Abstract
In Bacillus licheniformis 749/I, BlaP β-lactamase is induced by the presence of a β-lactam antibiotic outside the cell. The first step in the induction mechanism is the detection of the antibiotic by the membrane-bound penicillin receptor BlaR1 that is composed of two functional domains: a carboxy-terminal domain exposed outside the cell, which acts as a penicillin sensor, and an amino-terminal domain anchored to the cytoplasmic membrane, which works as a transducer-transmitter. The acylation of BlaR1 sensor domain by the antibiotic generates an intramolecular signal that leads to the activation of the L3 cytoplasmic loop of the transmitter by a single-point cleavage. The exact mechanism of L3 activation and the nature of the secondary cytoplasmic signal launched by the activated transmitter remain unknown. However, these two events seem to be linked to the presence of a HEXXH zinc binding motif of neutral zinc metallopeptidases. By different experimental approaches, we demonstrated that the L3 loop binds zinc ion, belongs to Gluzincin metallopeptidase superfamily and is activated by self-proteolysis.
Introduction
During evolution, the most common resistance mechanism acquired by eubacteria to resist β-lactam antibiotic action is the production of a β-lactamase that degrades these antibiotics by opening their β-lactam ring [1], [2]. The cleaved antibiotic is then unable to acylate its membrane-bound D,D-peptidase targets, which are involved in the peptidoglycan cross-linking or remodeling. These enzymes inhibited by β-lactam antibiotics are named penicillin-binding proteins (PBPs).
In Bacillus licheniformis and Staphylococcus aureus, the production of an inducible class A β-lactamase, BlaP and BlaZ respectively, is regulated by the BlaI repressor that maintains β-lactamase production at a low level in the absence of a β-lactam antibiotic outside the cell. In presence of a β-lactam antibiotic at sublethal concentration, the BlaI repressor is inactivated by a protein relay including the membrane-bound penicillin-sensory transducer BlaR1 and the BlaR2 protein, yet to be identified [3], [4], [5]. The regulatory genes, blaI and blaR1 are located downstream the blaP/blaZ genes and are divergently transcribed as a polycistronic mRNA [6].
In S. aureus, another related mechanism that mediates resistance to β-lactam antibiotics has been described. It involves the induction of the low-affinity PBP2a or MecA capable to replace the inhibited constitutive PBPs [7]. The mecA expression is under the control of MecI and MecR proteins that are homologous to S. aureus and B. licheniformis BlaI and BlaR1, and play the same role. Furthermore, MecI and BlaI from S. aureus are interchangeable, but BlaR1 and MecR are not [8], [9].
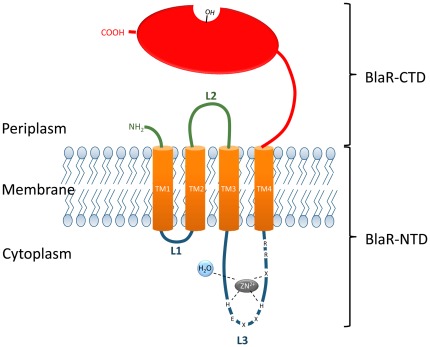
The BlaR1 and MecR membrane-bound penicillin-sensory transducers are composed of two domains, an amino-terminal domain (BlaR/MecR-NTD) anchored into the cytoplasmic membrane and a carboxy-terminal domain (BlaR/MecR-CTD) exposed outside the cell. This latter domain contains in its primary structure, the conserved motifs of the serine-penicillin recognizing enzymes (SPRE) and acts as a penicillin sensor [10], [11]. The BlaR-CTD from B. licheniformis and from S. aureus can be produced as soluble domains and their crystal structures exhibit the conserved SPRE 3D-fold [12], [13], [14]. The membrane topology of B. licheniformis BlaR-NTD (Met1-Pro339) has been experimentally determined: it contains four transmembrane segments (TM1, TM2, TM3, TM4) connected by three loops named L1 (Lys27-Thr35), L2 (Pro53-Ser115) and L3 (Tyr134-Arg322). L1 and L3 are cytoplasmic, whereas L2 is exposed outside the cell (Figure 1) [15]. Although very little is known concerning the mechanism of signal transduction by BlaR1/MecR, the first step is the acylation of BlaR/MecR-CTD by the β-lactam antibiotic outside the cell. The acylation of this domain by the antibiotic causes a modification of the interaction between BlaR-CTD and L2 and gives rise to the activation of the L3 cytoplasmic loop via a conformation change of the four transmembrane segments. The activated L3 would launch a cytoplasmic signal whose target is BlaI/MecI [10], [15], [16], [17], [18]. In this signaling pathway, L2 and the four transmembrane segments would act as a signal transducer and L3 as a cytoplasmic signal amplifier. In S. aureus, the BlaR1 L3 activation results in its single-point cleavage between the Arg293 and Arg294 residues [19]. This cleavage site is conserved in B. licheniformis BlaR1 and partially conserved in S. aureus MecR (Figure 2). In S. aureus, the activated L3 loop generates a cytoplasmic signal that leads to the inactivation of the BlaI repressor by proteolysis whereas in B. licheniformis it has been demonstrated that BlaI is not inactivated by proteolysis but by the presence of a coactivator coming from peptidoglycan turnover [20]. Regardless of the route of BlaI inactivation, activated L3 acts as a protease cleaving either BlaI or a pro-coactivator [20], [21], [22]. The hypothesis of L3 protease activity is supported by the presence within the BlaR1/MecR L3 primary structures, of a HEXXH motif characteristic of the zinc-binding signature of the neutral zinc metallopeptidases (M4 superfamily) such as thermolysin (Figure 1) [23]. In S. aureus BlaR1, the mutation of the conserved glutamic acid residue to alanine gives rise to a non-inducible β-lactamase phenotype [19].
Figure 1. Membrane topology of the penicillin-sensory transducer, BlaR1.
This receptor contains two domains, an extracellular domain: BlaR-CTD and a transmembrane domain: BlaR-NTD. BlaR-CTD exhibits the three motifs of the penicillin binding protein family (S*402TYK, Y476GN, K539TG, where S402 is the active serine). BlaR-NTD includes four transmembrane segments (TM1, TM2, TM3, TM4) connected by three loops (L1, L2, L3). The cytoplasmic L3 loop contains the H212EXXH motif, characteristic to zinc-metalloproteases.
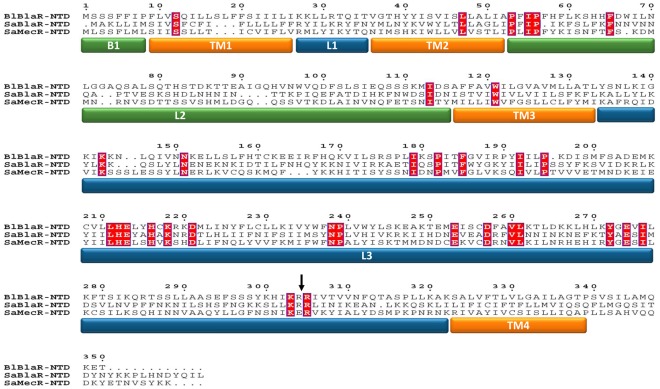
Figure 2. Sequence alignment of the amino-terminal domain of the proteins B. licheniformis 749/I BlaR1 (BlBlaR-NTD) and S. aureus RN4220 BlaR1 (SaBlaR-NTD) and S. aureus MRSA252 MecR (SaMecR-NTD).
The conserved residues are highlighted in red and the loop and transmembrane segments are marked: the transmembrane segments (TM1 to 4) in orange; the extracellular loops B1 (M1-P8) and L2 (P53-S115) in green and the cytoplasmic loops L1 (K27-T35) and L3 (Y134-K322) in blue. The site of cleavage, situated between the R293 and R294 in S. aureus BlaR1, is indicated by an arrow.
In this paper, we performed site-directed mutagenesis experiments combined with zinc-blot, Western blot and β-lactamase induction analyses to demonstrate that the cytoplasmic BlaR1 L3 loop belongs to Gluzincin metallopeptidase superfamily and that its cleavage during BlaR1 activation occurs by self-proteolysis.
Results
L3 Conserved Residues
A multiple sequence alignment of BlaR1/MecR N-terminal domains of B. licheniformis and S. aureus (Figure 2) revealed 32 strictly conserved residues in the three aligned sequences corresponding to 9.4% of identity when the length of B. licheniformis BlaR-NTD sequence is used as reference ([34/339]×100 = 9.4%). Out of these 32 conserved residues, 24 are located within the L3 loops ([24/187]×100 = 12.8%) highlighting the importance of this conserved loop for intracellular signaling. Conserved residues include the zinc-binding signature HEXXH of neutral zinc metalloproteases. Thermolysin is considered as the canonical enzyme of this protease family, its catalytically essential zinc ion is coordinated by the two histidine residues of the motif (H142EXXH, mature thermolysin numbering), the E166 and a water molecule. The catalytic mechanism involves a general base mechanism including E143 of the H142EXXH motif and the protonated H231 [24]. Within the strictly conserved residues present in L3 loop consensus, 4 acidic residues (D: 2 and E: 2) could play the role of the third zinc ligand or could be involved in the general base relay. No third conserved histidine residue able to play the catalytic role of H231 in thermolysin is present in the L3 loop. The best candidate for this role is a conserved tyrosine residue (Y272 in B. licheniformis BlaR-NTD).
Conserved L3 Residues Important for Signal Transduction or Zn Binding
To highlight if the conserved E, D, H and Y residues are important for the zinc binding or for the catalytic activity in the activated L3 loop, the mutants listed in Table 1 were constructed. The resulting plasmids (pDML1268-78, Table 2) were introduced into B. subtilis 168 and the recombinant resulting strains tested for their ability to induce BlaP β-lactamase in presence of the inducer. In all cases, the mutated receptor expression in non-induced or induced membranes has been detected by Western blotting analysis (Figure 3) showing that the introduced mutations do not lead to the production of an unstable, proteolysis susceptible receptor.
Table 1. Induction of BlaPβ-lactamase of the BlaR1 L3 loop mutants.
| Mutation | Plasmids | Induction factor* (3h) | |
| Wild type | pDML995 | 37.8±1.5 | |
| Mutants in H212EXXH motif | H212A/E213A/H216A | pDML1268 | 1.6±0.1 |
| E213A | pDML1269 | 1.2±0.1 | |
| H212A/H216A | pDML1270 | 1.2±0.1 | |
| H216A | pDML1271 | 1.27±0.01 | |
| E213D | pDML1279 | 1.07±0.02 | |
| E213Q | pDML1280 | 1.6±0.2 | |
| Mutants in putative Zn++ ligands | D221A | pDML1277 | 1.2±0.1 |
| E253A | pDML1273 | 1.2±0.3 | |
| D257A | pDML1274 | 1.9±0.1 | |
| E274A | pDML1275 | 33.9±0.9 | |
| Mutants in putative general base | Y 272A | pDML1278 | 1.1±0.1 |
| Mutants in R304R cleavage site | R304A/R305A | pDML3045 | 7.6±1.7 |
The induction factor corresponds to the ratio between the β-lactamase quantity/A600 for induced culture and the β-lactamase quantity/A600 for uninduced culture.
Table 2. Bacterial strains and plasmids used in this study.
| Strain or plasmid | Relevant characteristic(s) | Source |
| Bacillus licheniformis 749/I | β-lactamase inducible strain, wild-type | |
| Bacillus subtilis 168 | Recipient cell for E. coli/Bacillus shuttle vectors | ATCC 23857 |
| Escherichia coli GI724 | Host cell for heterologous expression of BlaR1 L3, F-, lamda-, lacPL8, ampC::Ptrpcl | Invitrogen |
| pUCBM20 | Cloning plasmid, Apr | Boehringer |
| pCR-ScriptTMSK(+) | Cloning vector for PCR fragments, Apr | Stratagene |
| pLEX | Expression vector from inducible P-lambda promoter, Apr | Novagen |
| pDML995 | A derivative of the Bacillus/E. coli shuttle vector pMK4 [39] carrying the wild-type B. licheniformis 749I bladivergon, Apr in E.coli and Cmr in Bacillus | A. Brans (unpublished) |
| pDML1251 | pUCBM20 derivative carrying the SstI/EcoRIblaR1 fragment | This study |
| pDML1255 | pDML1251 derivative carrying BlaR1H212A/E213A/H216A mutation | This study |
| pDML1256 | pDML1251 derivative carrying BlaR1 E213A mutation | This study |
| pDML1257 | pDML1251 derivative carrying BlaR1 H212A/H216A mutation | This study |
| pDML1258 | pDML1251 derivative carrying BlaR1 H216A mutation | This study |
| pDML1260 | pDML1251 derivative carrying BlaR1 E253A mutation | This study |
| pDML1261 | pDML1251 derivative carrying BlaR1 D257A mutation | This study |
| pDML1262 | pDML1251 derivative carrying BlaR1 E274A mutation | This study |
| pDML1264 | pDML1251 derivative carrying BlaR1 D221A mutation | This study |
| pDML1265 | pDML1251 derivative carrying BlaR1 Y272A mutation | This study |
| pDML1266 | pDML1251 derivative carrying BlaR1 E213D mutation | This study |
| pDML1267 | pDML1251 derivative carrying BlaR1 E213Q mutation | This study |
| pDML1268 | pDML995 derivative carrying BlaR1 H212A/E213A/H216A mutation | This study |
| pDML1269 | pDML995 derivative carrying BlaR1 E213A mutation | This study |
| pDML1270 | pDML995 derivative carrying BlaR1 H212A/H216A mutation | This study |
| pDML1271 | pDML995 derivative carrying BlaR1 H216A mutation | This study |
| pDML1273 | pDML995 derivative carrying BlaR1 E253A mutation | This study |
| pDML1274 | pDML995 derivative carrying BlaR1 D257Amutation | This study |
| pDML1275 | pDML995 derivative carrying BlaR1 E274A mutation | This study |
| pDML1277 | pDML995 derivative carrying BlaR1 D221A mutation | This study |
| pDML1278 | pDML995 derivative carrying BlaR1 Y272A mutation | This study |
| pDML1279 | pDML995 derivative carrying BlaR1 E213D mutation | This study |
| pDML1280 | pDML995 derivative carrying BlaR1 E213Q mutation | This study |
| pDML3045 | pDML995 derivative carrying BlaR1 R304A/R305A mutation | This study |
| pDML1283 | pCR-ScriptTMSK(+) derivative carrying BlaR1 L3 loop coding sequence | This study |
| pDML1284 | pDML1283 derivative carrying BlaR1 L3 E213A mutation | This study |
| pDML1285 | pDML1283 derivative carrying BlaR1 L3 H212A/H216A mutation | This study |
| pDML1286 | pDML1283 derivative carrying BlaR1 L3 D257A mutation | This study |
| pDML1287 | pDML1283 derivative carrying BlaR1 L3 D221A mutation | This study |
| pDML1293 | pDML1283 derivative carrying BlaR1 L3 E253A mutation | This study |
| pDML1288 | pLEX derivative allowing the expression of BlaR1 L3 loop | This study |
| pDML1289 | pLEX derivative allowing the expression of BlaR1 L3 carrying E213A mutation | This study |
| pDML1290 | pLEX derivative allowing the expression of BlaR1 L3 carrying H212A/H216A mutation | This study |
| pDML1291 | pLEX derivative allowing the expression of BlaR1 L3 carrying D257A mutation | This study |
| pDML1292 | pLEX derivative allowing the expression of BlaR1 L3 carrying D221A mutation | This study |
| pDML1294 | pLEX derivative allowing the expression of BlaR1 L3 carrying E253A mutation | This study |
Apr: ampicillin resistance.
Cmr: chloramphenicol resistance.
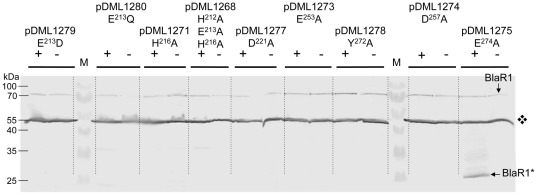
Figure 3. Western blot analysis of membrane-bound BlaR1 produced by B. subtilis strains carrying plasmids harboring blaR1 mutants.
Membrane proteins from induced (+) or uninduced (-) cultures were separatedon on SDS-PAGE. Purified BlaR-CTD antibodies were used for the Western blotting. Pre-stained protein molecular weight markers were used (M). The  pinpoints an intense and non-specific band (for details see Figure 6). BlaR1 and BlaR1* point out, respectively, the full size and the activated B. licheniformis BlaR1 receptor. Except for mutant E274A, all other mutants exhibit non-inducible β-lactamase phenotype. The E274A mutant has the same profile as the wild-type (to compare see Figure 6). For details see Tables 1 and 2 and Experimental procedures.
pinpoints an intense and non-specific band (for details see Figure 6). BlaR1 and BlaR1* point out, respectively, the full size and the activated B. licheniformis BlaR1 receptor. Except for mutant E274A, all other mutants exhibit non-inducible β-lactamase phenotype. The E274A mutant has the same profile as the wild-type (to compare see Figure 6). For details see Tables 1 and 2 and Experimental procedures.
In order to determine the importance of the HEXXH motif in L3 activity, alanine scanning mutagenesis was used to generate the following mutants: H212A/E213A/H216A, E213A, H212A/H216A and H216A. Analyses of these mutants indicate that all of these substitutions give rise to a non-inducible phenotype (Table 1) with a β-lactamase production similar to that obtained for a non-induced wild type strain (data not shown).
Neutral Zinc proteases catalyse cleavage of peptide bonds via a general-base type mechanism in which the glutamate residue of the conserved motif acts as catalytic base. The mutation of this glutamate residue to aspartate (E213D) or to glutamine (E213Q) prevents β-lactamase induction (Table 1, Figure 3). These results demonstrate that all the residues of this conserved motif play an essential role in the induction process.
The sequence alignment of the protein BlaR1 from B. licheniformis with the proteins BlaR1 and MecR from S. aureus, highlights four conserved residues that could act as the third zinc ligand (D221, E253, D257and E274; see above and Figure 2). Site-directed mutagenesis of these residues in alanine has been done and the resulting plasmids carrying these mutations (pDML1273-74-75-77) were introduced in B. subtilis 168 to probe the β-lactamase induction (Table 2). The D221, E253 and D257A mutants show a non-inducible phenotype while the E274A mutant retains an inducible phenotype similar to the wild type receptor (Table 1, Figure 3). This result indicates that E274 cannot be the third zinc ligand. The three other conserved residues could play an important role either in the enzymatic activity, in the Zinc coordination and/or in the folding of BlaR1 L3 loop. Finally, the mutation of the conserved tyrosine residue in alanine (Y272A) also leads to the loss of the β-lactamase induction (Table 1, Figure 3) suggesting that this residue could be part of the relay system involved in the acid-base catalysis.
L3 Overexpression and Zinc-binding Analysis
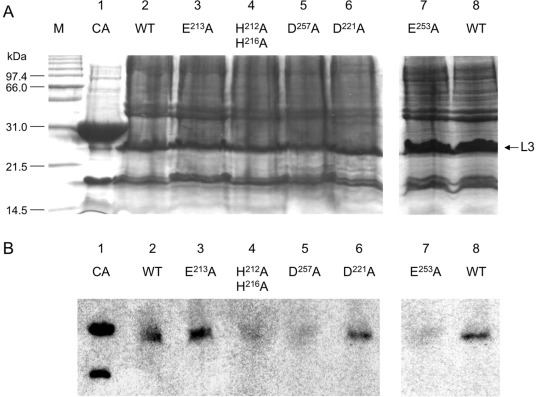
The DNA sequence coding for the B. licheniformis L3 loop was introduced into the pLex vector in which l3 was under the control of the inducible PL promoter from lambda phage. The resulting plasmid, pDML1288 (Table 2) was introduced in E. coli GI724. In the induced recombinant strain, the L3 loop was overproduced as inclusions bodies. After partial purification and SDS solubilization of inclusion bodies, proteins were separated by SDS-PAGE (Figure 4 A) and transferred onto a nitrocellulose membrane. The blotted proteins were refolded in presence of radioactive zinc chloride solution and a specific radioactive band of 25 kDa was highlighted by autoradiography (Figure 4 B, lane 2). In the utilized technique, it is well accepted that, after protein refolding, a fraction of enzyme macromolecules are correctly folded and active. This band is compatible with the expected molecular mass of L3 (22.116 kDa). The recombinant strains harboring plasmids with mutated l3 gene encoding for H212A/H216A, E213A, D221A, E253A or D257A (Table 2) has been also analyzed by Zinc blot analysis after solubilization of the inclusion bodies (Figure 4 B). As shown in the Figure 4 B, the H212A/H216A, E253A and D257A have lost their ability to bind Zn++ whereas E213A and D221A could be detected as zinc-binding proteins, highlighting the role of H212, H216, E253A and D257 residues in Zn binding.
Figure 4. Coomassie Blue-stained SDS-PAGE of partially purified inclusion bodies of wild-type and L3 loop mutants (A) and Zinc blot analysis (B).
In A and B: 1: bovine carbonic anhydrase (AC) (∼30 kDa; 5 µg), was used as positive control and 2 to 6 are, respectively, wild type (WT), E213A, H212A/H216A, D257 A, D221 A and E253 A L3 loop mutants. For each gel 40 µg of inclusion bodies were loaded. The fluorographies were exposed at -70°C for 72 hours. M: molecular weight marker. The arrow indicates L3 loops.
B. licheniformis L3 is Activated by Autocleavage
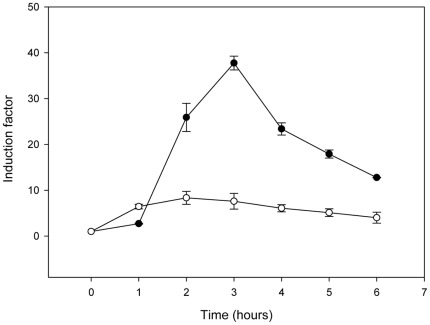
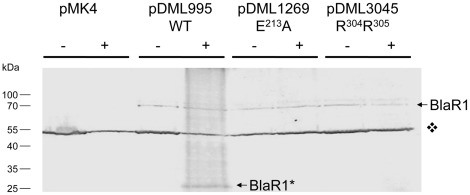
To avoid inclusion bodies and/or misfolded proteins due to overexpression of recombinant membrane protein, the fate of BlaR1 in presence or not of β-lactam (respectively induced or uninduced) was carried out by Western blot analysis of B. subtilis-pDML995 membrane fractions with purified anti-BlaR-CTD antibodies. These experiments revealed the presence of two induced bands: a major band of 33 kDa and a minor one of 68 kDa (Figure 5). The band of low molecular mass is compatible with the cleaved and activated BlaR1 C-terminal domain whereas the other of high molecular mass should be the full-length receptor. These results are in agreement with a cleavage of B. licheniformis BlaR1 between the R304 and R305 residues as in the case of S. aureus BlaR1 cleavage between R294 and R295 (Figure 2, [19]). To confirm these results in B. licheniformis BlaR1, the putative residues of the cleavage site has been mutated to R304A/R305A and the modified receptor was assayed for its ability to induce BlaP production. In presence of β-lactam antibiotic, R304A/R305A mutants lightly induces BlaP β-lactamase (IF = 7.6, Table 1 and Figure 5). Furthermore, Western blot analysis of membrane fractions from non- and induced cells with purified anti-BlaR-CTD antibodies did not show any L3 cleavage (Figure 6). To explore if the cleavage of BlaR1 observed in induced B. subtilis/pDML995 could be the result of L3 proteolytic activity, we analyzed the fate of BlaR1 HA213XXH in induced B. subtilis/pDML1269 membrane fractions by Western blotting as described above. In this mutant, the catalytic activity of L3 loop is abolished due to the mutation of the conserved glutamic residue of the HEXXH motif involved in the neutral Zinc metallopeptidase family acid-base relay. As shown in Figure 6, no cleavage of BlaR1 HA213XXH could be detected in presence of the inducer suggesting that the receptor should be activated via self-proteolysis.
Figure 5. Induction of β-lactamase synthesis in presence (induced) or not (non-induced) of inducer (2.5 µg ml−1 cephalosporin C) for the B. subtilis strains transformed with pDML995 (wild-type divergeon; •) or pDML3045 (mutation R304A/R305A;○).
The level of the induction is expressed as an induction factor (IF, the ratio between the β-lactamase quantity/A600 for induced culture and the β-lactamase quantity/A600 for uninduced culture).
Figure 6. Western blot analysis of membranes of B. subtilis transformed with pMK4 (negative control), pDML995 (wild type), pDML1269 (E213A mutant) or pDML3045 (R304A/R305A mutant).
Membrane proteins from induced (+) or uninduced (−) cultures were separatedon on SDS-PAGE. Purified BlaR-CTD antibodies were used for the Western blotting. Pre-stained protein molecular weight markers were used (M). The  indicates an intense and non-specific band. BlaR1 and BlaR1* highlight, respectively, the full size and the activated B. licheniformis BlaR1 receptor.
indicates an intense and non-specific band. BlaR1 and BlaR1* highlight, respectively, the full size and the activated B. licheniformis BlaR1 receptor.
Discussion
Structural studies on thermolysin and homologous enzymes (Thermolysin-like proteinases, TLPs) have revealed two short conserved motifs in their active sites: HEXXH and EXXXD [25]. The two histidine residues of the HEXXH sequence serve as ligands to zinc, whereas the glutamic acid is involved in the catalytic mechanism by transferring hydrogen atoms and polarizing a zinc-bound water molecule for nucleophilic attack on the scissile peptide bond of the bound substrate. The third zinc ligand is the glutamic acid residue of the sequence motif EXXXD located 20–60 residues downstream of the second histidine of the first motif. For this reason, members of thermolysin enzyme family are named gluzincins [26], [27]. The second histidine residue of the HEXXH motif was found to bridge both the zinc ion and the carboxylate side chain of the aspartic acid residue within the second conserved motif. The exact role of this conserved triad, Zn-His-Asp, is not well understood but is crucial for the catalytic activity [26], [27]. In the absence of a substrate, a water molecule acts as an additional fourth ligand to zinc and is clamped between the catalytic glutamic acid residue of the HEXXH motif and the metal. During TLP catalysis, the catalytic glutamic acid residue accepts a proton from the zinc-bound water nucleophile and transfers it to the leaving group of the substrate. Further stabilization of the transition state is provided by the side-chain of a conserved histidine residue (H231 in thermolysin) [28].
Site-directed mutagenesis of selected conserved residues in BlaR1/MecR L3 loops postulated to be involved in catalysis or zinc chelation, combined with zinc-blot experiments and β-lactamase induction assay have confirmed all the features present in the active sites of TLPs. We show that the two histidine residues of the motif H212EXXH (B. licheniformis BlaR1 numbering) effectively coordinate a Zinc ion and are essential for cytoplasmic signalling. The observed non-inducible β-lactamase phenotype, the conservation of the Zinc binding capacity and the loss of self-proteolysis exhibited by H(E213A)XXH mutant are in agreement with a glutamic acid residue acting as catalytic residue and a peptidase activity of L3 loop. The E253A and D257A mutants, which exhibited the same non-inducible phenotype (Table 1), showed that these residues are crucial for the L3 enzymatic activity and that they are good candidates to be included in the second conserved motif of TLPs. In BlaR1/MecR L3 loops, the EXXXD motif is located 37 residues downstream HEXXH motif, a distance similar to that observed in TLPs, which varies from 20 residues in thermolysin to 59 in mammalian neutral endopeptidase. These new observations allow us to classify BlaR1/MecR L3 loops as new members of TLPs or gluzincins. According to the properties of this enzyme family, the glutamic residue of the EXXXD motif is the third zinc ligand and the aspartic acid residue forms a carboxylate-histidine-zinc interaction crucial for TLP activity (see above). Indeed, as shown by 3D structures from several different enzymes included in the TLP family, this hydrogen bonding stabilizes the coordination of the zinc ion by one of the histidine residues of the HEXXH sequence [29]. In rat dipepeptidyl peptidase, the mutation of the EXXXD motif to EXXXA gives rise to the loss of the zinc ion and concomitantly the loss of enzyme activity [26] as it is the case for L3 EXXXA257 mutant (figure 4 and Table 1). By analogy, we postulate that the D257 residue of L3 loop is involved in zinc binding by orienting the imidazole ring of the H212 residue and/or by enhancing the basicity of the histidine-zinc ligand. To complete the zinc-coordination polyhedron, a water molecule, in interaction with the glutamic acid residue of the HEXXH motif, should be the fourth zinc ligand.
The conserved residues, D221 and Y272 (B. licheniformisBlaR1 numbering) are crucial for the penicillin receptor signalling. The most likely role for Y272 is to be involved in the stabilization of the tetrahedral intermediate complex during the catalysis of the activated BlaR1/MecR L3 loop. Indeed, carboxypeptidase A, which shares a common topology with thermolysin for its active site, possesses anY248 positioned at the same spatial position of H231 in thermolysin [30]. As D221 side chain is negatively charged, it is difficult to postulate that this side chain could directly interact with the negatively charged tetrahedral intermediate. Another hypothesis is that this conserved aspartic acid could be essential for substrate binding. As our results are consistent with a self-activation and as the cleavage site includes at least the positively charged side chain of one arginine residue, this hypothesis is plausible.
Similar to many other proteolytic enzymes, TLPs are synthesized as precursors with a propeptide. Up to 70% of known TLPs possess a propeptide containing two distinguished domains: the first one is an intramolecular chaperone (FTP domain) essential for the folding of the catalytic domain whereas the second one (PepSY domain) acts as an inhibitor to prevent detrimental TLP activity [31]. The cleavage of the propeptide occurs through autoprocessing and is mediated by the propeptide [26]. In some TLPs, C-terminal extensions, fused to the Zinc catalytic domain, mediate their binding to insoluble substrates. These C-terminal extensions can be autoprocessed and their cleavage modulates the enzyme specificity and activity [32].
BlaR1/MecR L3 loop activation results in a cascade of events originated by the acylation of the C-teminal sensor by the penicillin and finishing by selfproteolysis of the TLP receptor domain. It has been postulated that the cleavage of the L3 domain is triggered by a conformational change of the transmembrane segments due to a change in the interaction between BlaR/MecR-CTD and BlaR/MecR-NTD in presence of penicillin [16]. The results presented in this paper seem to show that propeptide inhibiting the BlaR1/MecR TLP domain is present and would be located at the C-terminal end of TLP domain. The cleavage site should be included in the K303(E/R)↓R conserved motif (Figure 2), in which “↓” indicates the cleaved peptide bond, instead of R304↓R motif as previously described [19]. The presence of a C-terminal propeptide inhibiting L3 TLP activity and cleaved by self-proteolysis to give the fully active L3 TLP would be unique to BlaR1/MecR in the TLP superfamily.
The RR/AA cleavage site mutant also sheds new light on the BlaR1/MecR intramolecular transduction mechanism. Indeed, in presence of β-lactam antibiotic, the acylation of the mutated BlaR1 receptor results in a slight induction of BlaP β-lactamase production (about 20% of the wild type) but without L3 cleavage. In the uncleaved protein, the C-terminal portion of the L3 loop acts as an inhibitor of its TLP domain. Cleavage of the KR↓R (both BlaR1 proteins) or KE↓R (S. aureusMecR) results in an activation of the peptidase activity, but it is not known whether the C-terminal peptide remains associated or not with the active domain. In fact, in this mutant, we have decoupled the L3 activation due to BlaR-CTD acylation from that caused by L3 selfproteolysis. The BlaP induction results indicate that in this case, the uncleaved L3 loop exhibits a residual activity which seems unable to perform a successful autocleavage but is able to produce a sufficient amount of coactivator to partially inactivate BlaI. This finding is in agreement with a selfproteolysis mechanism in which it is necessary that BlaR-CTD acylation slightly activates L3 loop to allow its own proteolysis and its full activation.
In conclusion BlaR1/MecR L3 loop is a gluzincin and a new member of the TLP superfamily that has diverged very early from canonical TLPs, since no significant identity can be detected between BlaR1/MecR L3 loops and TLPs by using comparison algorithms such as BLAST. The L3 signalling activity of BlaR1/MecR receptor would be repressed by the combination of two intramolecular interactions: one between L3 and its C-terminal propeptide and the second between L2 and BlaR-CTD. Similarly the full L3 activation triggered BlaR-CTD acylation would be the consequence of two successive events described above.
Materials and Methods
Bacterial Strains, Plasmids and DNA Manipulations
The strains and plasmids used in this study are described in Table 2. To introduce point mutations into BlaR1 or L3 loop, mutagenesis was carried out using the Quick Change Site-Directed mutagenesis procedure (Stratagene). For all mutations, except BlaR1 R304A/R305A mutation, the plasmids pDML1251 and 1283 were used as the templates in polymerase chain reactions (PCR) for blaR1 and l3 loop site-directed mutagenesis, respectively. For BlaR1 R304A/R305A mutation, pDML995 was used as template. Synthetic oligonucleotides 23–35 bases long containing a mutated codon in the middle of their sequences (Table 3) were employed to mutagenize the original codons. Following the verification of mutations by DNA sequencing (GIGA-DNA sequencing plateform, University of Liège), the corresponding restricted fragments (blaR1: 672 bp SstI-EcoRI; l3 loop: 567 bp NdeI-SalI) were cloned into pDML995 and pLex, respectively.
Table 3. Oligonucleotides used in this study.
| Name | Nucleotide sequence | Utilities |
| L3up | 5′-TAACATATGGATCCTAGCAATCTAAAAATCGGC-3′ | L3 loop sequence |
| L3rp | 5′-AAGCTTGTCGACTTATTTCGCCTTTAGCAAAGGTG-3′ | amplification |
| E213Aup | 5′-GCTTCATGCACTGTACCATTGCAAAC-3′ | Mutation |
| E213Arp | 5′-GTTTGCAATGGTACAGTGCATGAAGC-3′ | E213A |
| H212AH216Aup | 5′-GAAATGTGTTTTGCTTGC TGAACTGTACGC TTGC-3′ | Mutation |
| H212AH216Arp | 5′-GCAAGCGTACAGTTCAGCAAGCAAAACACATTTC-3′ | H212A/H216A |
| H212AE213AH216Aup | 5′-GCTTGC TGCACTGTACGC TTGCCAAACG-3′ | Mutation |
| H212AE213AH216Arp | 5′-CGTTTGGCAAGCGTACAGTGCAGCAAGC-3′ | H212A/E213A/H212A |
| E253Aup | 5′-CGAAAACGGAGATGGCGATTTCTTGC-3′ | Mutation |
| E253Arp | 5′-GCAAGAAATCGCCATCTCCGTTTTCG -3′ | E253A |
| D257Aup | 5′-GATTTCTTGCGCCTTTGCCGTATTAAAAAC-3′ | Mutation |
| D257Arp | 5′-GTTTTTAATACGGCAAAGGCGCAAGAAATC-3′ | D257A |
| D221Aup | 5′-GCAAACGAAAAGCCATGCTTATCAAC-3′ | Mutation |
| D221Arp | 5′-GTTGATAAGCATGG CTTTTCGTTTGC-3′ | D221A |
| Y272Aup | 5′-GCACCTCAAAGC TGGCGAGGTG-3′ | Mutation |
| Y272Arp | 5′-CACCTCGCCAGCTTTGAGGTGC-3′ | Y272A |
| E274Aup | 5′-CCTCAAATATGGCGCGGTGATTTTAAAATTCAC-3′ | Mutation |
| E274Arp | 5′-GTGAATTTTAAAATCACCGCGCCATATTTGAGG-3′ | E274A |
| E213Qup | 5′-GTTTTGCTTCATC AGCTGTACCATTGC-3′ | Mutation |
| E213Qrp | 5′-GCAATGGTACAGC TGATGAAGCAAAAC-3′ | E213Q |
| E213Dup | 5′-GTTTTGCTTCATGACCTGTACCATTGC-3′ | Mutation |
| E213Drp | 5′-GCAAATGGTACAGG TCATGAAGCAAAAC-3' | E213D |
| R304AR305Aup | 5'-TACAAACATATCAAAGC AGCGATTGTTACAGTTGTCAAC-3' | Mutation |
| R304AR305Arp | 5'-GTTGACAACTGTAACAATCGCTGCTTTGATATGTTTGTA-3' | R304A/R305A |
Modified codons are underlined and mutagenised bases are highlighted.
All routine DNA isolation and manipulation were performed as described by Sambrook et al. [33] or following instructions of the manufacturer. Restriction enzymes and mini-prep kit to prepare small-scale plasmid DNA were purchased from Promega and Fermentas respectively.
Overexpression of B. licheniformis BlaR1 L3 Loop and Purification of Inclusion Bodies
The coding sequence corresponding to B. licheniformis BlaR1 L3 loop (residues Tyr134 to Arg322) was amplified by PCR from pDML995 plasmid and with KBJ1 and KBJ2 primers (Table 3) containing respectively restriction sites, NdeI and SalI at their 5′-ends. The resulting fragment (567 bp) was cloned into pCR-Script to give pDML1283 plasmid. After purification of the resulting plasmid and the sequencing of the insert to verify its integrity, the plasmid was digested with NdeI and SalI and the fragment corresponding to the B. licheniformis BlaR1 L3 loop was ligated into the pLex vector digested with the same restriction endonucleases to give pDML1288. The pDML1289-94 plasmids are pDML1288 derivatives carrying respectively the mutation E213A, H212H/H216H, E253A, D257A or D221A (Table 2). These mutations were introduced as described above.
The E. coli GI724/pDML1288-94 strains were grown over night at 30°C in RM medium (40 mM Na2HPO4, 20 mM KH2PO4, 10 mM NaCl, 200 mM NH4Cl, 2% casamino acids, 1% glycerol, 1 mM MgCl2, 100 µg ml−1 ampicillin). The culture was then diluted 100 times in IM medium (40 mM Na2HPO4, 20 mM KH2PO4, 10 mMNaCl, 200 mM NH4Cl, 2% casamino acids, 0.5% glucose, 1 mM MgCl2, 100 µg ml−1 ampicilline) and cultivated at 30°C. When the absorbance at 550 nm reached 0.6, L3 loop expression was induced by both the addition of tryptophan (final concentration: 100 µg ml−1) and a temperature shift to 37°C. After 3 hours of induction, the cells were harvested by centrifugation, washed with 50 mM HEPES-NaOH (pH 7.5) and suspended in 50 mM Tris-HCl (pH 7.5). Bacterial cells were disrupted using a Basic Z Model disintegrator (Warwick, UK). After the addition of Benzonase (final concentration: 20 µg ml−1) and an incubation of 30 min at 4°C, the insoluble cell fraction containing inclusion bodies, was separated from the soluble fraction by centrifugation at 18000 g for 30 min at 4°C. The insoluble fraction was suspended in 50 mM HEPES-NaOH (pH 7.5), 0.5 mM NaCl, 5 mM DTT. After addition of Triton X100 (final concentration: 0.1%), the suspension was incubated under agitation at room temperature for 1 hour. The inclusion bodies containing the L3 loop were recovered by centrifugation at 18000 g, for 30 min and at 4°C.
Zinc Binding Assay
Soluble or insoluble Zinc binding proteins were detected by Zinc blot assay [34], [35], [36]. Purified inclusion bodies were solubilized by sodium dodecyl sulfate (SDS, solubilization buffer: Tris-HCl 0.5M pH 6.8) and separated by SDS-PAGE. After electrophoresis, the gel was soaked for one hour at 37°C in 25 mM Tris-HCl (pH 8.3), 190 mM glycine, 0.1% SDS, 5% 2-β-Mercaptoéthanol. The proteins were electrophoretically transferred to a nitrocellulose membrane in 25 mM Tris-HCl (pH 8.3), 190 mM glycine, 0.1% SDS, 20% methanol.
The nitrocellulose membrane was first incubated for 1 hour at room temperature in 10 mM Tris-HCl (pH 7.5) to partially renature blotted proteins. The membrane was then soaked two times, 15 min per soaking, in 50 ml of binding buffer (10 mM Tris-HCl (pH 7.5), 0.1 M KCl) containing 1µCi 65ZnCl and washed 30 minutes with two changes of binding buffer. The blot was dried one hour at room temperature and analyzed by autoradiography using X-OMAT™ film (Eastman Kodak Company) and an exposure time of 72 hours at −70°C.
β-lactamase Induction Assay
Bacillus subtilis 168 transformed by pDML995 or derivative plasmids was grown at 37°C in Luria-Bertani (LB) medium containing 7 µg ml−1 of chloramphenicol until A600 reached 0.2. Then, the culture is divided in two portions, one as a control (non induced cells) and the other for the addition of 5 µg ml−1 cephalosporin C (induced cells). In these conditions the concentration of inducer is below its minimal inhibitory concentration: 10 and 20 µg ml−1, respectively, for B. subtilis recombinant strains exhibiting non- and inducible β-lactamase phenotype (Ana Amoroso, unpublished data).
The incubation was continued and samples were withdrawn at hourly intervals for 6 hours. For each sample, the A600 was measured to monitor the cell density as well as the β-lactamase activity. This latter activity was spectrophotometrically determined by using nitrocefin as previously describred [17]. The β-lactamase quantity by cell [Et] was calculated by using the following equation: v0/(cellular density) = ΔA482/(Δt. ε. A600) = (kcat × [Et] × [S])/(Km + [S]) were v0: the initial velocity; ΔA482and Δt are absorbance and time variations, respectively; kcat: catalytic constant (470 s−1) [37]; [S]: substrate concentration (100 µM); Km: 40 µM [37] and ε: nitrocefin coefficient of molar extinction at 482 nm (15000 M−1 cm−1). The β-lactamase induction level was expressed as “induction factor” (IF), determined as: FI = ([Et] of induced culture)/([Et] of uninduced culture).
Anti-BlaR-CTD Antibodies Production and Western Blotting
A polyclonal antiserum directed against the BlaR1 sensor (BlaR-CTD), was generated by immunizing New Zealand rabbits with purified BlaR-CTD [17]. Polyclonal antibodies were purified against BlaR-CTD immobilized on Sepharose 4B as described in reference [38]. The purified primary antibodies were used in immunoblotting at a final dilution of 1∶1000.
Ten milliliters of induced or uninduced recombinant B. subtilis strains were harvested by centrifugation and resuspended in 200 µl of 50 mM Tris-HCl (pH 7.5). Cells were disrupted by sonication in a Branson ultrasonic disintegrator at amplitude of 6 µm for tree 30 s bursts. Membranes fraction, containing the BlaR1 receptor were obtained as insoluble fraction by centrifugation of the lysates. Membrane proteins were solubilized by SDS (solubilization buffer: Tris-HCl 0.5 M pH 6.8) and separated by SDS-PAGE (15% acrylamide) and transferred onto a nitrocellulose membrane (Millipore). Incubation of the membrane with primary antibodies was followed by incubation with alkaline phosphatase-conjugated anti-rabbit antibodies. The immune complexes were detected by a color reaction with nitroblue tetrazolium and 5-bromo-4-chloro-3-indoyl phosphate, as recommended by the manufacturer (Roche Applied Science).
Acknowledgments
We warmly thank Dr Jean-Marie Frère for fruitful discussions.
Footnotes
Competing Interests: The authors have declared that no competing interests exist.
Funding: This research was supported in part by Belgian Program on Interuniversity Poles of Attraction initiated by the Belgian State (Grant P6/19), the Actions de Recherche Concertées (Grant 03/08-297), the Fonds de la Recherche Fondamentale Collective (Grant 6 2.4511.06 and 2.4524.03), the Fonds de la recherche en Sciences Medicales (Grant 7 3.4586.05) and the Fonds pour la formation à la Recherche dans l’Industrie et dans l’Agriculture (FRIA). Stéphanie Berzigotti is a fellow of FRIA. The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
References
- 1.Philippon A, Dusart J, Joris B, Frère JM. The diversity, structure and regulation of beta-lactamases. Cell Mol Life Sci. 1998;54:341–346. doi: 10.1007/s000180050161. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Frère JM. Beta-lactamases and bacterial resistance to antibiotics. Mol Microbiol. 1995;16:385–395. doi: 10.1111/j.1365-2958.1995.tb02404.x. [DOI] [PubMed] [Google Scholar]
- 3.Clarke SR, Dyke KG. The signal transducer (BlaRI) and the repressor (BlaI) of the Staphylococcus aureus beta-lactamase operon are inducible. Microbiology. 2001;147:803–810. doi: 10.1099/00221287-147-4-803. [DOI] [PubMed] [Google Scholar]
- 4.Kobayashi T, Zhu YF, Nicholls NJ, Lampen JO. A second regulatory gene, blaR1, encoding a potential penicillin-binding protein required for induction of beta-lactamase in Bacillus licheniformis. J Bacteriol. 1987;169:3873–3878. doi: 10.1128/jb.169.9.3873-3878.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Sherratt DJ, Collins JF. Analysis by transformation of the penicillinase system in Bacillus licheniformis. J Gen Microbiol. 1973;76:217–230. doi: 10.1099/00221287-76-1-217. [DOI] [PubMed] [Google Scholar]
- 6.Salerno AJ, Lampen JO. Differential transcription of the bla regulatory region during induction of beta-lactamase in Bacillus licheniformis. FEBS Lett. 1988;227:61–65. doi: 10.1016/0014-5793(88)81414-5. [DOI] [PubMed] [Google Scholar]
- 7.Stapleton PD, Taylor PW. Methicillin resistance in Staphylococcus aureus: mechanisms and modulation. Sci Prog. 2002;85:57–72. doi: 10.3184/003685002783238870. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Lewis RA, Dyke KG. MecI represses synthesis from the beta-lactamase operon of Staphylococcus aureus. J Antimicrob Chemother. 2000;45:139–144. doi: 10.1093/jac/45.2.139. [DOI] [PubMed] [Google Scholar]
- 9.McKinney TK, Sharma VK, Craig WA, Archer GL. Transcription of the gene mediating methicillin resistance in Staphylococcus aureus (mecA) is corepressed but not coinduced by cognate mecA and beta-lactamase regulators. J Bacteriol. 2001;183:6862–6868. doi: 10.1128/JB.183.23.6862-6868.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Zhu Y, Englebert S, Joris B, Ghuysen JM, Kobayashi T, et al. Structure, function, and fate of the BlaR signal transducer involved in induction of beta-lactamase in Bacillus licheniformis. J Bacteriol. 1992;174:6171–6178. doi: 10.1128/jb.174.19.6171-6178.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Joris B, Ledent P, Kobayashi T, Lampen JO, Ghuysen JM. Expression in Escherichia coli of the carboxy terminal domain of the BLAR sensory-transducer protein of Bacillus licheniformis as a water-soluble Mr 26,000 penicillin-binding protein. FEMS Microbiol Lett. 1990;58:107–113. doi: 10.1016/0378-1097(90)90111-3. [DOI] [PubMed] [Google Scholar]
- 12.Kerff F, Charlier P, Colombo ML, Sauvage E, Brans A, et al. Crystal structure of the sensor domain of the BlaR penicillin receptor from Bacillus licheniformis. Biochemistry. 2003;42:12835–12843. doi: 10.1021/bi034976a. [DOI] [PubMed] [Google Scholar]
- 13.Wilke MS, Hills TL, Zhang HZ, Chambers HF, Strynadka NC. Crystal structures of the Apo and penicillin-acylated forms of the BlaR1 beta-lactam sensor of Staphylococcus aureus. J Biol Chem. 2004;279:47278–47287. doi: 10.1074/jbc.M407054200. [DOI] [PubMed] [Google Scholar]
- 14.Birck C, Cha JY, Cross J, Schulze-Briese C, Meroueh SO, et al. X-ray crystal structure of the acylated beta-lactam sensor domain of BlaR1 from Staphylococcus aureus and the mechanism of receptor activation for signal transduction. Journal of the American Chemical Society. 2004;126:13945–13947. doi: 10.1021/ja044742u. [DOI] [PubMed] [Google Scholar]
- 15.Hardt K, Joris B, Lepage S, Brasseur R, Lampen JO, et al. The penicillin sensory transducer, BlaR, involved in the inducibility of beta-lactamase synthesis in Bacillus licheniformis is embedded in the plasma membrane via a four-alpha-helix bundle. Mol Microbiol. 1997;23:935–944. doi: 10.1046/j.1365-2958.1997.2761642.x. [DOI] [PubMed] [Google Scholar]
- 16.Hanique S, Colombo ML, Goormaghtigh E, Soumillion P, Frère JM, et al. Evidence of an intramolecular interaction between the two domains of the BlaR1 penicillin receptor during the signal transduction. J Biol Chem. 2004;279:14264–14272. doi: 10.1074/jbc.M313488200. [DOI] [PubMed] [Google Scholar]
- 17.Duval V, Swinnen M, Lepage S, Brans A, Granier B, et al. The kinetic properties of the carboxy terminal domain of the Bacillus licheniformis 749/I BlaR penicillin-receptor shed a new light on the derepression of beta-lactamase synthesis. Mol Microbiol. 2003;48:1553–1564. doi: 10.1046/j.1365-2958.2003.03520.x. [DOI] [PubMed] [Google Scholar]
- 18.Llarrull LI, Toth M, Champion MM, Mobashery S. Activation of BlaR1 protein of methicillin-resistant Staphylococcus aureus, its proteolytic processing, and recovery from induction of resistance. The Journal of biological chemistry. 2011;286:38148–38158. doi: 10.1074/jbc.M111.288985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Zhang HZ, Hackbarth CJ, Chansky KM, Chambers HF. A proteolytic transmembrane signaling pathway and resistance to beta-lactams in staphylococci. Science. 2001;291:1962–1965. doi: 10.1126/science.1055144. [DOI] [PubMed] [Google Scholar]
- 20.Amoroso A, Boudet J, Berzigotti S, Duval V, Teller N, et al. A Peptidoglycan Fragment Triggers beta-lactam Resistance in Bacillus licheniformis. PLoS pathogens. 2012;8:e1002571. doi: 10.1371/journal.ppat.1002571. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Filée P, Benlafya K, Delmarcelle M, Moutzourelis G, Frère JM, et al. The fate of the BlaI repressor during the induction of the Bacillus licheniformis BlaP beta-lactamase. Mol Microbiol. 2002;44:685–694. doi: 10.1046/j.1365-2958.2002.02888.x. [DOI] [PubMed] [Google Scholar]
- 22.Gregory PD, Lewis RA, Curnock SP, Dyke KG. Studies of the repressor (BlaI) of beta-lactamase synthesis in Staphylococcus aureus. Mol Microbiol. 1997;24:1025–1037. doi: 10.1046/j.1365-2958.1997.4051770.x. [DOI] [PubMed] [Google Scholar]
- 23.Rawlings ND, Barrett AJ. Evolutionary families of metallopeptidases. Methods Enzymol. 1995;248:183–228. doi: 10.1016/0076-6879(95)48015-3. [DOI] [PubMed] [Google Scholar]
- 24.Pelmenschikov V, Blomberg MR, Siegbahn PE. A theoretical study of the mechanism for peptide hydrolysis by thermolysin. J Biol Inorg Chem. 2002;7:284–298. doi: 10.1007/s007750100295. [DOI] [PubMed] [Google Scholar]
- 25.Hooper NM. Families of zinc metalloproteases. FEBS letters. 1994;354:1–6. doi: 10.1016/0014-5793(94)01079-x. [DOI] [PubMed] [Google Scholar]
- 26.Gao X, Wang J, Yu DQ, Bian F, Xie BB, et al. Structural basis for the autoprocessing of zinc metalloproteases in the thermolysin family. Proc Natl Acad Sci U S A. 2010;107:17569–17574. doi: 10.1073/pnas.1005681107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Le Moual H, Devault A, Roques BP, Crine P, Boileau G. Identification of glutamic acid 646 as a zinc-coordinating residue in endopeptidase-24.11. J Biol Chem. 1991;266:15670–15674. [PubMed] [Google Scholar]
- 28.Lipscomb WN, Strater N. Recent Advances in Zinc Enzymology. Chemical reviews. 1996;96:2375–2434. doi: 10.1021/cr950042j. [DOI] [PubMed] [Google Scholar]
- 29.Fukasawa KM, Hata T, Ono Y, Hirose J. Journal of amino acids 2011: 574816; 2011. Metal preferences of zinc-binding motif on metalloproteases.574816. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Hausrath AC, Matthews BW. Redetermination and refinement of the complex of benzylsuccinic acid with thermolysin and its relation to the complex with carboxypeptidase A. The Journal of biological chemistry. 1994;269:18839–18842. doi: 10.2210/pdb1hyt/pdb. [DOI] [PubMed] [Google Scholar]
- 31.Demidyuk IV, Gasanov EV, Safina DR, Kostrov SV. Structural organization of precursors of thermolysin-like proteinases. The protein journal. 2008;27:343–354. doi: 10.1007/s10930-008-9143-2. [DOI] [PubMed] [Google Scholar]
- 32.Del Papa MF, Hancock LE, Thomas VC, Perego M. Full activation of Enterococcus faecalis gelatinase by a C-terminal proteolytic cleavage. Journal of bacteriology. 2007;189:8835–8843. doi: 10.1128/JB.01311-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Sambrook J, EF Fritsch, T Maniatis. NY: Cold Spring Harbor Laboratory Press; 2001. Molecular cloning: a laboratory manual. [Google Scholar]
- 34.Gong Z, Hew CL. Zinc and DNA binding properties of a novel LIM homeodomain protein Isl-2. Biochemistry. 1994;33:15149–15158. doi: 10.1021/bi00254a026. [DOI] [PubMed] [Google Scholar]
- 35.Mazen A, Gradwohl G, de Murcia G. Zinc-binding proteins detected by protein blotting. Anal Biochem. 1988;172:39–42. doi: 10.1016/0003-2697(88)90408-3. [DOI] [PubMed] [Google Scholar]
- 36.McEuen AR, Edwards B, Koepke KA, Ball AE, Jennings BA, et al. Zinc binding by retroviral integrase. Biochemical and biophysical research communications. 1992;189:813–818. doi: 10.1016/0006-291x(92)92275-3. [DOI] [PubMed] [Google Scholar]
- 37.Matagne A, Misselyn-Bauduin AM, Joris B, Erpicum T, Granier B, et al. The diversity of the catalytic properties of class A beta-lactamases. Biochem J. 1990;265:131–146. doi: 10.1042/bj2650131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Sambrook J, EF Fritsch, T Maniatis. Molecular cloning: a laboratory manual. 2008/02/12 ed. NY: Cold Spring Harbor Laboratory Press; 2001. Purification of antibodies. pp. 18.11–18.18. [Google Scholar]
- 39.Sullivan MA, Yasbin RE, Young FE. New shuttle vectors for Bacillus subtilis and Escherichia coli which allow rapid detection of inserted fragments. Gene. 1984;29:21–26. doi: 10.1016/0378-1119(84)90161-6. [DOI] [PubMed] [Google Scholar]