SUMMARY
Autophagy defects have been recently associated with chromosomal instability (CIN), a hallmark of human cancer. However, the functional specificity and mechanism of action of autophagy-related factors in genome stability remain elusive. Here we report that UVRAG, an autophagic tumor suppressor, plays a dual role in chromosomal stability, surprisingly independent of autophagy. We establish that UVRAG promotes DNA double-strand-breaks repair by directly binding and activating DNA-PK in non-homologous end-joining. Disruption of UVRAG increases genetic instability and sensitivity of cells to irradiation. Furthermore, UVRAG was found also localized at centrosomes and physically associated with CEP63, an integral component of centrosomes. Disruption of the association of UVRAG with centrosomes causes centrosome instability and aneuploidy. UVRAG thus represents an autophagy-related molecular factor that also has a convergent role in patrolling both the structural integrity and proper segregation of chromosomes, which may confer autophagy-independent tumor suppressor activity.
Keywords: UVRAG, genomic integrity, DNA damage repair, centrosome, DNA-PK
INTRODUCTION
The accurate partition of genetic information during cell division and the protection of cellular genome against various insults are of prime importance for proper cell function (Holland and Cleveland, 2009). Any defects in these pathways give rise to chromosomal instability (CIN), a conspicuous feature of many tumors, which drives cancer progression and is also associated with poor prognoses (Fukasawa, 2007; Holland and Cleveland, 2009). Despite its importance, the mechanisms leading to CIN are still poorly understood.
DNA double-strand breaks (DSBs) constitute formidable challenge to the stability of genetic materials and are particularly involved in chromosomal rearrangement that potentiates oncogenesis (Negrini et al.). Of the two major pathways for DSBs repair, homologous recombination (HR) and non-homologous end joining (NHEJ), NHEJ is considered to be a preferential route responding to DSBs in mammalian cells, since NHEJ rejoins the broken DNA ends without the requirement of a homologous template and functions thereof throughout the cell cycle (Lieber et al., 2003). During NHEJ, the DNA-end binding Ku70/80 heterodimer recognizes and binds DSB ends and recruits the catalytic subunit of the DNA-dependent protein kinase (DNA-PK) complex, DNA-PKcs, leading to the assembly and activation of DNA-PK holoenzyme as well as other cofactors for end-processing and subsequent ligation (Lieber et al., 2003). Disruption of any component of DNA-PK leads to extensive chromosomal instability (CIN), hypersensitivity to irradiation, and accelerated tumor development (Lieber et al., 2004). Autophagy has been proposed to limit DNA damage in response to metabolic stress (Mathew et al., 2007b). The basic tenet of autophagy is outlined as a lysosome-dependent bulk degradation and recycling of cytoplasmic materials, which allows cells to respond appropriately to stress (Levine and Kroemer, 2008; Liang and Jung). Recent findings demonstrate that cells with defective autophagy are prone to genomic instability with increased DNA damage and aneuploidy (Karantza-Wadsworth et al., 2007; Mathew et al., 2007b). However, evidence in support of autophagy as a straight genome protector is limited mostly to a phenotypic association between loss of autophagy-related gene(s) and gain of CIN in cells. Thus, the precise mechanism(s) underlying autophagy- or autophagy factor-mediated genome protection is still unknown.
In addition to DNA damage repair, faithful chromosome segregation during cell division represents an equally important aspect of genomic stability, which largely depends on the proper assembly of a bipolar spindle by centrosomes during mitosis (Fukasawa, 2007; Holland and Cleveland, 2009). The centrosome is composed of a pair of centrioles surrounded by pericentriolar material (PCM) (Bornens, 2002 and Nigg, 2004). To fulfill its critical task in assembling the spindle apparatus, the single interphase centrosome duplicates once and only once per cell cycle (Nigg, 2002). Almost inevitably, centrosome amplification results in erroneous chromosomal segregation, with dire consequences for genomic instability and aneuploidy (Fukasawa, 2007). Indeed, centrosome aberrations are considered as a major contributing factor to CIN in cancer cells (Bornens, 2002; Fukasawa, 2007).
UV-irradiation-resistance-associated gene (UVRAG) maps to a tumor susceptibility locus on human chromosome 11q13 that is frequently implicated in common human cancers, including breast, colorectal, and gastric cancers (Bekri et al., 1997; Goi et al., 2003; Ionov et al., 2004; Kim et al., 2008; Perelman et al., 1997). We and others have previously shown that UVRAG associates with Beclin1 and activates PI(3) kinase class III (PI(3)KC3) kinase in autophagy (Liang et al., 2006; Matsunaga et al., 2009; Takahashi et al., 2007b; Zhong et al., 2009). Gain-of-function of UVRAG which induces autonomous activation of autophagy inhibited cancer cell proliferation in our study, suggesting that UVRAG may control cell growth at least in part through its regulation of PI(3)KC3 signaling (Liang et al., 2006; Takahashi et al., 2007a). However, emerging evidence indicates that other functions unrelated to autophagy may exist. For instance, UVRAG has been found not exclusively present in autophagosomes (Liang et al., 2008). Moreover, the endosome-associated UVRAG promotes endocytic trafficking by interacting with the class C Vps complex (Liang et al., 2008). Importantly, aberrant levels of UVRAG exist in many cancers, but a clear correlation between impaired autophagy and carcinogenesis as a result of UVRAG deficiency has not been observed (Knaevelsrud et al.). Therefore, additional activities of UVRAG beyond autophagy might exist in the cancer-related cellular processes.
Here, we demonstrate that UVRAG plays a crucial role in maintaining chromosomal stability through mechanisms that are in addition to its role in autophagy. Loss of UVRAG results in defects in DSBs repair and hypersensitivity of cells to radiation. We found that UVRAG helps the assembly of DNA-PK and activates DNA-PK to maintain the stability of chromosomes through modulation of the NHEJ repair. Moreover, UVRAG is found to be associated with centrosomes by its interaction with CEP63. Disturbance of the UVRAG-centrosome interaction destabilizes centrosomes, resulting in extensive aneuploidy. Our findings thus reveal a tumor suppressor function for UVRAG, while at the same time unfolding mechanisms whereby the autophagy-related UVRAG acts to patrol chromosomal stability in an autophagy-independent manner.
RESULTS
UVRAG Deficiency Leads to an Accumulation of DSBs
Given that UVRAG is monoallelically mutated in cancer cells (Ionov et al., 2004; Liang et al., 2006), we attempted to determine whether downregulation of UVRAG affects any of the aspects of genomic stability that was shown previously to be associated with autophagy (Mathew et al., 2007a). We first analyzed the incidence of DNA damage in UVRAG wild-type (UVRAG+/+) mouse embryonic stem (ES) cells and cells with allelic loss of UVRAG (UVRAG+/−) that are available by immunostaining for the Ser139-phosphorylated form of histone H2AX (γ-H2AX), a sensitive DSBs marker (Rogakou et al., 1998). UVRAG+/− cells showed reduced levels of UVRAG expression than that in UVRAG+/+ cells (Figure 1A). Intriguingly, we found that the intensity and number of γ-H2AX foci per cell as well as the percentage of γ-H2AX foci-positive cells remarkably increased in UVRAG+/−, compared to the wild-type (Figure 1A). Likewise, the levels of γ-H2AX were also elevated in UVRAG+/− (Figure 1A). Re-introducing UVRAG clearly suppressed the levels of DSBs in these cells, indicating the specific effect of UVRAG in limiting DSBs in cells (Figure S1A–B). Similar results were obtained in human dermal fibroblasts (HDFα) (Figure S1F–H) and primary MEF (Figure S1C–E). To further confirm this, we used a neutral comet assay to detect the occurrence of DSBs in individual cells (Illuzzi et al., 2009) and found a marked increase in both the comet tail length and tail moment upon UVRAG depletion (Figure 1B–D). These results indicate that there is increased DNA-strand breakage when UVRAG is underexpressed.
Figure 1. UVRAG is Required for Efficient DNA DSB Repair.
(A) UVRAG deficiency leads to the accumulation of DNA DSBs. UVRAG+/+ and UVRAG+/− ES cells were stained with antibody against γ-H2AX (red) and DNAs were stained by DAPI (blue). Representative images of γ-H2AX foci are shown (left panel). The percent distribution of cells with different γ-H2AX foci is determined (middle panel) and western blot shows the level of endogenous UVRAG and γ-H2AX expression in these cells (right panel). Data represents mean ± s.d. from three independent experiments. Bars, 10 μm. See also Figure S1A–B.
(B–D) Neutral comet analysis of HDFα cells after UVRAG depletion. Representative comet images from control shRNA- and UVRAG shRNA-treated cells are illustrated in (B). Type I denotes cells with no comet tails; Type II denotes cells with a tail length less than 20 μm; Type III denotes cells with a tail length longer than 20 μm. Quantification of the distribution of cells with different type of comet tail moments is shown in (C). Quantification of percentage of cells with a tail moment is shown in (D). Data shown represent mean ± s.d. for data combined from three independent experiments. P < 0.001 (Wilcoxon signed-rank test).
(E–G) UVRAG deficiency impedes DSBs repair. Control shRNA- and UVRAG shRNA-treated HDFα cells (E–F), or UVRAG+/+ and UVRAG+/− ES cells (G), were treated with 2 Gy of ionizing radiation (IR) and recovered for a period of time as indicated. DSBs were determined by immunostaining of γ-H2AX foci formation. Representative images are shown in (E). Bar, 10 μm. (F–G) Immunoblotting analysis of the levels of γ-H2AX and UVRAG in HDFα (F) and ES (G) cells as indicated. See also Figure S1I.
(H) Depletion of UVRAG promotes DSBs in Atg3-knockout MEFs. Atg3+/+ and Atg3−/− immortalized MEF cells were treated with control shRNA or UVRAG-specific shRNA for 72 h and stained for γ-H2AX. Representative images of DSBs in these cells are shown in the left panel. The percentage of γ-H2AX-positive cells was quantified (middle panel). Western blot showing the levels of UVRAG, γ-H2AX, and Atg3 in these cells with actin serving as a loading control (right panel). Data shown represent mean ± s.d. for data combined from three independent experiments. Bar, 50 μm. See also Figure S1J–L.
UVRAG is Required for Efficient DNA Repair Independently of Autophagy
To determine the observed accumulation of DSBs reflects more of DNA damage accrual or of impaired DNA repair, we used the comet assay to measure the unrepaired DSBs after ionizing radiation (IR). We found that radiation induced comparable levels of DNA damage in both control and in UVRAG-knockdown 293T cells (1 h post IR in Figure S1I). However, at 24 h post-radiation, a highly persistence of comet tails was observed in UVRAG-depleted cells (Figure S1I). Consistent with this result, insufficiency of UVRAG prolonged the persistence of γ-H2AX foci and the expression of γ-H2AX in UVRAG-knockdown HDFα cells as well as in UVRAG+/− ES cells after IR, although the initial induction of γ-H2AX was similar to that of wild-type cells (Figure 1E–G). These data indicate that UVRAG suppression affects the rapid repair of DSBs and genomic stability.
To further explore whether the damage-protecting role of UVRAG might be related to autophagy, or instead reflects an independent event, we examined the effect of UVRAG on DNA damage in an autophagy-deficient background using Atg3- or Atg5-deficient immortalized MEFs (iMEF) (Mizushima et al.; Sou et al., 2008). We found that UVRAG depletion by itself induced a marked increase in the levels γ-H2AX and the γ-H2AX-foci staining in cells regardless of their autophagy status (Figure 1H and S1J–L). These data indicate that UVRAG-mediated suppression of DNA damage does not necessarily require functional autophagy machinery. While these observations do not exclude a role for autophagy in DNA damage, such events would appear to be downstream of a critical UVRAG-dependent step in maintaining genome stability. Therefore, UVRAG may directly regulate DNA damage repair through a mechanism independent of autophagy.
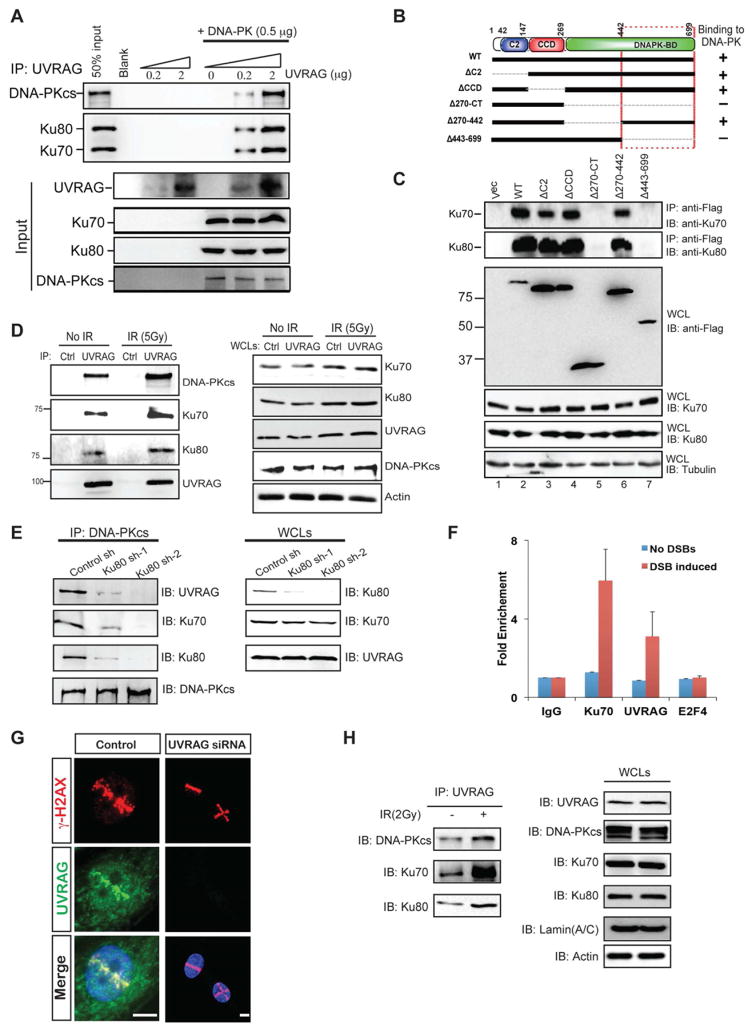
UVRAG Interacts Directly with the DNA-PK Complex in vitro and in vivo
To elucidate the mechanism by which UVRAG acts in DSB repair, we immunoaffinity-purified HA-UVRAG-containing complexes before and after exposure to IR, and the identity of UVRAG-interacting molecules induced by IR was determined by mass spectrometry analysis (Figure S2A). Peptides corresponding to Ku80 and DNA-PKcs were identified in independent assays of IR treatment with high confidence (Figure S2A). Ku80 is well known to pair with Ku70 to form a heterodimer for DNA end-binding, which recruits DNA-PKcs to form the DNA-PK complex, initiating NHEJ repair (Lieber et al., 2003). Using purified recombinant UVRAG and DNA-PK components, we established that UVRAG interacts directly with DNA-PK in a dose-dependent manner in vitro, independently of DNA (Figure 2A). Moreover, the carboxy terminus of UVRAG (residues 443-699) mediates this interaction (Lane 7 in Figure 2C), which is independent from its association with Beclin1 through the CCD (Figure 2B–C). We further verified physical interactions of the endogenous UVRAG and DNA-PK proteins, which was intensified after the treatment of IR or bleomycin (Figure 2D and S2B). Furthermore, knockdown of Ku70 or Ku80 reduced interaction of UVRAG with DNA-PKcs, suggesting that Ku70/80 facilitates UVRAG association with DNA-PK (Figure 2E and S2C). The strong induction of UVRAG-DNA-PK interaction upon radiation suggests that an increased number of UVRAG molecules is recruited to DNA-PK complex to facilitate the DNA repair process.
Figure 2. Functional Interaction between UVRAG and the DNA-PK Complex.
(A) Purified UVRAG directly interacts with recombinant DNA-PK complex in vitro. Indicated amount of purified UVRAG and DNA-PK complex were mixed and subjected to immunoprecipitation with the UVRAG-specific antibody followed by IB for Ku70, Ku80, and DNA-PKcs. All three subunits were co-immunoprecipitated with UVRAG in vitro.
(B) Schematic representation of UVRAG wild-type (WT) and its deletion mutants, and summary of their interactions with DNA-PK. Interaction was determined by coimmunoprecipitation of Flag-UVRAG with endogenous Ku70 and Ku80 from 293T cell lystates. C2, calcium-dependent lipid-binding domain; CCD, coiled-coil domain. +, strong binding; −, no binding.
(C) UVRAG C-terminal 443-699 region interacts with DNA-PK. 293T cells were transfected with Flag-UVRAG or its mutant derivatives and whole cell lysates (WCLs) were subjected to immunoprecipitation (IP) with an anti-Flag antibody, followed by immunoblotting (IB) with anti-Ku70 and anti-Ku80. WCLs were also used for IB with the indicated antibodies to show expression.
(D) Interaction between endogenous UVRAG and subunits of the DNA complex under basal condition and ionizing radiation (IR) treatment. WCLs of 293T cells were used for IP with control serum (control) or an anti-UVRAG antibody, followed by IB with the indicated antibodies. The bottom panel shows endogenous protein expression. See also Figure S2B.
(E) Ku80 knockdown inhibits UVRAG interaction with the DNA-PK complex. 293T cells were transfected with control shRNA or Ku80-specific shRNA for 72 h, followed by IP with a DNA-PKcs antibody and IB for UVRAG and the DNA-PK complex subunits. The right panels show endogenous protein expression. See also Figure S2C.
(F) Recruitment of UVRAG to I-SceI-induced DSBs. Enrichment of UVRAG and Ku70 around DNA damage site was quantified by anti-UVRAG and anti-Ku70 ChIP analysis, respectively, followed by real-time PCR using primers adjacent to DNA damage site. ChIP analyses using control IgG and E2F4 antibodies serve as controls.
(G) Recruitment of UVRAG to DSBs. Left panel: representative images showing the recruitment/accumulation of UVRAG to laser-induced DNA damage stripes. DNA damage was generated by laser microirradiation followed by immunofluorescence (1 h after damage) with the indicated antibodies against UVRAG, γ-H2AX, and DAPI. γ-H2AX marks the ‘laser stripes’ containing damaged DNA. Bar, 10μm. Right panel: same assay was performed in cells treated with UVRAG siRNA. See also Figure S2E-F.
(H) Nuclear fraction of UVRAG interacts with DNA-PK. A co-IP experiment was performed with isolated nuclear fraction of untreated or IR (2 Gy)-treated 293T cells using anti-UVRAG antibody followed by IB with the indicated antibodies of the DNA-PK complex.
To test whether UVRAG is recruited to the damaged sites of DSBs, we used an established model HEK293 cell system in which a unique I-SceI site has been integrated into the cellular genome (Bennardo et al., 2008). Using this system, recruitment of repair factors to the endonuclease-induced DSB site can be quantified using targeted chromatin immunoprecipitation (ChIP) analysis followed by real-time PCR with specific primers adjacent to the I-SceI DSB sites (Bennardo et al., 2008; Jirawatnotai et al., 2011). Indeed, we observed specific recruitment of both Ku and UVRAG to the I-SceI-induced DSB sites (Figure 2F). Moreover, UVRAG, similar to as has been observed for Ku (Mari et al., 2006) and DNA-PKcs (Uematsu et al., 2007), was enriched at sites of laser-induced DNA damage stripes containing γ-H2AX (Figure 2G). This UVRAG enrichment disappeared upon shRNA-mediated knockdown, consistent with specific staining of the endogenous protein (Figure 2G). We have also found that DNA-PKcs deficiency in M059J cells reduced recruitment of UVRAG to laser-induced damage stripes (Figure S2E) and to radiation-induced γ-H2AX foci (Figure S2F), whereas this was not the case in DNA-PKcs-competent M059K cells. This result indicates that DNA-PK is involved in the recruitment process of UVRAG. As expected, more co-IP complex of UVRAG with DNA-PK was recovered after radiation using the isolated nuclear extracts from 293T cells (Figure 2H). Collectively, these data suggest that UVRAG is recruited to DNA damage sites and interacts directly with DNA-PK.
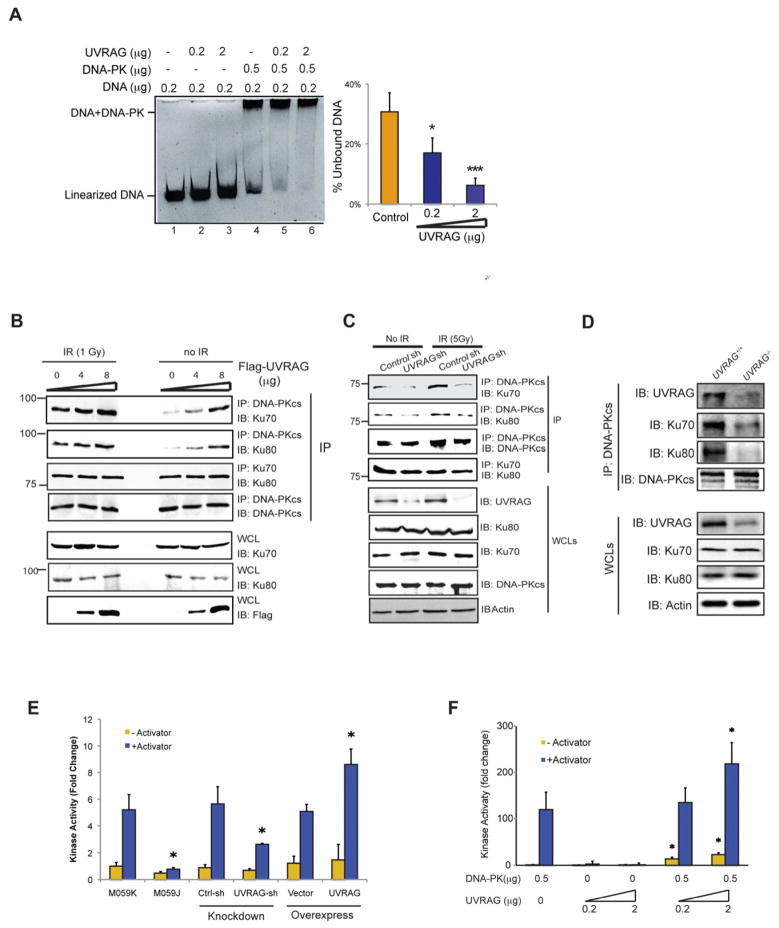
UVRAG Activates the Formation of the DNA-PK Complex and DNA-PKcs Activity
To investigate how UVRAG facilitates DNA repair, we examined the DNA end-binding, Ku-DNA-PKcs complex formation, and kinase activity of DNA-PK, three steps that are required for DNA-PK to perform end-joining repair. Using electrophoretic mobility shift assay (EMSA) (Muller et al., 2001) with purified DNA-PK, UVRAG, and dsDNA fragments, we observed a concentration-dependent sharp increase in the DNA-binding capacity of DNA-PK induced by UVRAG (17% and 6% unbound; Lane 5–6 in Figure 3A), although no apparent shift was detected for UVRAG per se (Lane 2–3 in Figure 3A). This data suggests that UVRAG enhances the affinity of DNA-PK to double-stranded DNA ends. We next asked whether UVRAG may further coordinate the complex assembly of DNA-PK. We found that more DNA-PKcs coimmunoprecipitated with Ku proteins in cells over-expressing UVRAG (Figure 3B), whereas depletion of UVRAG severely hindered the interaction of DNA-PKcs with Ku70/80 (Figure 3C). This data suggests that UVRAG promotes the efficient assembly of the Ku/DNA-PKcs complex, although it did not evidently affect the Ku heterodimer formation (Figure 3B–C). Such effect of UVRAG on DNA-PK became more evident upon IR treatment (Figure 3B–C). In agreement, more DNA-PK complex was recovered from UVRAG+/+ cells rather than UVRAG+/− cells after radiation (Figure 3D).
Figure 3. UVRAG Activates DNA-PK During DSB Repair.
(A) UVRAG promotes the DNA end-binding ability of DNA-PK in vitro. Indicated amount of purified UVRAG and DNA-PK complex were mixed with linearized DNA for 1 h, and then subjected to EMSA assay. The DNA end-binding ability was quantified by the percentage of unbound DNA (right). Data represented as mean ± s.d. from three independent experiments. *, P < 0.01; ***, P < 0.001.
(B) Transient expression of UVRAG facilitates the assembly of the DNA-PK complex. 293T cells transfected with increasing amounts of Flag-UVRAG were treated with IR (5 Gy). Co-IP was performed using DNA-PKcs or Ku70 antibody. Western Blot analyzed the amount of DNA-PKcs, Ku70, Ku80, and UVRAG within the complex.
(C) UVRAG knockdown impairs the DNA-PK complex formation. 293T were transfected with control shRNA or UVRAG shRNA for 72 h followed by IR (5 Gy) treatment. WCLs were used for IP with anti-DNA-PKcs followed by IB with the indicated antibody. Bottom panel showed endogenous protein expressions in cells.
(D) DNA-PK complex formation in UVRAG+/+ and UVRAG+/− ES cells. The ES cells were treated with IR (2 Gy). WCLs were used for IP with anti-DNA-PKcs followed by IB with the indicated antibody.
(E) UVRAG enhances DNA-PK activity. DNA-PK activity was measured using the SignaTECT DNA-PK Assay Kit in 293T cell expressing an empty vector, Flag-UVRAG, control shRNA or UVRAG specific shRNA. M059J and M059K cells were used as controls. Data represented as mean ± s.d. from three independent experiments. *, P < 0.05.
(F) in vitro kinase assay with purified UVRAG and DNA-PK complex was performed. Data represented as mean ± s.d. from three independent experiments. *, P < 0.05.
Since the assembly of DNA-PK at DNA damage sites stimulates the kinase activity of DNA-PKcs, we investigated further the effects of UVRAG on the enzymatic activity of DNA-PK. As a control to validate this assay, no obvious kinase activity was observed in the radiated DNA-PKcs-deficient M059J cells, whereas robust induction of DNA-PK was detected in the radiated M059K cells, akin to previous reports (Burma and Chen, 2004) (Figure 3E). Remarkably, UVRAG knockdown led to a more than 50% suppression of DNA-PK activity in comparison with control shRNA-treated cells, while expression of UVRAG strikingly stimulated DNA-PK (Figure 3E). When examined in vitro, we found that the kinase activity of recombinant DNA-PK was mobilized significantly with increasing amounts of purified UVRAG (Figure 3F). Collectively, these results are consistent with the model that UVRAG is recruited to DNA damage sites through a direct UVRAG-DNA-PK interaction. This interaction in turn facilitates the formation of the DSB-repair-promoting DNA-PK complex and increases Ku-associated DNA-PKcs activity, leading to efficient DNA repair.
UVRAG Is Required for Efficient NHEJ Repair and Protects Cells From DSBs Assaults
Our observation suggests that UVRAG may function in the DNA-PK-mediated NHEJ. Using a NHEJ repair reporter, the EJ5-GFP system (Bennardo et al., 2008), we found that expression of UVRAG markedly enhanced the rate of NHEJ repair, whereas depletion of UVRAG resulted in a consistent 30% reduction in NHEJ (Figure 4A). This observation was further confirmed in an independent assay that measures the rejoining of a linearized mCherry-reporter plasmid in vivo as described (Sharma and Raghavan). As shown in Figure 4C, despite their equivalent transfection efficiency marked by pTracer-GFP, the percentage of GFP-positive cells possessing rejoined cherry signals was significantly higher in the presence of UVRAG expression. Notably, expression of the UVRAGΔ443-699 mutant which is deficient in DNA-PK binding failed to promote efficient end-joining (Figure 4D–E). In contrast, expression of the ΔCCD and Δ270-442 mutants that are capable of DNA-PK interaction resulted in efficient DNA repair as seen with wild-type UVRAG (Figure 4D–E). These data indicate that UVRAG and its interaction with DNA-PK are required for an efficient DNA end-joining process. Consistent with this notion, UVRAG depletion or allelic loss of UVRAG (Figure S3A–B) sensitized the cells to radiation by showing reduced rates of clonogenic survival, in concordance with the findings that cells defective in NHEJ are hypersensitive to DSBs-inducing agents (Lieber et al., 2003). We also found that depletion of UVRAG has no observable effect on DNA HR repair rate (Figure S3C).
Figure 4. UVRAG Is Required for Efficient NHEJ Repair.
(A–B) NHEJ assay in HEK293 cells using EJ5-GFP reporter system. HEK293 cells stably expressing the NHEJ reporter were transfected with an empty vector, Flag-UVRAG, control shRNA or UVRAG-specific shRNA before the induction of DSBs by SceI transfection. The NHEJ repair activities as assessed by the reconstituted GFP signal were quantified by FACS. Data shown represent mean ± s.d. for data combined from three independent experiments. The expression levels of UVRAG were analyzed by Western blot in (B).
(C–E) UVRAG overexpression promotes DNA end-joining. 293T cells were sequentially transfected with pTracer-vector, pTracer-UVRAG or its mutant derivatives, and linearized pmCherry DNA. 2 days after transfection, cells were fixed and imaged. GFP is an indicator of transfection, while mCherry is an indicator of precise DNA end-joining. Cells from (C) were analyzed by FACS and the DNA end-joining activity was calculated based on the percentage of mCherry-positive cells in the population of GFP-positive cells in (D). Data shown represent mean ± s.d. for data combined from three independent experiments. (E) Immunoblotting shows UVRAG and its mutants expression. Actin serves as a loading control. Bar, 10 μm.
(F) Colony survival assay of Atg5+/+ and Atg5−/− iMEFs expressing control shRNA (control sh) or UVRAG shRNA (UVRAG sh), after an increasing dosage of IR. Values in the graphs represent means ± s.d. of triplicate samples and the experiment was repeated at least twice. See also Figure S3D.
(G) IR-sensitivity of M059J and M059K cells upon UVRAG knockdown. M059J and M059K cells depleted of UVRAG for 72 h were treated with an increasing dosage of IR and colony-forming ability was measured. Values in the graphs represent means ± s.d. of triplicate samples and the experiment was repeated at least twice. See also Figure S3A–B.
To further test whether UVRAG-associated cell survival upon radiation is related to autophagy, we evaluated the impact of UVRAG on the radiosensitivity of Atg5−/− cells. Despite that loss of Atg5 sensitized cells to the IR-induced cell death, as noted previously (Jin and White, 2007), downregulation of UVRAG in Atg5−/− cells caused a significant loss (approximately 10-fold) of clonogenicity in response to IR (Figure 4F), as also seen when cells were treated with bleomycin (Figure S3D). In contrast, UVRAG had minimal effect on the sensitivity of DNA-PK-deficient M059J cells to IR (Figure 4G). These results indicate that this increased radiosensitivity induced by UVRAG depletion manifests an inefficient DNA repair rate beyond autophagy. Taken together, UVRAG is necessary for efficient NHEJ repair through a direct UVRAG-DNA-PK interaction, which is required for genetic stability and thus optimal cell survival following DNA damage.
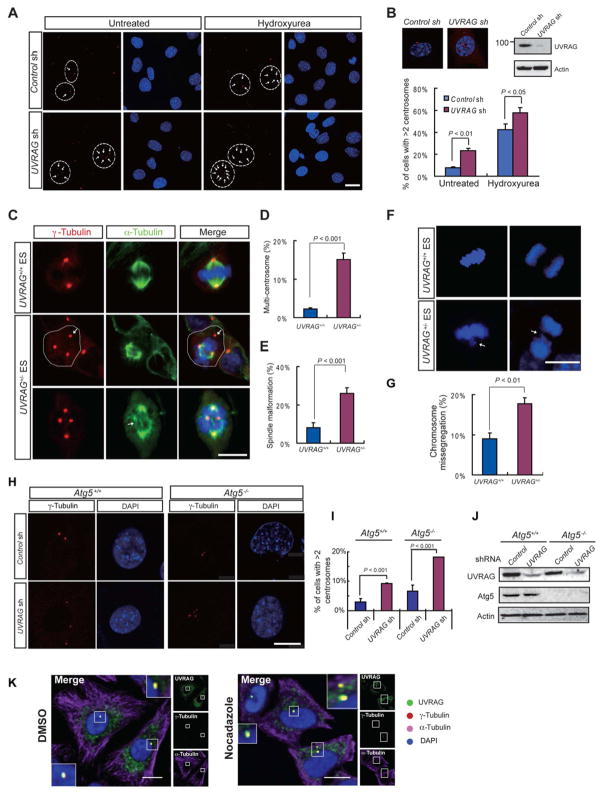
UVRAG Is Required for Centrosome Stability and Proper Chromosome Segregation
In addition to DNA damage, another feature of genomic instability associated with autophagy is centrosome abnormalities and aneuploidy (Mathew et al., 2007b). To test whether UVRAG also plays a possible role in centrosome, we examined centrosome function in mouse fibroblasts (MEF) and HeLa cells with efficient depletion of UVRAG. Intriguingly, we found that there was a profound increase in cells containing excess centrosomes when UVRAG was silenced (Figure 5A–B and S4A). Moreover, most of the UVRAG-depleted cells with supernumerary centrosomes were mononucleated, indicating that the accumulation of surplus centrosomes is not caused by aborted cell division (Figure 5A and S4A). Hydroxyurea (Hu) treatment, which dissociates centrosome duplication events from DNA synthesis, further increased centrosome numbers in the UVRAG-depleted cells (Figure 5A–B; S4A). These results indicate that UVRAG deficiency has a permissive effect for centrosome overduplication.
Figure 5. UVRAG Deficiency Results in Centrosome Amplification, Spindle Malformation, and Chromosomal Missegregation.
(A–B) Depletion of UVRAG leads to centrosome amplification in MEFs. MEF cells were transfected with control shRNA or UVRAG-specific shRNA, and either exposed to hydroxyurea for 48 h or left untreated. (A) Centrosomes were scored by immunostaining for γ-tubulin (red). Nuclei were stained with DAPI (blue). Arrows mark centrosomes. Cell borders are delineated where necessary to distinguish centrosome clusters belonging to adjacent cells. The percentages of cells in (A) with centrosome amplification (>2 centrosome per cell) were determined by counting over 100 cells and shown in (B), together with insets showing representative images of γ-tubulin foci in individual cells upon control or UVRAG knockdown (upper left) and the expression of UVRAG in treated MEF 72 h after transfection (upper right). Data represents mean ± s.d. from three independent experiments. Bar, 10 μm. See also Figure S4A.
(C–E) Centrosome amplification and spindle malformation in UVRAG+/− ES cells. Representative images of centrosomes and spindles in mitotic UVRAG+/+ and UVRAG+/− ES cells are shown in (C). Cells were co-immunostained with anti-γ-tubulin (red) for centrosome labeling and anti-α-tubulin (green) for the mitotic asters. Cell border is delineated where necessary to distinguish centrosome clusters belonging to adjacent cells. The percentages of UVRAG+/+ and UVRAG+/− ES cells with more than two centrosomes (D), or with disorganized spindle (E), were quantified in over 100 mitotic cells. Data are mean ± s.d. from three independent experiments. Bar, 10 μm. See also Figure S4B.
(F–G) Chromosome missegregation in UVRAG+/− ES cells. Representative images of chromosome segregation in UVRAG+/+ and UVRAG+/− ES cells are shown in (F). Arrows indicate a misaligned chromosome(s) at metaphase or a lagging chromosome(s) at telophase. Chromosomes were stained with DAPI, Bar, 10 μm. The percentages of UVRAG+/− ES cells with abnormal chromosome segregation were quantified in (G).
(H–J) UVRAG deficiency leads to centrosome amplification in autophagy-defective cells. Atg5+/+ and Atg5−/− immortalized MEF cells were transfected with either control shRNA or UVRAG-specific shRNA. Centrosomes were stained as in (A). Representative images are shown in (H). The percentage of cells with supernumerary centrosomes were quantified (I). Whole cell lysates prepared in parallel were analyzed for the expression of UVRAG and Atg5 by immunoblotting with actin as a loading control (J). Bar, 10 μm.
(K) UVRAG localizes in centrosomes. HeLa cells were fixed and immunostained with antibody against UVRAG (green), γ-tubulin (red), and α-tubulin (purple), after treatment with DMSO or nocadazole (200 ng/ml) for 2 h. Co-localization between UVRAG and γ-tubulin was observed as highlighted by the insets, regardless of the treatment. Bars, 10 μm. See also Figure S4C–E.
Extra centrosomes almost inevitably cause spindle malformation and erroneous chromosomal segregation (Ganem et al., 2009). As expected, we found that the proportion of cells with aberrant spindles was clearly increased after UVRAG knockdown in MEF cells (Figure S4B). Similarly, over 25% of UVRAG+/− ES cells exhibited disorganized spindles and supernumerary centrosomes as compared to 7% in control cells (Figure 5C–E). In accord, there was a distinct two-fold increase in the occurrence of chromosome mis-alignment (at metaphase) and mis-segregation (at anaphase) in UVRAG+/− cells (Figure 5F–G), which was largely restored by the expression of wild-type UVRAG (see later in Figure 6H). These results demonstrate that UVRAG constitutes an important factor for centrosome stability and is thus required for proper chromosome segregation.
Figure 6. Association of UVRAG with CEP63 in Centrosomes is essential for Centrosome Stability.
(A) UVRAG interaction with CEP63. 293T cells were co-transfected with Flag-UVRAG and HA-CEP63. WCLs were IP with anti-Flag, followed by IB with anti-HA.
(B) Interaction between endogenous UVRAG and CEP63. WCLs of 293T cells were immunoprecipiated with control IgG, or anti-CEP63 (left panel), or anti-UVRAG (middle panel), followed by immunoblotting with anti-CEP63 or anti-UVRAG. Right panel shows the expression of endogenous UVRAG and CEP63 proteins.
(C) Colocalization between endogenous UVRAG and CEP63 at the centrosome. HeLa cells were co-stained with antibodies against UVRAG (green), CEP63 (purple), and γ-tubulin (red), followed by confocal microscopy. Arrows highlight the colocalization. Bar, 10 μm. See also Figure S5A.
(D) Confocal microscopy analysis of the colocalization of endogenous UVRAG and γ-tubulin in CEP63+/+ and CEP63−/− DT40 cells. Arrows denote colocalized UVRAG and γ-tubulin in CEP63+/+, whereas insets highlight relative distribution of UVRAG with the γ-tubulin-labeled centrosomes in CEP63−/− cells. Bars, 10 μm.
(E) UVRAG C-terminal region interacts with CEP63. 48 h posttransfection with HA-CEP63 together with Flag-UVRAG or its mutants, 293T WCLs were IP with anti-Flag followed by IB with anti-HA. WCLs were also used for IB with the indicated antibodies to show expression. See also Figure S5C.
(F) Schematic representation of UVRAG wild-type (WT) and its deletion mutants, and summary of their interactions with CEP63. Interaction was determined by coimmunoprecipitation of Flag-UVRAG with HA-CEP63 from 293T cell lysates. +, strong binding; −, no binding.
(G–H) UVRAG association with centrosomes is required for centrosome integrity and proper chromosomal segregation. (G) UVRAG+/− ES cells reconstituted with empty vector (1st row), Flag-UVRAG (2nd row), or Flag-UVRAGΔ270-442 (3rd row) were stained with anti-γ-tubulin (red) for centrosomes, anti-α-tubulin (red) for mitotic asters, and DAPI for chromosomes (blue). Arrows denote over-duplicated centrosomes and abnormal spindle (left panel) or mis-segregated chromosomes in mitosis (right panel). Bar, 10 μm. Quantification of abnormal centrosome amplification, spindle malformation, and chromosomal missegregation from these cells is shown in (H). Data represents mean ± s.d. from three independent experiments. ***, P <0.001. See also Figure S5E–G.
(I–J) UVRAG association with centrosomes is required for chromosomal stability. The frequency of the numerical aberration of chromosomes from UVRAG+/− ES cells reconstituted as in (G) with empty vector, Flag-UVRAG, or Flag-UVRAGΔ270-442 was quantified (I) and reconstituted UVRAG expression was confirmed by immunoblotting (J).
To further test whether this effect of UVRAG on centrosomes involves an autophagy mechanism, we used autophagy-defective, Atg5−/−-deficient iMEF and found that there was a marked increase in the percentage of cells harboring supernumerary centrosomes when UVRAG was depleted regardless of Atg5 expression (Figure 5H–J) This data thus indicates that UVRAG has an autophagy-independent function to maintain centrosome stability.
UVRAG Is a Centrosome–associated Protein
Given the centrosome amplification induced by UVRAG depletion, we examined the distribution of UVRAG in comparison to centrosome markers. Irrespective of the fixation method and the cellular context, a portion of endogenous UVRAG or transiently expressed Flag-UVRAG concentrated in the proximity of nuclei and was co-stained with γ-tubulin, CEP170, and the centriolar marker, centrin, which was not perturbed by nocodazole (Figure S4C and 5K). Furthermore, endogenous UVRAG co-migrated with γ-tubulin in the centrosomal fraction (Figure S4D). Using phosphorylated-histone 3 (Phospho-H3) as a mitotic marker (Ajiro et al., 1996), we established that UVRAG continues to associate with centrosomes throughout most of the cell cycle, with the signal enriched at spindle poles at metaphase (Figure S4E). Thus, we identify UVRAG as the autophagy-related factor physically associated with centrosomes.
CEP63 Targets UVRAG to the Centrosome
To investigate how UVRAG functions at centrosomes, we conducted a yeast two-hybrid screen to identify UVRAG-interacting proteins that are centrosome-related. CEP63, an evolutionarily conserved centrosome protein for spindle organization (Andersen et al., 2003; Smith et al., 2009), was identified as a prominent candidate with nine clones recovered in our screening. This interaction was confirmed by co-immunoprecipitation (co-IP) in 293T cells transiently expressing Flag-UVRAG and HA-CEP63 (Figure 6A). Moreover, endogenous CEP63 and UVRAG co-immunoprecipitated with each other, suggesting their complex formation in vivo (Figure 6B). Consistent with the fact of being centrosome-associated, significant overlapping staining was observed between UVRAG (exogenous and endogenous) and CEP63 at γ-tubulin dots (Figure 6C and S5A). Additionally, both UVRAG and CEP63 co-fractionated with γ-tubulin (Figure S4D). These findings indicate that UVRAG primarily associates with CEP63 at centrosomes.
We next examined whether CEP63 is required for the centrosomal targeting of UVRAG using CEP63-knockout DT40 cells. We found that UVRAG was largely displaced from the centrosome in CEP63−/− cells but not in control cells, although its expression was not affected by CEP63-knockout (Figure 6D). Of note, ablation of CEP63 is sufficient to induce centrosome amplification (Figure S5B) (Smith et al., 2009). These data indicate that CEP63 is required for centrosome localization of UVRAG. By examining a series of UVRAG deletion mutants, we identified that the C-terminal region of residues 270-442 (Lane 6 in Figure 6E) conferred the major interaction with CEP63 in vivo, but not the C2, or the CCD that is known to mediate an interaction with Beclin1 (Liang et al., 2006), nor the region of residues 443-699 that binds DNA-PK as aforenoted (Figure 6E–F and S5C). Notably, the Δ270-442 mutant of UVRAG, albeit defective in CEP63 binding, preserved efficient interaction with the Beclin1-PI(3)KC3 complex (data not shown) and DNA-PK (Figure 2C), suggesting that the UVRAG association with centrosomes is independent from its autophagy-related and DNA repair-related interaction. Furthermore, the removal of autophagy-related CCD which was not required for CEP63-bindng, or the 443-699 region of UVRAG that abolishes UVRAG’s ability in DNA repair did not alter the centrosomal localization of UVRAG (Figure S5D). In contrast, the UVRAGΔ270-442 mutant which was unable to interact with CEP63 also lost the ability to localize to centrosomes (Figure S5D), though it has little effects on UVRAG-mediated end-joining repair (Figure 4D). These data thus indicate that UVRAG association with centrosomes is structurally and functionally distinguishable from the UVRAG-elicited autophagy and DNA repair activities.
Physical Association of UVRAG with Centrosomes is Necessary for Centrosome Stability and Chromosome Integrity
To further address whether centrosomal association of UVRAG is required for centrosome stability and chromosomal segregation, we reconstituted UVRAG+/− ES cells with either wild-type or the centrosome-binding-defective UVRAGΔ270-442 mutant. As described above, although allelic loss of UVRAG resulted in excess centrosomes, accompanied by significant spindles malformation and chromosomes missegregation, these defects were reversed by the expression of wild-type, but not by UVRAGΔ270-442 mutant expression (Figure 6G–H). Similarly, expression of the shRNA-resistant wild-type UVRAG rescued centrosome defects in UVRAG-depleted HeLa cells, but the Δ270-442 mutant failed to do so (Figure S5E–F). Furthermore, the cells expressing UVRAGΔ270-442 had high rates of aneuploidy as that of UVRAG+/− cells, whereas the expression of wild-type UVRAG suppressed aneuploidy (Figure 6I). These results thus indicate that centrosome association of UVRAG is sufficient and necessary for proper chromosome segregation to maintain genomic stability.
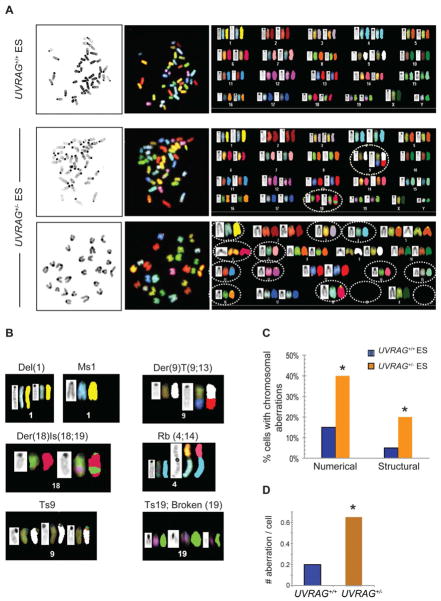
UVRAG is A Guardian of Chromosomes
Given the dual role of UVRAG in DNA repair and centrosome stability and the link between UVRAG deficiency and genomic stability, we examined the consequences of downregulation of UVRAG to chromosomes. Metaphase spreads prepared from UVRAG+/+ and UVRAG+/− cells were analyzed by multicolor spectral karotyping (SKY). We found that control ES cells were largely uniform and mostly diploid in karyotype with a few chromosomal aberrations (Figure 7A, 1st row; Figure S6A). In contrast, UVRAG+/− cells were highly heterogeneous with respect to both structural (Figure 7A, 2nd row) and numerical aberrations (Figure 7A, 3rd row), including aneuploidy, chromosome breaks, translocations, and fusions (Figure 7 and S6A). While no chromosomal translocations of any kind were found in UVRAG+/+ cells, complex translocations involving multiple chromosomes, such as Robertsonian centromere fusion (Rb), were often detected in UVRAG+/− cells (Figure 7B). Moreover, there was a greater than two-fold increase in aneuploidy (2n ± x) in UVRAG+/− compared to control cells (40% versus 15%; Figure 7C), with one spread of UVRAG+/− (AC-10 in Figure S6A) showing massive chromosomal loss. The total number of chromosomal aberrations detected in UVRAG+/− and UVRAG+/+ cells differed by 3-fold (0.6 per metaphase for UVRAG+/− versus 0.2 per metaphase for UVRAG+/+) (Figure 7D). Notably, re-expression of UVRAG effectively suppressed chromosomal abnormalities in UVRAG+/− cells (Figure S6B). Taken together, these results demonstrate that UVRAG is crucially needed for both the numerical and structural chromosomal stability, which may explain its important function in cancer development.
Figure 7. UVRAG Is Required for Chromosome Stability.
(A) Spectral karyotyping (SKY) analysis of UVRAG+/− (1st row) and UVRAG+/− (2nd and 3rd rows) mouse ES cells. Each representative metaphase spread is shown with DAPI-stained chromosomes (left panel), spectral-based display (middle panel), and the corresponding karyotype table (right panel) depicting individual chromosome in DAPI staining (left), spectral-based color (center) and classified color (right). The chromosomes are arranged in numerical order from left to right and top to bottom. UVRAG+/+ cells show normal diploid SKY karyotype (1st row). Structural (2nd row; 40XY, Der(9)T(9;13), Der(18)Is(18;19)) and numerical (3rd row; 26X, (-Y, -1, -3, -4, -6, -7, -11, -12, -14, -15, -16, -18, -19, -19)) chromosomal abnormalities were observed in UVRAG+/− ES cells and denoted by dotted lines.
(B) Representative examples of chromosomal abnormalities in UVRAG+/− ES cells. The DAPI-banded (left), display colors-banded (middle), and the spectrally classified chromosomes (right) are shown. Del, Deletion; Der, Derivative Chromosome; T, Translocation; Rb, Robertsonian translocation; Is, Insertion; Ms, Monosomy; Ts, Trisomy.
(C–D) Quantification of chromosomal aberrations in UVRAG+/+ and UVRAG+/− ES cells. Frequencies of structural and numerical aberrations (C) and the average events of chromosomal aberrations per cell (D) were determined from 20–40 metaphase spreads (pooled from two independent experiments) of UVRAG+/+ and UVRAG+/− ES cells. *, P <0.05, Wilcoxon Signed-rank Test. See also Figure S6.
DISCUSSION
The maintenance of chromosomal stability is a fundamental biological process and a critical aspect of cancer. Herein, we demonstrate that the autophagic tumor suppressor, UVRAG, plays a pivotal role in maintaining genomic stability through mechanisms involving DSBs repair and centrosome stability, independently of autophagy. To the best of our knowledge, these findings represent the description of an autophagy-related factor directly functioning in centrosomes and DNA repair to guard genome integrity, which thereby reveals a broader functional scope for UVRAG and also provides mechanistic insights into the role of UVRAG in cancer development.
The current understanding of UVRAG function is primarily focused on its activation of the autophagy-related Beclin1-PI(3)KC3 complex (Liang et al., 2006; Liang et al., 2008; Zhong et al., 2009). We herein establish that UVRAG depletion eventually leads to DSBs accumulation and thereof cells’ hypersensitivity to radiation, highlighting a second important mechanism for UVRAG in cancer-associated genomic instability. It is possible that aneuploidy associated with UVRAG deficiency may cause secondary mutations of other genes, whose products could facilitate DSB repair. In this scenario, loss of UVRAG may indirectly affect DSBs. However, we observed that re-introduction of wild-type UVRAG into UVRAG+/− cells significantly lessens DNA damage, underscoring a direct control of UVRAG on DSBs and a minor effect of putative secondary mutations that might arise due to UVRAG deficiency. Consistent with this notion, we prove that UVRAG acts directly on the step-wise activation of DNA-PK upon DSBs induction, from the initial recruitment of DNA-PK to broken DNA ends, the Ku/DNA-PKcs complex assembly thereafter, to the eventual kinase activation of DNA-PK, suggesting that UVRAG is a key regulator for the mammalian NHEJ apparatus. Since NHEJ factors help guarantee genomic integrity through the proper repair of DNA lesions, our data of UVRAG acting on NHEJ thus provide an important view that dysfunction of UVRAG may accumulate DNA damage and facilitate tumor progression.
In addition to the dysfunction of DNA repair that undermines the fidelity of genetic stability, we also identified UVRAG as a novel centrosome-associated protein, ensuring centrosome stability. Inhibition of UVRAG causes severe centrosome amplification with profound consequences on spindle malformation and chromosomal missegregation. Furthermore, the region of UVRAG responsible for centrosome-targeting is mapped to the C-terminal region, separable from its autophagy-related interaction with Beclin1. Further evidence for an autophagy-independent effect of UVRAG is provided by the observation that autophagy loss could not forestall centrosome amplification induced by UVRAG deficiency. Notably, a mutant version of UVRAG that lacks the centrosome-binding ability, while remaining competent for autophagy and DNA repair, induced centrosome amplification and consequent aneuploidy as a relic of UVRAG deficiency, suggesting that the association of UVRAG with centrosomes is important for centrosomal stability. It is also important to note that the UVRAG-associated centrosome stability and DNA repair represent two distinct aspects of autophagy-independent functions of UVRAG. As mentioned above, a mutation in UVRAG that prevents centrosome targeting generally has minor effects on DNA repair, but pronounced effects on centrosome. In contrast, a mutation in UVRAG that prevents DNA-PK association behaves in an opposite manner. These results imply that UVRAG functions through two distinct activities in genomic stability: one involving CEP63 in centrosome integrity and the other involving DNA-PK in DSBs repair.
A role for UVRAG in tumor suppression has been recognized for long, but the means by which UVRAG achieves this task remains elusive. Our work uncovered an unexpected role for UVRAG in promoting chromosomal stability through distinct mechanisms. UVRAG constitutes a direct mediator of DNA repair machinery to affect overall responsiveness of cells to DSB assault and presumably also affect the responsive of certain groups of cancer patients toward radiotherapy and DNA-damaging chemotherapies. Intriguingly, the genome-protecting function of UVRAG can be executed in an autophagy-independent manner. Moreover, UVRAG patrols centrosome stability and chromosome segregation. While centrosome abnormality may be due to an insufficient action of UVRAG in DNA repair, the physical presence of UVRAG in centrosomes and its suppression of centrosome amplification in the absence of DNA damage suggest a direct involvement of this protein in centrosome function. In the same vein, the DNA repair activity of UVRAG is not a straight result from its effect on centrosomes, instead it occurs in a nuclear NHEJ-dependent manner. An intriguing question is how UVRAG coordinates these distinct activities including autophagy. How are these complex processes coupled mechanistically? Future studies on these topics will undoubtedly illuminate new views on UVRAG as a protector of the genome and also hold a therapeutic potential for cancer research. Nevertheless, functional interactions between the pro-autophagic protein, UVRAG, the DNA repair factor, DNA-PK, as well as the centrosomal component, represent a potentially important point of convergence of the autophagy, centrosome, and DNA repair machinery in chromosomal stability, which may represent a fundamental role for UVRAG in cancer.
EXPERIMENTAL PROCEDURES
In vivo DNA DSB Repair
To measure the DNA DSB repair activities, a GFP-based chromosomally-integrated reporter was utilized (Bennardo et al., 2008). In brief, the HEK293 cells stably expressing EJ5-GFP reporter were transfected with UVRAG-specific shRNA or a scrambled control shRNA. Two days later, a secondary transfection was performed with the same shRNA plus an I-SceI expression vector (pCBASce), together with pmCherry as a transfection indicator. Cells were harvested after another 48 hours, and analyzed with a standard flow cytometry method. UVRAG expression was verified with western blotting. The repair activity of DSBs generated by I-SceI was calculated by the percentage of GFP-positive (repaired) cells in the mCherry-positive cells (transfected).
Neutral Comet Assay
Neutral comet assay was performed using CometAssay® kit (Trevigen) following the manufacturer’s instruction. Briefly, 10 μl of cell suspension (105 cells/ml) was mixed well with 90 μl of molten LMAgarose. After solidification, slides were immersed in Lysis Solution at 4°C for 1 h, and equilibrated in chilled neutral electrophoresis buffer for 30 min. Electrophoresis was performed in neutral electrophoresis buffer for 1 h with a electric field of 1 volt/cm. Slides were further treated with DNA Precipitation Solution followed by 70% ethanol for 30 min each at room temperature. After air dry, cells were stained with SYBR Green (1 μg/ml) or Propidium Iodide (1 μg/ml). Comet images were captured using an epifluorescence microscope (Nikon Eclipse C1). To analyze, cells were first scored into 3 categories based on tail length (no tails, tail length shorter than 20 μm, tail length longer than 20 μm), and quantified.
DNA-PK Kinase Assay
DNA-PK kinase activity is measured with the SignaTECT DNA-PK Assay System (Promega) following the manufacturer’s instruction. Briefly, nuclear extracts from 293T cells were adjusted to a protein concentration of 1mg/ml; 10 μl of extract was mixed with biotinylated peptide substrate and [γ-32P]-ATP in a reaction buffer containing 0.1mM ATP, 0.1mg/ml BSA. The reaction mix was incubated at 37°C for 15 min, either in the absence or presence of DNA-PK activator, followed by termination with 12.5 μl termination buffer. 10 μl of the reaction mix was then spotted onto a square of the SAM2® Membrane. After air dry, the membrane squares were extensively washed with 2M NaCl and 1% H3PO4 in 2M NaCl, then subjected to scintillation counting. Alternatively, nuclear extract was replaced by a mixture of 0.2–2 μg of purified UVRAG and 0.5 μg of purified DNA-PK.
Nuclear Fractionation
For the subcellular fractionation study of UVRAG in response to DNA damage, 107 cells were harvested and resuspended in the prechilled hypertonic buffer. After 15 min incubation, the cells were mixed with 25 μl Detergent Solution, vortexed for 10 seconds, and then centrifuged for 30 seconds. The supernatant was collected as cytoplasmic fraction, the nuclear pellet was resuspended in 50–100 ml Complete Lysis Buffer by pipetting and vortex. After 1 h incubation on ice, the nuclear fraction was centrifuged for 10 min, and the supernatant was collected as nuclear extract. Alternatively, the nuclear pellets were dissolved in RIPA buffer with protease inhibitors and sonicated on ice to directly achieve total nuclear fraction.
Centrosome Isolation
Centrosome isolation was performed based on standard procedures (Cold Spring Harbor). In brief, ~108 293T cells were treated with Nocodazole (10 μg/ml) and Cytochalasin D (2 μg/ml) for 90 min, collected and lysed in 5 ml of solution with 1mM Hepes, pH 7.2, 0.5% NP-40, 0.5mM MgCl2, 1mM DTT supplemented with protease inhibitor and phosphatase inhibitor cocktails (Roche). WCLs were centrifuged at 2,500x g for 10 min, and the supernatant was treated with DNase I (2 unit/ml, NEB) for 30 min on ice. The centrosomes were enriched from that of the WCLs by centrifugation at 10,000 × g for 30 min onto a underlaid cushion of 0.5 ml 60% sucrose in 10 mM Pipes, pH 7.2, 0.1% Triton X-100, 1mM DTT. 1.5 ml from the bottom was taken and fractionated at 350,000x g for 4 h with a self-forming gradient (30% iodixanol, OptiPrepTM Axis-shield). After centrifugation, 10 fractions were collected and diluted with 1ml 10mM Pipes buffer. Centrosomes were recovered by centrifugation at 15,000 rpm for 15 min, and dissolved in SDS sample buffer, followed by SDS-PAGE analysis.
Chromosomal Analysis by SKY
SKY analysis of ES cells was performed as described previously (Padilla-Nash et al., 2006). Briefly, metaphase chromosome were prepared from exponentially growing cells after treatment with colcemid (KaryoMAX, GIBCO) at 0.1 μg/ml for 1 h using a standard procedure (Padilla-Nash et al., 2006). Cells were swollen in prewarmed 0.56% KCl for 10 min at 37°C, then carefully fixed in methanol: acetic acid (3:1) overnight and kept at −20°C. Metaphase spreads were prepared by dropping cells in the fixative onto chilled Superfrost glass slides (Fisher Scientific) at 25°C and 60% of humidity. After air dry and pepsin digestion, slides were denatured at 80°C for 5 min, hybridization was performed using SKY probe (Applied Spectral Imaging, San Diego) and fluorescence-conjugated secondary antibodies in accordance with the manufacturer’s specification. Metaphase images were captured and analyzed using a SpectraCube® imaging system and software (Applied Spectral Imaging). At lease 20 metaphases from each cell line were scored for chromosomal aberration
Statistical Analysis
All experiments were independently repeated at least three times. Data are presented as mean ± s.d. The statistical significance was calculated using the Student’s t test or oneway ANOVA test, unless otherwise stated. A P value of ≤ 0.05 was considered statistically significant.
Supplementary Material
HIGHTLIGHTS.
UVRAG protects the genome from double-strand-break insults
UVRAG activates DNA-PK and promotes NHEJ repair to maintain genetic stability
Centrosome-associated UVRAG patrols centrosome stability and chromosome segregation
UVRAG maintains genomic stability independent of autophagy
Acknowledgments
The authors wish to acknowledge the NIH-sponsored Mutant Mouse Regional Resource Center (MMRRC) National System as the source of genetically altered UVRAG+/+ (E14TG2a.4) and UVRAG+/− (AC0571) mouse embryonic stem cells for use in this study. We thank Victoria Bedell and the Cytogenetics Core of City of Hope (Duarte, CA) for the SKY analysis. We also thank Ross Tomaino for Mass Spectrometry Analysis. We acknowledge the USC Broad CIRM Center Flow Cytometry Core Facility, supported by the National Cancer Institute Cancer Center Shared Grant #5 P30 CA014089 and the USC Provost Funds in BCC CU Accounts, for flow cytometry study. We thank Drs. J.U. Jung, M. Liber, B. Levine, S. Virgin, S. Field, T. Yoshimori, and Y. Ohsumi for providing reagents. This work is supported by the Baxter Foundation, American Cancer Society (RSG-11-121-01-CCG to C. Liang), National Institutes of Health grants (R01 CA140964 and R21 AI083841 to C. Liang; R01 CA120954 to JMS), and core services performed through grant NIAID U19AI083025 and Norris Cancer Center Grant P30CA014089-34 (C. Liang).
Footnotes
Supplemental information includes six supplementary figures and supplemental experimental procedures.
COMPETING FINANCIAL INTEREST STATEMENT
The authors declare that they have no competing financial interests.
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Ajiro K, Yoda K, Utsumi K, Nishikawa Y. Alteration of cell cycle-dependent histone phosphorylations by okadaic acid. Induction of mitosis-specific H3 phosphorylation and chromatin condensation in mammalian interphase cells. J Biol Chem. 1996;271:13197–13201. doi: 10.1074/jbc.271.22.13197. [DOI] [PubMed] [Google Scholar]
- Andersen JS, Wilkinson CJ, Mayor T, Mortensen P, Nigg EA, Mann M. Proteomic characterization of the human centrosome by protein correlation profiling. Nature. 2003;426:570–574. doi: 10.1038/nature02166. [DOI] [PubMed] [Google Scholar]
- Bekri S, Adelaide J, Merscher S, Grosgeorge J, Caroli-Bosc F, Perucca-Lostanlen D, Kelley PM, Pebusque MJ, Theillet C, Birnbaum D, et al. Detailed map of a region commonly amplified at 11q13-->q14 in human breast carcinoma. Cytogenet Cell Genet. 1997;79:125–131. doi: 10.1159/000134699. [DOI] [PubMed] [Google Scholar]
- Bennardo N, Cheng A, Huang N, Stark JM. Alternative-NHEJ is a mechanistically distinct pathway of mammalian chromosome break repair. PLoS Genet. 2008;4:e1000110. doi: 10.1371/journal.pgen.1000110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bornens M. Centrosome composition and microtubule anchoring mechanisms. Curr Opin Cell Biol. 2002;14:25–34. doi: 10.1016/s0955-0674(01)00290-3. [DOI] [PubMed] [Google Scholar]
- Burma S, Chen DJ. Role of DNA-PK in the cellular response to DNA double-strand breaks. DNA Repair (Amst) 2004;3:909–918. doi: 10.1016/j.dnarep.2004.03.021. [DOI] [PubMed] [Google Scholar]
- Fukasawa K. Oncogenes and tumour suppressors take on centrosomes. Nat Rev Cancer. 2007;7:911–924. doi: 10.1038/nrc2249. [DOI] [PubMed] [Google Scholar]
- Ganem NJ, Godinho SA, Pellman D. A mechanism linking extra centrosomes to chromosomal instability. Nature. 2009;460:278–282. doi: 10.1038/nature08136. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goi T, Kawasaki M, Yamazaki T, Koneri K, Katayama K, Hirose K, Yamaguchi A. Ascending colon cancer with hepatic metastasis and cholecystolithiasis in a patient with situs inversus totalis without any expression of UVRAG mRNA: report of a case. Surg Today. 2003;33:702–706. doi: 10.1007/s00595-002-2567-y. [DOI] [PubMed] [Google Scholar]
- Holland AJ, Cleveland DW. Boveri revisited: chromosomal instability, aneuploidy and tumorigenesis. Nat Rev Mol Cell Biol. 2009;10:478–487. doi: 10.1038/nrm2718. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Illuzzi J, Yerkes S, Parekh-Olmedo H, Kmiec EB. DNA breakage and induction of DNA damage response proteins precede the appearance of visible mutant huntingtin aggregates. J Neurosci Res. 2009;87:733–747. doi: 10.1002/jnr.21881. [DOI] [PubMed] [Google Scholar]
- Ionov Y, Nowak N, Perucho M, Markowitz S, Cowell JK. Manipulation of nonsense mediated decay identifies gene mutations in colon cancer Cells with microsatellite instability. Oncogene. 2004;23:639–645. doi: 10.1038/sj.onc.1207178. [DOI] [PubMed] [Google Scholar]
- Jin S, White E. Role of autophagy in cancer: management of metabolic stress. Autophagy. 2007;3:28–31. doi: 10.4161/auto.3269. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jirawatnotai S, Hu Y, Michowski W, Elias JE, Becks L, Bienvenu F, Zagozdzon A, Goswami T, Wang YE, Clark AB, et al. A function for cyclin D1 in DNA repair uncovered by protein interactome analyses in human cancers. Nature. 2011;474:230–234. doi: 10.1038/nature10155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Karantza-Wadsworth V, Patel S, Kravchuk O, Chen G, Mathew R, Jin S, White E. Autophagy mitigates metabolic stress and genome damage in mammary tumorigenesis. Genes Dev. 2007;21:1621–1635. doi: 10.1101/gad.1565707. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim MS, Jeong EG, Ahn CH, Kim SS, Lee SH, Yoo NJ. Frameshift mutation of UVRAG, an autophagy-related gene, in gastric carcinomas with microsatellite instability. Hum Pathol. 2008 doi: 10.1016/j.humpath.2007.11.013. [DOI] [PubMed] [Google Scholar]
- Knaevelsrud H, Ahlquist T, Merok MA, Nesbakken A, Stenmark H, Lothe RA, Simonsen A. UVRAG mutations associated with microsatellite unstable colon cancer do not affect autophagy. Autophagy. 2010;6:863–870. doi: 10.4161/auto.6.7.13033. [DOI] [PubMed] [Google Scholar]
- Levine B, Kroemer G. Autophagy in the pathogenesis of disease. Cell. 2008;132:27–42. doi: 10.1016/j.cell.2007.12.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liang C, Feng P, Ku B, Dotan I, Canaani D, Oh BH, Jung JU. Autophagic and tumour suppressor activity of a novel Beclin1-binding protein UVRAG. Nat Cell Biol. 2006;8:688–699. doi: 10.1038/ncb1426. [DOI] [PubMed] [Google Scholar]
- Liang C, Jung JU. Autophagy genes as tumor suppressors. Curr Opin Cell Biol. 2010;22:226–233. doi: 10.1016/j.ceb.2009.11.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liang C, Lee JS, Inn KS, Gack MU, Li Q, Roberts EA, Vergne I, Deretic V, Feng P, Akazawa C, et al. Beclin1-binding UVRAG targets the class C Vps complex to coordinate autophagosome maturation and endocytic trafficking. Nat Cell Biol. 2008;10:776–787. doi: 10.1038/ncb1740. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lieber MR, Ma Y, Pannicke U, Schwarz K. Mechanism and regulation of human non-homologous DNA end-joining. Nat Rev Mol Cell Biol. 2003;4:712–720. doi: 10.1038/nrm1202. [DOI] [PubMed] [Google Scholar]
- Lieber MR, Ma Y, Pannicke U, Schwarz K. The mechanism of vertebrate nonhomologous DNA end joining and its role in V(D)J recombination. DNA Repair (Amst) 2004;3:817–826. doi: 10.1016/j.dnarep.2004.03.015. [DOI] [PubMed] [Google Scholar]
- Mari PO, Florea BI, Persengiev SP, Verkaik NS, Bruggenwirth HT, Modesti M, Giglia-Mari G, Bezstarosti K, Demmers JA, Luider TM, et al. Dynamic assembly of end-joining complexes requires interaction between Ku70/80 and XRCC4. Proc Natl Acad Sci U S A. 2006;103:18597–18602. doi: 10.1073/pnas.0609061103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mathew R, Karantza-Wadsworth V, White E. Role of autophagy in cancer. Nat Rev Cancer. 2007a;7:961–967. doi: 10.1038/nrc2254. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mathew R, Kongara S, Beaudoin B, Karp CM, Bray K, Degenhardt K, Chen G, Jin S, White E. Autophagy suppresses tumor progression by limiting chromosomal instability. Genes Dev. 2007b;21:1367–1381. doi: 10.1101/gad.1545107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matsunaga K, Saitoh T, Tabata K, Omori H, Satoh T, Kurotori N, Maejima I, Shirahama-Noda K, Ichimura T, Isobe T, et al. Two Beclin 1-binding proteins, Atg14L and Rubicon, reciprocally regulate autophagy at different stages. Nat Cell Biol. 2009;11:385–396. doi: 10.1038/ncb1846. [DOI] [PubMed] [Google Scholar]
- Mizushima N, Yoshimori T, Levine B. Methods in mammalian autophagy research. Cell. 2010;140:313–326. doi: 10.1016/j.cell.2010.01.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Muller C, Monferran S, Gamp AC, Calsou P, Salles B. Inhibition of Ku heterodimer DNA end binding activity during granulocytic differentiation of human promyelocytic cell lines. Oncogene. 2001;20:4373–4382. doi: 10.1038/sj.onc.1204571. [DOI] [PubMed] [Google Scholar]
- Negrini S, Gorgoulis VG, Halazonetis TD. Genomic instability--an evolving hallmark of cancer. Nat Rev Mol Cell Biol. 2010;11:220–228. doi: 10.1038/nrm2858. [DOI] [PubMed] [Google Scholar]
- Nigg EA. Centrosome aberrations: cause or consequence of cancer progression? Nat Rev Cancer. 2002;2:815–825. doi: 10.1038/nrc924. [DOI] [PubMed] [Google Scholar]
- Padilla-Nash HM, Barenboim-Stapleton L, Difilippantonio MJ, Ried T. Spectral karyotyping analysis of human and mouse chromosomes. Nat Protoc. 2006;1:3129–3142. doi: 10.1038/nprot.2006.358. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Perelman B, Dafni N, Naiman T, Eli D, Yaakov M, Feng TL, Sinha S, Weber G, Khodaei S, Sancar A, et al. Molecular cloning of a novel human gene encoding a 63-kDa protein and its sublocalization within the 11q13 locus. Genomics. 1997;41:397–405. doi: 10.1006/geno.1997.4623. [DOI] [PubMed] [Google Scholar]
- Rogakou EP, Pilch DR, Orr AH, Ivanova VS, Bonner WM. DNA double-stranded breaks induce histone H2AX phosphorylation on serine 139. J Biol Chem. 1998;273:5858–5868. doi: 10.1074/jbc.273.10.5858. [DOI] [PubMed] [Google Scholar]
- Sharma S, Raghavan SC. Nonhomologous DNA end joining in cell-free extracts. J Nucleic Acids. 2010;2010 doi: 10.4061/2010/389129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith E, Dejsuphong D, Balestrini A, Hampel M, Lenz C, Takeda S, Vindigni A, Costanzo V. An ATM- and ATR-dependent checkpoint inactivates spindle assembly by targeting CEP63. Nat Cell Biol. 2009;11:278–285. doi: 10.1038/ncb1835. [DOI] [PubMed] [Google Scholar]
- Sou YS, Waguri S, Iwata J, Ueno T, Fujimura T, Hara T, Sawada N, Yamada A, Mizushima N, Uchiyama Y, et al. The Atg8 conjugation system is indispensable for proper development of autophagic isolation membranes in mice. Mol Biol Cell. 2008;19:4762–4775. doi: 10.1091/mbc.E08-03-0309. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takahashi Y, Coppola D, Matsushita N, Cualing HD, Sun M, Sato Y, Liang C, Jung JU, Cheng JQ, Mul JJ, et al. Bif-1 interacts with Beclin 1 through UVRAG and regulates autophagy and tumorigenesis. Nat Cell Biol. 2007a;9:1142–1151. doi: 10.1038/ncb1634. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takahashi Y, Coppola D, Matsushita N, Cualing HD, Sun M, Sato Y, Liang C, Jung JU, Cheng JQ, Mule JJ, et al. Bif-1 interacts with Beclin 1 through UVRAG and regulates autophagy and tumorigenesis. Nat Cell Biol. 2007b;9:1142–1151. doi: 10.1038/ncb1634. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Uematsu N, Weterings E, Yano K, Morotomi-Yano K, Jakob B, Taucher-Scholz G, Mari PO, van Gent DC, Chen BP, Chen DJ. Autophosphorylation of DNA-PKCS regulates its dynamics at DNA double-strand breaks. J Cell Biol. 2007;177:219–229. doi: 10.1083/jcb.200608077. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhong Y, Wang QJ, Li X, Yan Y, Backer JM, Chait BT, Heintz N, Yue Z. Distinct regulation of autophagic activity by Atg14L and Rubicon associated with Beclin 1-phosphatidylinositol-3-kinase complex. Nat Cell Biol. 2009;11:468–476. doi: 10.1038/ncb1854. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.