Abstract
Introduction:
The role of the nicotinic acetylcholine receptor gene cluster on chromosome 15q24-25 in the etiology of nicotine dependence (ND) is still being defined. In this study, we included all 15 tagging single nucleotide polymorphisms (SNPs) within the CHRNA5-CHRNA3-CHRNB4 cluster and tested associations with 30 smoking-related phenotypes.
Methods:
The study sample was ascertained from the Finnish Twin Cohort study. Twin pairs born 1938–1957 and concordant for a history of cigarette smoking were recruited along with their family members (mainly siblings), as part of the Nicotine Addiction Genetics consortium. The study sample consisted of 1,428 individuals (59% males) from 735 families, with mean age 55.6 years.
Results:
We detected multiple novel associations for ND. DSM-IV ND symptoms associated significantly with the proxy SNP Locus 1 (rs2036527, p = .000009) and Locus 2 (rs578776, p = .0001) and tolerance factor of the Nicotine Dependence Syndrome Scale (NDSS) showed suggestive association to rs11636753 (p = .0059), rs11634351 (p = .0069), and rs1948 (p = .0071) in CHRNB4. Furthermore, we report significant association with DSM-IV ND diagnosis (rs2036527, p = .0003) for the first time in a Caucasian population. Several SNPs indicated suggestive association for traits related to ages at smoking initiation. Also, rs11636753 in CHRNB4 showed suggestive association with regular drinking (p = .0029) and the comorbidity of depression and ND (p = .0034).
Conclusions:
We demonstrate novel associations of DSM-IV ND symptoms and the NDSS tolerance subscale. Our results confirm and extend association findings for other ND measures. We show pleiotropic effects of this gene cluster on multiple measures of ND and also regular drinking and the comorbidity of ND and depression.
Introduction
Genes are known to be involved in the etiology of nicotine dependence (ND) with heritability estimates ranging from 40% to 75% (Rose, Broms, Korhonen, Dick, & Kaprio, 2009). Similarly, moderate to high heritability estimates have been reported for smoking initiation (32%–78%; Rose et al., 2009), first smoking experiences (37%; Haberstick, Ehringer, Lessem, Hopfer, & Hewitt, 2011), cigarettes smoked per day (CPD; 40%–56%; Rose et al., 2009), and smoking cessation (11%–86%; Rose et al., 2009). However, studies on smoking and ND based on linkage scans and candidate gene approach has yielded somewhat inconsistent results (Han, Gelernter, Luo, & Yang, 2010), even though multiple pathways and neurotransmitter systems have been implicated (Wang & Li, 2010).
The most established finding involves the CHRNA5-CHRNA3-CHRNB4 nicotinic acetylcholine receptor (nAChR) gene cluster on chromosome 15q24-25. A number of studies have implicated the consistent role of this gene cluster in a wide range of smoking-related phenotypes: ever smoking (smoked 100 cigarettes or more in lifetime; Erlich et al., 2010), CPD (Berrettini et al., 2008; Caporaso et al., 2009; Erlich et al., 2010; Keskitalo et al., 2009; Li et al., 2010; Liu et al., 2010; Saccone et al., 2010; Spitz, Amos, Dong, Lin, & Wu, 2008; Stevens et al., 2008; Thorgeirsson et al., 2008, 2010; Tobacco and Genetics Consortium, 2010), persistent smoking (Bierut et al., 2008), heavy smoking (Stevens et al., 2008), number of quitting attempts (Erlich et al., 2010), age at smoking initiation (Grucza et al., 2010; Schlaepfer et al., 2008; Weiss et al., 2008), pleasurable early smoking experience (Sherva et al., 2008), physical effects reported after smoking first experimental cigarette (Hoft, Stitzel, Hutchison, & Ehringer, 2011), as well as serum cotinine levels (Keskitalo et al., 2009). The CHRNA5-CHRNA3-CHRNB4 gene cluster has been associated with ND defined by the Fagerström Test for Nicotine Dependence (FTND; Heatherton, Kozlowski, Frecker, & Fagerström, 1991) (Bierut et al., 2007; L. S. Chen et al., 2009; X. Chen et al., 2009; Erlich et al., 2010; Johnson et al., 2010; Kim et al., 2011; Li et al., 2010; Saccone et al., 2009; Saccone, Hinrichs, et al., 2007a; Spitz et al., 2008; Weiss et al., 2008; Wessel et al., 2010), as well as the Heaviness of Smoking Index (HSI) consisting of two key items of FTND (Li et al., 2010; Marques-Vidal et al., 2011). In addition, association has been found with the DSM-IV ND diagnosis (American Psychiatric Association, 1994) in a Black sample (Sherva et al., 2010) and in an Islandic sample defining ND as a score of 4 or more on the FTND or endorsement of 3 or more DSM-IV ND criteria (Thorgeirsson et al., 2008). Furthermore, haplotypes within the gene cluster were associated with the multidimensional ND scale, the Wisconsin Inventory of Smoking Dependence Motives, (WISDM-68) among individuals with early smoking initiation (Baker et al., 2009).
The CHRNA5-CHRNA3-CHRNB4 gene cluster codes for the α5, α3, and β4 nAChR subunits. Nicotinic acetylcholine receptors are the primary targets for nicotine and initiate the brain and peripheral responses to smoking. It is thus biologically plausible that genetic variants in the genes coding for the nAChR subunits influence smoking intensity and ND. Supporting this hypothesis, functional studies have shown that a common nonsynonymous variant (rs16969968) in CHRNA5 affects receptor function (Bierut et al., 2008; Kuryatov, Berrettini, & Lindstrom, 2011). Furthermore, rs588765 and correlates are associated with CHRNA5 mRNA levels in brain tissue (Wang et al., 2009). In addition, Chrna5 knockout mice have shown reduced sensitivity to nicotine-induced hypolocomotion and seizures (Salas et al., 2003).
Variation in genes coding for nAChRs has an established role in ND but may also play a role in alcohol dependence (Sherva et al., 2010; Wang et al., 2009), early alcohol use (Schlaepfer et al., 2008), cocaine dependence (Sherva et al., 2010), and opioid dependence (Erlich et al., 2010). The effects of different drugs share biological mechanisms, most importantly the increase of dopamine release from limbic brain areas. The α4α5β2 nAChR subtype is involved in nicotine-stimulated dopamine release (Salminen et al., 2004). Recent work highlights the role of the medial habenula, with high density of α4α5β2 receptors, in responses to nicotine (Fowler, Lu, Johnson, Marks, & Kenny, 2011; Salas, Sturm, Boulter, & De Biasi, 2009).
The CHRNA5-CHRNA3-CHRNB4 gene cluster exhibits extensive linkage disequilibrium (LD). CHRNA5 and CHRNA3 are oriented in opposite directions and share part of their 3′UTR; thus, coordinated expression of these two genes may occur (Solda et al., 2005). Previous studies support the existence of multiple distinct smoking behavior loci within the CHRNA5-CHRNA3-CHRNB4 region (Liu et al., 2010; Saccone et al., 2010; Thorgeirsson et al., 2010; Tobacco and Genetics Consortium, 2010). The large meta-analysis on CPD (Saccone et al., 2010) identified three statistically distinct loci: Locus 1 tagged by rs16969968, Locus 2 tagged by rs578776, and Locus 3 tagged by rs588765. For each loci, a list of so-called proxy single nucleotide polymorphisms (SNPs; Saccone et al., 2010), that is, adjacent SNPs in high LD with these variants, has been provided to ease future study comparisons and meta-analyses.
Both depression and alcohol use are known to co-occur with smoking and ND (Dani & Harris, 2005; Durazzo & Meyerhoff, 2007; Korhonen et al., 2007). Twin and family studies show that significant genetic correlations underlie this co-occurrence (Rose et al., 2009), suggesting shared genetic predisposition. As nAChRs have an important role in mediating the effect of nicotine on the dopaminergic pathway (Benowitz, 2010), it is reasonable to consider that variation in nicotine receptor genes may have pleiotropic effects and potentially associates not exclusively to ND but also to alcohol use and depression. Furthermore, the various ND measurements are suggested to capture slightly different aspects of ND, and by including several measures, we attempt to comprehensively portray the dimensions of ND. Here, the study sample utilized was specifically enriched for smoking and ND, with detailed phenotype profiles including not only assessment of CPD and ND but also age at smoking initiation, diagnoses and symptoms of depression, as well as of alcohol use, abuse, and dependence. The aim of this study was to utilize detailed phenotype information to more comprehensively clarify the involvement of the CHRNA5-CHRNA3-CHRNB4 gene cluster in the etiology of ND and other smoking related traits as well as co-occurring phenotypes.
Methods
Study Sample
The sample collection has been previously described in detail (Broms et al., 2007; Loukola et al., 2008; Saccone, Pergadia, et al., 2007b). Briefly, the study sample was ascertained from the Finnish Twin Cohort study consisting of adult twins born between 1938 and 1957. Based on earlier health questionnaires, the twin pairs concordant for ever smoking were identified and recruited along with their family members (mainly siblings) for the Nicotine Addiction Genetics Finland study (N = 2,265), as part of the consortium including Finland, Australia, and United States. Data collection took place between 2001 and 2005. Because of the relatively old age of the siblings, very few parents were available for the study. The study sample consisted of 1,428 individuals (59% males) from 735 families, with mean age 55.6 years, who smoked on average 19.7 (SD = 9.9) CPD. Ninety-four percent had smoked 100 or more cigarettes over lifetime. Sample characteristics are presented in Table 1. The study was approved by the Ethics committee of the Hospital District of Helsinki and Uusimaa, Finland and by the IRB of Washington University, St. Louis, Missouri, USA.
Table 1.
Basic Characteristics of the Study Sample
| Families | Individuals | MZ twins | DZ twins | Regular smoker | CPD ≥ 20 | DSM-IV NDa | FTND ≥ 4a | M age (years) | |
| Total | 735 | 1,428 | 140 | 970 | 1,347 | 649 | 735 | 687 | 55.6 |
| Males | 845 (59%) | 98 | 589 | 818 | 456 | 440 | 437 | 55.2 | |
| Females | 583 (41%) | 42 | 381 | 529 | 193 | 295 | 250 | 56.1 |
Notes. CPD = cigarettes per day; DSM-IV = Diagnostic and Statistical Manual of Mental Disorders, 4th edition (American Psychiatric Association, 1994); DZ = dizygotic; FTND = Fagerström Test for Nicotine Dependence (Heatherton et al., 1991); MZ = monozygotic; ND = nicotine dependence.
Conditioned for regular smoking.
In order to gain further information on the LD structure, we utilized a total of 679 genome-wide association (GWA) samples ascertained from the population-based FinnTwin12 study (unselected for smoking; Kaprio, 2006).
Phenotypes
Participants were telephone interviewed using the diagnostic Semi-Structured Assessment for the Genetics of Alcoholism (Bucholz et al., 1994) protocol, including an additional section on smoking behavior and ND adapted from the Composite International Diagnostic Interview (Cottler et al., 1991). The customized computer-assisted telephone interviews included more than 100 questions on smoking behavior. All participants provided written informed consent forms by mail. All phenotypes used in the analyses are based on the interview data (survey for the Nicotine Dependence Syndrome Scale [NDSS]; Shiffman, Waters, & Hickcox, 2004). Phenotype definitions are presented in Table 2. The examined binary, continuous, and categorical smoking-related phenotypes are divided into five groups: (a) amount smoked, (b) smoking initiation, (c) ND, (d) DSM-IV–based lifetime major depression, and (e) alcohol use, abuse, and dependence.
Table 2.
Phenotypes Used in the Study
| Phenotype | Definition | Continuous/categorical: min–max (M), binary: prevalence (%) |
| I Amount smoked | ||
| (1) Regular smoker | Smoked at least 100 cigarettes in lifetime and at least once a week for at least two consecutive months. | 94.7% |
| (2) CPD | Number of cigarettes smoked per day during month of heaviest smoking (eight categories: 1–2, 3–5, 6–10, 11–15, 16–19, 20–25, 26–39, ≥40)a | 1–2 to ≥40 (19.7)b |
| (3) Maximum CPD | Maximum number of cigarettes ever smoked during one day (24-hr period). | 2–98 (29.8) |
| (4) Heavy smoker | Smoked 20 cigarettes or more daily during heaviest period of smoking or, 40 or more in a single day. | 51.1% |
| II Smoking initiation | ||
| (5) Age at first puff | Age (years) at first puff of cigarette (“How old were you the very first time you smoked even a puff of a cigarette?”). | 3–54 (14.1) |
| (6) Age at first cigarette | Age (years) when first whole cigarette smoked (“How old were you the first time you smoked a whole cigarette?”). | 4–54 (15.8) |
| (7) Immediacy | Tried smoking again on same or next day (“After you tried smoking a cigarette for the first time, how soon did you try smoking again?”). | 18.3% |
| (8) Second cigarette | Second cigarette smoked same or next day after first one (“After you first smoked a whole cigarette, how long was it before you smoked your second whole cigarette?”). | 51.6% |
| (9) Age of onset of weekly smoking | Age (years) when started to smoke weekly (“How old were you when you first smoked a cigarette at least once a week for at least two months in a row?”). | 6–54 (17.5) |
| (10) Age of onset of daily smoking | Age (years) when one started to smoke daily (“How old were you when you first smoked cigarette every day or nearly every day for at least two months in a row?”). | 7–54 (18.4) |
| (11) First time sensation | Sensation felt after smoking the first cigarette or first puffs (“While smoking your very first cigarettes, did you (1) like the taste or smell of the cigarette, (2) cough, (3) feel dizzy or light headed, (4) feel more relaxed, (5) get a headache, (6) feel a pleasurable rush or buzz, (7) feel your heart racing, (8) feel nauseated, like vomiting, (9) feel your muscles tremble or become jittery, (10) feel burning in your throat”). Sum score of 10 questions (items #1, #4, and #6 were reverse scored before summation): 0 points if answered “No,” 1 = “A little bit,” 2 = “Some,” 3 = “Quite a bit,” and 4 = “A great deal.” Cronbach’s alpha = .70. | 3.6–15.8 (10.1) |
| III Nicotine dependence | ||
| (12) DSM-IV ND | ND by DSM-IV diagnosis (three symptoms or more of seven occurring within a year). | 51.5% |
| (13) DSM-IV ND symptoms | Number of DSM-IV ND symptoms from 0 to 7. | 0–7 (3.0) |
| (14) FTND score | FTND score: 0–10 points. | 0–10 (3.7) |
| (15) FTND (≥4) | Nicotine dependent if 4 or more of 10 points in FTND. | 48.1% |
| (16) FTND TTF | TTF in the morning (one item of the FTND scale). Five categories: 0–5 min, 6–15 minc, 16–30 minc, 31–60 min, and >60 min. | 1–5 (3.1)b |
| (17) NDSS sum score | NDSSd sum score as a continuous variable from 0 to 56. | 0–56 (21.0) |
| (18) NDSS drive/priority factore | Drive reflects craving, withdrawal, and smoking compulsions; priority reflects preference for smoking over other reinforces. | −2.0–3.3(−0.1) |
| (19) NDSS stereotypy/continuity factore | Continuity reflects the regularity of smoking rate; stereotypy reflects the invariance of smoking. | −2.6–2.3(−0.3) |
| (20) NDSS tolerance factore | Tolerance reflects reduced sensitivity to the effects of smoking. | −2.5–2.7(−0.1) |
| (21) DSM-IV ND and HSI | Co-occurrence of DSM-IV ND diagnosis and HSI (Heatherton et al. 1989; HSI ≥ 4 of 6 points). | 17.9% |
| IV Depression | ||
| (22) DSM-IV depression | Lifetime Major Depressive Disorder DSM-IV diagnosis (five symptoms or more of nine, including insufficiency). | 17.2% |
| (23) DSM-IV depression symptoms | Number of DSM-IV defined depression symptoms ranging from 0 to 9. | 0–9 (1.6) |
| (24) Comorbidity of depression and ND | Fulfilling the criteria of depression and nicotine dependence (either DSM-IV ND criteria or FTND ≥ 4). | 13.8% |
| V Alcohol use | ||
| (25) Regular drinker | Drinks at least one alcoholic drink at least once a week. | 67.2% |
| (26) Heavy drinker | Drinks at least five or more alcoholic drinks once a week. | 50.8% |
| (27) DSM-IV alcohol dependence | DSM-IV alcohol dependence (three or more of seven symptoms occurring within a year). | 25.6% |
| (28) Binge drinker | Drinks at least five alcoholic drinks at least three days per year. | 79.4% |
| (29) Comorbidity of binge drinking and ND | Comorbidity of binge drinking (five or more alcoholic drinks three or more days per year) and FTND (≥4 points). | 41.5% |
| (30) Maximum drinks | Maximum number of alcoholic drinks ever drunk during one day (24-hr period). | 1–72 (15.4) |
Notes. The 30 examined binary, continuous, and categorical phenotypes are presented in five groups: (I) amount smoked, (II) smoking initiation, (III) ND, (IV) DSM-IV–based lifetime major depression, and (V) alcohol use, abuse, and dependence. CPD = cigarettes per day; DSM-IV = Diagnostic and Statistical Manual of Mental Disorders, 4th edition; FTND = Fagerström Test for Nicotine Dependence; HSI = Heaviness of Smoking Index; ND = nicotine dependence; TTF = time to first cigarette.
In the statistical analyses of the CPD variable, original categorical observations (1–8) were replaced with class means of CPD (1.5, 3.5, 8, 13, 17.5, 22.5, 32.5, and 45 CPD, respectively). Regression coefficients can therefore be interpreted as the average change in number of cigarettes smoked per day when the number of minor allele is increased by one.
Weighted arithmetic mean, where weights determined by class frequencies.
Categorization differs from original four categories (Heatherton et al., 1991), that is, 6–30 min is split into 6–15 min and 16–30 min. In our dataset, 46% of smokers belong to the group of 6–30 min, and from the smoking behavior point of view, there is a significant difference whether one smokes the first cigarette within 6 min or 30 min from waking up. In this dataset, 22% of smokers belong to the 6–15 min and 24% to the 16–30 min group.
NDSS = Nicotine Dependence Syndrome Scale (Shiffman et al., 2004).
Factor structure of the NDSS scale with 31 items conditioned for current smoking and CPD more than 10 generated a three-factor solution (as reported also earlier in Broms et al., 2007). Factors accounted for 92% of the common variance among items: 1. drive/priority factor, 11 items (Cronbach’s alpha (α) = .88, accounted for 33%); 2. stereotypy/continuity factor, 9 items (α = .89, accounted for 33%); and 3. tolerance factor, 5 items (α = .82, accounted for 26%).
Prevalences and correlations of phenotypes were calculated with Stata 11.1 statistical software (StataCorp, 2009). Phenotype correlations are presented in Supplementary Table 1.
Genotyping
Participants were mailed a blood sample kit, which they took to the nearest health center laboratory for phlebotomy, and the venous blood samples were returned to the National Public Health Institute by mail. DNA was extracted by standard methods.
A total of 303 individuals were genotyped using Sequenom’s homogeneous Mass Extend (hME) and iPLEX Gold technology (Sequenom, San Diego, CA, USA). Tagging SNPs were selected based on the HapMap Project (http://www.hapmap.org) and NCBI (http://www.ncbi.nlm.nih.gov) databases. The selected variants were biallelic and had a minor allele frequency (MAF) for more than 10% in the Caucasian population. The ability to amplify the flanking regions of each SNP was determined by using the applications SNPper (http://www.snpper.chip.org) and RealSNP (http://www.realsnp.com), which define, respectively, the most reliable regions for designing primers and the quality of the amplicons. All tagging SNPs failing during the procedure were replaced by newly generated tagging SNPs proposed by Haploview (Barrett, Fry, Maller, & Daly, 2005). The polymerase chain reaction (PCR) and extension primers were designed using Sequenom’s MassARRAY Assay Design software (version 2.0). SNPs were genotyped in 384-well plates according to manufacturer’s instructions. For quality controls, each plate contained at least eight water controls and 22 duplicate samples. PCR reactions were performed in a total reaction volume of 5 μl using 20 ng of genomic DNA. The alleles were automatically called by Sequenom's MassARRAY Typer Analyzer software and verified by two independent persons. Further, marker-specific quality controls included a call rate more than 80% and a Hardy–Weinberg equilibrium (HWE) p value > .01 (estimated using unrelated individuals). Mendelian errors were excluded using PedCheck (O’Connell & Weeks, 1998).
For 1,125 individuals, genotypes for the same 15 SNPs were derived from GWA data. Genotyping was performed at the Welcome Trust Sanger Institute (Hinxton, UK) on the Human670-QuadCustom Illumina BeadChip (Illumina, Inc., San Diego, CA, USA). The data were checked for minor allele frequency (>1%), genotyping success rate per SNP and per individual (>95%), HWE (p > 1 × 10−6), gender, and heterozygosity. In addition, to check whether any individuals were unexpectedly related to each other, an multidimensional scaling plot (using a pairwise-IBS matrix) with only one member of each known family was created. After the pedigree was reassured to be correctly formulated, the basic filters (MAF, genotyping success, HWE) were reapplied to the data. Seven of the markers were genotyped and eight were imputed using the software IMPUTE v2.1.0 (Howie et al., 2009). The reference panel used in the imputation was HapMap rel#24 CEU—NCBI Build 36 (dbSNP b126), which is available on the IMPUTE website (https://mathgen.stats.ox.ac.uk/impute/impute_v1.html#Using_IMPUTE_with_the_HapMap_Data). The posterior probability threshold for “best-guess” imputed genotype was .9. Genotypes below the threshold were set to missing.
Marker quality controls are presented in Supplementary Table 2.
Statistical Analyses
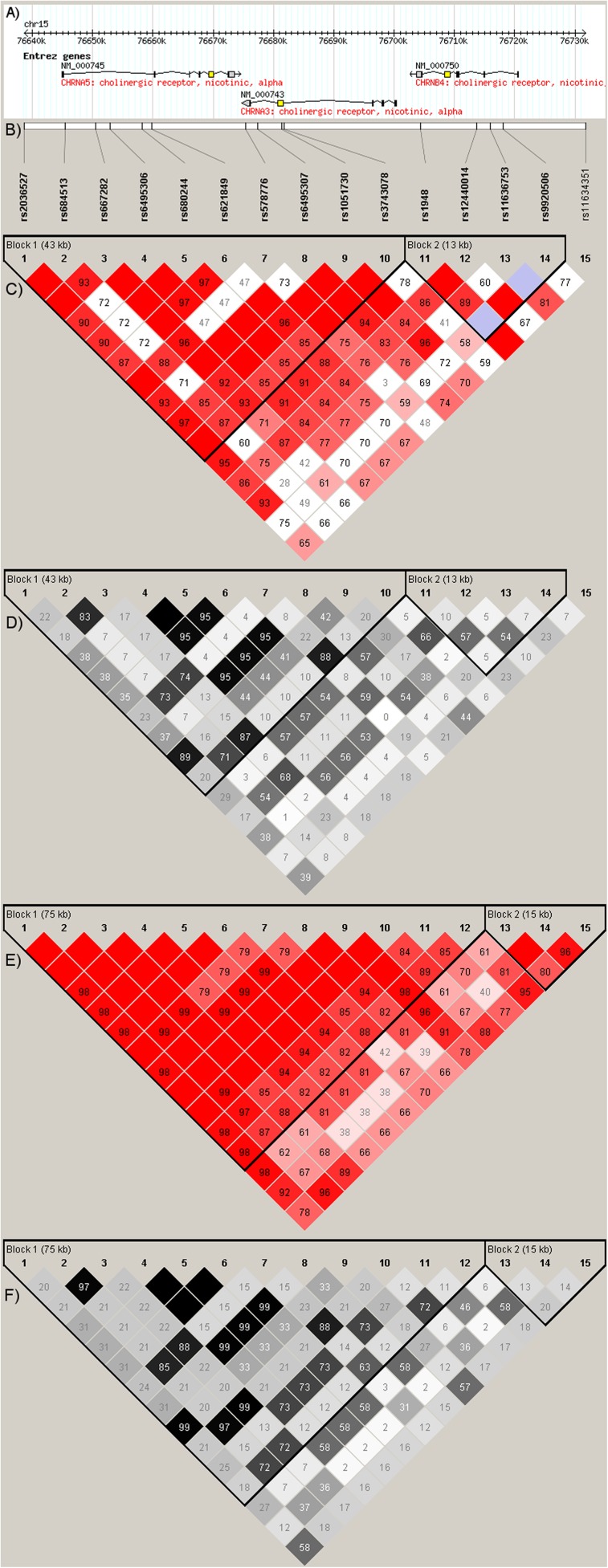
The LD between SNPs was estimated among nonrelated individuals (one per family) by using Haploview 4.2 (Barrett et al., 2005). The pairwise comparisons of markers more than 500 kb apart were excluded. Haplotype blocks were defined according to the “solid spine of LD” algorithm by using the default threshold values for block estimation (Figure 1).
Figure 1.
(A) Gene structures in the CHRNA5-CHRNA3-CHRNB4 region, (B) genotyped single nucleotide polymorphisms, (C) D′ in the HapMap CEPH data (NCBI Build 36), (D) r2 in the HapMap CEPH data, (E) D′ in the study sample (nonrelated individuals; one per family), (F) r2 in the study sample.
The associations between discrete phenotypes and candidate SNPs (qualitative association) were estimated with pseudomarker (Goring & Terwilliger, 2000), which performs separate and joint linkage and LD analyses, testing each marker locus against a phenotype-based “pseudomarker” locus. This likelihood-based estimation method is numerically equivalent to model-free analysis and efficiently uses data on all family types. Both recessive and dominant models (default parameters) were fitted. We report uncorrected p values minimized over “LD given linkage,” “LD given no linkage,” and “LD and linkage” (joint test) as well as dominant and recessive models.
The associations between continuous phenotypes and candidate SNPs (quantitative association) were performed with quantitative transmission disequilibrium test (QTDT; Abecasis, Cardon, & Cookson, 2000). In the analysis, the proportion of alleles shared identically by descent (IBD) was estimated by multipoint computation of MERLIN (Abecasis, Cherny, Cookson, & Cardon, 2002) to extract maximal inheritance information from the pedigrees. Prior to analysis, we estimated our sample not being stratified with the appropriate “population stratification” model of QTDT, which compares the between and within family components of association (Abecasis et al., 2000). The total association model was used, allowing powerful analysis of the sample, including incomplete families. In the analysis, the variance components “polygenic,” “nonshared environment” (environmental effects unique to each family member), “common environment” (environmental effects shared by all related individuals), “nuclear family environment” (environmental effects shared by all members of a nuclear family), and “twin environment” (environmental effects shared only by twins) were used to model the phenotypic similarities between the pedigree members. In the analysis of traits related to age of initiation (age at first puff, age at first cigarette, age of onset of weekly smoking, age of onset of daily smoking), sex was included as a covariate, whereas both sex and age at recruitment were used as covariates for all other continuous traits.
To account for multiple testing we used a modified Bonferroni correction to set p value thresholds for significant and suggestive association signals. Since neither analyzed markers nor traits are independent, we utilized an established methodology to evaluate the numbers of corresponding independent markers and traits with the programs SNPSpD and matSpD, respectively (Cheverud, 2001; Li & Ji, 2005; Nyholt, 2004), and their MeffLi and VeffLi estimates (Li & Ji, 2005) were used as they were smaller than Meff and Veff, respectively, as recommended by the author (http://gump.qimr.edu.au/general/daleN/SNPSpD/). The trait “regular smoker” was not accounted for when estimating the number of independent traits, as it is the ascertainment criteria for our families. In our dataset, the number of independent markers was 6.0022 and the number of independent traits was 16.956. A p value threshold of .0005 for significant association was achieved by dividing p = .05 by the product of the number of independent markers and the number of independent traits. A p value threshold of .0083 for suggestive association was achieved by dividing p = .05 by the number of independent markers.
Nonnormally distributed continuous variables (kurtosis and/or skewness >1 or <−1) were transformed with square root or 10-base logarithm, whichever provided most normal-like distribution. Trait values above 30 years considered as outliers were removed from the analysis for phenotypes measuring the age at first puff (seven individuals), age at first cigarette (nine individuals), age of onset of weekly smoking (20 individuals), and age of onset of daily smoking (28 individuals).
To estimate effect sizes for all SNPs showing significant or suggestive p values in the genetic association analyses, we performed regression analyses with Stata 11.1 statistical software (StataCorp, 2009) by using an additive model. Odds ratios (OR) and beta coefficients (β) with 95% CIs were reported for binary and quantitative traits, respectively. Regression analyses were adjusted for sex and age. Furthermore, individuals were clustered based on the possible lack of statistical independence of observations of subjects who came from the same family (Williams, 2000).
Results
In the study sample, the LD blocks were similar to those in the HapMap CEPH data (Figure 1) and the somewhat stronger LD between markers is in agreement with previous findings from the Finnish population (Service et al., 2006). In the HapMap CEPH data, CHRNB4 SNPs lie outside the CHRNA5–CHRNA3 LD block, whereas in the study sample, the LD block extends to rs1948 and rs12440014 in CHRNB4 (Figure 1). The LD blocks were almost identical between the study sample and the FinnTwin12 sample (data not shown).
We detected significant association with a variety of traits measuring ND, the strongest signal emerging for DSM-IV ND symptoms in Locus 1 (rs2036527 p = .000009, rs1051730 p = .00002) and Locus 2 (rs578776 p = .0001, rs667282 p = .0002). A significant association was observed with DSM-IV ND diagnosis in Locus 1 (rs2036527 p = .0003, rs1051730 p = .0004). Altogether we detected significant or suggestive p values for six traits measuring ND (DSM-IV ND, DSM-IV ND symptoms, FTND score, NDSS tolerance factor, Time to first cigarette (TTF), and DSM-IV ND + HSI; Table 3).
Table 3.
Association Analyses Results for Traits Showing Significant (p < .0005, Bold and Underlined) or Suggestive (.0005 < p < .008, Bold) Association
| Amount smoked |
Smoking initiation |
Nicotine dependence |
Depression | Alcohol use | |||||||||
| Marker | Gene | CPD | Maximum CPD | Age of onset of weekly smoking | Age of onset of daily smoking | DSM-IV ND | DSM-IV ND symptoms | FTND score | FTND TTF | NDSS tolerance factor | DSM-IV ND and HSI | Comorbidity of depression and ND | Regular drinker |
| rs2036527 L1 | CHRNA5 | 0.0073 | 0.0710 | 0.9188 | 0.2158 | 0.0003 | 0.000009 | 0.0004 | 0.0061 | 0.0844 | 0.0047 | 0.0676 | 0.3381 |
| rs684513 | CHRNA5 | 0.0336 | 0.0118 | 0.0034 | 0.0065 | 0.0609 | 0.0005 | 0.0447 | 0.0567 | 0.9696 | 0.0531 | 0.4674 | 0.2136 |
| rs667282 L2 | CHRNA5 | 0.0273 | 0.0120 | 0.0070 | 0.0032 | 0.0137 | 0.0002 | 0.0254 | 0.0848 | 0.9959 | 0.0370 | 0.4252 | 0.0189 |
| rs6495306 L3 | CHRNA5 | 0.7143 | 0.4308 | 0.0588 | 0.2134 | 0.4361 | 0.5403 | 0.2601 | 0.4519 | 0.0480 | 0.6088 | 0.0303 | 0.1686 |
| rs680244 L3 | CHRNA5 | 0.7607 | 0.4795 | 0.0460 | 0.1794 | 0.3855 | 0.5335 | 0.2753 | 0.4555 | 0.0468 | 0.5717 | 0.0203 | 0.0088 |
| rs621849 L3 | CHRNA5 | 0.7060 | 0.3919 | 0.0450 | 0.1254 | 0.4102 | 0.5538 | 0.2617 | 0.3998 | 0.0271 | 0.3757 | 0.0250 | 0.1685 |
| rs578776 L2 | CHRNA3 | 0.0125 | 0.0125 | 0.0054 | 0.0077 | 0.0217 | 0.0001 | 0.0047 | 0.0132 | 0.8518 | 0.0315 | 0.7507 | 0.0411 |
| rs6495307 L3 | CHRNA3 | 0.6800 | 0.4627 | 0.0581 | 0.1932 | 0.4648 | 0.5274 | 0.2621 | 0.3762 | 0.0544 | 0.5503 | 0.0228 | 0.1713 |
| rs1051730 L1 | CHRNA3 | 0.0077 | 0.0711 | 0.5518 | 0.1354 | 0.0004 | 0.00002 | 0.0005 | 0.0091 | 0.0138 | 0.0094 | 0.0726 | 0.3098 |
| rs3743078 | CHRNA3 | 0.0198 | 0.0084 | 0.0062 | 0.0033 | 0.0118 | 0.0001 | 0.0108 | 0.0545 | 0.7586 | 0.0632 | 0.3510 | 0.0619 |
| rs1948 | CHRNB4 | 0.8906 | 0.7394 | 0.0156 | 0.1698 | 0.1662 | 0.4901 | 0.4708 | 0.7041 | 0.0071 | 0.6646 | 0.0861 | 0.0615 |
| rs12440014 | CHRNB4 | 0.0722 | 0.0074 | 0.0957 | 0.0287 | 0.1264 | 0.0040 | 0.0411 | 0.1014 | 0.5114 | 0.1347 | 0.4426 | 0.3351 |
| rs11636753 | CHRNB4 | 0.0909 | 0.4171 | 0.4022 | 0.8948 | 0.3541 | 0.2331 | 0.0603 | 0.0830 | 0.0059 | 0.0403 | 0.0034 | 0.0029 |
| rs9920506 | CHRNB4 | 0.6322 | 0.9541 | 0.9561 | 0.4164 | 0.3091 | 0.1238 | 0.8613 | 0.8019 | 0.6695 | 0.5209 | 0.0672 | 0.3220 |
| rs11634351 | CHRNB4 | 0.1672 | 0.5042 | 0.4820 | 0.0902 | 0.0377 | 0.0011 | 0.0535 | 0.3443 | 0.0069 | 0.0939 | 0.0813 | 0.0168 |
Note. L1 = Locus 1 (tagged by rs16969968): proxy SNP identifier from Saccone et al. (2010) and by personal communication by S. M. Hartz. L2 = Locus 2 (tagged by rs578776): proxy SNP identifier from Saccone et al. (2010) and by personal communication by S. M. Hartz. L3 = Locus 3 (tagged by rs588765): proxy SNP identifier from Saccone et al. (2010) and by personal communication by S. M. Hartz. CPD = cigarettes per day; DSM-IV = Diagnostic and Statistical Manual of Mental Disorders, 4th edition; FTND = Fagerström Test for Nicotine Dependence; HSI = Heaviness of Smoking Index; ND = nicotine dependence; NDSS = Nicotine Dependence Syndrome Scale; TTF = time to first cigarette.
We detected suggestive association with the categorized variable of CPD in Locus 1 (rs1051730 p = .0077, rs2036527 p = .0073) and maximum CPD at rs12440014 (p = .0074) (Table 3). Additionally, four SNPs (rs578776 and rs667282 at Locus 2, rs684513, rs3743078) indicated suggestive association for the age of onset of both weekly and daily smoking (Table 3).
Furthermore, rs11636753 in CHRNB4 showed suggestive association with both regular drinking (p = .0029) and the comorbidity of depression and ND (p = .0034) (Table 3).
Association analysis results for all phenotypes are presented in Supplementary Table 3.
For those SNPs showing significant associations, notable effect sizes were detected (Supplementary Table 4). In Locus 1, DSM-IV ND diagnosis showed an OR of 1.25 (p = .0085), and corresponding effect size for DSM-IV ND symptoms was a beta coefficient of 0.30 (p < .0001). Similarly, Locus 1 showed a beta coefficient of 0.33 (p = .0017) for the FTND score. Locus 2 showed plausible protective effect for DSM-IV ND symptoms (β = −0.28, p = .0001). For SNPs showing suggestive association, modest effect sizes were detected. As an exception, a notable protective effect (OR = 0.76, p = .0239) was detected for rs11636753 in the comorbidity of depression and ND. The effect size for the regular drinker phenotype failed to meet statistical significance (p > .05; Supplementary Table 4).
Discussion
Multiple distinct loci at the CHRNA5-CHRNA3-CHRNB4 locus on 15q24-25 appear to affect smoking behavior (Liu et al., 2010; Saccone et al., 2010; Thorgeirsson et al., 2010; Tobacco and Genetics Consortium, 2010). In a previous meta-analysis including the current study sample (Saccone et al., 2010), three SNPs in the CHRNA5-CHRNA3-CHRNB4 gene cluster (rs16969968, rs578776, and rs588765) showed association with CPD (heavy i.e., CPD > 20 vs. light i.e., CPD ≤ 10). Presuming the previous evidence as an “a priori” hypothesis, we set out to extend and deepen the analyses of these three loci by utilizing previously established proxy SNPs (Saccone et al., 2010; S. M. Hartz [personal communication, May 30, 2011]) as well as all detected tagging SNPs within this gene cluster. We performed association analyses in a Finnish family sample of twins and their siblings ascertained specifically for smoking and ND using a broad spectrum of phenotypes covering aspects of smoking behavior, ND, and depression, as well as alcohol use, abuse, and dependence.
In agreement with previous evidence from extensive studies with over 1,40,000 participants (Liu et al., 2010; Saccone et al., 2010; Thorgeirsson et al., 2008, 2010; Tobacco and Genetics Consortium, 2010), we detected some association with CPD. Larger samples may be needed to detect significant association with CPD, as shown by a study of over 13,000 Icelandic smokers (Thorgeirsson et al., 2008) in which variation in Locus 1 (rs1051730) accounted for only about 1% of the variance in CPD, the average effect per allele being about 1 CPD (Thorgeirsson et al., 2008). In agreement with this, an effect size of a beta coefficient 1.05 was detected in our sample, roughly corresponding to an increment of one cigarette per day for each allele of the locus.
The involvement of the CHRNA5-CHRNA3-CHRNB4 gene cluster in FTND has been well documented in the literature, whereas studies reporting findings for DSM-IV ND are sparse. It is unclear whether this is due to lack of positive findings, publication bias, challenges in reliably creating DSM-IV ND diagnosis, or the predominance of FTND in measuring ND, possible due to ease of assessment. Here, we report a significant novel association with the number of DSM-IV ND symptoms, with Locus 1 conferring risk (β = 0.30) and Locus 2 showing plausible protective effect (β = −0.28), in agreement with a previous study reporting similar effects for CPD (Saccone et al., 2010). We also report the first significant association with DSM-IV ND diagnosis in a Caucasian sample, replicating previous findings in Blacks (Sherva et al., 2010). Further, we replicate association with the FTND as a continuous trait (X. Chen et al., 2009; Erlich et al., 2010; Kim et al., 2011; Wessel et al., 2010). These two widely used ND measures are suggested to extract partly different dimensions of ND (Moolchan et al., 2002; Piper, McCarthy, & Baker, 2006; Piper et al., 2008). DSM-IV predominantly measures loss of control in terms of smoking behavior, including cognitive, behavioral, and physiological symptoms, resulting in tolerance, withdrawal, and compulsive drug-taking behavior (American Psychiatric Association, 1994). FTND aims to measure the construct of physical dependence, associating with cessation outcome, predicting smoking relapse, and having the key component of difficulty to stand reduced nicotine levels (Haddock, Lando, Klesges, Talcott, & Renaud, 1999; Heatherton et al., 1991).
We demonstrate the first evidence of genetic association of the NDSS tolerance factor, TTF, as well as the combination of DSM-IV ND diagnosis and HSI (a combination of CPD and TTF) in the CHRNA5-CHRNA3-CHRNB4 locus. Functional evidence for the involvement of the CHRNA5-CHRNA3-CHRNB4 gene cluster in tolerance includes knockout mice showing association with alpha4beta2 nAChR subunits and the development of tolerance (McCallum, Collins, Paylor, & Marks, 2006; Tapper, McKinney, Marks, & Lester, 2007). Further, chronically treated mice lacking the beta4 subunit show an increased tolerance to an acute dose of nicotine (E. E. Meyers and M. J. Marks [personal communication, May 31, 2011]). Tolerance to nicotine is defined by the ability to smoke increased amounts of cigarettes without experiencing toxic effects (Piper et al., 2006). Repeated exposure to nicotine leads to tolerance (neuroadaptation), and as neuroadaptation develops, the number of binding sites on the nAChRs in the brain increases, probably in response to nicotine-mediated desensitization of receptors (Benowitz, 2010).
The gene cluster has previously been associated with the WISDM (Piper et al., 2004) tolerance factor among 886 early-onset smokers from the United States (Baker et al., 2009). NDSS and WISDM are measuring somewhat different aspects of ND and have a slightly different focus on the tolerance dimension as well (Piper et al., 2008). The use of multidimensional scales such as NDSS and WISDM potentially aids in deciphering the construct and nature of ND (Piper et al., 2006). Our results confirm the CHRNA5-CHRNA3-CHRNB4 association to tolerance, one of the dimensions of ND, which supposedly also is embedded within the traditional unidimensional scales, DSM-IV and FTND.
Twin studies suggest that genetic influences on age at smoking initiation are correlated with the amount smoked (Broms, Silventoinen, Madden, Heath, & Kaprio, 2006; Morley et al., 2007). Several SNPs within the CHRNA5-CHRNA3-CHRNB4 locus associate with multiple phenotypes measuring age at onset of smoking, consistent with previous evidence (Schlaepfer et al., 2008). Previous results have been inconsistent, with findings supporting CHRNA5-CHRNA3-CHRNB4 variation underlying ND in individuals with early age at smoking initiation (≤16 years; Weiss et al., 2008) and CHRNA5 association with later age at smoking initiation (≥17 years; Grucza et al., 2010). Adolescence clearly is a crucial period of vulnerability for the development of substance use disorders (Chambers, Taylor, & Potenza, 2003; Crews, He, & Hodge, 2007), and also in the current study, we found that later starters were less dependent (see Supplementary Table 1) as previously reported (Breslau & Peterson, 1996; Broms, Silventoinen, Lahelma, Koskenvuo, & Kaprio, 2004; Chassin, Presson, Rose, & Sherman, 1996; J. Chen & Millar, 1998; Kandel, Hu, Griesler, & Schaffran, 2007; Lando et al., 1999). This study covered four stages of smoking initiation, allowing a comprehensive evaluation of the genetic factors contributing to this process.
Depression commonly co-occurs with smoking and ND (Chaiton, Cohen, O’Loughlin, & Rehm, 2009; Korhonen et al., 2007; Morrell & Cohen, 2006; Rose et al., 2009), recent evidence showing that ND predicts depression (Boden, Fergusson, & Horwood, 2010) but the causal nature of this association is unknown. Under certain conditions, nicotine can act as an antidepressant (Picciotto, Brunzell, & Caldarone, 2002), whereas chronic nicotine use can lead to neuroadaptation resulting in increase of depressed mood (Picciotto et al., 2002). On the other hand, depressed mood may promote continued nicotine intake to maintain desensitization of nAChRs (Mineur & Picciotto, 2009). So far, the effects of specific genes on the comorbidity of smoking and depression have been investigated in a limited number of studies with small sample sizes (Rose et al., 2009), mostly for dopaminergic genes (Audrain-McGovern, Lerman, Wileyto, Rodriguez, & Shields, 2004; Lerman & Niaura, 2002; Lerman et al., 1998). By stratifying on smoking status, it has been shown that genetic predisposition for depression actually run in families of smokers (Pergadia et al., 2011). The involvement of nAChRs in depression is a biologically plausible hypothesis. Preclinical studies show antidepressant-like effects in drugs targeting nAChRs for example, α4β2 nAChR modulators, such as varenicline (Philip, Carpenter, Tyrka, & Price, 2010). Interestingly, in our study, rs11636753 in CHRNB4 showed suggestive association with the comorbidity of depression and ND, to our best knowledge a novel finding. It is noteworthy that the very same rs11636753 was associated also with the NDSS tolerance subscale. Such pleiotropic effect could be explained so that tolerance encompasses neuroadaptation, that is, quantitative and qualitative changes in nAChRs, being further associated with withdrawal symptoms, such as depressed mood (Benowitz, 2010).
Comorbidities for cigarette smoking and ND are alcohol use and dependence (Dani & Harris, 2005; Durazzo & Meyerhoff, 2007), shared genetic vulnerability being supported by twin studies (Madden & Heath, 2002; True et al., 1999). Genetic variation within the CHRNA5-CHRNA3-CHRNB4 gene cluster has been associated with DSM-IV alcohol dependence (Sherva et al., 2010; Wang et al., 2009), DSM-IV alcohol symptoms (X. Chen et al., 2009), and early alcohol use onset (Schlaepfer et al., 2008). Interestingly, Locus 1 variants (rs16969968 and rs1051730) are associated with both ND and alcohol abuse, however, with opposite risk alleles (X. Chen et al., 2009), possibly reflecting the different actions of the two substances: alcohol being a depressant and nicotine a stimulant. In our sample ascertained for smoking, we detected suggestive association between rs11636753 and regular drinking. Heavy drinking or alcohol dependence did not show any association, although those traits likely are more genetically relevant extreme phenotypes. In the Finnish drinking culture, regular drinking defined as one or more drinks per week reflects a social drinking pattern, slightly surmising our association findings. Furthermore, the regular drinker phenotype showed statistically nonsignificant results (p > .05) in the effect size analysis (Supplementary Table 4).
Our study is based on a strong a priori hypothesis for the involvement of this locus in ND and smoking quantity. We set up to scrutinize a vast array of phenotypes to gain comprehensive information on the involvement of the 15q24-25 region and to study potential pleiotrophic effects in traits related to or co-occurring with ND. Although acknowledging the issue of increased number of tests, we chose to implement all relevant phenotypes within a single study rather than dividing them into separate entities. In order to account for multiple testing, we used a modified Bonferroni correction, utilizing estimated numbers of independent markers and traits, to set p value thresholds for significant and suggestive association signals. As the included markers and traits are correlated, standard procedures for the correction for multiple testing would certainly be overly conservative. The estimation of independent markers, based on LD matrixes, is rather straightforward. However, the number of independent traits is difficult to estimate and depends on what that information is used for. One criterion could be informativeness in predicting cessation or disease outcomes, which could be tested in a multivariate predictive model (logistic regression or survival models). In the current study, we used a statistical estimate based on the correlation/covariance matrix, resulting in a sample-based estimate that may fluctuate when applied to novel independent population samples. The use of estimated numbers of independent markers and traits in adjusting p value thresholds likely is quite conservative but nevertheless successful in reducing the type I error rate. We acknowledge that many of our findings are suggestive and need to be further confirmed in independent replication samples.
Although our study sample was of moderate size (n = 1,428), it harbors a number of advantages. Our sample of twins and their siblings was ascertained specifically for smoking and ND, being initially drawn from the population-based Finnish twin cohort with extensive phenotypic profiles. The population of Finland represents a well-established isolate with minuscule population admixture. In isolates, the genetic drift may lead to an overabundance of morbid alleles for particular disorders, and a high proportion of patients are likely to share these alleles IBD. Although the effect is strongest for rare disease alleles, isolates are also advantageous for genetic studies of common disorders (Peltonen, Palotie, & Lange, 2000).
The inclusion of other variants besides the well-established loci (tagged by rs16969968, rs578776, and rs588765; Saccone et al., 2010) proved beneficial. Although our strongest association signals emerged from Locus 1 (rs1051730 and rs2036527; in full LD with the functional and most well-known Locus 1 proxy SNP, rs16969968), we also detected association with SNPs not included in the proxy lists. Although we aimed to study haplotype effects, the construction of haplotypes in our family sample with virtually all parental genotypes missing proved to be a challenging task. We tested several softwares including PHASE 2.1.1 (Stephens & Scheet, 2005; Stephens, Smith, & Donnelly, 2001; disregarding family structures) and FBAT 2.0.2c (Laird, Horvath, & Xu, 2000; including nuclear family information), but these led to either very inconsistent and unreliable haplotypes or the exclusion of virtually all pedigrees due to missing founder genotypes.
To conclude, we report the novel associations of DSM-IV ND symptoms, NDSS tolerance subscale, TTF, the combined phenotype of DSM-IV ND and HSI, as well as regular drinking and the comorbidity of ND and depression to the CHRNA5-CHRNA3-CHRNB4 gene cluster. Our results confirm and extend the association findings for DSM-IV ND, FTND, CPD, and age at smoking initiation. Using detailed phenotype profiles, we show a broad involvement of this gene cluster on various traits measuring or co-occurring with ND.
Supplementary Material
Supplementary Tables 1–4 can be found online at http://www.ntr.oxfordjournals.org
Funding
This work was supported for data collection by Academy of Finland grants to JK and a National Institutes of Health (NIH) grant (grant numbers DA12854 to PAFM, DA019951 to MLP). This work was supported by Doctoral Programs of Public Health (UB), the Yrjö Jahnsson Foundation (UB), the Juho Vainio Foundation for Post-Doctoral research (UB), Academy of Finland Post-Doctoral Fellowship (AL), Finnish Cultural Foundation (TK, JW), Finnish Medical Foundation (JW), Helsinki Biomedical Graduate School (JW), a NIH grant (DA019951 to MLP), a Global Research Awards for Nicotine Dependence funded by Pfizer Inc. (to JK), and by the Academy of Finland Center of Excellence in Complex Disease Genetics (grant numbers: 213506, 129680 to JK).
Declaration of Interests
Dr. Kaprio has served as a consultant to Pfizer in 2008 and 2011. Dr. Broms has served as a consultant to Pfizer in 2008. Dr. Korhonen has served as a consultant to Pfizer in 2011.
Supplementary Material
Acknowledgments
We warmly thank the participating twin pairs and their family members for their contribution. We would like to express our appreciation to the skilled study interviewers A.-M. Iivonen, K. Karhu, H.-M. Kuha, U. Kulmala-Gråhn, M. Mantere, K. Saanakorpi, M. Saarinen, R. Sipilä, L.Viljanen, and E. Voipio. M. Levander, L. Arala, O. Törnwall, and M. Jussila are acknowledged for their skillful technical assistance. E. E. Meyers, M. J. Marks, and S. M. Hartz are acknowledged for personal communication. We are ever grateful to the late Academician Leena Peltonen for her indispensable contribution throughout the years of the study.
References
- Abecasis GR, Cardon LR, Cookson WO. A general test of association for quantitative traits in nuclear families. American Journal of Human Genetics. 2000;66:279–292. doi: 10.1086/302698. doi:10.1086/302698. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Abecasis GR, Cherny SS, Cookson WO, Cardon LR. Merlin—Rapid analysis of dense genetic maps using sparse gene flow trees. see comment. Nature Genetics. 2002;30:97–101. doi: 10.1038/ng786. doi:10.1038/ng786. [DOI] [PubMed] [Google Scholar]
- American Psychiatric Association. Diagnostic and statistical manual of mental disorders: DSM-IV. 4th ed. Washington, DC: Author; 1994. [Google Scholar]
- Audrain-McGovern J, Lerman C, Wileyto EP, Rodriguez D, Shields PG. Interacting effects of genetic predisposition and depression on adolescent smoking progression. American Journal of Psychiatry. 2004;161:1224–1230. doi: 10.1176/appi.ajp.161.7.1224. [DOI] [PubMed] [Google Scholar]
- Baker TB, Weiss RB, Bolt D, von Niederhausern A, Fiore MC, Dunn DM, et al. Human neuronal acetylcholine receptor A5-A3-B4 haplotypes are associated with multiple nicotine dependence phenotypes. Nicotine & Tobacco Research. 2009;11:785–796. doi: 10.1093/ntr/ntp064. doi:10.1093/ntr/ntp064. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barrett JC, Fry B, Maller J, Daly MJ. Haploview: Analysis and visualization of LD and haplotype maps. Bioinformatics. 2005;21:263–265. doi: 10.1093/bioinformatics/bth457. doi:10.1093/bioinformatics/bth457. [DOI] [PubMed] [Google Scholar]
- Benowitz NL. Nicotine addiction. New England Journal of Medicine. 2010;362:2295–2303. doi: 10.1056/NEJMra0809890. doi:10.1056/NEJMra0809890. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Berrettini W, Yuan X, Tozzi F, Song K, Francks C, Chilcoat H, et al. Alpha-5/alpha-3 nicotinic receptor subunit alleles increase risk for heavy smoking. Molecular Psychiatry. 2008;13:368–373. doi: 10.1038/sj.mp.4002154. doi:10.1038/sj.mp.4002154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bierut LJ, Madden PA, Breslau N, Johnson EO, Hatsukami D, Pomerleau OF, et al. Novel genes identified in a high-density genome wide association study for nicotine dependence. Human Molecular Genetics. 2007;16:24–35. doi: 10.1093/hmg/ddl441. doi:10.1093/hmg/ddl441. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bierut LJ, Stitzel JA, Wang JC, Hinrichs AL, Grucza RA, Xuei X, et al. Variants in nicotinic receptors and risk for nicotine dependence. American Journal of Psychiatry. 2008;165:1163–1171. doi: 10.1176/appi.ajp.2008.07111711. doi:10.1176/appi.ajp.2008.07111711. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boden JM, Fergusson DM, Horwood LJ. Cigarette smoking and depression: Tests of causal linkages using a longitudinal birth cohort. British Journal of Psychiatry: The Journal of Mental Science. 2010;196:440–446. doi: 10.1192/bjp.bp.109.065912. doi:10.1192/bjp.bp.109.065912. [DOI] [PubMed] [Google Scholar]
- Breslau N, Peterson EL. Smoking cessation in young adults: Age at initiation of cigarette smoking and other suspected influences. American Journal of Public Health. 1996;86:214–220. doi: 10.2105/ajph.86.2.214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Broms U, Madden PA, Heath AC, Pergadia ML, Shiffman S, Kaprio J. The nicotine dependence syndrome scale in Finnish smokers. Drug & Alcohol Dependence. 2007;89:42–51. doi: 10.1016/j.drugalcdep.2006.11.017. doi:10.1016/j.drugalcdep.2006.11.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Broms U, Silventoinen K, Lahelma E, Koskenvuo M, Kaprio J. Smoking cessation by socioeconomic status and marital status: The contribution of smoking behavior and family background. Nicotine & Tobacco Research. 2004;6:447–455. doi: 10.1080/14622200410001696637. doi:10.1080/14622200410001696637. [DOI] [PubMed] [Google Scholar]
- Broms U, Silventoinen K, Madden PA, Heath AC, Kaprio J. Genetic architecture of smoking behavior: A study of finnish adult twins. Twin Research & Human Genetics. 2006;9:64–72. doi: 10.1375/183242706776403046. [DOI] [PubMed] [Google Scholar]
- Bucholz KK, Cadoret R, Cloninger CR, Dinwiddie SH, Hesselbrock VM, Nurnberger JI, Jr, et al. A new, semi-structured psychiatric interview for use in genetic linkage studies: A report on the reliability of the SSAGA. Journal of Studies on Alcohol. 1994;55:149–158. doi: 10.15288/jsa.1994.55.149. [DOI] [PubMed] [Google Scholar]
- Caporaso N, Gu F, Chatterjee N, Sheng-Chih J, Yu K, Yeager M, et al. Genome-wide and candidate gene association study of cigarette smoking behaviors. PLoS One. 2009;4:e4653. doi: 10.1371/journal.pone.0004653. doi:10.1371/journal.pone.0004653. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chaiton MO, Cohen JE, O’Loughlin J, Rehm J. A systematic review of longitudinal studies on the association between depression and smoking in adolescents. BMC Public Health. 2009;9:356. doi: 10.1186/1471-2458-9-356. doi:10.1186/1471-2458-9-356. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chambers RA, Taylor JR, Potenza MN. Developmental neurocircuitry of motivation in adolescence: A critical period of addiction vulnerability. American Journal of Psychiatry. 2003;160:1041–1052. doi: 10.1176/appi.ajp.160.6.1041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chassin L, Presson CC, Rose JS, Sherman SJ. The natural history of cigarette smoking from adolescence to adulthood: Demographic predictors of continuity and change. Health Psychology. 1996;15:478–484. doi: 10.1037//0278-6133.15.6.478. doi:10.1037/0278-6133.15.6.478. [DOI] [PubMed] [Google Scholar]
- Chen J, Millar WJ. Age of smoking initiation: Implications for quitting. Health Reports. 1998;9 39–46(Eng); 39–48(Fre) [PubMed] [Google Scholar]
- Chen LS, Johnson EO, Breslau N, Hatsukami D, Saccone NL, Grucza RA, et al. Interplay of genetic risk factors and parent monitoring in risk for nicotine dependence. Addiction. 2009;104:1731–1740. doi: 10.1111/j.1360-0443.2009.02697.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen X, Chen J, Williamson VS, An SS, Hettema JM, Aggen SH, et al. Variants in nicotinic acetylcholine receptors alpha5 and alpha3 increase risks to nicotine dependence. American Journal of Medical Genetics. Part B, Neuropsychiatric Genetics. 2009;150B:926–933. doi: 10.1002/ajmg.b.30919. doi:10.1002/ajmg.b.30919. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cheverud JM. A simple correction for multiple comparisons in interval mapping genome scans. Heredity. 2001;87:52–58. doi: 10.1046/j.1365-2540.2001.00901.x. [DOI] [PubMed] [Google Scholar]
- Cottler LB, Robins LN, Grant BF, Blaine J, Towle LH, Wittchen HU, et al. The CIDI-core substance abuse and dependence questions: Cross-cultural and nosological issues. The WHO/ADAMHA field trial. British Journal of Psychiatry. 1991;159:653–658. doi: 10.1192/bjp.159.5.653. [DOI] [PubMed] [Google Scholar]
- Crews F, He J, Hodge C. Adolescent cortical development: A critical period of vulnerability for addiction. Pharmacology, Biochemistry, and Behavior. 2007;86:189–199. doi: 10.1016/j.pbb.2006.12.001. doi:10.1016/j.pbb.2006.12.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dani JA, Harris RA. Nicotine addiction and comorbidity with alcohol abuse and mental illness. Nature Neuroscience. 2005;8:1465–1470. doi: 10.1038/nn1580. doi:10.1038/nn1580. [DOI] [PubMed] [Google Scholar]
- Durazzo TC, Meyerhoff DJ. Neurobiological and neurocognitive effects of chronic cigarette smoking and alcoholism. Frontiers in Bioscience: A Journal and Virtual Library. 2007;12:4079–4100. doi: 10.2741/2373. doi:10.1093/alcalc/agm020. [DOI] [PubMed] [Google Scholar]
- Erlich PM, Hoffman SN, Rukstalis M, Han JJ, Chu X, Linda Kao WH, et al. Nicotinic acetylcholine receptor genes on chromosome 15q25.1 are associated with nicotine and opioid dependence severity. Human Genetics. 2010;128:491–499. doi: 10.1007/s00439-010-0876-6. doi:10.1007/s00439-010-0876-6. [DOI] [PubMed] [Google Scholar]
- Fowler CD, Lu Q, Johnson PM, Marks MJ, Kenny PJ. Habenular alpha5 nicotinic receptor subunit signalling controls nicotine intake. Nature. 2011;471:597–601. doi: 10.1038/nature09797. doi:10.1038/nature09797. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goring HH, Terwilliger JD. Linkage analysis in the presence of errors IV: Joint pseudomarker analysis of linkage and/or linkage disequilibrium on a mixture of pedigrees and singletons when the mode of inheritance cannot be accurately specified. American Journal of Human Genetics. 2000;66:1310–1327. doi: 10.1086/302845. doi:10.1086/302845. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grucza RA, Johnson EO, Krueger RF, Breslau N, Saccone NL, Chen LS, et al. Incorporating age at onset of smoking into genetic models for nicotine dependence: Evidence for interaction with multiple genes. Addiction Biology. 2010;15:346–357. doi: 10.1111/j.1369-1600.2010.00220.x. doi:10.1111/j.1369-1600.2010.00220.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haberstick BC, Ehringer MA, Lessem JM, Hopfer CJ, Hewitt JK. Dizziness and the genetic influences on subjective experiences to initial cigarette use. Addiction. 2011;106:391–399. doi: 10.1111/j.1360-0443.2010.03133.x. doi:10.1111/j.1360-0443.2010.03133.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haddock CK, Lando H, Klesges RC, Talcott GW, Renaud EA. A study of the psychometric and predictive properties of the fagerstrom test for nicotine dependence in a population of young smokers. Nicotine & Tobacco Research. 1999;1:59–66. doi: 10.1080/14622299050011161. [DOI] [PubMed] [Google Scholar]
- Han S, Gelernter J, Luo X, Yang BZ. Meta-analysis of 15 genome-wide linkage scans of smoking behavior. Biological Psychiatry. 2010;67:12–19. doi: 10.1016/j.biopsych.2009.08.028. doi:10.1016/j.biopsych.2009.08.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heatherton TF, Kozlowski LT, Frecker RC, Fagerström K.-O. The Fagerström Test for Nicotine Dependence: A revision of the Fagerstrom Tolerance Questionnaire. British Journal of Addiction. 1991;86:1119–1127. doi: 10.1111/j.1360-0443.1991.tb01879.x. [DOI] [PubMed] [Google Scholar]
- Heatherton TF, Kozlowski LT, Frecker RC, Rickert W, Robinson J. Measuring the heaviness of smoking: Using self-reported time to the first cigarette of the day and number of cigarettes smoked per day. British Journal of Addiction. 1989;84:791–799. doi: 10.1111/j.1360-0443.1989.tb03059.x. [DOI] [PubMed] [Google Scholar]
- Hoft NR, Stitzel JA, Hutchison KE, Ehringer MA. CHRNB2 promoter region: Association with subjective effects to nicotine and gene expression differences. Genes, Brain, and Behavior. 2011;10:176–185. doi: 10.1111/j.1601-183X.2010.00650.x. doi:10.1111/j.1601-183X.2010.00650.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Howie BN, Donnelly P, Marchini J. A flexible and accurate genotype imputation method for the next generation of genome-wide association studies. PLoS Genetics. 2009;5:e1000529. doi: 10.1371/journal.pgen.1000529. doi:10.1371/journal.pgen.1000529. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johnson EO, Chen LS, Breslau N, Hatsukami D, Robbins T, Saccone NL, et al. Peer smoking and the nicotinic receptor genes: An examination of genetic and environmental risks for nicotine dependence. Addiction. 2010;105:2014–2022. doi: 10.1111/j.1360-0443.2010.03074.x. doi:10.1111/j.1360-0443.2010.03074.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kandel DB, Hu MC, Griesler PC, Schaffran C. On the development of nicotine dependence in adolescence. Drug & Alcohol Dependence. 2007;91:26–39. doi: 10.1016/j.drugalcdep.2007.04.011. doi:10.1016/j.drugalcdep.2007.04.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaprio J. Twin studies in Finland 2006. Twin Research and Human Genetics. 2006;9:772–777. doi: 10.1375/183242706779462778. doi:10.1375/183242706779462778. [DOI] [PubMed] [Google Scholar]
- Keskitalo K, Broms U, Heliovaara M, Ripatti S, Surakka I, Perola M, et al. Association of serum cotinine level with a cluster of three nicotinic acetylcholine receptor genes (CHRNA3/CHRNA5/CHRNB4) on chromosome 15. Human Molecular Genetics. 2009;18:4007–4012. doi: 10.1093/hmg/ddp322. doi:10.1093/hmg/ddp322. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim DK, Hersh CP, Washko GR, Hokanson JE, Lynch DA, Newell JD, et al. Epidemiology, radiology, and genetics of nicotine dependence in COPD. Respiratory Research. 2011;12:9. doi: 10.1186/1465-9921-12-9. doi:10.1186/1465-9921-12-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Korhonen T, Broms U, Varjonen J, Romanov K, Koskenvuo M, Kinnunen T, et al. Smoking behaviour as a predictor of depression among Finnish men and women: A prospective cohort study of adult twins. Psychological Medicine. 2007;37:705–715. doi: 10.1017/S0033291706009639. doi:10.1017/S0033291706009639. [DOI] [PubMed] [Google Scholar]
- Kuryatov A, Berrettini W, Lindstrom J. Acetylcholine receptor (AChR) alpha5 subunit variant associated with risk for nicotine dependence and lung cancer reduces (alpha4beta2)alpha5 AChR function. Molecular Pharmacology. 2011;79:119–125. doi: 10.1124/mol.110.066357. doi:10.1124/mol.110.066357. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Laird NM, Horvath S, Xu X. Implementing a unified approach to family-based tests of association. Genetic Epidemiology. 2000;1(Suppl. 1):S36–S42. doi: 10.1002/1098-2272(2000)19:1+<::AID-GEPI6>3.0.CO;2-M. doi:2-M. [DOI] [PubMed] [Google Scholar]
- Lando HA, Thai DT, Murray DM, Robinson LA, Jeffery RW, Sherwood NE, et al. Age of initiation, smoking patterns, and risk in a population of working adults. Preventive Medicine. 1999;29:590–598. doi: 10.1006/pmed.1999.0590. doi:10.1006/pmed.1999.0590. [DOI] [PubMed] [Google Scholar]
- Lerman C, Caporaso N, Main D, Audrain J, Boyd NR, Bowman ED, et al. Depression and self-medication with nicotine: The modifying influence of the dopamine D4 receptor gene. Health Psychology. 1998;17:56–62. doi: 10.1037//0278-6133.17.1.56. doi:10.1037/0278-6133.17.1.56. [DOI] [PubMed] [Google Scholar]
- Lerman C, Niaura R. Applying genetic approaches to the treatment of nicotine dependence. Oncogene. 2002;21:7412–7420. doi: 10.1038/sj.onc.1205801. doi:10.1038/sj.onc.1205801. [DOI] [PubMed] [Google Scholar]
- Li J, Ji L. Adjusting multiple testing in multilocus analyses using the eigenvalues of a correlation matrix. Heredity. 2005;95:221–227. doi: 10.1038/sj.hdy.6800717. doi:10.1038/sj.hdy.6800717. [DOI] [PubMed] [Google Scholar]
- Li MD, Xu Q, Lou XY, Payne TJ, Niu T, Ma JZ. Association and interaction analysis of variants in CHRNA5/CHRNA3/CHRNB4 gene cluster with nicotine dependence in African and European Americans. American Journal of Medical Genetics. Part B, Neuropsychiatric Genetics. 2010;153B:745–756. doi: 10.1002/ajmg.b.31043. doi:10.1002/ajmg.b.31043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu JZ, Tozzi F, Waterworth DM, Pillai SG, Muglia P, Middleton L, et al. Meta-analysis and imputation refines the association of 15q25 with smoking quantity. Nature Genetics. 2010;42:436–440. doi: 10.1038/ng.572. doi:10.1038/ng.572. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Loukola A, Broms U, Maunu H, Widen E, Heikkila K, Siivola M, et al. Linkage of nicotine dependence and smoking behavior on 10q, 7q and 11p in twins with homogeneous genetic background. Pharmacogenomics Journal. 2008;8:209–219. doi: 10.1038/sj.tpj.6500464. doi:10.1038/sj.tpj.6500464. [DOI] [PubMed] [Google Scholar]
- Madden PA, Heath AC. Shared genetic vulnerability in alcohol and cigarette use and dependence. Alcoholism, Clinical and Experimental Research. 2002;26:1919–1921. doi: 10.1097/01.ALC.0000040960.15151.30. doi:10.1097/01.ALC.0000040960.15151.30. [DOI] [PubMed] [Google Scholar]
- Marques-Vidal P, Kutalik Z, Paccaud F, Bergman S, Waeber G, Vollenweider P, et al. Variant within the promoter region of the CHRNA3 gene associated with FTN dependence is not related to self-reported willingness to quit smoking. Nicotine & Tobacco Research. 2011;13:833–839. doi: 10.1093/ntr/ntr084. doi:10.1093/ntr/ntr084. [DOI] [PubMed] [Google Scholar]
- McCallum SE, Collins AC, Paylor R, Marks MJ. Deletion of the beta 2 nicotinic acetylcholine receptor subunit alters development of tolerance to nicotine and eliminates receptor upregulation. Psychopharmacology. 2006;184:314–327. doi: 10.1007/s00213-005-0076-6. doi:10.1007/s00213-005-0076-6. [DOI] [PubMed] [Google Scholar]
- Mineur YS, Picciotto MR. Biological basis for the co-morbidity between smoking and mood disorders. Journal of Dual Diagnosis. 2009;5:122–130. doi: 10.1080/15504260902869964. doi:10.1080/15504260902869964. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moolchan ET, Radzius A, Epstein DH, Uhl G, Gorelick DA, Cadet JL, et al. The Fagerstrom Test for Nicotine Dependence and the Diagnostic Interview Schedule: Do they diagnose the same smokers? Addictive Behaviors. 2002;27:101–113. doi: 10.1016/s0306-4603(00)00171-4. [DOI] [PubMed] [Google Scholar]
- Morley KI, Lynskey MT, Madden PA, Treloar SA, Heath AC, Martin NG. Exploring the inter-relationship of smoking age-at-onset, cigarette consumption and smoking persistence: Genes or environment? Psychological Medicine. 2007;37:1357–1367. doi: 10.1017/S0033291707000748. doi:10.1017/S0033291707000748. [DOI] [PubMed] [Google Scholar]
- Morrell HER, Cohen LM. Cigarette smoking, anxiety, and depression. Journal of Psychopathology & Behavioral Assessment. 2006;28:281–295. doi:10.1007/s10862-005-9011-8. [Google Scholar]
- Nyholt DR. A simple correction for multiple testing for single-nucleotide polymorphisms in linkage disequilibrium with each other. American Journal of Human Genetics. 2004;74:765–769. doi: 10.1086/383251. doi:10.1086/383251. [DOI] [PMC free article] [PubMed] [Google Scholar]
- O’Connell JR, Weeks DE. PedCheck: A program for identification of genotype incompatibilities in linkage analysis. American Journal of Human Genetics. 1998;63:259–266. doi: 10.1086/301904. doi:10.1086/301904. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peltonen L, Palotie A, Lange K. Use of population isolates for mapping complex traits. Nature Reviews. Genetics. 2000;1:182–190. doi: 10.1038/35042049. doi:10.1038/35042049. [DOI] [PubMed] [Google Scholar]
- Pergadia ML, Glowinski AL, Wray NR, Agrawal A, Saccone SF, Loukola A, et al. A 3p26-3p25 genetic linkage finding for DSM-IV major depression in heavy smoking families. American Journal of Psychiatry. 2011;168:848–852. doi: 10.1176/appi.ajp.2011.10091319. doi:10.1176/appi.ajp.2011.10091319. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Philip NS, Carpenter LL, Tyrka AR, Price LH. Nicotinic acetylcholine receptors and depression: A review of the preclinical and clinical literature. Psychopharmacology. 2010;212:1–12. doi: 10.1007/s00213-010-1932-6. doi:10.1007/s00213-010-1932-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Picciotto MR, Brunzell DH, Caldarone BJ. Effect of nicotine and nicotinic receptors on anxiety and depression. Neuroreport. 2002;13:1097–1106. doi: 10.1097/00001756-200207020-00006. [DOI] [PubMed] [Google Scholar]
- Piper ME, McCarthy DE, Baker TB. Assessing tobacco dependence: A guide to measure evaluation and selection. Nicotine & Tobacco Research. 2006;8:339–351. doi: 10.1080/14622200600672765. doi:10.1080/14622200600672765. [DOI] [PubMed] [Google Scholar]
- Piper ME, McCarthy DE, Bolt DM, Smith SS, Lerman C, Benowitz N, et al. Assessing dimensions of nicotine dependence: An evaluation of the Nicotine Dependence Syndrome Scale (NDSS) and the Wisconsin Inventory of Smoking Dependence Motives (WISDM) Nicotine & Tobacco Research. 2008;10:1009–1020. doi: 10.1080/14622200802097563. doi:10.1080/14622200802097563. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Piper ME, Piasecki TM, Federman EB, Bolt DM, Smith SS, Fiore MC, et al. A multiple motives approach to tobacco dependence: The Wisconsin Inventory of Smoking Dependence Motives (WISDM-68) Journal of Consulting & Clinical Psychology. 2004;72:139–154. doi: 10.1037/0022-006X.72.2.139. doi:10.1037/0022-006X.72.2.139. [DOI] [PubMed] [Google Scholar]
- Rose JE, Broms U, Korhonen T, Dick DM, Kaprio J. Genetics of smoking behavior. In: Kim YK, editor. Handbook of behavior genetics. 1st ed. New York: Springer; 2009. pp. 411–432. [Google Scholar]
- Saccone NL, Culverhouse RC, Schwantes-An TH, Cannon DS, Chen X, Cichon S, et al. Multiple independent loci at chromosome 15q25.1 affect smoking quantity: A meta-analysis and comparison with lung cancer and COPD. PLoS Genetics. 2010;6:e1001053. doi: 10.1371/journal.pgen.1001053. doi:10.1371/journal.pgen.1001053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saccone NL, Wang JC, Breslau N, Johnson EO, Hatsukami D, Saccone SF, et al. The CHRNA5-CHRNA3-CHRNB4 nicotinic receptor subunit gene cluster affects risk for nicotine dependence in African-Americans and in European-Americans. Cancer Research. 2009;69:6848–6856. doi: 10.1158/0008-5472.CAN-09-0786. doi:10.1158/0008-5472.CAN-09-0786. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saccone SF, Hinrichs AL, Saccone NL, Chase GA, Konvicka K, Madden PA, et al. Cholinergic nicotinic receptor genes implicated in a nicotine dependence association study targeting 348 candidate genes with 3713 SNPs. Human Molecular Genetics. 2007a;16:36–49. doi: 10.1093/hmg/ddl438. doi:10.1093/hmg/ddl438. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saccone SF, Pergadia ML, Loukola A, Broms U, Montgomery GW, Wang JC, et al. Genetic linkage to chromosome 22q12 for a heavy-smoking quantitative trait in two independent samples. American Journal of Human Genetics. 2007b;80:856–866. doi: 10.1086/513703. doi:10.1086/513703. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Salas R, Orr-Urtreger A, Broide RS, Beaudet A, Paylor R, De Biasi M. The nicotinic acetylcholine receptor subunit alpha 5 mediates short-term effects of nicotine in vivo. Molecular Pharmacology. 2003;63:1059–1066. doi: 10.1124/mol.63.5.1059. doi:10.1124/mol.63.5.1059. [DOI] [PubMed] [Google Scholar]
- Salas R, Sturm R, Boulter J, De Biasi M. Nicotinic receptors in the habenulo-interpeduncular system are necessary for nicotine withdrawal in mice. Journal of Neuroscience. 2009;29:3014–3018. doi: 10.1523/JNEUROSCI.4934-08.2009. doi:10.1523/JNEUROSCI.4934-08.2009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Salminen O, Murphy KL, McIntosh JM, Drago J, Marks MJ, Collins AC, et al. Subunit composition and pharmacology of two classes of striatal presynaptic nicotinic acetylcholine receptors mediating dopamine release in mice. Molecular Pharmacology. 2004;65:1526–1535. doi: 10.1124/mol.65.6.1526. doi:10.1124/mol.65.6.1526. [DOI] [PubMed] [Google Scholar]
- Schlaepfer IR, Hoft NR, Collins AC, Corley RP, Hewitt JK, Hopfer CJ, et al. The CHRNA5/A3/B4 gene cluster variability as an important determinant of early alcohol and tobacco initiation in young adults. Biological Psychiatry. 2008;63:1039–1046. doi: 10.1016/j.biopsych.2007.10.024. doi:10.1016/j.biopsych.2007.10.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Service S, DeYoung J, Karayiorgou M, Roos JL, Pretorious H, Bedoya G, et al. Magnitude and distribution of linkage disequilibrium in population isolates and implications for genome-wide association studies. Nature Genetics. 2006;38:556–560. doi: 10.1038/ng1770. doi:10.1038/ng1770. [DOI] [PubMed] [Google Scholar]
- Sherva R, Kranzler HR, Yu Y, Logue MW, Poling J, Arias AJ, et al. Variation in nicotinic acetylcholine receptor genes is associated with multiple substance dependence phenotypes. Neuropsychopharmacology. 2010;35:1921–1931. doi: 10.1038/npp.2010.64. doi:10.1038/npp.2010.64. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sherva R, Wilhelmsen K, Pomerleau CS, Chasse SA, Rice JP, Snedecor SM, et al. Association of a single nucleotide polymorphism in neuronal acetylcholine receptor subunit alpha 5 (CHRNA5) with smoking status and with ‘pleasurable buzz’ during early experimentation with smoking. Addiction. 2008;103:1544–1552. doi: 10.1111/j.1360-0443.2008.02279.x. doi:10.1111/j.1360-0443.2008.02279.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shiffman S, Waters A, Hickcox M. The nicotine dependence syndrome scale: A multidimensional measure of nicotine dependence. Nicotine & Tobacco Research. 2004;6:327–348. doi: 10.1080/1462220042000202481. [DOI] [PubMed] [Google Scholar]
- Solda G, Boi S, Duga S, Fornasari D, Benfante R, Malcovati M, et al. In vivo RNA-RNA duplexes from human alpha3 and alpha5 nicotinic receptor subunit mRNAs. Gene. 2005;345:155–164. doi: 10.1016/j.gene.2004.12.005. doi:10.1016/j.gene.2004.12.005. [DOI] [PubMed] [Google Scholar]
- Spitz MR, Amos CI, Dong Q, Lin J, Wu X. The CHRNA5-A3 region on chromosome 15q24-25.1 is a risk factor both for nicotine dependence and for lung cancer. Journal of the National Cancer Institute. 2008;100:1552–1556. doi: 10.1093/jnci/djn363. doi:10.1093/jnci/djn363. [DOI] [PMC free article] [PubMed] [Google Scholar]
- StataCorp. Stata statistical software: Release 11. College Station, TX: StataCorp LP; 2009. [Google Scholar]
- Stephens M, Scheet P. Accounting for decay of linkage disequilibrium in haplotype inference and missing-data imputation. American Journal of Human Genetics. 2005;76:449–462. doi: 10.1086/428594. doi:10.1086/428594. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stephens M, Smith NJ, Donnelly P. A new statistical method for haplotype reconstruction from population data. American Journal of Human Genetics. 2001;68:978–989. doi: 10.1086/319501. doi:10.1086/319501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stevens VL, Bierut LJ, Talbot JT, Wang JC, Sun J, Hinrichs AL, et al. Nicotinic receptor gene variants influence susceptibility to heavy smoking. Cancer Epidemiology, Biomarkers & Prevention: A Publication of the American Association for Cancer Research, Cosponsored by the American Society of Preventive Oncology. 2008;17:3517–3525. doi: 10.1158/1055-9965.EPI-08-0585. doi:10.1158/1055-9965.EPI-08-0585. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tapper AR, McKinney SL, Marks MJ, Lester HA. Nicotine responses in hypersensitive and knockout alpha 4 mice account for tolerance to both hypothermia and locomotor suppression in wild-type mice. Physiological Genomics. 2007;31:422–428. doi: 10.1152/physiolgenomics.00063.2007. doi:10.1152/physiolgenomics.00063.2007. [DOI] [PubMed] [Google Scholar]
- Thorgeirsson TE, Geller F, Sulem P, Rafnar T, Wiste A, Magnusson KP, et al. A variant associated with nicotine dependence, lung cancer and peripheral arterial disease. Nature. 2008;452:638–642. doi: 10.1038/nature06846. doi:10.1038/nature06846. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thorgeirsson TE, Gudbjartsson DF, Surakka I, Vink JM, Amin N, Geller F, et al. Sequence variants at CHRNB3-CHRNA6 and CYP2A6 affect smoking behavior. Nature Genetics. 2010;42:448–453. doi: 10.1038/ng.573. doi:10.1038/ng.573. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tobacco and Genetics Consortium. Genome-wide meta-analyses identify multiple loci associated with smoking behavior. Nature Genetics. 2010;42:441–447. doi: 10.1038/ng.571. doi:10.1038/ng.571. [DOI] [PMC free article] [PubMed] [Google Scholar]
- True WR, Xian H, Scherrer JF, Madden PA, Bucholz KK, Heath AC, et al. Common genetic vulnerability for nicotine and alcohol dependence in men. Archives of General Psychiatry. 1999;56:655–661. doi: 10.1001/archpsyc.56.7.655. [DOI] [PubMed] [Google Scholar]
- Wang J, Li MD. Common and unique biological pathways associated with smoking initiation/progression, nicotine dependence, and smoking cessation. Neuropsychopharmacology. 2010;35:702–719. doi: 10.1038/npp.2009.178. doi:10.1038/npp.2009.178. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang JC, Grucza R, Cruchaga C, Hinrichs AL, Bertelsen S, Budde JP, et al. Genetic variation in the CHRNA5 gene affects mRNA levels and is associated with risk for alcohol dependence. Molecular Psychiatry. 2009;14:501–510. doi: 10.1038/mp.2008.42. doi:10.1038/mp.2008.42. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weiss RB, Baker TB, Cannon DS, von Niederhausern A, Dunn DM, Matsunami N, et al. A candidate gene approach identifies the CHRNA5-A3-B4 region as a risk factor for age-dependent nicotine addiction. PLoS Genetics. 2008;4:e1000125. doi: 10.1371/journal.pgen.1000125. doi:10.1371/journal.pgen.1000125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wessel J, McDonald SM, Hinds DA, Stokowski RP, Javitz HS, Kennemer M, et al. Resequencing of nicotinic acetylcholine receptor genes and association of common and rare variants with the Fagerstrom Test for Nicotine Dependence. Neuropsychopharmacology. 2010;35:2392–2402. doi: 10.1038/npp.2010.120. doi:10.1038/npp.2010.120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Williams RL. A note on robust variance estimation for cluster-correlated data. Biometrics. 2000;56:645–646. doi: 10.1111/j.0006-341x.2000.00645.x. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.