Abstract
Background
Cerebral autosomal dominant arteriopathy with subcortical infarcts and leukoencephalopathy (CADASIL) is an inherited disease leading to recurrent ischemic stroke and vascular dementia. Numerous mutations in the 23 exons of the NOTCH3 gene have been reported to cause CADASIL in Caucasian populations, but the full spectrum of genetic changes leading to this disease is yet to be known and, especially, very few reports are available on CADASIL in Asian populations.
Methods and Results
We genotyped members of a 5-generational Han Chinese family with CADASIL patients and identified an R133C mutation in the NOTCH3 gene. Clinical analysis demonstrated that the penetrance of the mutation was not complete. Five of the mutation carriers, not exposed to the known vascular risk factors, did not show any clinical feature of CADASIL, suggesting the importance of environmental factors to the development of this disease.
Conclusions
Members of a 5-generational Han Chinese family with CADASIL patients had an R133C mutation in the NOTCH3 gene but only individuals exposed to known vascular risk factors developed CADASIL.
Introduction
Cerebral autosomal dominant arteriopathy with subcortical infarcts and leukoencephalopathy (CADASIL) is an inherited disease with mutations in the NOTCH3 gene [1], [2], [3]. This disorder has been found in many race-ethnicities, with most reported cases coming from European Caucasian families [2], [4], [5]. To date, very few cases in Asian families have been reported [6], which however may not necessarily indicate that the disease is rare in Asia.
The main clinical feature of the disease is the disfunctioning of the central nervous system (CNS), characterized by recurrent ischemic attacks or strokes, migraine, cognitive impairment, dementia and psychiatric disturbances [7]. The mean onset age is around 45 years old, ranging from 30 to 70 [2], [4], [5]. About 85% of symptomatic CADASIL patients have ischemic attacks or stroke and 22–64% show migraine, which may begin early during childhood or adolescence but mostly during the third decade [2], [4], [6]. Many CADASIL patients also show cognitive decline, dementia and psychiatric symptoms [6]. In addition to these common CNS symptoms and signs, some less frequent manifestations of the disease have also been reported, such as epilepsy, transient disturbances of consciousness, visual impairment, and hemorrhagic strokes [7], [8], [9], [10].
A large number of mutations in the 23 exons of the NOTCH3 gene have been reported to be associated with CADASIL [1], [5], [7], [11], [12], [13], [14]. However, the full spectrum of genetic changes leading to this disease is yet to be known and, especially, very few reports are available on CADASIL in Asian populations. Here, we report an R133C mutation on exon 4 of the NOTCH3 gene in members of a 5-generational Han Chinese family and describe the unusual clinical manifestations of the disease in this family.
Results
Clinical Data
The proband was a 60-year-old male (ΙΙ:5; Figure 1), whose symptoms began at the age of 47. The main clinical manifestations included mild dysarthria and left central facial and tongue paralysis. Lower jaw reflex was brisk, bilateral palm-chin reflex was brisk, bilateral gag reflex was slow, limb tendon reflexes were brisk, with the lower limbs being pronounced. Left rotation movement was clumsy, and the Romberg sign was brisk. The patient had a right hemiparesis.
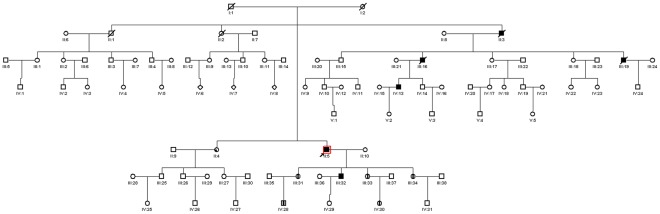
Figure 1. Pedigree of a 5-generational Chinese Han family affected with CADASIL.
Squares and circles indicate males and females, respectively. Filled symbols denote affected status. Normal individuals are shown as empty symbols. Mutation carriers without evident clinical features are indicated by a circle with a vertical line in the middle (III:31, III:33, III:34, IV:30) or a square with a vertical line in the middle (IV:28). II:4 had a large area cerebral infarction but was not a CADASIL patient.
The MRI examination results showed long T1 and long T2 signals on the white matter around the ventricles, and punctate long T1 and long T2 signals in the brainstem (Figure 2). His intelligence score was normal. Urine routine test, blood glucose, blood lipids, liver and kidney functions, blood homocysteine levels, ECG and abdominal ultrasound results were all normal. This 5-generational family included 6 affected individuals, 72 unaffected individuals, 5 mutation carriers who did not show symptoms and one large area cerebral infarction patient (ΙΙ:4), who was not a CADASIL patient and did not have the mutation. The main clinical features of the proband and the other affected individuals in the family were summarized in Table 1.
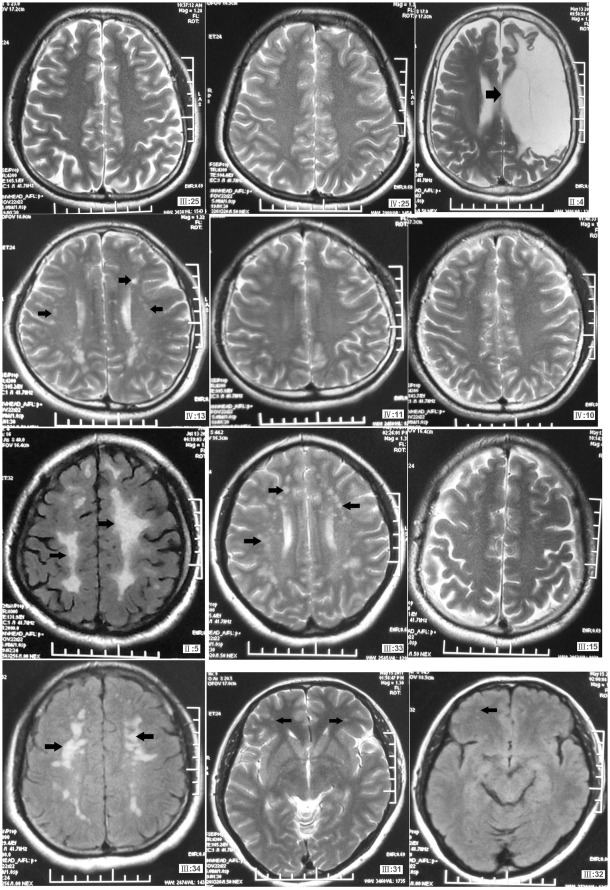
Figure 2. Cerebral MRI examination in the CADASIL family members.
ΙΙ:5, the proband, was a 60-year-old male affected family member. ΙΙΙ:31, ΙΙΙ:32, ΙΙΙ:33 and ΙΙΙ:34 were the children of the proband (ΙΙ:5). ΙΙ:5 and ΙΙΙ:32 showed evident clinical features, but ΙΙΙ:31, ΙΙΙ:33 and ΙΙΙ:34 did not show any clinical feature, although they were all mutation carriers (see the long T1 and T2 signals). ΙΙΙ:25, ΙΙΙ:15, ΙV:25, ΙV:10 and ΙV:11 were healthy core family members. ΙΙΙ:4 is a patient with l massive cerebral infarction, who did not have the R133C mutation.
Table 1. Clinical features of the affected individuals.
| Patient | Sex | Age | Onset Age | Migraine | Memory | MRI Examination | Vascular Risk Factors | Other Symptom |
| ΙΙ:4 | F | 67 | 48 | – | Severe | Gliosis in the lefttemporal lobe, foreheadand parietallobe; massivecerebralinfarction | – | Ineffective activity ofright limbs accompaniedby verbal clumsinessfor over 20 years;obvious memorydecline; nodysphagia or dysuria. |
| ΙΙ:5 | M | 60 | 47 | – | Mild | Long T1 andT2 signals shownon the whitematter around theventricles; punctuatelong T1 andlong T2signals in the brainstem;CADASIL patient. | Smoking | Unsteady gait for2 months, gettingworse for1 week; numbnessin hands andfeet; languageunclear; ataxia;lower extremityweakness. |
| ΙΙΙ:32 | M | 40 | 30 | + | Mild | Punctate longT1 and longT2 signals onthe temporal lobes;bilateral basal gangliaand corona radiateand high-intensitysignals of FLAIRsequence; CADASIL patient. | Smoking | Numbness in handsand feet; nosymptoms ofineffective activityof the limbs,memory declineor headache. |
| ΙV:13 | M | 31 | 20 | + | Mild | Punctate long T1and T2 signals onright hemisphere ofcerebellum, bilateral temporallobes, bilateral basalganglia and coronaradiate, high-intensitysignals of FLAIRsequence; CADASILpatient. | Hypertension | Drowsinessaccompanied byparoxysmal hemicrania forover 10 years;headache during thecourse of thedisease; MMSEscore 29. |
| ΙΙΙ:31 | F | 41 | – | – | – | Abnormal punctate longT1 and T2signals on bilateralfrontal lobes, slightlyhigher signal intensityof lesions onFLAIR;CADASIL patient. | – | – |
| ΙΙΙ:34 | F | 36 | – | – | – | Abnormal punctate long T1 and long T2 signal shown on bilateral frontal lobe; CADASIL patient. | – | – |
| ΙΙΙ:33 | F | 36 | – | – | – | Punctate long T1and long T2signals on bilateraltemporal lobes,bilateral basal gangliaand corona radiate;CADASIL patient. | – | – |
NOTCH3 Mutation
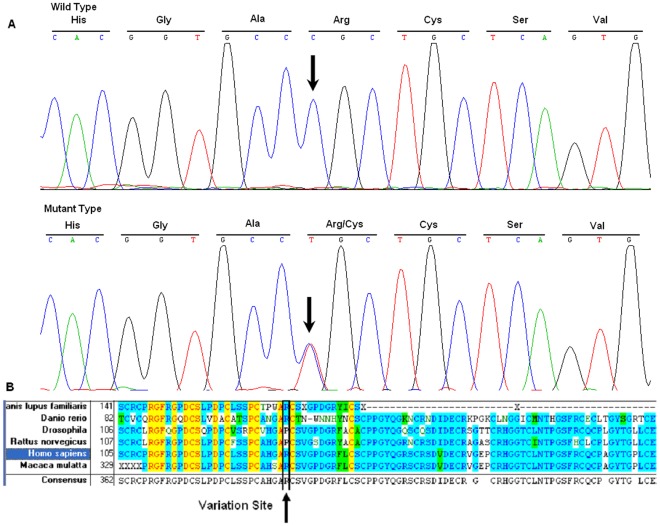
Sanger sequencing of the amplified fragments of NOTCH3 in all affected individuals identified a single base alteration, 475C→T (Figure 3A, B), in exon 4 of the NOTCH3 gene (GI:4854) located at 19p13.2-p13.1, resulting in the substitution of Arg to Cys at codon 133 (R133C). The remainder of the coding sequence showed no other changes. Further sequence analysis revealed that all affected members in this family carried the 475C→T mutation, although 5 mutation carriers did not show symptoms. The other individuals in this family did not carry this mutation, and the mutation was not present in 100 normal controls.
Figure 3. Analysis of the DNA and protein sequences.
DNA sequence chromatogram of the R133C mutation in NOTCH3 and multiple-sequence alignment of NOTCH3 protein family. The C→T transition at position 475 resulting in the R133C mutation is located within a highly conserved region. A: DNA sequence chromatogram of the unaffected family members and ΙΙΙ:4; B: DNA sequence chromatogram of the affected family members and ΙΙΙ:31 and ΙΙΙ:33; C: multiple-sequence alignment of NOTCH3 protein family.
Conservation of the Protein in Evolution
Comparison of NOTCH3 protein sequences from six mammalian species by multiple-sequence alignment analysis showed that the 133Arg residue was located in a highly conserved region of the protein (Figure 3C).
Discussion
In this study, we identified a mutation, 475C→T (R133C), in the NOTCH3 gene in a 5-generational Han Chinese family with CADASIL patients. This mutation co-segregated with the disease phenotype in all affected individuals except ΙΙΙ:4, who was a patient with massive cerebral infarction but not a CADASIL patient. The mutation was not present in 100 normal control subjects. Five members of the family, ?:31, ?:33, ?:34,108 ?28 and ?30, were mutation carriers but did not show any clinical feature of CADASIL. The result of multiple-sequence alignments showed that Arg133, mutation of which was previously described by Joutel [15] and Mykkanen [16], was a conserved residue, indicating its importance for normal NOTCH3 function.
In this family, the penetrance rate was not 100% in the mutation carriers, but MRI examination showed that all the mutation carriers had long T1 and T2 signals within the temporal lobes and high-intensity signals of FLAIR sequence. However, ΙΙΙ:31, ΙΙΙ:33 and ΙΙΙ:34 did not show any clinical feature of CADASIL. One fact worth noting is that these three family members had not been exposed to any known vascular risk factors, such as smoking, drinking, and hypertension. On the other hand, ΙΙΙ:5, ΙΙΙ:32 and ΙV:13, who showed very evident clinical features, had obviously contacted vascular risk factors, such as smoking or hypertension. The genotype–phenotype correlation in CADASIL has not been clarified. Although some data support genotype–phenotype correlation in CADASIL [7], [13], [17], [18], the mutation found in patients of the family did not have specific effects on the expressivity of the disease. Some authors report that vascular risk factors [19], [20] or other unexplored factors may influence the phenotypic variability and lead to atypical features of the CADASIL patients, which may explain why ΙΙΙ:31, ΙΙΙ:33 and ΙΙΙ:34 did not show any clinical feature of CADASIL. However, correlations between vascular risk factors and expressivity of the disease would require larger scales of study involving many more patients and controls for a conclusion.
NOTCH3 (N3) is one member of the Notch receptor superfamily, which regulates cell fate during embryonic development [21] and is predominantly expressed in vascular smooth muscle cells (VSMC) in adulthood [1], [22]. Appropriately half of identified CADASIL-related mutations are located on exons 2–4 [7], [11], which encode the extracellular domain of N3 (N3ECD) within epidermal growth factor-like (EGF-like) repeat domains [15], [23], [24]. Modular structures show that six highly conserved cysteine residues within these domains stabilize the domain [25]. Dichgans and colleagues, by blocking or facilitating disulfide bridge formation, found that multimerization of N3, at least in part, depends on disulfide bridges and unpaired cysteine residue might make CADASIL-mutated N3ECD more susceptible to multimerization in higher order complexes [26].
In summary, this study identified the 475C→T (R133C) mutation in the NOTCH3 gene in a 5-generational Han Chinese family with CADASIL. Five family members were not exposed to any vascular risk factors and they did not show any clinical feature of CADASIL. Vascular risk factors may play a vital role in the development of CADASIL.
Materials and Methods
Affected and Unaffected Individuals in the Family
We ascertained a 5-generational Chinese Han family with non-syndromic CADASIL (see Figure 1) at the Second Affiliated Hospital of Harbin Medical University, Harbin, China. Informed consent was obtained from each participant, consistent with the Declaration of Helsinki. We recorded their medical history in detail. Physical and MRI examination was carried on each of the family members. Genomic DNA was extracted from peripheral blood leukocytes using standard protocols.
DNA sequencing
Individual exons of NOTCH3 were amplified by PCR using primer pairs shown in Table S1. The PCR products were sequenced on an ABI3130 Automated Sequencer.
Multiple Sequence Alignment
From the NCBI website (http://www.ncbi.nlm.nih.gov/), the NOTCH3 protein sequence of various species were obtained and, by using the Vector NTI software, multiple-sequence alignments of NOTCH3 proteins were carried out.
Supporting Information
PCR primers and PCR product sizes for NOTCH3 sequence analysis.
(DOC)
Acknowledgments
The authors thank the patients and the family members for their cooperation and participation in this study.
Footnotes
Competing Interests: The authors have declared that no competing interests exist.
Funding: This work was supported by a grant of National Natural Science Foundation of China (NSFC30970078) and a grant of Natural Science Foundation of Heilongjiang Province of China to GRL; a grant from Heilongjiang Innovation Research Foundation for Graduate Studies (YJSCX2011-335HLJ) to ZXT; and a start-up grant from Harbin Medical University, grants of National Natural Science Foundation of China (NSFC30870098, 30970119, 81030029), and a Specialized Research Fund for the Doctoral Program of Higher Education (No.20092307110001) to SLL. The funders had no role in study design, data collection and analysis, decision to publish or preparation of the manuscript.
References
- 1.Joutel A, Corpechot C, Ducros A, Vahedi K, Chabriat H, et al. Notch3 mutations in CADASIL, a hereditary adult-onset condition causing stroke and dementia. Nature. 1996;383:707–710. doi: 10.1038/383707a0. [DOI] [PubMed] [Google Scholar]
- 2.Dichgans M, Mayer M, Uttner I, Bruning R, Muller-Hocker J, et al. The phenotypic spectrum of CADASIL: clinical findings in 102 cases. Ann Neurol. 1998;44:731–739. doi: 10.1002/ana.410440506. [DOI] [PubMed] [Google Scholar]
- 3.Opherk C, Peters N, Herzog J, Luedtke R, Dichgans M. Long-term prognosis and causes of death in CADASIL: a retrospective study in 411 patients. Brain. 2004;127:2533–2539. doi: 10.1093/brain/awh282. [DOI] [PubMed] [Google Scholar]
- 4.Chabriat H, Vahedi K, Iba-Zizen MT, Joutel A, Nibbio A, et al. Clinical spectrum of CADASIL: a study of 7 families. Cerebral autosomal dominant arteriopathy with subcortical infarcts and leukoencephalopathy. Lancet. 1995;346:934–939. doi: 10.1016/s0140-6736(95)91557-5. [DOI] [PubMed] [Google Scholar]
- 5.Markus HS, Martin RJ, Simpson MA, Dong YB, Ali N, et al. Diagnostic strategies in CADASIL. Neurology. 2002;59:1134–1138. doi: 10.1212/wnl.59.8.1134. [DOI] [PubMed] [Google Scholar]
- 6.Tang SC, Lee MJ, Jeng JS, Yip PK. Arg332Cys mutation of NOTCH3 gene in the first known Taiwanese family with cerebral autosomal dominant arteriopathy with subcortical infarcts and leukoencephalopathy. J Neurol Sci. 2005;228:125–128. doi: 10.1016/j.jns.2004.10.019. [DOI] [PubMed] [Google Scholar]
- 7.Chabriat H, Joutel A, Dichgans M, Tournier-Lasserve E, Bousser MG. Cadasil. Lancet Neurol. 2009;8:643–653. doi: 10.1016/S1474-4422(09)70127-9. [DOI] [PubMed] [Google Scholar]
- 8.Choi JC, Kang SY, Kang JH, Park JK. Intracerebral hemorrhages in CADASIL. Neurology. 2006;67:2042–2044. doi: 10.1212/01.wnl.0000246601.70918.06. [DOI] [PubMed] [Google Scholar]
- 9.Rufa A, De Stefano N, Dotti MT, Bianchi S, Sicurelli F, et al. Acute unilateral visual loss as the first symptom of cerebral autosomal dominant arteriopathy with subcortical infarcts and leukoencephalopathy. Arch Neurol. 2004;61:577–580. doi: 10.1001/archneur.61.4.577. [DOI] [PubMed] [Google Scholar]
- 10.Singhal S, Bevan S, Barrick T, Rich P, Markus HS. The influence of genetic and cardiovascular risk factors on the CADASIL phenotype. Brain. 2004;127:2031–2038. doi: 10.1093/brain/awh223. [DOI] [PubMed] [Google Scholar]
- 11.Tikka S, Mykkanen K, Ruchoux MM, Bergholm R, Junna M, et al. Congruence between NOTCH3 mutations and GOM in 131 CADASIL patients. Brain. 2009;132:933–939. doi: 10.1093/brain/awn364. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Escary JL, Cecillon M, Maciazek J, Lathrop M, Tournier-Lasserve E, et al. Evaluation of DHPLC analysis in mutational scanning of Notch3, a gene with a high G-C content. Hum Mutat. 2000;16:518–526. doi: 10.1002/1098-1004(200012)16:6<518::AID-HUMU9>3.0.CO;2-Q. [DOI] [PubMed] [Google Scholar]
- 13.Dotti MT, Federico A, Mazzei R, Bianchi S, Scali O, et al. The spectrum of Notch3 mutations in 28 Italian CADASIL families. J Neurol Neurosurg Psychiatry. 2005;76:736–738. doi: 10.1136/jnnp.2004.048207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Federico A, Bianchi S, Dotti MT. The spectrum of mutations for CADASIL diagnosis. Neurol Sci. 2005;26:117–124. doi: 10.1007/s10072-005-0444-3. [DOI] [PubMed] [Google Scholar]
- 15.Joutel A, Vahedi K, Corpechot C, Troesch A, Chabriat H, et al. Strong clustering and stereotyped nature of Notch3 mutations in CADASIL patients. Lancet. 1997;350:1511–1515. doi: 10.1016/S0140-6736(97)08083-5. [DOI] [PubMed] [Google Scholar]
- 16.Mykkanen K, Savontaus ML, Juvonen V, Sistonen P, Tuisku S, et al. Detection of the founder effect in Finnish CADASIL families. Eur J Hum Genet. 2004;12:813–819. doi: 10.1038/sj.ejhg.5201221. [DOI] [PubMed] [Google Scholar]
- 17.Desmond DW, Moroney JT, Lynch T, Chan S, Chin SS, et al. The natural history of CADASIL: a pooled analysis of previously published cases. Stroke. 1999;30:1230–1233. doi: 10.1161/01.str.30.6.1230. [DOI] [PubMed] [Google Scholar]
- 18.Pescini F, Bianchi S, Salvadori E, Poggesi A, Dotti MT, et al. A pathogenic mutation on exon 21 of the NOTCH3 gene causing CADASIL in an octogenarian paucisymptomatic patient. J Neurol Sci. 2008;267:170–173. doi: 10.1016/j.jns.2007.10.017. [DOI] [PubMed] [Google Scholar]
- 19.Pantoni L, Sarti C, Pescini F, Bianchi S, Bartolini L, et al. Thrombophilic risk factors and unusual clinical features in three Italian CADASIL patients. Eur J Neurol. 2004;11:782–787. doi: 10.1111/j.1468-1331.2004.00915.x. [DOI] [PubMed] [Google Scholar]
- 20.Adib-Samii P, Brice G, Martin RJ, Markus HS Clinical spectrum of CADASIL and the effect of cardiovascular risk factors on phenotype: study in 200 consecutively recruited individuals. Stroke. 41:630–634. doi: 10.1161/STROKEAHA.109.568402. [DOI] [PubMed] [Google Scholar]
- 21.Artavanis-Tsakonas S, Rand MD, Lake RJ. Notch signaling: cell fate control and signal integration in development. Science. 1999;284:770–776. doi: 10.1126/science.284.5415.770. [DOI] [PubMed] [Google Scholar]
- 22.Joutel A, Andreux F, Gaulis S, Domenga V, Cecillon M, et al. The ectodomain of the Notch3 receptor accumulates within the cerebrovasculature of CADASIL patients. J Clin Invest. 2000;105:597–605. doi: 10.1172/JCI8047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Dichgans M, Ludwig H, Muller-Hocker J, Messerschmidt A, Gasser T. Small in-frame deletions and missense mutations in CADASIL: 3D models predict misfolding of Notch3 EGF-like repeat domains. Eur J Hum Genet. 2000;8:280–285. doi: 10.1038/sj.ejhg.5200460. [DOI] [PubMed] [Google Scholar]
- 24.Peters N, Opherk C, Bergmann T, Castro M, Herzog J, et al. Spectrum of mutations in biopsy-proven CADASIL: implications for diagnostic strategies. Arch Neurol. 2005;62:1091–1094. doi: 10.1001/archneur.62.7.1091. [DOI] [PubMed] [Google Scholar]
- 25.Downing AK, Knott V, Werner JM, Cardy CM, Campbell ID, et al. Solution structure of a pair of calcium-binding epidermal growth factor-like domains: implications for the Marfan syndrome and other genetic disorders. Cell. 1996;85:597–605. doi: 10.1016/s0092-8674(00)81259-3. [DOI] [PubMed] [Google Scholar]
- 26.Opherk C, Duering M, Peters N, Karpinska A, Rosner S, et al. CADASIL mutations enhance spontaneous multimerization of NOTCH3. Hum Mol Genet. 2009;18:2761–2767. doi: 10.1093/hmg/ddp211. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
PCR primers and PCR product sizes for NOTCH3 sequence analysis.
(DOC)