. Subsequently, the polyphenolic compound used as inhibitor was spotted on the conjugated RNA oligonucleotide and the SARS-CoV N protein. After incubation for 1 hour at 25°C, the chip was then washed three times with phosphate-buffered saline (pH 7.2) for 1 minute. The chip was analyzed by a confocal laser scanning microscope LSM 510 META (Carl Zeiss, Jena, Germany). The signal intensity was determined by software for the LSM 510 (LSM Image Browser; Carl Zeiss). A histogram of the intensity was achieved from the region of the spotted chip. The value of signal intensity was achieved by calculating and expressing it as the mean intensity.
Results and discussion
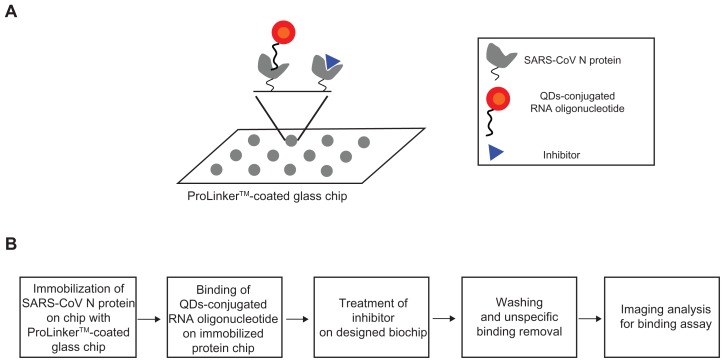
Scheme for inhibitor screening of SARS-CoV N protein on chip
For the inhibitor screening of the SARS-CoV N protein, we designed the QDs-conjugated specific RNA oligonucleotide for specific SARS-CoV N protein targeting: first, the SARS-CoV N protein (1 μL) was immobilized on a glass chip; second, QDs-conjugated RNA oligonucleotide conjugates (1 μL) were bound on an immobilized chip; third, inhibitor treatment was performed on the conjugated RNA oligonucleotide and SARS-CoV N protein; fourth, washing and unspecific binding removal was done; fifth, detection was achieved to show directly the specific recognition of the inhibition effect of SARS-CoV N protein on the biochip. The schematic design of the inhibitor screening for effective monitoring of SARS-CoV N protein is illustrated in Figure 1. To accomplish the feasibility of targeting and imaging, we used QD605 conjugates having an RNA oligonucleotide for SARS-CoV N protein with an emission wavelength as the optical imaging probe.
Figure 1.
A representative scheme for the inhibitor screening of SARS-CoV N protein using QDs-conjugated RNA oligonucleotide on biochip.
Abbreviations: N, nucleocapsid; SARS-CoV, severe acute respiratory syndrome-associated coronavirus; QDs, quantum dots.
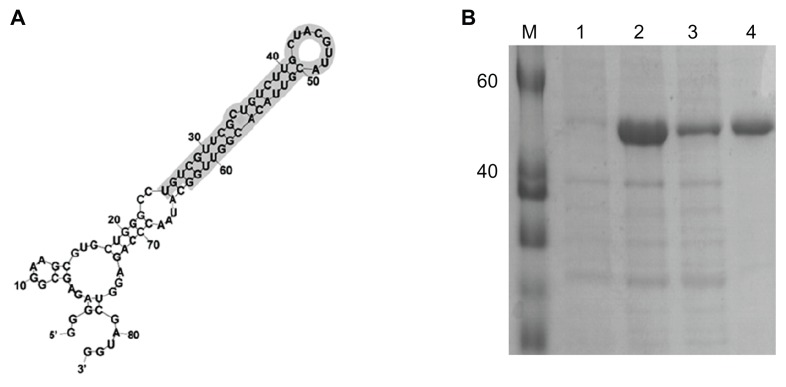
Secondary structure of RNA oligonucleotide and expression and purification of SARS-CoV N protein
Figure 2A presents the secondary structure of RNA oligonucleotide that binds to SARS-CoV N protein. The RNA secondary structure of the used RNA oligonucleotide was analyzed using the Mfold program.36 In order to improve the solubility of the overexpressed recombinant SARS-CoV N protein, the coexpression of N protein with E. coli molecular chaperone GroES/EL was performed. The SARS-CoV N protein was purified by a single chromatography step on a Ni2+ affinity column. The C-terminal his-tagged SARS-CoV N protein was visualized with a molecular mass of approximately 48 kDa on a SDS-PAGE (Figure 2B). The sequence is 5′-gggagagcggaagcgugcugggccugucgguucgcugucuugcuacguuacguuacacgguuggcauaacccagaggucgauggaucccccc-3′.
Figure 2.
(A) Sequence and secondary structure of RNA oligonucleotide that binds to N protein. (B) Purification of SARS-CoV N protein, 12% SDS-PAGE gel showing SARS-CoV N protein with his-tag. M, protein marker; lane 1, before induction form of SARS-CoV N protein; lane 2, total form of SARS-CoV N protein; lane 3, soluble form of SARS-CoV N protein; lane 4, his-tag form of SARS-CoV N protein.
Abbreviations: N, nucleocapsid; SARS-CoV, severe acute respiratory syndrome-associated coronavirus; SDS-PAGE, sodium dodecyl sulphate-polyacrylamide gel electrophoresis; QDs, quantum dots.
Quantum dots (QDs)-conjugated RNA oligonucleotide
The QDs-supported RNA oligonucleotide was conjugated in the reaction for the amide formation from the coupling of 5′-end-amine-modified RNA oligonucleotide at the surface of QDs displaying carboxyl groups via standard EDC coupling. The QDs-conjugated RNA oligonucleotide was confirmed on a 2.5% agarose gel at 100 V in TAE buffer. Agarose gel electrophoresis showed a band with mobility having a different band pattern between the free QDs and the QDs-conjugated RNA oligonucleotide, which confirmed the formation of QDs-conjugated RNA oligonucleotide. The mobility shifts were compared (data not shown). On agarose gel, the QDs-conjugated RNA oligonucleotide showed a lower mobility shift than free QDs, thus demonstrating the amide formation between QDs and RNA oligonucleotide for conjugation.
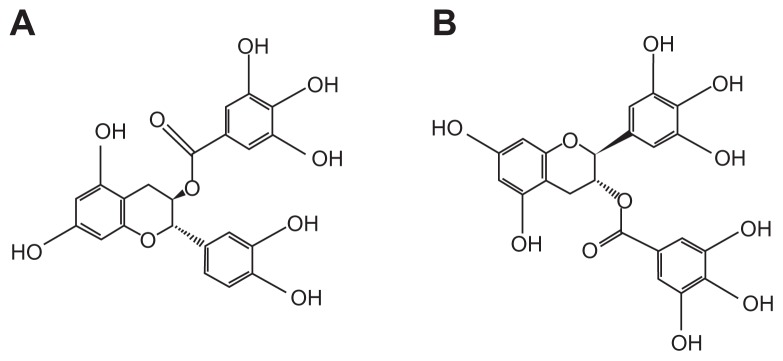
Inhibitory effect of SARS-CoV N protein
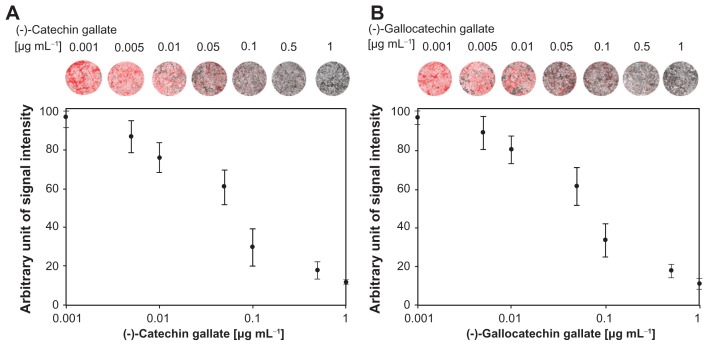
In Table 1, the effects of polyphenolic compounds on the inhibition of the SARS-CoV N protein used in this study are described. Among the polyphenolic compounds screened, (−)-catechin gallate and (−)-gallocatechin gallate showed high anti-SARS-CoV N protein activity. The chemical structures of (−)-catechin gallate and (−)-gallocatechin gallate are shown in Figure 3. Figure 4 shows our elucidation of (−)-catechin gallate; (−)-gallocatechin gallate showed high inhibition activity in a concentrated manner against SARS-CoV N protein. (−)-catechin gallate and (−)-gallocatechin gallate, at 0.005 μg mL−1 or more, concentration-dependently attenuated the binding affinity as evidenced by QDs-RNA oligonucleotide on the designed biochip (Figure 4A and B). At a concentration of 0.05 μg mL−1, (−)-catechin gallate and (−)-gallocatechin gallate showed more than 40% inhibition activity on a QDs-RNA oligonucleotide biochip platform. As shown in Figure 4A and B, (−)-catechin gallate and (−)-gallocatechin gallate showed a similar pattern when comparing the concentration-dependent anti-SARS activity. The half-maximal inhibitory concentration (IC50) values of (−)-catechin gallate and (−)-gallocatechin gallate were found to be approximately 0.05 μg mL−1, respectively (Figure 4A and B). Other polyphenolic compounds to the inhibition of SARS-CoV N protein on the nanoparticle-based RNA oligonucleotide biochip system were detected as nearly similar to the background signal, due to the high affinity with the QDs-conjugated aptamer-SARS-CoV N protein (data not shown). To perform high-throughput screening of the inhibitors, it would be efficient to be able to measure the anti-SARS activity from optical images of a biochip containing multiple reaction compounds. To demonstrate the feasibility of the inhibitor assays, the latter were carried out on a biochip within 60 minutes at 37°C, and optical images were obtained from the reaction mixtures on a plate using a QDs-based imaging system. The inhibition of the anti-SARS activity from (−)-catechin gallate and (−)-gallocatechin gallate were clearly illustrated, and dose dependency was distinctly observable in the optical images.
Table 1.
The effects of polyphenolic compounds on the inhibition of SARS-CoV N protein used in this study
| Compounds | Inhibition activity |
|---|---|
| Quercetin | − |
| Acacetin | − |
| Apigenin | − |
| Baicalein | − |
| Hesperidin | − |
| Morin | − |
| Rutin | − |
| Naringin | − |
| Naringenin | − |
| (−)-Catechin | − |
| (−)-Catechin gallate | + |
| (−)-Gallocatechin gallate | + |
| Diosmin | − |
| Daidzein | − |
| Genistein | − |
| Glycitein | − |
| Kaempferol | − |
| Luteolin | − |
| Myricetin | − |
| Silibinin | − |
| Silymarin | − |
| Orientin | − |
| Oroxylin A | − |
Notes: + indicates inhibition activity on SARS-CoV N protein; − indicates no inhibition activity on SARS-CoV N protein.
Abbreviations: N, nucleocapsid; SARS-CoV, severe acute respiratory syndrome-associated coronavirus.
Figure 3.
Chemical structures of (A) (−)-catechin gallate and (B) (−)-gallocatechin gallate.
Figure 4.
Inhibitory effect of (A) (−)-catechin gallate and (B) (−)-gallocatechin gallate to SARS-CoV N protein.
Abbreviations: N, nucleocapsid; SARS-CoV, severe acute respiratory syndrome-associated coronavirus.
Conclusion
We demonstrated inhibitor screening on a biochip platform using a QDs-conjugated RNA oligonucleotide. We discovered a novel function of (−)-catechin gallate and (−)-gallocatechin gallate as anti-SARS agents. The discovery of anti-SARS agents has been of considerable interest in developing an efficient and effective methodology for high-throughput screening. Our main goal in this study is to demonstrate a proof-of-concept that SARS-CoV N protein can be inhibited and detected with remarkable simplicity and speed. Regarding its application, this designed platform for a novel inhibitor assay possesses significant potential as a target screening. In particular, it is a promising method for inhibitor screening because of its high sensitivity, low cost, rapid response, compatibility for miniaturization, and low labor-intensity. In addition, this platform is expected to be applicable to the inhibitor screening of other types of diseases.
Acknowledgments
This research was supported by the Basic Science Research Program through the National Research Foundation of Korea (NRF), which is funded by the Ministry of Education, Science and Technology (grant no 2011-0010634).
Footnotes
Disclosure
The author reports no conflicts of interest in this work.
References
- 1.Roh C, Jo SK. Quantitative and sensitive detection of SARS coronavirus nucleocapsid protein using quantum dots-conjugated RNA aptamer on chip. J Chem Technol Biotech. 2011;86:1475–1479. doi: 10.1002/jctb.2721. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Peiris JSM, Lai ST, Poon LL, et al. Coronavirus as a possible cause of severe acute respiratory syndrome. Lancet. 2003;361:1319–1325. doi: 10.1016/S0140-6736(03)13077-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Ksiazek TG, Erdman D, Goldsmith CS, et al. A novel coronavirus associated with severe acute respiratory syndrome. N Engl J Med. 2003;348:1953–1966. doi: 10.1056/NEJMoa030781. [DOI] [PubMed] [Google Scholar]
- 4.Drosten C, Gunther S, Preiser W, et al. Identification of a novel coronavirus in patients with severe acute respiratory syndrome. N Engl J Med. 2003;348:1967–1976. doi: 10.1056/NEJMoa030747. [DOI] [PubMed] [Google Scholar]
- 5.Du L, He Y, Zhou Y, et al. The spike protein of SARS-CoV – a target for vaccine and therapeutic development. Nat Rev Microbiol. 2009;7:226–236. doi: 10.1038/nrmicro2090. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Abbott A. Are drugs for SARS on the horizon? Nature. 2003;424:125–126. [Google Scholar]
- 7.World Health Organization. Severe acute respiratory syndrome (SARS): multi-country outbreak. 2003. [Accessed 16 April, 2012]. Available from: http://www.who.int/csr/don/2003_04_11/en/
- 8.Marra MA, Jones SJ, Astell CR, et al. The genome sequence of the SARS-associated coronavirus. Science. 2003;300:1399–1404. doi: 10.1126/science.1085953. [DOI] [PubMed] [Google Scholar]
- 9.Rota PA, Oberste MS, Monroe SS, et al. Characterization of a novel coronavirus associated with severe acute respiratory syndrome. Science. 2003;300:1394–1399. doi: 10.1126/science.1085952. [DOI] [PubMed] [Google Scholar]
- 10.Gorbalenya AE, Koonin EV, Donchenko AP, et al. Coronavirus genome: prediction of putative functional domains in the non-structural polyprotein by comparative amino acid sequence analysis. Nucleic Acids Res. 1989;17:4847–4861. doi: 10.1093/nar/17.12.4847. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Ruan YJ, Wei CL, Ee AL, et al. Comparative full-length genome sequence analysis of 14 SARS coronavirus isolates and common mutations associated with putative origins of infection. Lancet. 2003;361:1779–1785. doi: 10.1016/S0140-6736(03)13414-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Hatakeyama S, Matsuoka Y, Ueshiba H, et al. Dissection and identification of regions required to form pseudoparticles by the interaction between the nucleocapsid (N) and membrane (M) proteins of SARS coronavirus. Virology. 2008;380:99–108. doi: 10.1016/j.virol.2008.07.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Hurst KR, Kuo L, Koetzner CA, et al. A major determinant for membrane protein interaction localizes to the carboxy-terminal domain of the mouse coronavirus nucleocapsid protein. J Virol. 2005;79:13285–13297. doi: 10.1128/JVI.79.21.13285-13297.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Drosten C, Gunther S, Preiser W, et al. Identification of a novel coronavirus in patients with severe acute respiratory syndrome. N Engl J Med. 2003;348:1967–1976. doi: 10.1056/NEJMoa030747. [DOI] [PubMed] [Google Scholar]
- 15.Grant PR, Garson JA, Tedder RS, et al. Detection of SARS coronavirus in plasma by real-time RT-PCR. N Engl J Med. 2003;349:2468–2469. doi: 10.1056/NEJM200312183492522. [DOI] [PubMed] [Google Scholar]
- 16.Poon LLM, Leung CSW, Tashiro M, et al. Rapid detection of severe acute respiratory syndrome (SARS) coronavirus by a loop-mediated isothermal amplification assay. Clin Chem. 2004;50:1050–1052. doi: 10.1373/clinchem.2004.032011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Woo PC, Lau SK, Wong BH, et al. Detection of specific antibodies to severe acute respiratory syndrome (SARS) coronavirus nucleocapsid protein for serodiagnosis of SARS coronavirus pneumonia. J Clin Microbiol. 2004;42:2306–2309. doi: 10.1128/JCM.42.5.2306-2309.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Lau SK, Woo PC, Wong BH, et al. Detection of SARS coronavirus nucleocapsid protein in SARS patients by enzyme-linked immunosorbent assay. J Clin Microbiol. 2004;42:2884–2889. doi: 10.1128/JCM.42.7.2884-2889.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Dixon RA, Ferreira D. Molecules of interest genistein. Phytochem. 2002;60:205–211. doi: 10.1016/s0031-9422(02)00116-4. [DOI] [PubMed] [Google Scholar]
- 20.Middleton JE, Kandaswami C. Effects of flavanoids on immune and inflammatory cell functions. Biochem Pharm. 1992;43:1167–1179. doi: 10.1016/0006-2952(92)90489-6. [DOI] [PubMed] [Google Scholar]
- 21.Heinonen SM, Waehaelae HK, Adlercreutz H. Metabolism of isoflavones in human subjects. Phytochem Rev. 2002;1:175–182. [Google Scholar]
- 22.Galeotti F, Barile E, Curir P, Dolci M, Lanzotti V. Flavonoids from carnation (Dianthus caryophyllus) and their antifungal activity. Phytochem Lett. 2008;1:44–48. [Google Scholar]
- 23.Yamamoto Y, Gaynor RB. Therapeutic potential of inhibition of the NF-κB pathway in the treatment of inflammation and cancer. J Clin Invest. 2001;107:135–142. doi: 10.1172/JCI11914. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Cushnie TPT, Lamb AJ. Antimicrobial activity of flavonoids. Int J Antimicrob Agents. 2005;26:343–356. doi: 10.1016/j.ijantimicag.2005.09.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Spencer JP. Flavonoids: modulators of brain function? Br J Nutr. 2008;99:ES60–ES77. doi: 10.1017/S0007114508965776. [DOI] [PubMed] [Google Scholar]
- 26.Katiyar S, Elmets CA, Katiyar SK. Green tea and skin cancer: photoimmunology, angiogenesis and DNA repair. J Nutr Biochem. 2007;18:287–296. doi: 10.1016/j.jnutbio.2006.08.004. [DOI] [PubMed] [Google Scholar]
- 27.Pyrko P, Schonthal AH, Hofman FM, et al. The unfolded protein response regulator GRP78/BiP as a novel target for increasing chemosensitivity in malignant gliomas. Cancer Res. 2007;67:9809–9816. doi: 10.1158/0008-5472.CAN-07-0625. [DOI] [PubMed] [Google Scholar]
- 28.Aktas O, Waiczies S, Zipp F. Neurodegeneration in autoimmune demyelination: recent mechanistic insights reveal novel therapeutic targets. J Neuroimmunol. 2007;184:17–26. doi: 10.1016/j.jneuroim.2006.11.026. [DOI] [PubMed] [Google Scholar]
- 29.Williamson MP, McCormick TG, Nance CL, et al. Epigallocatechin gallate, the main polyphenol in green tea, binds to the T-cell receptor, CD4: potential for HIV-1 therapy. J Allergy Clin Immunol. 2006;118:1369–1374. doi: 10.1016/j.jaci.2006.08.016. [DOI] [PubMed] [Google Scholar]
- 30.Hamza A, Zhan CG. How can (−)-epigallocatechin gallate from green tea prevent HIV-1 infection? Mechanistic insights from computational modeling and the implication for rational design of anti-HIV-1 entry inhibitors. J Phy Chem B. 2006;110:2910–2917. doi: 10.1021/jp0550762. [DOI] [PubMed] [Google Scholar]
- 31.Das A, Banik NL, Ray SK. Flavonoids activated caspases for apoptosis in human glioblastoma T98G and U87MG cells but not in human normal astrocytes. Cancer. 2009;116:164–176. doi: 10.1002/cncr.24699. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Hsieh TC, Wu JM. Targeting CWR22Rv1 prostate cancer cell proliferation and gene expression by combinations of the phytochemicals EGCG, genistein and quercetin. Anticancer Res. 2009;29:4025–4032. [PMC free article] [PubMed] [Google Scholar]
- 33.Philips BJ, Coyle CH, Morrisroe SN, et al. Induction of apoptosis in human bladder cancer cells by green tea catechins. Biomed Res. 2009;30:207–215. doi: 10.2220/biomedres.30.207. [DOI] [PubMed] [Google Scholar]
- 34.Laemmli UK. Cleavage of structural proteins during the assembly of the head of Bacteriophage T4. Nature. 1970;227:680–685. doi: 10.1038/227680a0. [DOI] [PubMed] [Google Scholar]
- 35.Bradford M. A rapid and sensitive method for the quantitation of microgram quantities of protein utilizing the principle of protein-dye binding. Anal Biochem. 1976;72:248–254. doi: 10.1016/0003-2697(76)90527-3. [DOI] [PubMed] [Google Scholar]
- 36.Jacobson AB, Zuker M. Structural analysis by energy dot plot of a large mRNA. J Mol Biology. 1993;233:261–269. doi: 10.1006/jmbi.1993.1504. [DOI] [PubMed] [Google Scholar]