Abstract
GL13NH2 is a bacteria-agglutinating peptide derived from the sequence of the salivary protein parotid secretory protein (PSP, BPIFA2, SPLUNC2, C20orf70). The peptide agglutinates both Gram negative and Gram positive bacteria, and shows anti-lipopolysaccharide activity in vitro and in vivo. However, GL13NH2 does not exhibit bactericidal activity. To generate a more cationic peptide with potential bactericidal activity, three amino acid residues were replaced with lysine residues to generate the peptide GL13K. In this report, the antibacterial and anti-inflammatory activities of GL13K were characterized. GL13K had lost the ability to agglutinate bacteria but gained bactericidal activity. Substitution of individual amino acids in GL13K with alanine did not restore bacterial agglutination. GL13K was bactericidal against Pseudomonas aeruginosa, Streptococcus gordonii and Escherichia coli but not Porphyromonas gingivalis. Unlike the agglutinating activity of GL13NH2, the bactericidal activity of GL13K against Pseudomonas aeruginosa was retained in the presence of saliva. Both GL13NH2 and GL13K exhibited anti-lipopolysaccharide activity. In GL13K, this activity appeared to depend on a serine hydroxyl group. GL13K protected mice from lipopolysaccharide- induced sepsis and the peptide exhibited a low level of hemolysis, suggesting that it may be suitable for in vivo application.
Keywords: Agglutination, Bactericidal, BPIFA2, Hemolysis, Lipopolysaccharide, LPS
1. Introduction
Mucosal surfaces, including the oral cavity and upper airways are exposed to a wide variety of microorganisms. As an example, the oral cavity harbors more than 1000 major bacterial species [17]. These bacteria represent a balance of pathogens and commensals, which may become opportunistic pathogens when host immunity is weakened and cause infection. To manage the resident flora and combat new infections, the mucosal innate immune complex, including saliva, exhibits at least 45 different antimicrobial proteins, that belong to six different functional classes [14]. It is thought that this diversity is required to effectively control the wide array of infectious agents that can enter the oral cavity and upper respiratory tract.
Surfactant and agglutinating proteins are prominent in mucosal secretions where they can modulate bacterial adhesion and clearance. Human Parotid Secretory Protein (PSP; BPIFA2)3 was identified as an agglutinating protein and a potential new oral host-defense protein [1, 13, 15]. PSP belongs to the BPI-fold superfamily [3–5, 26]. The BPI-fold family is named for its predicted structural similarity to bactericidal/permeability-increasing protein (BPI) and includes lipopolysaccharide (LPS)-binding protein (LBP), cholesteryl-ester transport protein (CETP), phospholipid transport protein (PLTP) and the palate, lung and nasal epithelium clone (PLUNC) proteins [3, 4, 26]. Surfactant properties have been attributed to PLUNC proteins: PLUNC (BPIFA1) [10] and latherin (BPIFA4) [2, 21]. In addition, recombinant PSP can reduce colony forming units (CFU) of Pseudomonas aeruginosa in vitro [13], possibly by agglutination of bacteria [15]. Consistent with this finding it has been reported that mouse PSP binds bacterial membranes [23] and recent evidence suggests that PLUNC can control P. aeruginosa infections in the lungs of transgenic mice [18].
Based on the predicted structural similarity of PSP to other BPI-fold proteins, we designed the synthetic peptide GL13NH2 (PSP 141–153), which agglutinates both Gram negative and Gram positive bacteria, including the oral pathogen Aggregatibacter actinomycetemcomitans and the oral commensal Streptococcus gordonii [16]. GL13NH2 also shows anti-LPS activity in vitro [1] and may have anti-inflammatory activity in vivo [15].
GL13NH2 does not exhibit bactericidal activity. Since bactericidal peptides are typically cationic, we increased the net positive charge in this peptide by replacing amino acid residues at position 2 (glutamine), 5 (asparagines) and 11 (aspartic acid) with lysine residues to generate the peptide GL13K [15]. In this report, the antibacterial and anti-inflammatory activities of GL13K are characterized. While the anti-inflammatory activity of GL13NH2 is retained in GL13K, the latter peptide does not induce bacterial agglutination but instead exhibits bactericidal activity.
2. Material and Methods
2.1 Bacterial culture conditions
All bacterial media were from Difco/Becton Dickinson. Pseudomonas aeruginosa PAO1 was obtained from Dr. D.R. Demuth, University of Louisville and maintained on “Pseudomonas Isolation Agar”. Broth cultures of P. aeruginosa and Escherichia coli DH5-α (Invitrogen, Life Technologies, Grand Island, NY) were performed at 37° C in Luria Bertani (LB) medium. S. gordonii M5 was obtained from Dr. Demuth and cultured at 35° C in brain heart infusion medium containing 0.5% yeast extract (YBHI) under reduced oxygen condition. Porphyromonas gingivalis strains ATCC 53977, DPG3, and W50 were obtained from Dr. M. Costalonga, University of Minnesota and stored in 10% skim milk at −80°C. The bacteria were cultured at 37°C in an anaerobic chamber in Todd-Hewitt Base broth supplemented with 5 μg/ml hemin chloride (Calbiochem, La Jolla, CA), 0.5 μg/ml menadione (MP Biomedical) and 4% heat-inactivated fetal bovine serum. P. gingivalis colonies were cultured on Todd-Hewitt Base blood agar supplemented with hemin chloride/menadione and 5% defibrinated sheep blood (Gibson Laboratories).
2.2 Peptides
The peptides GK7NH2 [12], GL13NH2 [16] and GL13K [15] have been described previously. Their sequences are provided in Table 1. The positive control peptides LL-37 and polymyxin B (PMX) were obtained from Innovagen (Lund, Sweden) and Sigma-Aldrich (St. Louis, MO), respectively. To test the effect of individual amino acid changes in GL13K, 12 alanine-substituted peptides were obtained from the University of Minnesota peptide synthesis facility. Each peptide contained one amino acid substituted by alanine. The alanine residue in position 8 of GL13K was not altered.
Table 1.
Peptide sequences
| Peptide | Sequence | Net charge | Relative Hydrophobic moment |
|---|---|---|---|
| GK7NH2 | GQIINLK-NH2 | +2 | 0.33 |
| GL13NH2 | GQIINLKASLDLL-NH2 | +1 | 0.29 |
| GL13K | GKIIKLKASLKLL-NH2 | +5 | 0.29 |
| GL13K-R1 | IGIKLLKSKLKAL-NH2 | +5 | 0.49 |
| GL13K-R2 | KKKLSLKALIILG-NH2 | +5 | 0.04 |
The sequences of the PSP peptides used in this study are listed with calculated net charge (at pH 7) and mean hydrophobic moment.
Lysozyme (egg white) was obtained from Fisher Scientific (Pittsburgh, PA). All peptide stock solutions (1 mg/ml) were prepared in 0.01% acetic acid and aliquots stored at −20° C. Buffer control (blank) samples contained an equal volume of 0.01% acetic acid (HOAc).
2.3. Saliva Samples
Whole unstimulated saliva was self-collected into a centrifuge tube that was kept on ice and then centrifuged 30 min at 1000 × g. The resulting supernatant was diluted with 1 volume of sterile phosphate-buffered saline (PBS) and passed through a 0.2 μm sterile syringe filter (Corning) to remove endogenous bacteria. Filtered 50% saliva samples were stored at 4°C for up to 3 days.
2.4. Bacterial Agglutination Assay
Overnight cultures of E. coli DH5-α were pelleted (10 min, 3000 × g) and resuspended in PBS, pH 7.4 to a final OD600 of about 1.2. Two hundred micro-liters of bacterial solution were diluted with 250 μl of PBS and mixed with 50 μl of peptide stock solution for a final volume of 500 μl and peptide concentration of 100 μg/ml. Samples were incubated in a microcuvette at room temperature and the OD recorded every 15 minutes. At the end of some experiments, the bacteria were resuspended and the OD recorded as a measure of intact bacteria.
2.4.1 Saliva effect on agglutination
Overnight cultures of E. coli were centrifuged as above and resuspended in PBS or 50% saliva in PBS to an approximate OD=1.2. Bacteria (200 μl) were further diluted with 250 μl of PBS or 50% saliva in PBS and mixed with 50 μl of peptide (final concentration 100 μg/ml) or an equal volume of 0.01% acetic acid in a total sample volume of 500 μl. The final saliva concentration was 45%. The OD600 was recorded after mixing of the samples and again after incubation for 150 min. The samples were then incubated overnight at room temperature, mixed and the final OD600 recorded. In some experiments, 40 μg/ml lysozyme was used instead of 50% saliva.
2.5. Bacterial Killing Assay
Overnight cultures of E. coli, P. aeruginosa or S. gordonii were diluted to 1–2×105 CFU/ml in either 10 mM sodium phosphate, pH 7.4, PBS, or 50% saliva in PBS. Bacteria (450 μl) were incubated with 50 μl peptide solution (final peptide concentration 10 or 100 μg/ml) for 2 hours at 35°C. The bacterial samples were diluted and plated on LB agar plates (E. coli; P. aeruginosa) or YBHI agar plates (S. gordonii, under reduced oxygen conditions) and cultured at 35°C. For time course killing assays, P. aeruginosa (107 CFU) were exposed to 100 μg/ml peptide in 10 mM sodium phosphate, pH 7.4 at 35°C. Ten μl aliquots of this solution were spotted on a LB plate at selected time points.
Three strains of P. gingivalis (53977, DPG3, W50) were incubated with 200 μg/ml of each peptide for 2 ½ hours at 35°C. Surviving bacteria were plated on blood agar and cultured anaerobically at 37°C for 5–7 days. Following incubation, the surviving colonies were counted to determine CFU of each species. In some experiments, bacterial survival was determined by cellular ATP content using the Bactiter GLO assay (Promega, Madison, WI).
2.6. Determination of minimum inhibitory concentrations
To determine the minimum inhibitory concentration (MIC) of a peptide, overnight cultures of E. coli DH5α or P. aeruginosa PAO1 were pelleted (10 min, 3000 × g) and resuspended in MHB. P. gingivalis W50 was similarly incubated in Pg broth. The bacteria (100μl, 104 CFU) were incubated with 20μl of a two-fold serial dilution of each peptide (1mg/ml stock solution) in 10 mM sodium phosphate, pH 7.4. Control samples contained 20 μl of 0.01% acetic acid serially diluted in sodium phosphate. In some experiments, the bacteria were diluted in 50% saliva in broth. Bacterial growth was quantitated by OD at 570 nm after overnight incubation on a shaker at 35°C.
2.7 Lipopolysaccharide binding
LPS binding to LPS-binding protein and LPS-stimulated secretion of TNFα from RAW264.7 macrophage cells in the presence or absence of 180–200 μg/ml peptides was tested as previously described [1].
Free LPS was quantitated using a limulus amebocyte lysate chromogenic endpoint assay, following the manufacturer’s instructions (Hycult Biotech, Plymouth Meeting, PA). Peptides were tested at a concentration of 100 μg/ml against 10 ng/ml LPS. Control samples contained 100 μg/ml polymyxin B (0% free LPS) or 0.01% acetic acid (100% free LPS).
2.8 In vivo experiments
All animal studies were approved by the Institutional Animal Care and Use Committee at the University of Minnesota (IACUC protocol # 0910A73194). Male C57BL/6 mice (20–22 g) obtained from Charles River Laboratories were housed in standard cages and maintained in climate and light controlled rooms (12-hour dark/12-hour light cycle) with free access to food and water. The analgesic buprenorphine was administered subcutaneously (1μg/mouse) prior to each experiment (A pilot experiment had revealed that this treatment did not affect subsequent survival upon LPS injection). Then each mouse was injected intraperitoneally (i.p.) with 15 mg of D-galactosamine (Sigma Chemicals, St. Louis, MO). on right side and 100 ng of LPS or 100 μg of GL13K + 100 ng of LPS on the left side. The animals were observed for mortality for 30 h. Surviving animals were terminated by CO2 asphyxiation after 30 h. Survival curves were constructed using Graphpad Prism 5 (Graphpad Software, La Jolla, CA).
2.9. Hemolysis assay
Human red blood cells (Allcells, Emeryville, CA) were diluted 1:50 in PBS and incubated with 1 mg/ml of each peptide (10mg/ml stock solution, 0.01% acetic acid) in a total volume of 500 μl. The samples were incubated for 18 hours at 35°C and then centrifuged (5 min, 16,000 × g). Released hemoglobin was quantitated spectrophotometrically at 541 nm. Control samples were incubated with PBS (0% hemolysis) or dH2O (100% hemolysis).
2.10 Computational and statistical analyses
The net charge of peptides was calculated at pH 7.0 using Pepcalc.com provided by Innovagen AB, (Lund, Sweden). Mean hydrophobic moment was calculated according to CCS using a 100° alpha helix with an online calculator at http://www.bbcm.univ.trieste.it/~tossi/HydroCalc/HydroMCalc.html. The helical wheel representations of peptides were drawn with the program heliquest [11].
Statistical analyses were performed in Graphpad Prism 5. The data in Figure 1A–B were analyzed by repeated measures ANOVA with Dunnett’s multiple comparison post-test. Figure 4 was analyzed by one-way ANOVA with Dunnett’s multiple comparison post-test. Table 2 was analyzed by one-way ANOVA with Bonferroni’s multiple comparison post-test for select pairs. Mouse survival curves were compared by Mantel-Cox’ log-rank test. P<0.05 was considered statistically significant.
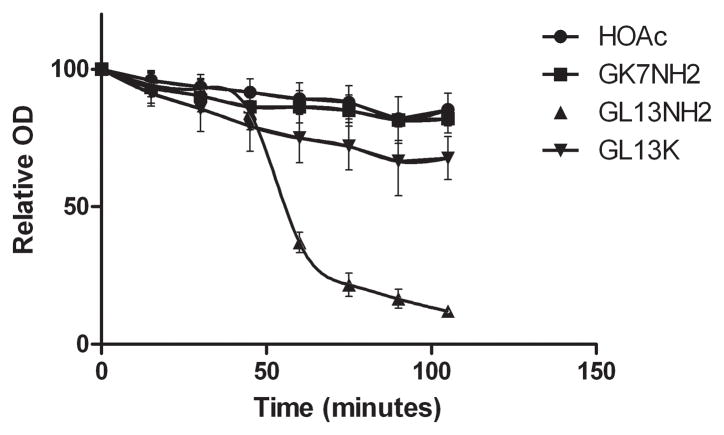
Figure 1. Agglutination of E. coli.
A: E. coli (105 CFU/ml) were incubated with GK7NH2, GL13NH2, GL13K or buffer containing 0.01% acetic acid (HOAc). The OD600 was monitored for 105 min and expressed relative to the starting OD for each sample. The data are presented as mean ± SEM from three independent experiments (N=3). The decrease in OD with GL13NH2 was different from buffer control (P<0.01).
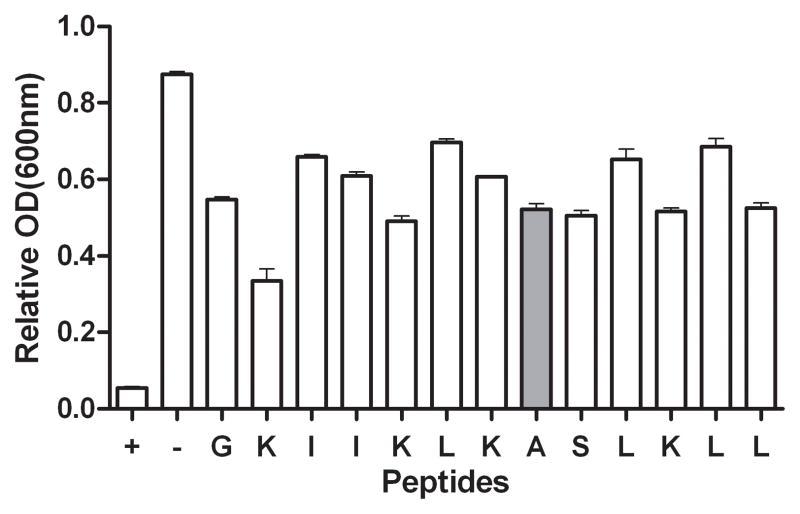
B: Agglutination of alanine-substituted GL13K. E. coli was incubated with each peptide for 3 h and the OD at 600 nm recorded and expressed relative to the starting OD for each sample. GL13NH2 was used as a positive control (+). Buffer alone was used as a negative control (−). The letters indicate the amino acid in the GL13K sequence that was replaced by alanine in each peptide. Ala8 is the original GL13K sequence (shaded bar). The data are shown as the mean ± range of two independent experiments (N=2).
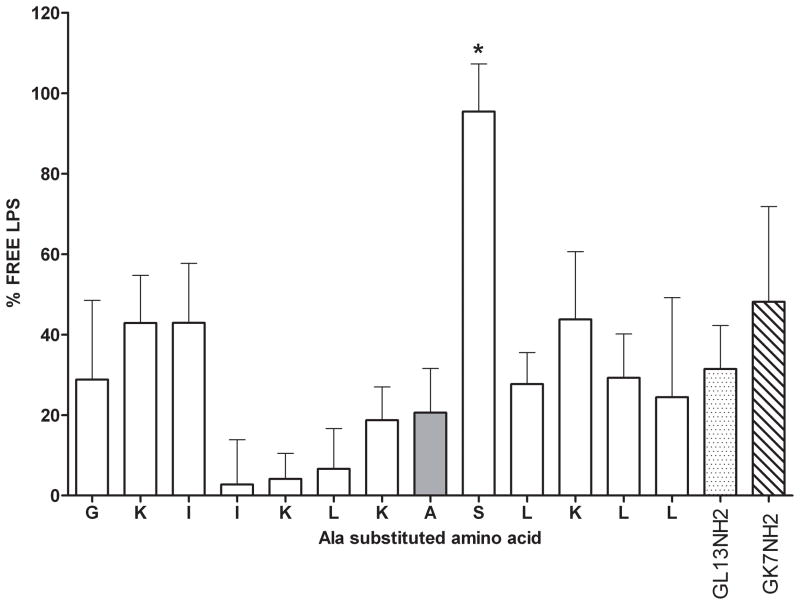
Figure 4. Inhibition of lipopolysaccharide binding by GL13K variants.
Twelve alanine substituted GL13K peptides (100 μg/ml) were tested for inhibition of LPS-binding activity. The amino acid, which was substituted with alanine in each peptide is shown. The unmodified GL13K peptide contains an Ala residue in position 8 (shaded bar). Free LPS was quantitated by the limulus amebocyte lysate assay and expressed relative to control samples that did not contain peptide (100% free LPS) and samples containing 100 μg/ml polymyxin B (0% free LPS). LPS binding to GL13NH2 or GK7NH2 is shown for comparison. The data from four independent experiments are shown as mean ± SEM (N=3–4). *) different from unmodified GL13K (A), P<0.001.
Table 2.
Minimum inhibitory concentrations.
| Peptide (μg/ml) | E. coli DHα5 | E.coli DHα5 50% saliva | P. aeruginosa PAO1 | P. gingivalis W50 |
|---|---|---|---|---|
| GL13K | 5 | 50 | 8 | >100 |
| LL-37 | 3 | >167 | 42 | 25 |
| GL13NH2 | >200 | ND | >167 | >167 |
| GK7NH2 | >200 | >167 | >167 | >167 |
ND: Not determined
The MIC (in μg/ml) of GL13K, LL-37, GL13NH2 and GK7NH2 were determined by broth dilution assay. Data for E. coli are the mean of five independent experiments; E. coli + saliva, two independent experiments; P. aeruginosa, 2–5 independent experiments, and P. gingivalis, seven independent experiments.
3. Results
3.1. Bacterial agglutination
To determine if GL13K agglutinates bacteria, E. coli DH5-α were incubated with GL13K or control peptides and the decrease in OD due to agglutination was recorded. Figure 1A shows that GL13NH2 agglutinated E. coli after 45 minutes and agglutination was essentially complete after 90 minutes. In contrast, neither GL13K nor the control peptide GK7NH2 agglutinated the bacteria, suggesting that the additional Lys residues in GL13K eliminated the agglutinating ability of GL13NH2. To test the effect of individual amino acid substitutions on agglutination, 12 alanine substituted GL13K variants were analyzed (Figure 1B). Most of these peptides exhibited less than 50% agglutination of E. coli although GL13K-A2, which contains an Ala residue in place of the Lys residue in position 2, showed a 70% decrease in OD after 180 minutes of incubation. In comparison, the positive control peptide GL13NH2 caused a 95% reduction of OD after 180 minutes.
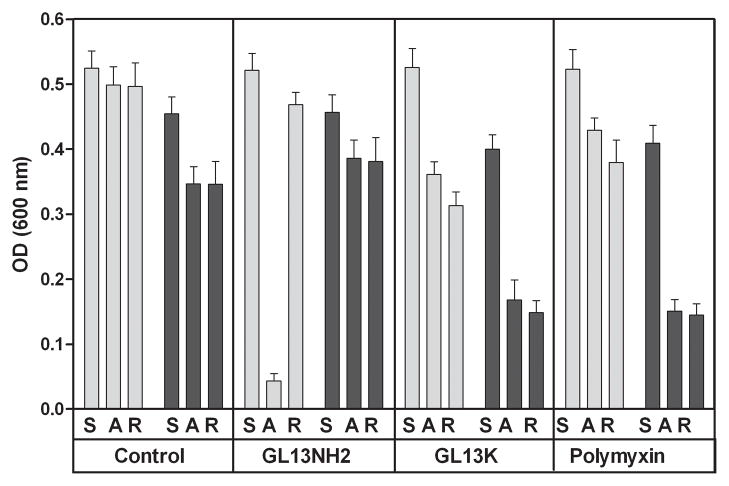
To determine the effect of saliva on bacterial agglutination, E. coli were incubated in the presence or absence of 45% saliva. Incubation of E. coli without saliva had no effect on the OD (Figure 2). In the presence of saliva, the OD was reduced 20% after incubation, suggesting either cell lysis or agglutination and precipitation of the bacteria. To distinguish between these two possibilities, the samples were mixed to redisperse any agglutinated cells. The finding that the optial density was no increased by mixing suggests that the bacteria were lysed in the presence of saliva. In the presence of GL13NH2, the samples exhibited a larger decrease in OD, which was reverted by mixing of the solution, consistent with bacterial agglutination and subsequent resuspension. The effect of GL13NH2 was lost in the presence of 45% saliva. The peptides GL13K and polymyxin, caused a decrease in OD, which was enhanced by 45% saliva and that was irreversible upon mixing, suggesting that it was due to bacterial lysis. Indeed, for GL13K and polymyxin the addition of 40 μg/ml lysozyme also resulted in a decreased OD that was not reversible by resuspension (not shown).
Figure 2. Effect of saliva on bacterial agglutination.
E. coli was incubated with PBS (control) or GL13NH2, GL13K or polymyxin (100 μg/ml) in the absence (light bars) or presence (dark bars) of 45% saliva in PBS. The OD 600 nm was recorded immediately after mixing the samples (Start - S), after a 150 min agglutination period (A) or after overnight incubation and resuspension of the samples (R). The data are shown as the mean ± SEM of five independent experiments (N=3–5).
3.2 Bactericidal activity
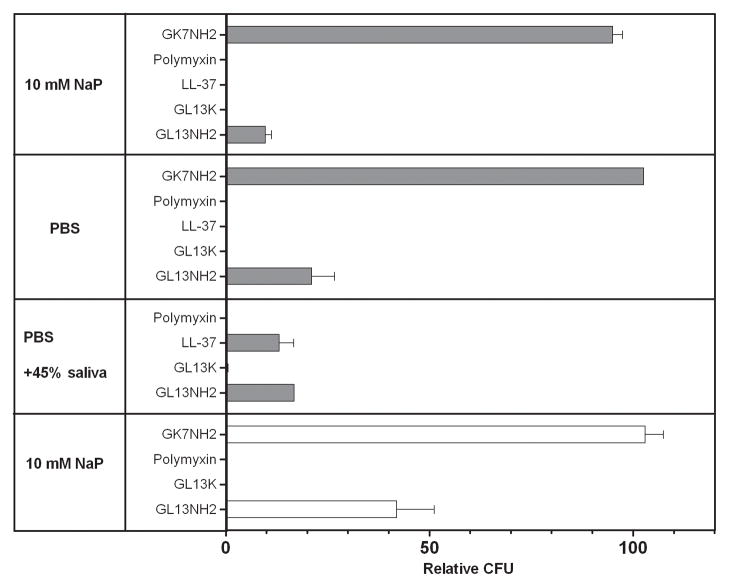
The finding that GL13K caused an irreversible decrease in OD, suggested that this peptide has bactericidal activity. To test this, bacteria were incubated with GL13K or control peptides for 2 hours and the CFU were enumerated. A negative control peptide GK7NH2 did not reduce CFU in 10 mM sodium phosphate, PBS or PBS with 45% saliva (Figure 3A). In contrast, GL13K effectively (99.8%) killed E. coli at peptide concentrations of 100 and 10 μg/ml. The effect was similar to that of the antimicrobial control peptides LL-37 and polymyxin in 10 mM sodium phosphate and PBS. GL13K and polymyxin retained bactericidal activity in the presence of 50% saliva, while LL-37 reduced CFUs less than 90% in 50% saliva (Figure 3A). The agglutinating peptide GL13NH2 reduced CFU 80–90% at 100 μg/ml and 40% at 10 μg/ml. This finding is consistent with a reduction of CFUs caused by bacterial agglutination [16]. The killing of E. coli was confirmed by quantitating ATP in live cells. In the concentration range from 10–168 μg/ml, GL13K reduced viable bacteria by at least 90%. GL13NH2 and GK7NH2 had no effect on bacterial viability in this concentration range (not shown).
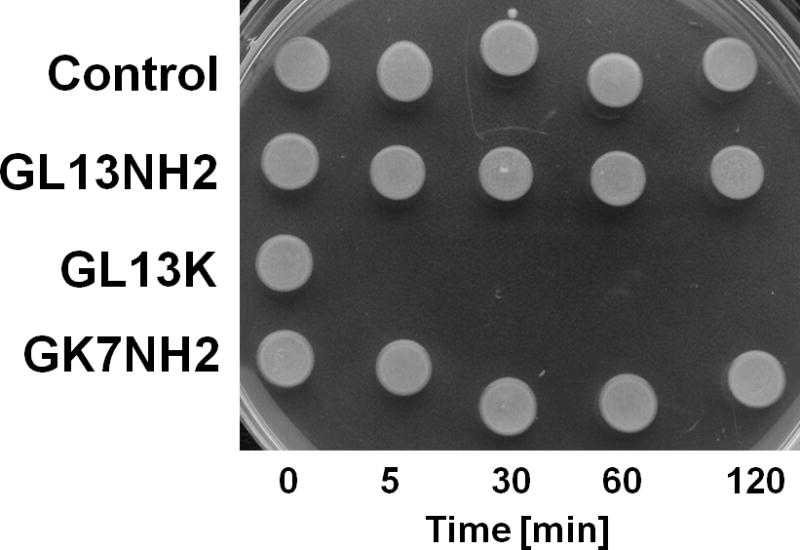
Figure 3. Bactericidal activity of GL13K.
A: Effect of buffer conditions. E. coli were incubated with GK7NH2, LL-37, polymyxin, GL13K or GL13NH2 (100 μg/ml -shaded bars; 10 μg/ml – open bars) for 2 hours in 10 mM sodium phosphate (NaP), PBS or PBS with 45% saliva. Surviving bacteria were enumerated as CFU and normalized to bacteria in samples incubated without peptide (100 Relative CFU).The data from 3–5 independent experiments are expressed as mean ± SEM (N=1–5).
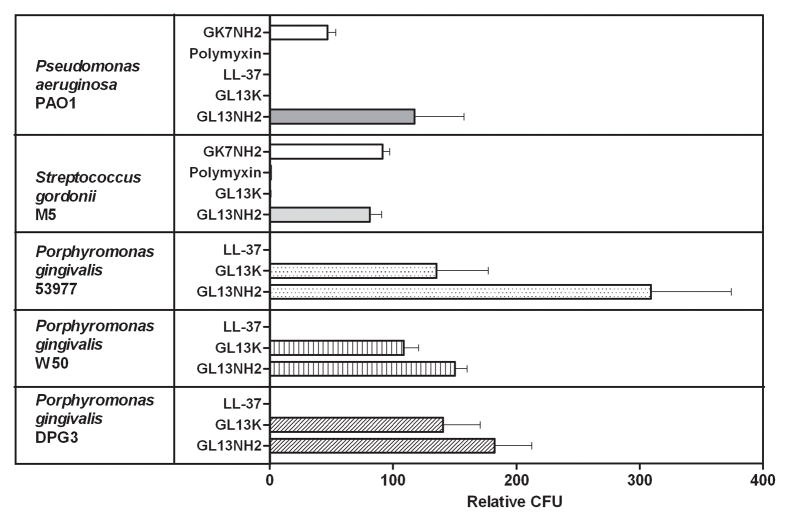
B: Bactericidal activity against different species. P. aeruginosa, S. gordonii or three stains of P. gingivalis were incubated with GK7NH2, LL-37, polymyxin, GL13K or GL13NH2 in 10 mM sodium phosphate. Surviving bacteria were enumerated as CFU and normalized to bacteria in samples incubated without peptide (100 Relative CFU).The data are expressed as mean ± SEM (P. aeruginosa, N=3–15; S. gordonii, N=3; P. gingivalis 53977, N=4; Pg W50, N=3; Pg DPG3, N=2 (mean ± range is shown)).
C: Time course of bacterial killing. P. aeruginosa were incubated without peptide (control) or with GL13NH2, GL13K, GK7NH2 (100 μg/ml) for 2 hours in 10 mM sodium phosphate. Ten μl aliquots were spotted on LB agar at the times indicated and cultured overnight.
Further bactericidal assays revealed that GL13K effectively killed the Gram negative bacterium P. aeruginosa and the Gram positive bacterium Streptococcus gordonii (Figure 3B). Time course experiments revealed that killing of P. aeruginosa (107 CFU) by GL13K (100 μg/ml) was complete in 5 minutes, whereas other peptides tested, including GL13NH2, did not show bactericidal activity in this assay (Figure 3C). In contrast, GL13K was ineffective against three strains of P. gingivalis that were killed by the antimicrobial peptide LL-37 (Figure 3B). In a control experiment, polymyxin B also proved ineffective against P. gingivalis (not shown), as previously reported by others [7]. As previously reported [16], GL13NH2 did not exhibit significant bactericidal activity against the bacterial strains tested. The increased number of CFU observed for P. gingivalis incubated with GL13 peptides was presumably due to the availability of these peptides as a carbon source during incubation. Consistent with this interpretation, the CFU of peptide treated samples were not elevated in a control experiment where the samples were incubated in Pg broth (not shown).
The MICs of GL13K and control peptides were determined in broth with or without 50% saliva (Table 2). GL13K showed a MIC of 5 μg/ml for E. coli without saliva and 50 μg/ml in the presence of saliva. A slightly lower MIC was determined for LL-37, but this peptide was not active in the presence of saliva. GL13K was highly effective against P. aeruginosa but the peptide did not exhibit a defined MIC for P. gingivalis.
3.3 Inhibiton of lipopolysaccharide activity by GL13K
The peptide GL13NH2 exhibits anti-LPS activity and blocks the binding of LPS to PSP, LPS-binding protein and inhibits LPS-stimulated activation of macrophages [1]. To test if the anti-LPS activity was retained by the modified peptide GL13K, the effects of GL13NH2 and GL13K on LPS binding to LBP and stimulation of TNFα secretion were compared (Table 3). The two peptides (180–200 μg/ml) reduced the effect of LPS in these assays by 60–70%, with no significant difference between the peptides. GL13K did not stimulate TNFα secretion from RAW 264.7 cells (not shown), suggesting lack of inflammatory activty, consistent with the results for GL13NH2 [1].
Table 3.
Lipopolysaccharide-blocking activity of GL13K
| Assay | LPS | LPS+GL13K | LPS+GL13NH2 |
|---|---|---|---|
| TNFα secretion | 100% | 26.1%±12%* (N=3)1 | 37.6%±6.5%* (N=21)2 |
| LBP binding | 100% | 45.3%±9.7% (N=3) | 35.3%±4.1%* (N=3)2 |
Comparison of anti-LPS activty of GL13NH2 and GL13K. The effects of 180–200 μg/ml GL13NH2 and GL13K on LPS-stimulated TNFα secretion from macrophages and LPS binding to LBP were compared. Data are shown as mean ± SEM.
) different from LPS alone, P<0.05. The peptide treated samples were not significantly different.
Data from [15]
Data from [1]
To further understand the effect of amino acid substitutions on anti-LPS activity of the GL13K peptide, each amino acid in GL13K was substituted with alanine and the resulting peptides (100 μg/ml) used to test LPS activity in the limulus amebocyte lysate assay (Figure 4). Eleven of the 12 alanine substituted peptides inhibited LPS activity similar to that of normal GL13K peptide. The control peptides GL13NH2 and GK7 likewise inhibited LPS activity, consistent with our previous results [1, 12]. In contrast, the serine residue in position 9 was found to be critical for anti-LPS activity (P<0.001), as loss of the serine hydroxyl group by substitution with alanine, led to complete loss of LPS-inhibitory activity.
3.4 GL13K activity in vivo
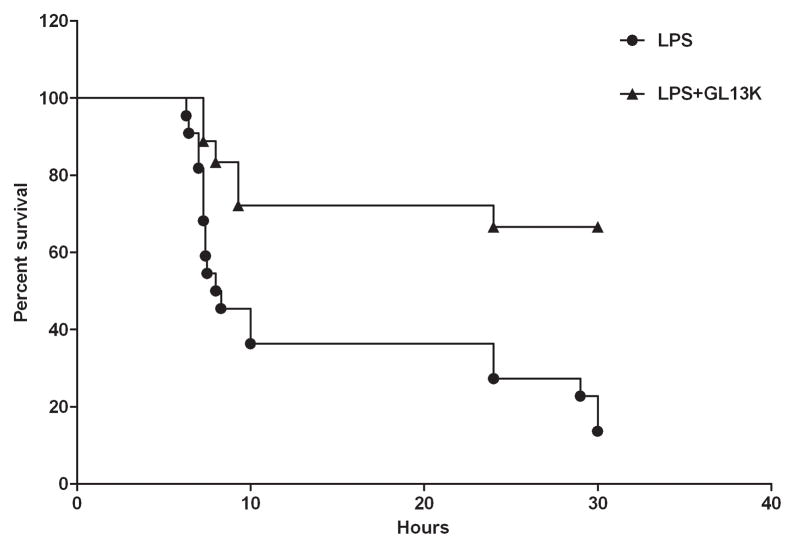
Preliminary data have indicated that GL13NH2 may be protective against LPS toxicity in a mouse sepsis model [15]. To determine if GL13K is protective in this model, mice were injected with 100 ng LPS alone or LPS that had been preincubated with 100 μg of GL13K. Figure 5 shows that GL13K significantly improved the survival of mice in this model. Thus, it is concluded that GL13K remains active and continues to block the binding of LPS to cellular receptors in vivo.
Figure 5. Mouse survival after LPS injection.
C57BL/6 mice were injected with LPS (filled circles) or LPS that was pre-incubated with GL13K (open squares) and survival was monitored for 30 hours. Percent survival was calculated at the times indicated. Survival was significantly increased in LPS+GL13K samples compared to LPS alone, P<0.02 (N=18–22).
3.5 Hemotoxicity assays
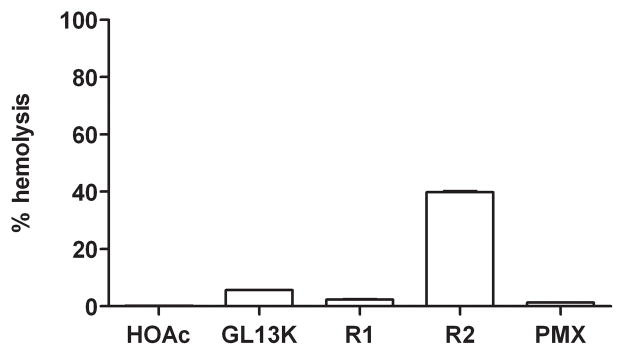
Unlike the peptides GL13NH2, GL13OH and GL13D/N [16], the modified peptide GL13K did not cause hemagglutination (not shown), consistent with the observed lack of bacterial agglutination for this peptide (Figure 1). GL13K (1 mg/ml) caused only minor lysis of human red blood cells (Figure 6) or sheep red blood cells (not shown) compared to buffer controls.
Figure 6. Hemolysis of human red blood cells.
Cells were incubated at 35°C in distilled water (dH2O) (100% hemolysis), PBS (0% hemolysis) or PBS with acetic acid (HOAc), GL13K, two randomized GL13K peptides (R1 and R2) or polymyxin B (PMX). The peptides were tested at a concentration of 1 mg/ml. Hemoglobin released from lysed cells was quantitated spectrophotometrically at 541 nm, expressed as mean ± range (N=2) or mean ± SEM (N=3–4). Data are from two independent experiments.
4. Discussion
The peptide GL13NH2 represents amino acids 141–153 of the PSP sequence. The peptide exhibits bacterial agglutinating activity, which is also found for recombinant PSP [15]. GL13NH2 contains several charged amino acid residues. In an attempt to achieve bactericidal activity, the residues in positions 2 (glutamine), 5 (asparagine) and 11 (aspartic acid) were replaced by lysine residues, thereby increasing the calculated net charge from +1 to +5 (pH 7.0) and the isoelectric point from 10.1 to 14, while retaining the relative hydrophobic moment (Table 1). The helical wheel representations predicted similar hydrophobic surfaces for the two peptides (Figure 7). However, GL13K contains two cationic surfaces while GL13NH2 contains a hydrophilic surface that is not strongly cationic. Indeed, the lysine residue in position 2 of GL13K, which is located in this hydrophilic side appears to be primarily disrupting agglutinating activity of the peptide. In addition, two charged residues, lysine and aspartic acid exhibit opposite charges near each other in GL13NH2 but two cationic residues are located here in GL13K (Figure 7). Thus, while single amino acid substitutions do not account for the observed transformation from an agglutinating peptide (GL13NH2) to a bactericidal peptide (GL13K), this change of activity appears to depend on a combinatorial effect of the amino acids. GL13K is active against both Gram negative and Gram positive bacteria. However, unlike LL-37, the peptide was not active against three strains of P. gingivalis. We speculate that bacterial gingipains caused the proteolysis of the peptide, as shown for the antimicrobial peptides human β-defensin 3 [[20] and BAR [8].
Figure 7. Helical wheel representation of peptide GL13NH2 and GL13K.
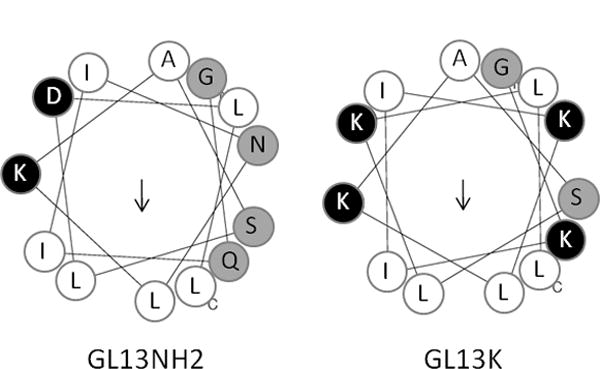
Open circles: hydrophobic residues. Black circles: charged residues; grey circles: hydrophilic residues and glycine. Glycine is the N-terminal residue in both peptides. The C-terminal leucine residues are labeled (c).
Preliminary data suggest that GL13NH2 causes bacterial agglutination by binding to membrane lipids in a carpet model mechanism of interaction (Balhara and DeWolf, personal communication). Preliminary data suggest that, unlike polymyxin B, GL13K does not permeabilize bacterial cells (Hirt and Gorr, unpublished observation). On the other hand, the bactericidal effect of GL13K is enhanced by saliva or lysozyme, supporting a role for bacterial lysis in the presence of salivary proteins. Thus, while lysis does not appear to be necessary for bacterial killing, the mechanisms behind the bactericidal activity of GL13K remains to be elucidated. The finding that GL13K remains active in the presence of saliva is promising for its use in the oral cavity or other mucosal surfaces.
Antimicrobial peptides often exhibit anti-LPS activity in addition to bactericidal activity [6, 9, 24]. Consistent with this, the GL13 peptides also exhibit dual activities. Interestingly, changing three amino acids in GL13NH2 to Lys residues to generate GL13K had a profound effect on the antibacterial activity, but did not significantly affect anti-LPS activity (Table 3). In fact, anti-LPS activity was strongly dependent on the serine residue at position 9, which was not affected by the changes introduced in GL13K. Replacing this serine residue with alanine eliminated LPS binding, suggesting that the hydroxyl group in serine is critical for this activity. As seen for saliva, GL13K remained active in the peritoneum and protected mice from the lethal effect of LPS. Similar results have previously been reported for GL13NH2 [15].
The higher survival of the GL13K treated mice also suggested that GL13K is not acutely toxic. Consistent with this, we found that the peptide caused neither hemagglutination nor hemolysis. We have previously reported low toxicity for the related peptide GL13NH2 [1, 15, 16]. A wide variety of peptides exhibit antimicrobial activity [25].Typically a combination of hydrophobic and cationic amino acid residues can exhibit antibacterial activity, but sometimes at the cost of toxicity to mammalian cells [19, 22]. The use of human peptide sequences to design antimicrobial peptides may overcome this problem. Thus, the GL13K sequence, which is based on the human protein PSP, shows low toxicity. In contrast a scrambled peptide with the same amino acid composition as GL13K exhibited 40% hemolysis. Together, these findings validate the approach of using a human salivary protein as the template for the design of antimicrobial peptides with low host toxicity.
Highlights.
Lysine substitution converts an agglutinating peptide to a bactericidal peptide
The peptide retains activity in saliva and in vivo
The peptide exhibits anti-lipopolysaccharide activity in vitro and in vivo
A serine hydroxyl is necessary for anti-LPS activity but not bactericidal activity
The peptide has low hemolytic activity
Acknowledgments
We thank Drs. Julie Sotsky and Anuradha Shelar, University of Louisville for contributing confirmatory data for select experiments and Dr. Paul B.M. Joyce, Concordia University, Montreal, Canada for critical reading of the manuscript. This work was funded by U.S. PHS grant R01DE17989 from the National Institute for Dental and Craniofacial Research. Institutional support from the University of Minnesota and University of Louisville Schools of Dentistry is gratefully acknowledged.
Footnotes
Abbreviations: BPI – bactericidal/permeability-increasing protein, CFU – colony forming units, LPS - lipopolysaccharide, MIC – minimum inhibitory concentration, OD – optical density, PBS – phosphate-buffered saline, PLUNC – palate, lung, nasal epithelium clone, PSP – parotid secretory protein
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Abdolhosseini M, Sotsky JB, Shelar A, Joyce PB, Gorr S-U. Human Parotid Secretory Protein is a lipopolysaccharide-binding protein: identification of an anti-inflammatory peptide domain. Mol Cell Biochem. 2011;359:1–8. doi: 10.1007/s11010-011-0991-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Beeley JG, Eason R, Snow DH. Isolation and characterization of latherin, a surface-active protein from horse sweat. Biochem J. 1986;235:645–50. doi: 10.1042/bj2350645. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Bingle CD, Craven CJ. PLUNC: a novel family of candidate host defence proteins expressed in the upper airways and nasopharynx. Human Mol Genet. 2002;11:937–43. doi: 10.1093/hmg/11.8.937. [DOI] [PubMed] [Google Scholar]
- 4.Bingle CD, Gorr S-U. Host defense in oral and airway epithelia: chromosome 20 contributes a new protein family. Intl J Biochem Cell Biol. 2004;36:2144–52. doi: 10.1016/j.biocel.2004.05.002. [DOI] [PubMed] [Google Scholar]
- 5.Bingle CD, Seal RL, Craven CJ. Systematic nomenclature for the PLUNC/PSP/BSP30/SMGB proteins as a subfamily of the BPI fold-containing superfamily. Biochem Soc Trans. 2011;39:977–83. doi: 10.1042/BST0390977. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Brandenburg K, Andrä J, Garidel P, Gutsmann T. Peptide-based treatment of sepsis. Appl Microbiol Biotechnol. 2011;90:799–808. doi: 10.1007/s00253-011-3185-7. [DOI] [PubMed] [Google Scholar]
- 7.Coats SR, To TT, Jain S, Braham PH, Darveau RP. Porphyromonas gingivalis resistance to polymyxin B is determined by the lipid A 4′-phosphatase, PGN_0524. Intl J Oral Sci. 2009;1:126–35. doi: 10.4248/IJOS.09062. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Daep CA, Novak EA, Lamont RJ, Demuth DR. Selective substitution of amino acids limits proteolytic cleavage and improves the bioactivity of an anti-biofilm peptide that targets the periodontal pathogen, Porphyromonas gingivalis. Peptides. 2010;31:2173–8. doi: 10.1016/j.peptides.2010.08.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Elsbach P, Weiss J. The bactericidal/permeability-increasing protein (BPI), a potent element in host-defense against gram-negative bacteria and lipopolysaccharide. Immunobiology. 1993;187:417–29. doi: 10.1016/S0171-2985(11)80354-2. [DOI] [PubMed] [Google Scholar]
- 10.Gakhar L, Bartlett JA, Penterman J, Mizrachi D, Singh PK, Mallampalli RK, et al. PLUNC is a novel airway surfactant protein with anti-biofilm activity. PLoS One. 2010;5:e9098. doi: 10.1371/journal.pone.0009098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Gautier R, Douguet D, Antonny B, Drin G. HELIQUEST: a web server to screen sequences with specific alpha-helical properties. Bioinformatics. 2008;24:2101–2. doi: 10.1093/bioinformatics/btn392. [DOI] [PubMed] [Google Scholar]
- 12.Geetha C, Venkatesh SG, Bingle L, Bingle CD, Gorr SU. Design and Validation of Anti-inflammatory Peptides from Human Parotid Secretory Protein. J Dent Res. 2005;84:149–53. doi: 10.1177/154405910508400208. [DOI] [PubMed] [Google Scholar]
- 13.Geetha C, Venkatesh SG, Fasciotto Dunn BH, Gorr S-U. Expression and anti-bacterial activity of human parotid secretory protein (PSP) Biochem Soc Trans. 2003;31:815–8. doi: 10.1042/bst0310815. [DOI] [PubMed] [Google Scholar]
- 14.Gorr S-U. Antimicrobial peptides of the oral cavity. Periodontol 2000. 2009;51:152–80. doi: 10.1111/j.1600-0757.2009.00310.x. [DOI] [PubMed] [Google Scholar]
- 15.Gorr S-U, Abdolhosseini M, Shelar A, Sotsky J. Dual host-defense functions of SPLUNC2/PSP and synthetic peptides derived from the protein. Biochem Soc Trans. 2011;39:1028–32. doi: 10.1042/BST0391028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Gorr S-U, Sotsky JB, Shelar AP, Demuth DR. Design of bacteria-agglutinating peptides derived from Parotid Secretory Protein, a member of the Bactericidal/Permeability Increasing-like protein family. Peptides. 2008;29:2118–27. doi: 10.1016/j.peptides.2008.09.019. [DOI] [PubMed] [Google Scholar]
- 17.Keijser BJF, Zaura E, Huse SM, van der Vossen JMBM, Schuren FHJ, Montijn RC, et al. Pyrosequencing analysis of the Oral Microflora of healthy adults. J Dent Res. 2008;87:1016–20. doi: 10.1177/154405910808701104. [DOI] [PubMed] [Google Scholar]
- 18.Lukinskiene L, Liu Y, Reynolds SD, Steele C, Stripp BR, Leikauf GD, et al. Antimicrobial Activity of PLUNC Protects against Pseudomonas aeruginosa Infection. J Immunol. 2011;187:382–90. doi: 10.4049/jimmunol.1001769. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Maher S, McClean S. Investigation of the cytotoxicity of eukaryotic and prokaryotic antimicrobial peptides in intestinal epithelial cells in vitro. Biochem Pharmacol. 2006;71:1289–98. doi: 10.1016/j.bcp.2006.01.012. [DOI] [PubMed] [Google Scholar]
- 20.Maisetta G, Brancatisano FL, Esin S, Campa M, Batoni G. Gingipains produced by Porphyromonas gingivalis ATCC49417 degrade human-β-defensin 3 and affect peptide’s antibacterial activity in vitro. Peptides. 2011;32:1073–7. doi: 10.1016/j.peptides.2011.02.003. [DOI] [PubMed] [Google Scholar]
- 21.McDonald RE, Fleming RI, Beeley JG, Bovell DL, Lu JR, Zhao X, et al. Latherin: a surfactant protein of horse sweat and saliva. PLoS One. 2009;4:e5726. doi: 10.1371/journal.pone.0005726. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.McInnes C, Kondejewski LH, Hodges RS, Sykes BD. Development of the Structural Basis for Antimicrobial and Hemolytic Activities of Peptides Based on Gramicidin S and Design of Novel Analogs Using NMR Spectroscopy. J Biol Chem. 2000;275:14287–94. doi: 10.1074/jbc.275.19.14287. [DOI] [PubMed] [Google Scholar]
- 23.Robinson CP, Bounous DI, Alford CE, Nguyen KH, Nanni JM, Peck AB, et al. PSP expression in murine lacrimal glands and function as a bacteria binding protein in exocrine secretions. Am J Physiol. 1997;272:G863–71. doi: 10.1152/ajpgi.1997.272.4.G863. [DOI] [PubMed] [Google Scholar]
- 24.Scott MG, Vreugdenhil ACE, Buurman WA, Hancock REW, Gold MR. Cutting Edge: Cationic Antimicrobial Peptides Block the Binding of Lipopolysaccharide (LPS) to LPS Binding Protein. J Immunol. 2000;164:549–53. doi: 10.4049/jimmunol.164.2.549. [DOI] [PubMed] [Google Scholar]
- 25.Wang G, Li X, Wang Z. APD2: the updated antimicrobial peptide database and its application in peptide design. Nucl Acids Res. 2009;37:D933–7. doi: 10.1093/nar/gkn823. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Weston WM, LeClair EE, Trzyna W, McHugh KM, Nugent P, Lafferty CM, et al. Differential display identification of plunc, a novel gene expressed in embryonic palate, nasal epithelium, and adult lung. J Biol Chem. 1999;274:13698–703. doi: 10.1074/jbc.274.19.13698. [DOI] [PubMed] [Google Scholar]