Abstract
Recently, researchers have focused on immunosuppression induced by rifampicin. Our previous investigation found that rifampicin was neuroprotective by inhibiting the production of pro-inflammatory mediators, thereby suppressing microglial activation. In this study, using 2-dimensional gel electrophoresis (2-DE) and mass spectrometry (MS), we discovered that 26S protease regulatory subunit 7 (MSS1) was decreased in rifampicin-treated microglia. Western blot analysis verified the downregulation of MSS1 expression by rifampicin. As it is indicated that the modulation of the ubiquitin-26S proteasome system (UPS) with proteasome inhibitors is efficacious for the treatment of neuro-inflammatory disorders, we next hypothesized that silencing MSS1 gene expression might inhibit microglial inflammation. Using RNA interference (RNAi), we showed significant reduction of IkBα degradation and NF-kB activation. The production of lipopolysaccharides-induced pro-inflammatory mediators such as inducible nitric oxide synthase (iNOS), nitric oxide, cyclooxygenase-2, and prostaglandin E2 were also reduced by MSS1 gene knockdown. Taken together, our findings suggested that rifampicin inhibited microglial inflammation by suppressing MSS1 protein production. Silencing MSS1 gene expression decreased neuroinflammation. We concluded that MSS1 inhibition, in addition to anti-inflammatory rifampicin, might represent a novel mechanism for the treatment of neuroinflammatory disorders.
Introduction
Microglial activation plays an important role in the pathophysiology of neurodegenerative diseases, and the suppression of microglial activation has been shown to prevent the progression of Alzheimer’s disease (AD), Parkinson’s disease (PD), trauma, multiple sclerosis, and cerebral ischemia [1]–[5].
Rifampicin is a macrocyclic antibiotic that is used extensively against Mycobacterium Tuberculosis and other mycobacterial infections [6]. It has been reported that rifampicin is immunosuppressive [7]–[10]. We previously found that rifampicin improved survival of catecholamine and α-synuclein-containing cells, which degenerate in PD, thus might be therapeutic in this disease [11]. Rifampicin suppressed the release of pro-inflammatory mediators including nitric oxide (NO), prostaglandin E2 (PGE2), tumor necrosis factor-α (TNF-α), and interleukin-1β (IL-1β) from BV2 microglial cells that were pre-treated with lipopolysaccharides (LPS). It acted as a neuroprotector to increase neuronal survival against microglia-induced neuron death [12]. Our results strongly supported rifampicin as a potential therapeutic for the treatment of neurodegenerative diseases. Despite the above findings, the mechanism through which rifampicin inhibits neuroinflammation is not completely understood.
NF-kB is an important transcription factor for the expression of pro-inflammatory mediators [13]. In unstimulated cells, nuclear factor-kappa B (NF-kB) binds to IkappaBalpha (IkBα) and its activity is inhibited. The activation of NF-kB is initiated by signal-induced degradation of IkBα proteins [14], which occurs primarily via the ubiquitin–proteasome pathway [15]. Proteasomes play a critical role in protein degradation and are essential to many intracellular processes [16]. The 26S proteasome, a multi-subunit enzyme complex, is a major cellular non-lysosomal protease. Modulation of the ubiquitin-26S proteasome system (UPS) with proteasome inhibitors has indicated possible efficacy for the treatment of neuroinflammatory disorders [17]. We used 2-dimensional gel electrophoresis (2-DE) and mass spectrometry (MS) to identify proteins affected by rifampicin in activated microglia. We uncovered that the expression of 26S protease regulatory subunit 7 (MSS1) was reduced. MSS1 localizes to both the nucleus and the cytoplasm. It functions as a chaperone-like subunit in the 19S regulatory complex and participates in intracellular proteasome events [18]. Based on the above evidence, we further hypothesized that rifampicin inhibited the expression of MSS1, thus suppressed IkBα degradation and the production of inflammatory mediators.
In this study, we examined the effect of rifampicin on the expression of MSS1 in LPS-stimulated BV2 microglia by western blot to confirm the results of proteomics experiments. We demonstrated that after silencing the expression of MSS1 gene via RNA interference (RNAi), IkBα protein degradation and NF-kB activity were both downregulated in LPS-stimulated BV2 microglia. We also showed that the production of inducible NO synthase (iNOS), NO, cyclooxygenase-2 (COX-2), and PGE2 were significantly decreased after MSS1 gene knockdown in LPS-activated BV2 microglia. Our results implied that rifampicin inhibited IkBα degradation by suppressing the expression of MSS1, therefore regulated the production of inflammatory mediators.
Materials and Methods
Chemicals and Reagents
Rifampicin (purity >98%), LPS and dimethyl sulfoxide (DMSO) were purchased from Sigma (St. Louis, MO). Rifampicin was dissolved in less than 0.1% of DMSO solution. Antibodies against iNOS, COX-2, and IkBα were obtained from Cell Signaling Tech (Beverly, MA). Mouse beta-actin antibody was purchased from Sigma. Dulbecco’s-modified Eagle’s medium (DMEM) containing L-arginine (200 mg/L), fetal bovine serum (FBS), and other tissue culture reagents were purchased from Gibco (Grand Island, NY).
Cell Culture
BV2-immortalized murine microglial cells were provided by the Cell Culture Center of the Chinese Academy of Medical Sciences (China). Cells were cultured in DMEM supplemented with 10% FBS, 100 units/ml penicillin, and 100 µg/ml streptomycin in a humidified atmosphere of 5% CO2 at 37°C [12]. To examine the effect of rifampicin on the expression of MSS1 in LPS-stimulated BV2 microglia, 3×105 cells per well were seeded in 6-well plates and pretreated with 150 µmol/L rifampicin for 2 hours (h) before the addition of LPS (1000 ng/ml).
2-dimensional Gel Electrophoresis and Image Analysis
LPS-treated cells were washed three times with ice-cold washing buffer (10 µM Tris-HCl, 250 µM sucrose, pH 7.0), collected in clean 1.5 ml eppendorf tubes. Lysis buffer [7 M urea, 2 M thiourea, 4% CHAPS (w/v), 1% dithiothreitol (DTT), 2% immobilized pH gradients (IPG) (v/v), pH 3–10 NL] was added, and samples were centrifuged at 13,200 g for 30 min at 4°C. The supernatant was subjected to 2-DE using an Amersham Biosciences IPGphor IEF System and Hoefer SE 600 (GE healthcare, Uppsala, Sweden) electrophoresis units (13 cm), according to manufacturer’s instructions and a previously described protocol [19]. Protein lysates and 2-DE gels were processed in parallel. Protein concentrations were determined using the Bradford assay. After 2-DE, the gels underwent silver nitrate staining according to a previously described protocol [20], then were scanned using an Image Scanner (GE Healthcare). The images were analyzed using the ImageMaster 2D Platinum (GE Healthcare).
Matrix-assisted Laser Desorption/ionization Time-of-flight Mass Spectrometry (MALDI-TOF-MS) and Database Search
Only protein spots that were consistently different in at least three independent experiments were considered to be significant for analysis by MALDI-TOF-MS. Protein spots were excised from the silver-stained gels and transferred into siliconized 1.5 ml eppendorf tubes. Tryptic in-gel digestion was performed as previously reported with slight modifications [19]. Molecular mass analysis of the tryptic peptides was performed using ABI 4800 plus a MALDI-TOF-MS mass spectrometer (Applied Biosystems, Foster City, CA). Spectra were interpreted and processed using the Global Protein Server Workstation (V3.6, Applied Biosystems) via the internal MASCOT search engine (V2.1, Matrix Science, London, UK) to analyze MALDI-TOF-MS and MS/MS data. Based on combined MALDI-TOF-MS and MS/MS spectra, MASCOT protein scores of greater than 65 were considered statistically significant (p<0.05). The individual MS/MS spectrum with the best ion score (based on MS/MS spectra) that was statistically significant (p<0.05) was also accepted. Searches were performed against the IPI mouse database (V3.36) with parameters as the following: the enzyme trypsin with one missed cleavage was allowed; variable modifications included acetamidation of cysteine and oxidation of methionine; peptide mass tolerance was set to 50 ppm and fragment ion mass tolerance was set to 0.2 Da; and only monoisotopic masses were included in the search.
MSS1 Gene silencing
Gene silencing was performed using small interference RNA (siRNA) targeting MSS1 mRNA for degradation. MSS1-specific siRNAs had the sense sequence of 5′-GUCGAACGCACAUCUUUAATT-3′, corresponding to a region that was 443–461 bases downstream of the first nucleotide of the start codon of mouse MSS1 cDNA (GenBank Accession Number: NC_000007.13). The sense sequence of scrambled siRNAs was 5′-UUCUCCGAACGUGUCACGUTT-3′. RNA duplexes were synthesized, purified and annealed by Dharmacon (Lafayette, CO). BV2 cells were transfected with targeting and scrambled RNA duplexes at a final concentration of 100 nM using Lipofectamine 2000 (Invitrogen, Grand Island, NY) in either 96-well or 6-well culture plates. The cells were assayed at 24 h post-transfection via western blotting.
Nitrite (Griess) Assay
The NO levels in the culture supernatants were determined by measuring nitrite levels using a Griess reaction [21]. Six wells of cells were treated with rifampicin per experiment. After the BV2 microglial cells were stimulated in 24-well plates for 24 h, 100 µl of the cell culture medium was taken out and mixed with the same volume of the Griess reagent [1% sulfanilamide, 0.1% N-(1-naphthyl)-ethylenediamine dihydrochloride, 2.5% H3PO4]. The nitrite concentration was determined by evaluating the absorbance at 540 nm using a 96-well microplate spectrophotometer and calculated by referring to a standard curve.
Enzyme-linked Immunosorbent Assay (ELISA)
The concentration of PGE2 in cell-conditioned culture medium was assessed using an ELISA kit (R&D Systems, Minneapolis, MN) according to the manufacturer’s instructions. Three wells of cells were treated per experiment.
Western Blot Analysis
The BV2 microglial cells were harvested from each group and followed by western blot analysis that was conducted as previously described [22]. Cell pellets were briefly lysed in RIPA buffer [1 mM ethylenediaminetetraacetic acid (EDTA), 150 mM NaCl, 1% igepal, 0.1% sodium dodecyl sulfate (SDS), 0.5% sodium deoxycholate, and 50 mM Tris-HCl, pH 8.0]. Equal amounts of cellular proteins were separated by 8–12% sodium dodecyl sulfate polyacrylamide gel electrophoresis (SDS-PAGE), transferred to polyvinylidene fluoride (PVDF) membranes, blocked with 5% nonfat milk for 2 h, and incubated with antibodies against iNOS (1∶1000), COX-2 (1∶1000), IkBα (1∶1000), and β-actin (1∶5000) at 4°C overnight. The next day, the membrane was washed by Tris-Buffered Saline Tween-20 (TBST) three times, 10 minutes each, and incubated with the corresponding secondary antibodies that were horseradish peroxidase-conjugated for 1 h at room temperature. Antibody interactions were detected using enhanced chemiluminescence (ECL) followed by exposure to film.
NF-kB Reporter Gene Assay
A total of 1×106 BV2 microglial cells were transfected with 2 µg NF-kB-Luciferase reporter plasmid and pCMV-gal control vector (Clontech, Mountain View, CA) using Lipofectamine reagents according to the manufacturer’s protocol (Invitrogen). BV2 microglial cells were plated in 96-well plates at a density of 1×104 cells per well and cultivated at 37°C overnight. Three wells of cells were treated per experiment. After incubation with the appropriate DNA-Lipofectamine mixtures, cells were pre-incubated with or without rifampicin for 2 h before the addition of LPS for 6 h. Cells were then washed, lysed, and centrifuged according to the manufacturer’s instructions (Promega, Madison, WI). 20 µl of cell extract was mixed with 100 µl of luciferase assay reagent at room temperature followed by the luciferase activity detection using a luminometer (Safire2, Tecan Instruments, Switzerland). Luciferase activity was normalized through dividing the mean luciferase relative light units (RLU) by the mean value of β-galactosidase RLU.
Statistical Analysis
Data were presented as the mean ± standard error of the mean (SEM) derived from three or more independent experiments. Comparisons between two groups were analyzed using Student’s t-test. A value of p<0.05 was deemed to be statistically significant.
Results
2-DE Maps and Protein Identification by MALDI-TOF-MS
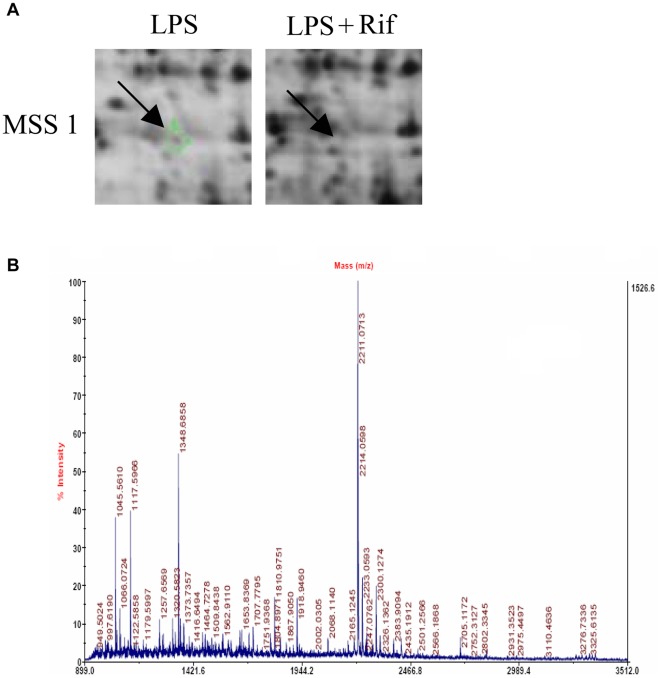
After matching, fifteen protein spots were extracted, digested, and submitted for identification by MALDI-TOF-MS. MSS1 protein was successfully identified. Its expression level was downregulated compared to the control (Figure 1). Detailed information about MSS1 proteins is listed in Table 1, including International Protein Index (IPI) accession number, molecular weight, pH indicated, and rifampicin treated-to-vehicle fluorescence ratios.
Figure 1. 2D-DIGE gel images of proteins isolated from LPS-stimulated BV2 microglia with or without rifampicin pretreatment.
Arrows indicate proteins that were differentially expressed in rifampicin-treated cells compared with non-treated controls. Peptide mass fingerprint spectra produced by MALDI-TOF-MS. Representative spectra from three independent experiments are shown. The x-axis represents mass-to-charge ratio (m/z), and the y-axis represents relative abundance. The peptide masses are labeled and annotated with corresponding m/z.
Table 1. Differential MSS1 protein expression identified by MALDI-TOF-MS.
| Accession Number | Name | Molecular Weight (Dalton) | pH Indicated | Ratio of Spot Density(Rifampicin/Vehicle) |
| IPI 00270326 | 26S proteasomeregulatory subunit 7 | 52,867 | 5.97 | −1,000,000 |
Rifampicin Significantly Suppressed the Expression of MSS1 in LPS-activated BV2 Microglia
To examine the effect of rifampicin on the expression of MSS1 in LPS stimulated BV2 microglia, we measured the protein levels of MSS1 in LPS-stimulated BV2 microglia. Rifampicin treatment greatly inhibited the LPS-induced MSS1 protein expression (Figure 2). The result suggested that rifampicin significantly reduced the expression of MSS1 in LPS-stimulated BV2 microglia.
Figure 2. Rifampicin significantly suppressed the expression of MSS1 in LPS-stimulated BV2 microglia.
Cells were treated with the indicated doses of rifampicin for 2 h prior to the addition of LPS (1000 ng/ml). At 24 h post-LPS incubation, cell lysates were analyzed for the protein production of MSS1 using western blot. Rifampicin significantly inhibited the LPS-induced MSS1 expression at protein levels. Data were obtained from three independent experiments with four to six replicates each. *p<0.05 compared with untreated cells and cells treated with LPS in the absence of rifampicin.
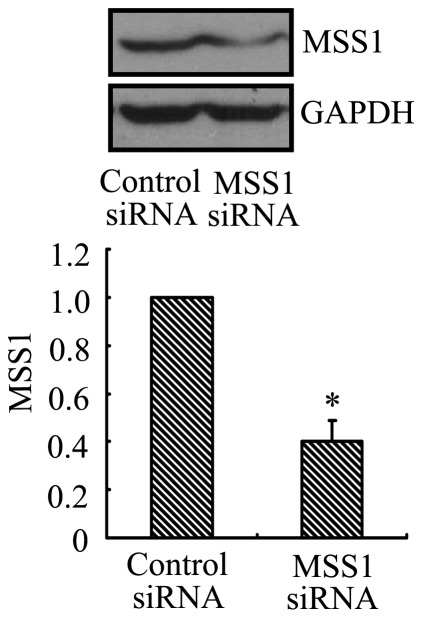
Verification of MSS1 Gene Silencing
We used western blot analysis to confirm the gene knockdown of MSS1 by its targeting siRNAs. After transfection with siRNAs, the expression of MSS1 protein was decreased to 40% compared with the negative control cells. The difference was statistically significant (p<0.01, Figure 3).
Figure 3. MSS1 gene knockdown reduced the expression of MSS1 at protein levels.
In order to assess the efficacy of gene silencing, western blot analysis was performed after transfection with siRNAs targeting MSS1. The specificity of MSS1 gene silencing was determined by comparing with cells transfected with the scrambled RNA duplex. The BV2 cells were transfected with either MSS1-specfic or control siRNAs. At 24 h post-incubation, cell lysates were analyzed for the protein expression of MSS1 using western blot. Compared with the negative control group, the expression of MSS1 was significantly reduced by incubation with MSS1-targeted siRNAs. Data were obtained from three independent experiments with four to six replicates each. *p<0.05 compared with the negative control group.
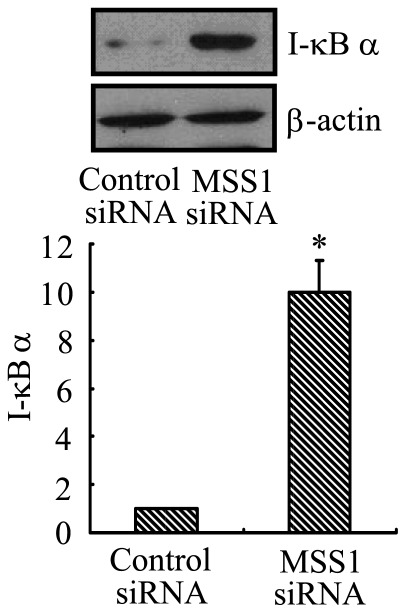
IkBα Protein Degradation was Significantly Reduced by MSS1 Gene Knockdown in LPS-stimulated BV2 Microglia
To examine the regulation IkBα degradation by MSS1 in LPS-activated microglia, the BV2 cells were transfected with either MSS1-specfic or control siRNAs for 24 h followed by LPS stimulation at 1000 ng/mL for 30 min. Cell lysates were analyzed for the protein expression of IkBα using western blot. Our results demonstrated that IkBα protein degradation was significantly reduced after MSS1 gene knockdown in LPS-stimulated BV2 microglia (Figure 4).
Figure 4. MSS1 gene silencing decreased IkBα protein degradation in LPS-activated microglia.
The BV2 cells were transfected with either MSS1-specfic or control siRNAs for 24 h, then cells were stimulated with LPS (1000 ng/mL) for 30 min before cell lysates were analyzed for IkBα expression using western blot. IkBα protein degradation was significantly reduced after the addition of siRNAs targeting MSS1 in LPS-induced BV2 microglia. Data were obtained from three independent experiments with four to six replicates each. *p<0.05 compared with the negative control group.
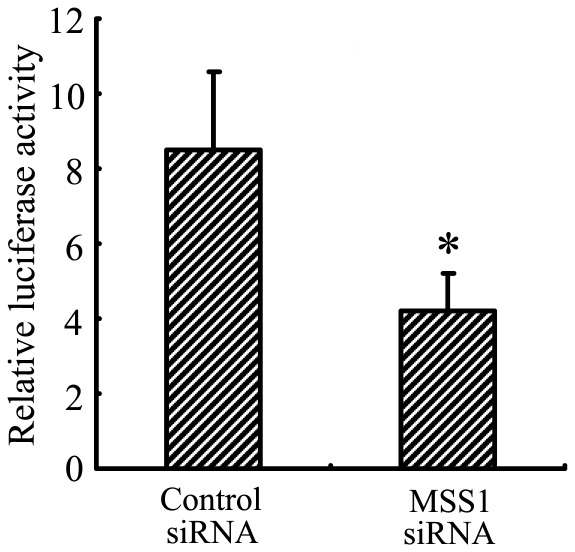
Downregulation of Microglial NF-kB Activation by MSS1 Gene Silencing in Response to LPS Stimulation
After MSS1 gene knockdown via RNAi, we assessed NF-kB activation using the NF-kB reporter gene assay. The BV2 cells were transfected with either MSS1-specfic or control siRNAs for 24 h. Cells were then incubated with LPS at 1000 ng/mL for 8 h. NF-kB activity was determined by measuring the relative luciferase activity. As shown in Figure 5, LPS markedly enhanced NF-kB activity, while transfection with MSS1-targeted siRNA significantly inhibited the enhancement.
Figure 5. MSS1 gene silencing inhibited microglial NF-kB activation in response to LPS stimulation.
The BV2 cells were transfected with either MSS1-specfic or control siRNA for 24 h, then cells were incubated with LPS at 1000 ng/mL for 8 h. After that, cells were transfected with NF-kB-luciferase reporter plasmid and pCMV-gal control vectors using Lipofectamine reagents. NF-kB activation was detected and expressed as relative luciferase activity. Compared with the negative control group, treatment with MSS1-targetd siRNA significantly suppressed the enhancement of NF-kB activity by LPS. Data were obtained from three independent experiments with four to six replicates each. *p<0.05 compared with the negative control group.
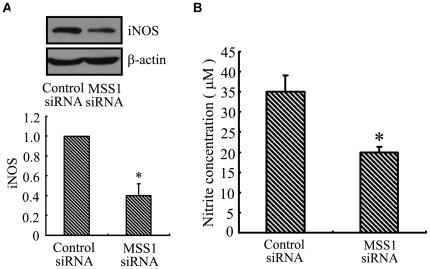
Decrease of iNOS Expression and NO Production by MSS1 Gene Knockdown in LPS-induced BV2 Microglia
To investigate the effect of MSS1 gene silencing on iNOS expression and NO production, we measured their protein levels as well as the accumulation of nitrite, a stable metabolite of NO, in LPS-stimulated BV2 microglia. Transfection using MSS1-specific siRNA greatly inhibited the LPS-induced iNOS protein expression (Figure 6). We next evaluated the NO production in culture supernatants by detecting nitrite levels using a Griess reaction. Consistent with the downregulation of iNOS, transfection with MSS1-targeted siRNAs reduced the LPS-mediated NO production in BV2 microglia (Fig. 6). Our results indicated that MSS1 gene silencing suppressed the production of pro-inflammatory NO by inhibiting the expression of iNOS in LPS-stimulated BV2 microglia.
Figure 6. Decreased iNOS expression and NO production by MSS1 gene silencing in LPS-activated microglia.
The BV2 cells were transfected with either MSS1-specfic or control siRNA for 24 h, then cells were stimulated for 24 h with LPS (1000 ng/mL). At 24 h post-LPS incubation, cell lysates were analyzed for the protein production of iNOS using western blot. The Griess assay was performed to measure the production of the NO metabolite, nitrite. Transfection with MSS1-specific siRNA suppressed the LPS-induced iNOS expression at protein levels, along with the production of nitrites Data were obtained from three independent experiments with four to six replicates each. *p<0.05 compared with the negative control group.
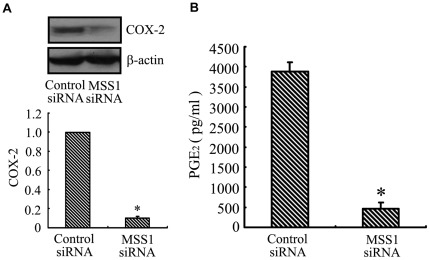
Inhibition of COX-2 Expression and PGE2 Production by MSS1 Gene Knockdown in LPS-activated BV2 Microglia
To address the effect of MSS1 gene silencing on COX-2 and PGE2 production, we assessed their expression in LPS-induced BV2 cells. Transfection using MSS1-targeted siRNAs significantly inhibited the LPS-induced COX-2 protein expression (Figure 7). We next collected the supernatant and analyzed the concentration of PGE2 using ELISA. Transfection with MSS1-specific siRNAs decreased the LPS-induced PGE2 production in BV2 microglia (Figure 7). These results implied that MSS1 gene silencing suppressed the production of pro-inflammatory PGE2 by inhibiting the expression of COX-2 in LPS-stimulated BV2 microglia.
Figure 7. Inhibition of COX-2 expression and PGE2 production by MSS1 gene knockdown in LPS-induced microglia.
The BV2 cells were transfected with either MSS1-specfic or control siRNA for 24 h, then cells were stimulated for 24 h with LPS (1000 ng/mL). At 24 h post-LPS incubation, cell lysates were analyzed for the protein production of COX-2 using western blot. We collected the supernatant and further analyzed the production of PGE2. MSS1 gene silencing suppressed the LPS-induced COX-2 expression at protein levels, as well as PGE2 production. Data were obtained from three independent experiments with four to six replicates each. *p<0.05 compared with the negative control group.
Discussion
The immunosuppressive properties of rifampicin have been discussed in the literature for 30 years [23]–[27]. Calleja et al. discovered that rifampicin activated the human glucocorticoid receptor (hGR), which regulated the expression of various genes including those that encoded interleukins [28]. Further investigations uncovered that rifampicin inhibited Toll-like receptor 2 (TLR2) via the suppression of the DNA binding of NF-kB, providing a novel mechanism contributing to the immunosuppression of rifampicin [29]. Our results showed that the anti-inflammatory, neuroprotective properties of rifampicin were mediated through the inhibition of signaling molecules, such as NF-kB and mitogen activated protein kinases (MAPKs) in LPS-activated BV2 microglial cells [12]. However, the mechanism by which rifampicin reduces microglial inflammation is not completely understood.
In this investigation, we used 2-DE and MALDI-TOF-MS to identify proteins affected by rifampicin in LPS-pretreated microglia. We successfully identified MSS1 protein and showed that its expression was downregulated (Figure 1A and 1B). The proteomic results were verified by western blot analysis, which also demonstrated the inhibition of the LPS-induced MSS1 expression by rifampicin (Figure 2). Our findings suggested that MSS1 was involved in microglial inflammation and its gene knockdown by RNAi alleviated neuroinflammation (Figure 3) in LPS-activated BV2 cells. Our results open the door to further study MSS1 as a potential therapeutic target for neuroinflammation.
UPS represent an ATP-dependent protein degradation mechanism in eukaryotic cells. The 26S proteasome is a multi-catalytic proteinase complex. It has a highly ordered structure composed of two complexes, a 20S core and a 19S regulator. The 19S regulator is composed of a base, which contains 6 ATPase subunits and 2 non-ATPase subunits, and a lid, which contains up to 10 non-ATPase subunits [30]. Proteasomes are distributed throughout eukaryotic cells at a high concentration and cleave peptides in an ATP/ubiquitin-dependent process in a non-lysosomal pathway [31]. Alterations in UPS are correlated with a variety of human pathologies, such as cancer, immunological disorders, inflammation, and neurodegeneration [31]–[33].
MSS1, also known as S7 or PSMC2, is a 433-amino-acid member of the AAA ATPase family. As a chaperone-like subunit of the 19S regulatory complex, MSS1 localizes in both the nucleus and the cytoplasm where it participates in proteasome events throughout the cell [18]. Additionally, MSS1 is thought to interact with several basal transcription factors and, via this interaction, play a role in transcriptional regulation [34]. Our data provided strong evidence for a novel mechanism of MSS1-mediated neuroinflammation. Additional investigations are needed to further elucidate this pathway and address the potential of MSS1 gene silencing as a therapy for neurodegenerative disorders.
NF-kB is an important transcription factor for the secretion of pro-inflammatory mediators [35]. LPS is shown to increase NF-kB activation, through IkB phosphorylation and the subsequent IkB degradation in macrophages [36]. In the canonical pathway of NF-kB induction, IkBs are phosphorylated at two amino-terminal serines, thus targeting them for polyubiquitination and the subsequent proteasomal degradation. IkB degradation enables NF-kB to translocate to the nucleus and bind to its target genes, including IkB. In addition, proteasomal degradation of transcriptionally active p65/RelA promotes the prompt termination of NF-kB responses [37]. The ubiquitin–proteasome pathway is considered pivotal to signal-induced IkB degradation [38]–[39]. In this study, we investigated the regulation of IkB and NF-kB signaling pathways by MSS1 gene silencing using a reporter gene assay and western blot analysis. Our results demonstrated that LPS caused rapid degradation of IkBα, while MSS1 gene knockdown significantly reduced IkBα degradation in LPS-stimulated BV2 microglia (Figure 4). We also found that LPS markedly enhanced NF-kB activity, whereas treatment with MSS1-specific siRNA significantly inhibited the enhancement (Figure 5). These results suggested that MSS1 gene knockdown suppressed NF-kB activation, likely through the blockage of IkBα degradation. The downregulation of IkBα provided a novel mechanism for MSS1’s immunomodulation in LPS-activated microglia.
Intranuclear blockage of NF-kB has been reported to suppress the expression of iNOS and COX-2 [40]. Our results indicated that transfection with MSS1-targeted siRNAs decreased the production of pro-inflammatory NO by inhibiting the expression of iNOS in LPS-stimulated BV2 microglia (Figure 6A and 6B). We also found that gene knockdown of MSS1 reduced the production of pro-inflammatory PGE2 through suppressing COX-2 gene expression in LPS-induced BV2 microglia (Figure 7A and 7B).
In conclusion, we demonstrated that rifampicin inhibited the expression of MSS1, which subsequently decreased IkBα degradation and the production of inflammatory mediators. Our results supported the potential application of MSS1 suppression, together with anti-inflammatory rifampicin, for the treatment of neuroinflammation and neurodegeneration.
Footnotes
Competing Interests: The authors have declared that no competing interests exist.
Funding: This work was funded by the Guangdong Provincial Department of Science and Technology (0400935505B33801003), the Ph.D. Programs Foundation of Ministry of Education of China (No. 20070558257), and the National Natural Science Foundation of China (81102449). The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
References
- 1.Liu B, Hong JS. Role of microglia in inflammation-mediated neurodegenerative diseases: mechamisms and strategies for therapeutic intervention. J Pharmacol Exp Ther. 2003;304:1–7. doi: 10.1124/jpet.102.035048. [DOI] [PubMed] [Google Scholar]
- 2.Eikelenboom P, van Gool WA. Neuroinflammatory perspectives on the two faces of Alzheimer’s disease. J Neural Transm. 2004;111:281–294. doi: 10.1007/s00702-003-0055-1. [DOI] [PubMed] [Google Scholar]
- 3.Koning N, Bö L, Hoek RM, Huitinga I. Downregulation of macrophage inhibitory molecules in multiple sclerosis lesions. Ann Neurol. 2007;62:504–514. doi: 10.1002/ana.21220. [DOI] [PubMed] [Google Scholar]
- 4.Krause DL, Müller N. Neuroinflammation, microglia and implications for anti-inflammatory treatment in Alzheimer’s disease. Int J Alzheimers Dis 2010. pii, 2010;732806 doi: 10.4061/2010/732806. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Qian L, Flood PM, Hong JS. Neuroinflammation is a key player in Parkinson’s disease and a prime target for therapy. J Neural Transm. 2010;117:971–979. doi: 10.1007/s00702-010-0428-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Blanchard JS. The ying and yang of rifampicin. Nat Med. 1998;4:92–96. doi: 10.1038/nm0198-014. [DOI] [PubMed] [Google Scholar]
- 7.Bellahsène A, Forsgren A. Effect of rifampin on the immune response in mice. Infect Immun. 1980;27:15–20. doi: 10.1128/iai.27.1.15-20.1980. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Tsankov N, Grozdev I, Kkzandjieva J. Old drug–new indication. Rifampicin in psoriasis. J Dermatolog Treat. 2006;17:18–23. doi: 10.1080/09546630500375643. [DOI] [PubMed] [Google Scholar]
- 9.Tsiskarishvili NV, Tsiskarishvili NI. The anti-tubercular drugs in the treatment of psoriasis.Georgian Med News. 2009;174:25–28. [PubMed] [Google Scholar]
- 10.Namazi HJ. Practice pearl: a novel use of rifampicin for treatment of carpal tunnel syndrome. Pain. 2008;9:380–381. doi: 10.1016/j.jpain.2008.01.325. [DOI] [PubMed] [Google Scholar]
- 11.Xu J, Wei C, Xu C, Bennett MC, Zhang G, et al. Rifampicin protects PC12 cells against MPP+-induced apoptosis and inhibits the expression of an alpha-Synuclein multimer. Brain Res. 2007;1139:220–225. doi: 10.1016/j.brainres.2006.12.074. [DOI] [PubMed] [Google Scholar]
- 12.Bi W, Zhu L, Wang C, Liang Y, Liu J, et al. Rifampicin inhibits microglial inflammation and improves neuron survival against inflammation. Brain Res. 2011;1395:12–20. doi: 10.1016/j.brainres.2011.04.019. [DOI] [PubMed] [Google Scholar]
- 13.Dai JN, Zong Y, Zhong LM, Li YM, Zhang W. Gastrodin inhibits expression of inducible NO synthase, cyclooxygenase-2 and pro-flammatory cytokines in cultured LPS-stimulated microglia via MAPK pathways. PLoS One. 2011;6:e21891. doi: 10.1371/journal.pone.0021891. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.S Vallabhapurapu, M Karin. Regulation and function of NF-kappaB transcription factors in the immune system, Annu. Rev. Immunol. 2009;27:693–733. doi: 10.1146/annurev.immunol.021908.132641. [DOI] [PubMed] [Google Scholar]
- 15.Zhang W, Wei Q. Calcineurin stimulates the expression of inflammatory factors in RAW 264.7 cells by interacting with proteasome subunit alpha type 6. Biochem Biophys Res Commun. 2011;407:668–673. doi: 10.1016/j.bbrc.2011.03.071. [DOI] [PubMed] [Google Scholar]
- 16.De Mot R, Nagy I, Walz J, Baumeister W. Proteasomes and other self-compartmentalizing proteases in prokaryotes. Trends Microbiol. 1999;7:88–92. doi: 10.1016/s0966-842x(98)01432-2. [DOI] [PubMed] [Google Scholar]
- 17.Williams AJ, Dave JR, Tortella FC. Neuroprotection with the proteasome inhibitor MLN519 in focal ischemic brain injury: relation to nuclear factor kappaB (NF-kappaB), inflammatory gene expression, and leukocyte infiltration. Neurochem Int. 2006;49:106–12. doi: 10.1016/j.neuint.2006.03.018. [DOI] [PubMed] [Google Scholar]
- 18.Yanagi S, Shimbara N, Tamura T. Tissue and cell distribution of a mammalian proteasomal ATPase, MSS1, and its complex formation with the basal transcription factors. Biochem Biophys Res Commun. 2000;279:568–573. doi: 10.1006/bbrc.2000.3969. [DOI] [PubMed] [Google Scholar]
- 19.Wang Y, Cheung YH, Yang Z, Chiu JF, Che CM, et al. Proteomic approach to study the cytotoxicity of dioscin (saponin). Proteomics. 2006;6:2422–2432. doi: 10.1002/pmic.200500595. [DOI] [PubMed] [Google Scholar]
- 20.Jessie K, Pang WW, Haji Z, Rahim A, Hashim OH. Proteomic analysis of whole human saliva detects enhanced expression of interleukin-1 receptor antagonist,thioredoxin and lipocalin-1 in cigarette smokers compared to non-smokers. International Journal of Molecular Sciences. 2010;11:4488–4505. doi: 10.3390/ijms11114488. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Tsikas D. Analysis of nitrite and nitrate in biological fluids by assays based on the Griess reaction: appraisal of the Griess reaction in the L-arginine/nitric oxide area of research. J Chromatogr B Analyt Technol Biomed Life Sci 15; 2007;851(1–2):51–70. doi: 10.1016/j.jchromb.2006.07.054. [DOI] [PubMed] [Google Scholar]
- 22.Woo MS, Jang PG, Park JS, Kim, WK, Jon TH, et al. Selective modulation of lipopolysaccharide-stimulated cytokine expression and mitogen-activated protein kinase pathways by dibutyryl-cAMP in BV2 microglial cells. Molecular Brain Res. 2003;113:86–96. doi: 10.1016/s0169-328x(03)00095-0. [DOI] [PubMed] [Google Scholar]
- 23.Dajani BM, Canadi MS, Thompson JS, Kasik JE. Rifampicin: an immunosuppressant? Lancet. 1972;2:1904. doi: 10.1016/s0140-6736(72)92390-2. [DOI] [PubMed] [Google Scholar]
- 24.Tsankov N, Angelova I. Rifampin in dermatology. Clin Dermatol. 2003;21:50–55. doi: 10.1016/s0738-081x(02)00328-0. [DOI] [PubMed] [Google Scholar]
- 25.Mlambo G, Sigola LB. Rifampicin and dexamethasone have similar effects on macrophage phagicytosis of zymosan, but differ in their effects on nitrite and TNF-alpha production. Int Immunopharmacol. 2003;3:513–522. doi: 10.1016/S1567-5769(03)00022-5. [DOI] [PubMed] [Google Scholar]
- 26.An N, Song Y, Zhang X, Ci X, Li H, et al. Pretreatment of mice with rifampicin prolongs survival of endotoxic shock by modulating the levels of inflammatory cytokines. Immunopharmacol Immunotoxicol. 2008;30:437–446. doi: 10.1080/08923970802135146. [DOI] [PubMed] [Google Scholar]
- 27.An N, Song Y, Zhang X, Ci X, Li H, et al. Group B streptococci exposed to rifampin or clindamycin (versus ampicillin or cefotaxime) stimulate reduced production of inflammatory mediators by murine macrophages. Pediatr Res. 2005;57:419–423. doi: 10.1203/01.PDR.0000153946.97159.79. [DOI] [PubMed] [Google Scholar]
- 28.Calleja C, Pascussi JM, Mani JC, Maurel P, Vilarem MJ. The antibiotic rifampicin is a nonsteroidal ligand and activator of the human glucocorticoid receptor. Nat Med. 1998;4:92–96. doi: 10.1038/nm0198-092. [DOI] [PubMed] [Google Scholar]
- 29.Kim SK, Kim YM, Yeum CE, Jin SH, Chae GT, et al. Rifampicin Inhibits the LPS-induced Expression of Toll-like Receptor 2 via the Suppression of NF-kappaB DNA-binding Activity in RAW 264.7 Cells. Korean J Physiol Pharmacol. 2009;13:475–482. doi: 10.4196/kjpp.2009.13.6.475. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Hershko A, Ciechanover A. The ubiquitin system. Annu Rev Biochem. 1998;67:425–479. doi: 10.1146/annurev.biochem.67.1.425. [DOI] [PubMed] [Google Scholar]
- 31.Finley D. Recognition and processing of ubiquitin-protein conjugates by the proteasome. Annu Rev Biochem. 2009;78:477–513. doi: 10.1146/annurev.biochem.78.081507.101607. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Goldberg AL. Protein degradation and protection against misfolded or damaged proteins. Nature. 2003;426:895–899. doi: 10.1038/nature02263. [DOI] [PubMed] [Google Scholar]
- 33.Sujashvili R. Ubiquitin-26s proteasome system. New therapeutic implications. Georgian Med News. 2008;161:48–51. [PubMed] [Google Scholar]
- 34.Yanagi S, Shimbara N, Tamur T. Tissue and cell distribution of a mammalian proteasomal ATPase, MSS1, and its complex formation with the basal transcription factors. Biochem Biophys Res Commun. 2000;279:568–573. doi: 10.1006/bbrc.2000.3969. [DOI] [PubMed] [Google Scholar]
- 35.Ozato K, Tsujimura H, Tamura T. Toll-like receptor signal in gand regulation of cytokine gene expression in the immune system. Biotechniques. 2002;66–72 [PubMed] [Google Scholar]
- 36.Kim WG, Mohney RP, Wilson B, Jeohn GH, Liu B, et al. Regional difference in susceptibility to lipopolysaccharide-induced neurotoxicity in the rat brain: role of microglia. J Neurosci. 2000;20:6309–6316. doi: 10.1523/JNEUROSCI.20-16-06309.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Saccani S, Marazzi I, Beg A, Natoli G. Degradation of promoter-bound p65/RelA is essential for the prompt termination of the nuclear factor kappaB response. J Exp Med. 2004;200:107–113. doi: 10.1084/jem.20040196. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Chen F, Lu Y, Kuhn DC, Maki M, Shi X, et al. Calpain Contributes to Silica-Induced IkBa Degradation and Nuclear Factor-kB Activation. ARCHIVES OF BIOCHEMISTRY AND BIOPHYSICS. 1997;342:383–388. doi: 10.1006/abbi.1997.0132. [DOI] [PubMed] [Google Scholar]
- 39.YmamotoY, Gaynor RB. IkappaB kinases: key regulators of the NF-kappaB Pathway. Trends Biochem Sci. 2004;29:72–79. doi: 10.1016/j.tibs.2003.12.003. [DOI] [PubMed] [Google Scholar]
- 40.Wang Y, Yu C, Pan Y, Li J, Zhang Y, et al. A novel compound c12 inhibits inflammatory cytokine production and protects from inflammatory injury in vivo. PLoS One. 2011;6:e24377. doi: 10.1371/journal.pone.0024377. [DOI] [PMC free article] [PubMed] [Google Scholar]