Abstract
The AddAB and RecBCD helicase-nucleases are related enzymes prevalent among bacteria but not eukaryotes and are instrumental in the repair of DNA double-strand breaks and in genetic recombination. Although these enzymes have been extensively studied both genetically and biochemically, inhibitors specific for this class of enzymes have not been reported. We developed a high-throughput screen based on the ability of phage T4 gene 2 mutants to grow in Escherichia coli only if the host RecBCD enzyme, or a related helicase-nuclease, is inhibited or genetically inactivated. We optimized this screen for use in 1536-well plates and screened 326,100 small molecules in the NIH molecular libraries sample collection for inhibitors of the Helicobacter pylori AddAB enzyme expressed in an E. coli recBCD deletion strain. Secondary screening used assays with cells expressing AddAB or RecBCD and a viability assay that measured the effect of compounds on cell growth without phage infection. From this screening campaign, 12 compounds exhibiting efficacy and selectivity were tested for inhibition of purified AddAB and RecBCD helicase and nuclease activities and in cell-based assays for recombination; seven were active in the 0.1 – 50 μM range in one or another assay. Compounds structurally related to two of these were similarly tested, and three were active in the 0.1 – 50 μM range. These compounds should be useful in further enzymatic, genetic, and physiological studies of these enzymes, both purified and in cells. They may also lead to useful antibacterial agents, since this class of enzymes is needed for successful bacterial infection of mammals.
Small-molecule inhibitors are exceptionally useful in enzymology and physiology, for they can allow immediate inactivation of an enzyme, either in the purified state or in cells. With multifunctional enzymes, such as those studied here, they can also halt an enzymatic reaction at an intermediate step of the reaction and permit, for example, determining the structure of reaction intermediates. Notable examples include the aminocoumarin and quinolone classes of DNA gyrase inhibitors and the camptothecin class of topoisomerase I inhibitors. In these cases, inhibitors were instrumental in showing that the proteins make covalent links with their DNA substrate 1, 2. The quinolone inhibitors, such as ciprofloxacin, are also useful antibacterial agents, since DNA gyrase is widely distributed in bacteria but closely related enzymes appear to be absent in eukaryotes.
The RecBCD class of enzymes and the closely related AddAB enzymes are bacterial helicase-nuclease complexes important for repair of broken DNA and for genetic recombination 3, 4, 5. Starting at a double-strand (ds) DNA end, these enzymes unwind DNA rapidly and highly processively while hydrolyzing ATP or another nucleoside triphosphate (Fig. 1). During unwinding, they also hydrolyze DNA by making endonucleolytic scissions at a rate dependent on the ratio of [ATP] to [Mg2+], both of which are required for the helicase and nuclease activities. The RecBCD enzyme of Escherichia coli makes endonucleolytic scissions at especially high frequency at Chi sites (5′ GCTGGTGG 3′), which as a consequence are hotspots of recombination 6. The RecBCD and AddAB enzymes from other species similarly act at other short nucleotide sequences 7. The single-stranded (ss) DNA resulting from unwinding is a potent substrate for the enzymes’ ATP-dependent ss nuclease, which, at least for the RecBCD enzyme of E. coli, produces a limit digest of primarily tetra- to hexanucleotides 8. Because this class of enzymes is important for the repair of DNA double-strand (ds) breaks, mutants lacking them are deficient in infecting mammals, likely because mammalian cells produce reactive oxygen species, such as hydrogen peroxide, that break the bacterial DNA upon infection 9, 10. Inhibitors of these enzymes may therefore be effective antibacterial drugs.
Figure 1.
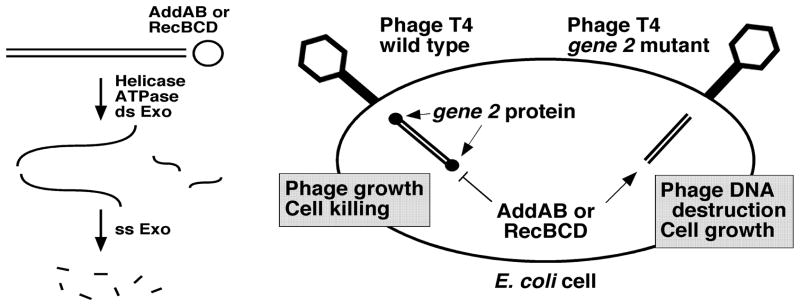
Principle of the cell-based screen for AddAB and RecBCD inihbitors. Left. Activities of RecBCD and AddAB helicase-nucleases. Both enzymes (open circle) are active on linear duplex DNA (double lines). ds exonuclease activity involves a combination of ATP-dependent DNA unwinding and endonucleolytic cuts. ss DNA intermediates are digested to short TCA-soluble oligonucleotides by the ss exonuclease activity. Right. RecBCD or AddAB nuclease activity blocks the growth of phage T4 gene 2 mutants. Upon injection into E. coli cells, wild-type T4 DNA is protected from AddAB and RecBCD nucleases by the gene 2 protein bound to the linear duplex DNA ends in the virion; phages grow and the cells are killed. Unprotected T4 gene 2 mutant DNA is digested by the nucleases; cells grow. Inhibition of AddAB or RecBCD is detected by lack of cell growth after T4 gene 2 mutant infection.
Although these complex enzymes have been studied by both biochemistry and genetics for over fifty years 5, 11, no small molecule inhibitors specific for them have, to our knowledge, been reported. Dziegielewska et al. 12 reported inhibition of RecBCD helicase activity by three compounds, but these compounds bind and alkylate DNA and likely would inhibit other helicases as well. Ca2+ and other divalent ions inhibit E. coli RecBCD and Bacillus subtilis AddAB nuclease 13, though not the helicase 14, 15, but Ca2+ inhibits many other enzymes. The Gam protein of phage λ also partially inhibits RecBCD 16, 17, perhaps by binding to the site at which DNA binds 18, 19. Non-specific inhibitors, such as EDTA and SDS, have also been used to stop the enzyme, but they have limited utility in studying reaction mechanisms.
To aid studies of RecBCD and AddAB and to find potential novel antibacterial agents, we have sought inhibitors of these enzymes by screening large libraries of small organic molecules. We developed a cell-based assay, so that compounds that do not enter E. coli would be immediately eliminated. The assay used here is based on the ability of phage T4 gene 2 mutants to grow in E. coli only if the RecBCD nuclease is inactivated by mutation 20 (Fig. 1). The nuclease, which resides in the RecB polypeptide 21, is active only if the RecD subunit is present and only if the RecB helicase is active 22, 23. The RecB helicase in turn is highly active only in the presence of the RecC subunit 24. In AddAB, each subunit contains a nuclease domain; only AddA appears to have an active helicase domain, but its inactivation blocks all detectable nuclease activity 13, 25, 26, 27. Consequently, small molecules that bind to any subunit of either enzyme might inhibit the nuclease, either directly or indirectly, and allow T4 gene 2 mutant phage to grow and thereby block the growth of E. coli. We report here five structural classes of inhibitors of the three-subunit E. coli RecBCD enzyme and the related two-subunit Helicobacter pylori AddAB enzyme.
RESULTS AND DISCUSSION
Rationale and Development of the Screen
Because phage T4 gene 2 mutants grow in E. coli mutants lacking RecBCD nuclease activity but not in E. coli wild-type or other nuclease-deficient mutants 20, RecBCD evidently is the only nuclease that blocks this mutant phage’s growth. Wild-type T4 phage are able to grow in wild-type E. coli presumably because the gene 2 protein binds to the ends of the ds DNA in the phage virions and, upon injection of the DNA into the host cell, blocks the action of RecBCD 20 (Fig. 1). Based on these observations, we developed conditions that allow E. coli recB21 null mutants but not recBCD+ cells to be lysed by T4 gene 2 mutant phage. After infection at a multiplicity of infection (MOI) of 0.01 in liquid culture, E. coli recB21 cells increase in OD for about 2 h and then cease growth, presumably when the phage have multiplied sufficiently to infect and begin to lyse most or all of the cells (Fig. S1). Under these conditions, recBCD+ cells grow about the same with or without phage infection. E. coli cells bearing a deletion of the recBCD genes and harboring a plasmid expressing the H. pylori addAB+ genes also grow about the same with or without phage infection 10. We reasoned that an inhibitor of RecBCD or AddAB nuclease would allow T4 gene 2 mutant phage to block the growth of recBCD+ (or addAB+) cells; an inhibitor specific for RecBCD or AddAB would not block the growth of uninfected cells. We used these criteria to screen for specific inhibitors of these two enzymes.
To screen large numbers of compounds in 0.1 ml cultures in 96-well plates, we found reproducible results by diluting freshly grown cells about 1:100 into LB broth containing compound and adding phage after 1 h of incubation. In every well recB21 cells were lysed or failed to grow and in nearly all of the wells recBCD+ cells grew to at least as high OD as without infection (e.g., Fig. S1); however, in about 2% of the wells recBCD+ cells were also lysed. Similar results were found with E. coli expressing H. pylori AddAB (SKA and LL, unpublished data). We found that these wells with lysed cells contained revertants or pseudorevertants of the gene 2 mutation, a nonsense mutation at codon 247 of 275 codons in the gene (SKA unpublished data) (NCBI NP_049754) 28. To circumvent this problem, we constructed a phage with three nonsense mutations, at codons 247, 248, and 249. [We were unable to construct a deletion of gene 2, presumably because part of the gene 2 protein is also required for packaging phage DNA 29.] We have not observed revertants of this triple nonsense phage, designated gene 2 am149, in >103 assays without a compound added.
Screen of 326,100 Compounds
To screen larger libraries, we converted the assay for use in 1536-well plates as detailed in Experimental Procedures and summarized in Table S1. The AddAB (strain V3605) phage assay was selected as the primary screen to test a total of 326,100 distinct chemical entities from the National Institutes of Health’s Molecular Libraries Small Molecules Repository (MLSMR). All compounds were tested at 12 μM in singlicate. Primary screen results were reviewed, and 937 compounds that appeared nominally active (“hits”) were advanced to secondary assays; 52 of these compounds were unavailable from the MLSMR. In secondary assays, the 885 available compounds were first retested in triplicate in the primary screening assay to confirm activity. The same compounds were also tested in triplicate for their effect on the viability of strain V3065 (i.e., in the absence of phage) and also screened in triplicate in strain V66 in the presence of phage to determine RecBCD inhibition. From these efforts, 225 hits that appeared active in either the RecBCD or AddAB inhibition assays were advanced to titration assays. In titration assays, compounds were tested in triplicate as 10-point titrations using the same protocols used for secondary assays; IC50 values were then determined. All HTS assays were determined to be robust, as each assay demonstrated Z’-scores greater than 0.8. A summary of the entire HTS effort, including summary statistics for all screening assays, is presented in Table S2.
Direct Enzyme Assays of 12 Compounds
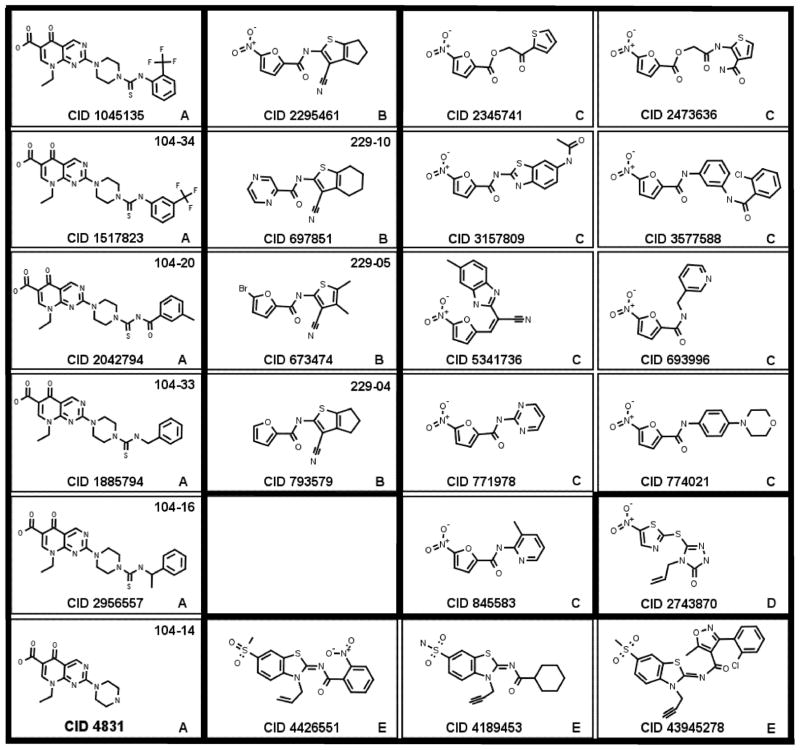
From the screens above, we chose the seven most potent compounds and five related compounds for direct tests with purified enzymes. These compounds, listed in Table 1 and diagramed in Fig. 2, form four structural classes, designated here “pyrimidopyridones” (group A), “cyanothiophenes” (group B), “nitrofurans” (group C), and “nitrothiazole” (group D with one member). For ease of identification, CID numbers are followed by the group designation in square brackets.
TABLE 1.
Summary of properties of 18 most active compounds found in the screens for AddAB and RecBCD inhibitors
| CID, (scaffold type)a | Inhibition (approximate IC50, μM) | Relative recombinant frequencyb | ||||||
|---|---|---|---|---|---|---|---|---|
| E. coli (paddA B) growth with T4 2− phage | AddA B nuclease | E. coli recBCD+ growth with T4 2− phage | RecBCD nuclease | Inhibition of RecBCD Chi cutting | E. coli Hfr recombination at 200 μM compound | Phage λ recombination at 200 μM compound | Chi hotspotc activity at 200 μM compound | |
| 1045135 [A] | 2.5 | 34 | >40 | 13 | 10 | 16, 15 | 9, 14 | 4.1, 4.6 |
| 4831 [A] | NDe | >100 | ND | >100 | ND | 0.2, 0.4 | 9, 11 | 3.1, 3.5 |
| 1517823 [A] | ND | 26 | ND | 5.1 | ND | 20, 10 | 18, 23 | 2.5, 2.1 |
| 2295461 [B] | 18 | 96 | >120 | 51 | 20 | 37, 21 | ND | ND |
| 697851 [B] | ND | 13 | ND | 33 | >500 | 118, 121d | ND | ND |
| 771978 [C] | 8.2 | >100 | >40 | >100 | >100 | 7, 5 | 0.4, 0.3 | 5.3, 5.2 |
| 693996 [C] | 9.7 | >100 | 86 | >100 | >500 | 6, 7 | 5, 6 | 4.9, 3.8 |
| 845583 [C] | 10 | >100 | >120 | >100 | 100 | 34, 42 | ND | ND |
| 5341736 [C] | 4.9 | >100 | >120 | >100 | 50 | 37, 43 | ND | ND |
| 3157809 [C] | >13 | >100 | >120 | >100 | >500 | 47, 51 | ND | ND |
| 2473636 [C] | 4.8 | >100 | >120 | >100 | >500 | 65, 73 | ND | ND |
| 3577588 [C] | 17 | >100 | >120 | >100 | 20 | 47, 55 | ND | ND |
| 2345741 [C] | 21 | >100 | >120 | >100 | >500 | 49, 63 | ND | ND |
| 774021 [C] | ND | >100 | ND | >100 | 200 | 49, 54 | ND | ND |
| 2743870 [D] | 12 | 130 | >120 | >100 | >500 | 0.6, 0.3 | 90, 50 | 3.1, 2.9 |
| 4189453 [E] | ND | ND | >100 | 10 | 50 | 64, 72d | ND | ND |
| 4426551 [E] | ND | ND | >100 | 80 | 50 | 78, 67d | ND | ND |
| 43945278 [E] | ND | ND | ND | 3 | 50 | 57, 47d | ND | ND |
See Figs. 2, S4, and S6. A, pyrimidopyridone; B, cyanothiophene; C, nitrofuran; D, nitrothiazole; E, iminobenzothiazole.
Data, from two experiments for each compound, are the recombinant frequencies as a percentage of that without compound. For Hfr recombination these were 8.7 and 8.9% His+ StrR per viable Hfr cell in the two experiments, respectively; for λ recombination these were 11.2% and 3.4% J+ R+, respectively.
Chi hotspot activity, measured in crosses between λ phages 1081 × 1082 and 1083 × 1084 as in 34, is the frequency of recombinants in an interval with Chi to that in the same interval without Chi. In the absence of compounds, values were 5.1 and 4.9 in the two experiments, respectively.
Compound was 100 μM; recombinant frequencies in the absence of compound were 7.6% and 8.4%.
ND, not determined.
Figure 2.
Structures of AddAB and RecBCD inhibitors. Compounds are grouped into “scaffolds” (A, B, C, D, and E in lower right corner or each panel) reflecting related structures. Trivial names, such as 104-34 and 229-10 in the upper right corner of some panels, refer to derivatives of the parent compound (see Figs. S4 and S6, which show additional compounds related to CID 1045135 [A] and CID 2295461 [B]). Heavy outlines separate compounds of different scaffold types (Fig. 2).
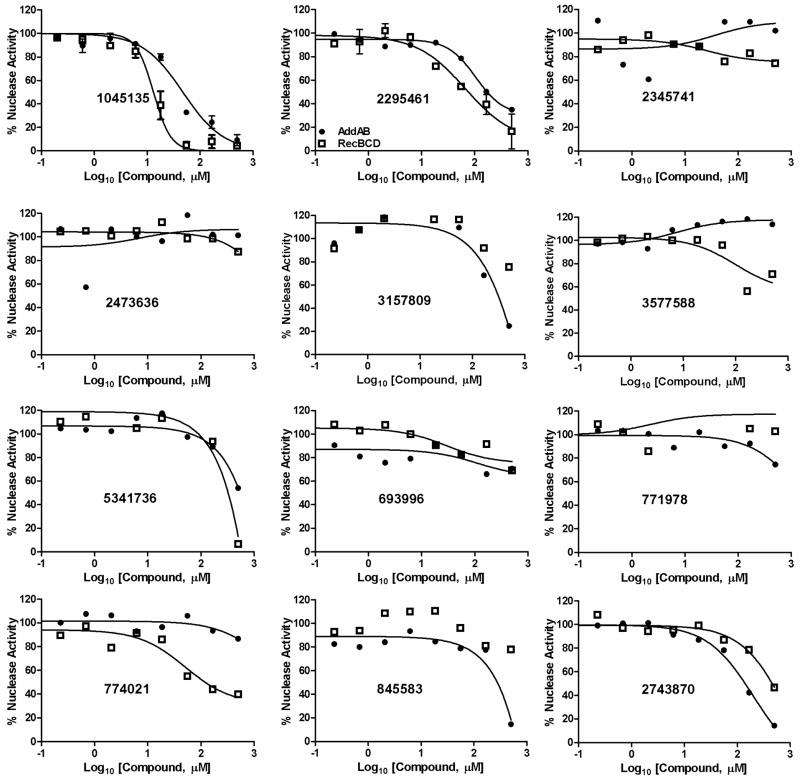
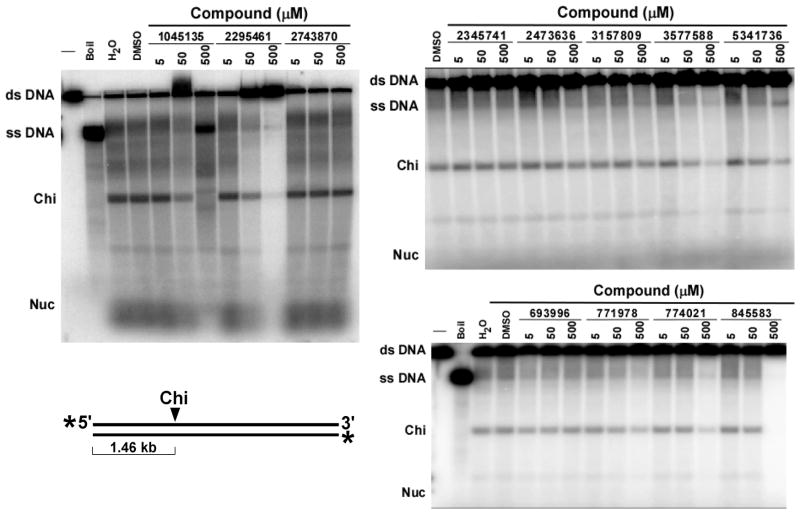
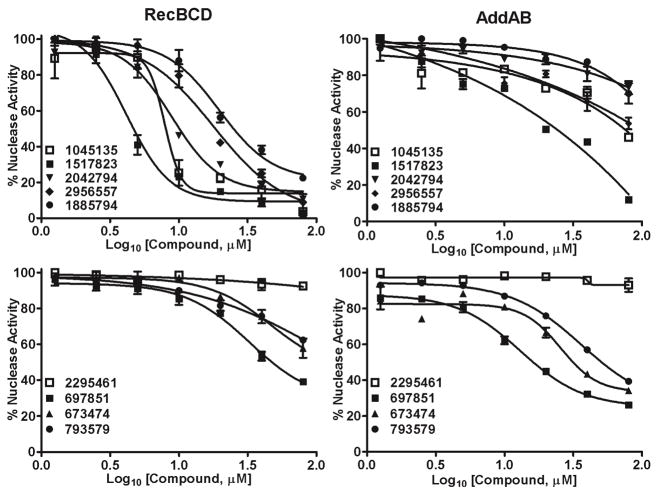
We first assayed the ability of the compounds to inhibit the ds exonuclease activity of purified RecBCD and AddAB enzymes. Compound concentrations from 0.2 μM to 500 μM were tested. IC50 values ranged from ~15 μM to >100 μM (Fig. 3 and Table 1). For both enzymes, CID 1045135 [A] (a pyrimidopyridone) and CID 2295461 [B] (a cyanothiophene) were the most potent. In helicase assays, these compounds did not significantly inhibit AddAB (Fig. S2), but several inhibited RecBCD’s helicase and Chi-cutting activities. CID 2295461 [B] inhibited both of these activities with an IC50 of ~20 μM (Fig. 4), and three nitrofurans (CID 3577588 [C], CID774021 [C], and CID 845583 [C]) inhibited with IC50 of <500 μM. CID 1045135 [A] appeared to inhibit in a biphasic way – it inhibited both helicase and Chi-cutting activities at 50 μM, but at 500 μM it appeared to stimulate the helicase and to change the position of specific cuts. Further investigations are required to determine the nature of the effect of CID 1045135 [A] on RecBCD.
Figure 3.
Inhibition of AddAB and RecBCD nuclease activities by the 12 most active compounds identified in the initial screen. ds exonuclease activity of AddAB (filled circles) and RecBCD (open squares) was measured in the presence of the indicated concentration of each compound (identified by CID number) and expressed as a percent of the activity in the absence of compound. Curves were fit by GraphPad software using the four-parameter logistic nonlinear regression model. For CID 1045135 [A] and CID 2295461 [B] data are means +/− SEM; n = 3 or 4; for other compounds data are from one experiment. Structures are in Fig. 2.
Figure 4.
Inhibition of RecBCD unwinding and Chi cutting activities by the 12 most active compounds identified in the initial screen. DNA unwinding and cutting at Chi hotspots by RecBCD enzyme was assayed in the presence of the indicated concentration of each compound (CID number shown). Unwinding is indicated by the amount of ss DNA and Chi cutting by the amount of Chi-dependent 1.46 kb ss DNA fragment (“Chi”) produced from the 4.36 kb ds DNA substrate. Structures are in Fig. 2.
Further Tests of Structurally Related Compounds
To find more potent compounds, we tested 22 compounds related to CID 1045135 [A] and 15 compounds related to CID 2295461 [B]. In the T4 gene 2 mutant-sensitization assay, used for the initial screen, derivative CID 1517823 [A] appeared RecBCD-specific and derivatives 104-29, 30, 31, and 34 appeared AddAB-specific; i. e., growth of cells with each enzyme was inhibited only if cells were infected with T4 gene 2−phage (Fig. S3). (See Fig. S4 for the structures and CID numbers corresponding to these trivial designations of group A compounds.) Derivative CID 4831 [A] inhibited growth of cells with either enzyme even without phage infection; thus, this compound appears to inhibit E. coli growth independently of RecBCD or AddAB. Derivatives 104-20, 21, 32, 33, and 35, and to a lesser extent 104-15 and16, also inhibited growth without phage infection, but only of cells with AddAB. This result may reflect the poorer growth of E. coli with AddAB than with RecBCD, perhaps because AddAB does not effectively utilize E. coli RecA but RecBCD does 25. With one exception, derivatives of CID 2295461 [B] did not greatly inhibit either enzyme in this cell-based assay; the exception was the parent compound, CID 2295461 [B], which slightly inhibited AddAB, though not as much as in the original screen at this concentration (50 μM).
In direct tests of inhibition of the purified enzymes, we found that most of the derivatives of CID 1045135 [A] inhibited RecBCD nuclease activity and several inhibited AddAB nuclease activity (Fig. S5). More derivatives may inhibit in these assays than in the cell-based assay because we were able to test a higher concentration with purified enzymes (100 μM) or because some compounds may not enter cells. Notably, however, derivative CID 1517823 [A] inhibited both enzymes in both cells-based and enzyme-based assays. Derivative CID 4831 [A] did not inhibit either purified enzyme, a result consistent with its inhibition of cell growth independent of RecBCD or AddAB (Fig. S3). Thus, these results validate the cell-based screen.
Three derivatives of CID 2295461 [B] (229-04, 05, and CID 697851 [B]) inhibited the nuclease activity of both of the purified enzymes with IC50 of <100 μM (Figs. 5 and S5), although they did not significantly inhibit in either cell-based assay using T4 gene 2 mutant infection. (See Fig. S6 for the structures and CID numbers corresponding to these trivial designations of group B compounds.) Thus, these compounds may not effectively enter E. coli cells. From these nuclease assays, compounds CID 1517823 [A] and CID 697851 [B] appear to be the most potent inhibitors identified in our screen. The IC50 of CID 1517823 [A] is ~5 μM for RecBCD and ~25 μM for AddAB, and that of CID 697851 [B] is ~30 μM for RecBCD and ~15 μM for AddAB (Fig. 5).
Figure 5.
Inhibition of AddAB and RecBCD nuclease activities by derivatives of CID 1045135 [A] and CID 2295461 [B] (dose responses). ds exonuclease activity was measured in the presence of the indicated concentration of each compound and expressed as a percent of the activity in the absence of compound. Structures are in Figs. 2, S4, and S6.
Assays of the RecBCD helicase and Chi-cutting activities showed that these two compounds inhibited these activities much like the parent compounds: CID 1517823 [A] inhibited in a biphasic way and CID 697851 [B] in a monophasic way (IC50 of ~5 and ~30 μM, respectively); neither compound inhibited AddAB unwinding activity (AK, unpublished data).
Inhibition of Intracellular Recombination
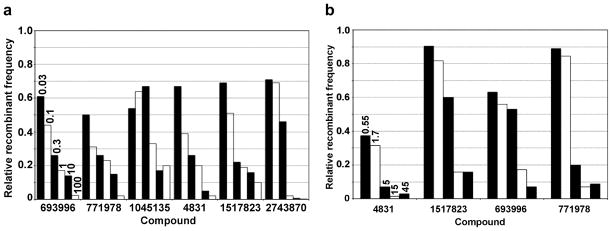
To explore further the ability of these compounds to inhibit RecBCD or AddAB in cells, we assayed the ability of the compounds to inhibit E. coli Hfr-based recombination and phage λ recombination. When tested at 100 or 200 μM, seven of the initial 12 compounds reduced Hfr recombinant frequencies by less than a factor of 2, but the remaining five compounds and four of the derivatives inhibited more, by factors up to ~200 (Table 1 and Figs. 6a and S7). For six of these latter nine, dose-response assays showed that CID 693996 [C], CID 771978 [C], CID 1045135 [A], CID 4831 [A], CID 1517823 [A], and CID 2743870 [D] inhibited with IC50 of <1 μM (Fig. 6a).
Figure 6.
(a) Inhibition of E. coli Hfr recombination by selected compounds. The frequency of His+ StrR recombinants in matings between strains V66 (F−recBCD +hisG4 rpsL +) and V1306 (Hfr PO44 rpsL31 his+) in the presence of compound (concentration in μM as indicated for CID 693996 [C] applies to all compounds) is expressed as a fraction of that in the absence of compound (9.3 % per viable Hfr cell). Structures are in Figs. 2, S4, and S6. Data are from one experiment; similar results were obtained in two others. (b)Inhibition of phage λ recombination by selected compounds. The mean frequency of J+ R+ recombinants in λ crosses (1081 × 1082 and 1083 × 1084) in strain V66 in the presence of the indicated compound (concentration in μM as indicated for CID 4831 [A] applies to all compounds) is expressed as a fraction of that without compound (6.9 ± 0.25 %; n = 4). Structures are in Figs. 2, S4, and S6.
For CID 693996 [C], CID 771978 [C], and CID 4831 [A] this outcome was surprising, since these compounds did not significantly affect RecBCD nuclease, unwinding or Chi-cutting activities (Figs. 3, 4, and S5). These compounds at ≥40 μM inhibit cell growth without T4 gene 2 mutant infection (Fig. S3; Table 1) and may inhibit some enzyme other than RecBCD required for recombination. For example, DNA gyrase, which is required for RecBCD pathway recombination 30 is inhibited by pyrimidopyridones such as CID 4831 [A] (pipemidic acid) 31, 32. In contrast CID 1045135 [A], CID 1517823 [A], and CID 2743870 [D] inhibited Hfr recombination with IC50 of <1 μM (Fig. 6a), as well as RecBCD nuclease, unwinding, or Chi-cutting activities, albeit at higher IC50 (Figs. 3, 4, 5, and S5). CID 1045135 [A] also inhibited Hfr recombination when AddAB replaced RecBCD in E. coli cells: 20 μM CID 1045135 [A] reduced the recombinant frequency to that of cells lacking AddAB or RecBCD (0.02% His+ StrR recombinants per Hfr donor cell), although this reduction was by a factor of only ~5 since AddAB does not confer as high recombination-proficiency as RecBCD 25. Inhibition by additional compounds of AddAB-dependent Hfr recombination was not tested because of instability of strains expressing both H. pylori addAB and recA from plasmids.
To determine if inhibition of Hfr recombination was specific to RecBCD, we tested the six compounds used in Fig. 6a for inhibition of Hfr recombination by two alternative pathways, called RecE and RecF, that are activated by mutations (sbc) that suppress the recombination-deficiency of recB recC null mutants 33. Each compound inhibited recombination by the RecBCD (wild-type) pathway to a greater extent than recombination by the RecE or RecF pathway (Table 2). Two compounds, CID 2743870 [D] and CID 4831 [A], significantly inhibited recombination by the latter pathways, by factors of 6 – 13, but the other four compounds inhibited recombination by these pathways only marginally if at all (by factors of ≤3). Thus, these data suggest that these six compounds are specific to RecBCD enzyme. They may, however, inhibit instead an enzyme required more stringently by the RecBCD pathway than by the other pathways, or both that enzyme and RecBCD. For example, DNA gyrase, which is inhibited by CID 4831 [A] 31, 32, may be the target of CID 4831 [A], as suggested above. Inhibition of another component, such as RecA protein, required differentially, at least quantitatively, may also explain the differential inhibitions seen by these compounds. Nevertheless, the effects of CID1045135 [A], CID 1517823 [A], CID 693996 [C], and CID 771978 [C] on purified RecBCD (Figs. 3, 4, 5, and S5) and on intracellular recombination (Tables 1 and 2; Figs. 6a and S7) suggest that at least these four compounds inhibit recombination by RecBCD in cells.
Table 2.
Pathway specificity of Hfr recombination inhibitors
| Pathway | Genotype | Fold-reduction in recombinant frequencya | ||||||
|---|---|---|---|---|---|---|---|---|
| Null mutantb | 2743870 [D] | 1045135 [A] | 4831 [A] | 1517823 [A] | 693996 [C] | 771978 [C] | ||
| RecBCD | recBCD+ | 1000 | 450 | 6.9 | 300 | 9 | 14 | 13 |
| RecF |
recB21 recC22 sbcB14, C(D) |
1000 | 13 | 1.4 | 13 | 1.8 | 1.0 | 1.2 |
| RecE |
recB21 recC22 sbcA23 |
100 | 11 | 2.0 | 5.6 | 1.8 | 2.7 | 3.1 |
Recipient strains were grown and mated with donor strain V1306 (Hfr PO44) in LB plus the indicated compound (100 μM). Structures are in Figs. 2 and S4. Data are the mean factor of reduction (n = 3) in recombinant frequencies compared to those of the untreated control, which were for V66 (recBCD+) 4.5, 10.2, and 11.1 %; for JC9387 (recB21 sbcB14 sbcC or D) 1.3, 2.2, 1.9%; and for JC8679 (recB21 sbcA23) 2.2, 2.1, 1.8%. SEM or range was <20% of the mean (n = 2 or 3). Strain JC9387 (recB21 sbcB14) presumably also carries an sbcC or sbcD mutation 52.
The factor of reduction in the frequency of recombinants observed in crosses between an Hfr donor strain and a recombination-deficient null mutant compared to that in the corresponding rec+ parent: for the RecBCD pathway V67 (recB21) compared to V66 37; for the RecF pathway JC8111 (recF143) compared to JC9387 53; for the RecE pathway N2510 (recN262) compared to JC8679 54.
To extend the intracellular assays, we tested the effects of the compounds on phage λ recombination dependent on RecBCD (the phages are red gam mutants). The most interesting results were obtained with CID 693996 [C], CID 771978 [C], CID 4831 [A], and CID 1517823 [A], which inhibited λ recombination with IC50 of <15, 5, 0.6, and 5 μM, respectively (Fig. 6b). These compounds also inhibited RecBCD-dependent Hfr recombination, as noted above. In λ crosses, the activity of the Chi hotspot, which regulates RecBCD activity 3, 4, 5, is measured as the ratio of the recombinant frequency in a genetic interval with Chi to that in the same interval without Chi; this ratio is ~5 in wild-type E. coli and 1 in recBCD null mutants, meaning that Chi is inactive in the absence of RecBCD 34. Three compounds, CID 4831 [A], CID 1517823 [A], CID 2743870 [D], significantly lowered Chi hotspot activity to a value of ~3 (Table 1). Since RecBCD directly interacts with Chi 6, this result suggests that the compounds directly inhibit RecBCD, but indirect effects on a step of recombination after that stimulated by Chi cannot be ruled out.
Another Class of Inhibitors Identified by Similarity to Other Helicase Inhibitors
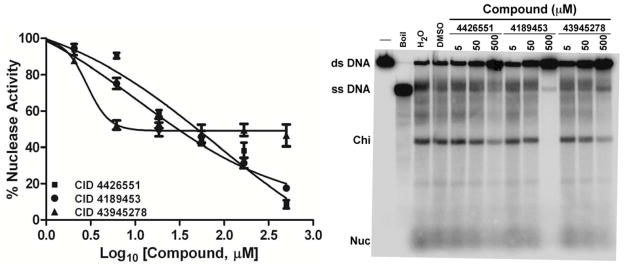
Large-scale screens for three DNA helicases have been reported on PubChem: AID 485395 for Staphylococcus aureus replicative helicase 35, AID 1945 for hepatitis C virus helicase NS3 helicase, and AID 2549 for human RecQ-like helicase 1. We noted that one class of compound, here called “iminobenzothiazoles,” appeared to be common among the reported inhibitors. We therefore tested 60 such compounds in a library from Life Chemicals. We tested these for inhibition of RecBCD nuclease activity and found three with IC50 of <100 μM (Figs. 7 and S8), but their dose responses were unexpected. CID 43945278 [E] detectably inhibited at concentrations as low as ~2 μM; inhibition was ~50% at ~5 μM and remained at that level at concentrations as high as 500 μM. As their concentrations were raised, CID 4426551 [E] and CID 4189453 [E] inhibited more gradually than expected for single-site inhibition: activity decreased from ~90% to ~10% over a range of ~2.5 log10 rather than over one log10 as expected for single-site inhibition.
Figure 7.
Inhibition of RecBCD nuclease, unwinding, and Chi cutting activities by “iminobenzothiazoles” [group E]. ds exonuclease activity (left panel) and unwinding and Chi cutting activities (right panel) were measured in the presence of the indicated concentration of each compound. ds exonuclease activity is expressed as a percent of the activity in the absence of compound. Unwinding is indicated by the amount of ss DNA, and Chi cutting by the amount of Chi-dependent ss DNA fragment (“Chi”). Structures are in Fig. 2.
These curious results may reflect differential inhibition of the two helicases in RecBCD, since DNA unwinding by both helicases is essential for full nuclease activity 22, 36. CID 43945278 [E] may inhibit only one of the helicases, with IC50 of ~2 μM, and this helicase may be responsible for only half of the nuclease activity measured. The other two compounds may inhibit this helicase with IC50 of ~10 μM and the other helicase with IC50 of ~100 μM, so that nuclease activity is inhibited only gradually as the concentration is raised. Further experiments are required to test this hypothesis.
DNA unwinding and Chi-cutting activities were also inhibited by these three compounds (Fig. 7). The IC50 values, most readily quantified for Chi cutting, were ~50 μM. In cell-based assays (sensitization to T4 gene 2 mutant infection and Hfr-dependent recombination) these compounds were inactive at the concentration tested (100 μM; Table S7), which may explain why this class of compounds was not identified in the initial screens.
Advantages of a cell-based screen
By using a cell-based assay, we would obviously study only compounds that entered E. coli cells sufficiently readily to inhibit the target enzyme, either the native RecBCD enzyme or the AddAB enzyme expressed from the H. pylori genes. In addition, these compounds must inhibit the enzyme in its natural environment, which might be markedly different from that of conditions normally employed to study the purified enzymes. Using these stringent criteria, we identified only about a dozen effective inhibitors from the 326,100 compounds initially tested, and four structurally different molecules (“scaffolds”) were identified. For two of these, we studied structurally related compounds and found six more potent inhibitors. Further structure-activity relationships (SAR) may reveal additional, highly potent inhibitors.
The assay used here is simple and inexpensive, since it employs only bacteria and phage, which are readily grown in large quantities, and reliable: Z′ factors of ~0.9 were routinely observed (Table S2). In principle this assay is specific for RecBCD or related nucleases, such as AddAB, since activity of the critical reagent used – phage T4 gene 2 mutants’ lysis of E. coli cells (Fig. 1) – is detected only in recBCD mutants 20. As anticipated, we found compounds that inhibit AddAB or RecBCD or both. The most potent compounds, CID 1045135 [A] (a “pyrimidopyridone”) and CID 2295461 [B] (a “cyanothiophene”), inhibited the ds exonuclease, DNA unwinding, and Chi-cutting activities of RecBCD and the ds exonuclease activity of AddAB (Figs. 3, 4, 5, and S5). Under the conditions used, neither compound inhibited the unwinding activity of AddAB (Fig. S2), but more sensitive assays under other conditions may reveal inhibition of AddAB unwinding activity. Thus, this screen indeed revealed compounds of the desired type.
An unexpected result of our screen, however, was the identification of CID 2743870 [D], which strongly inhibits Hfr recombination but appears to have little effect on purified RecBCD enzyme. Although Hfr recombination was inhibited at IC50 of ~0.3 μM (Fig. 6a), the nuclease was inhibited only at about 1000-fold higher concentration (IC50 of ~200 μM; Fig. 3) and unwinding and Chi-cutting were not detectably inhibited even at 500 μM (Fig. 4). Although differences in the cellular conditions vs. those used with the purified enzyme may account for this marked disparity, CID 2743870 [D] may affect another component required for Hfr recombination, since it also significantly inhibits Hfr recombination by the RecE and RecF pathways (Table 2). For example, it may affect RecA protein loading onto DNA or its DNA strand exchange activity, since RecA is required for all three pathways of recombination 33. The partial inhibition of Chi hotspot activity (Table 1) might also reflect inhibition of RecA, because RecA is required for the Chi–stimulated RecBCD pathway 33. If indeed CID 2743870 [D] does not inhibit RecBCD, its identification in our screen must be fortuitous, because RecA is not needed to block the growth of phage T4 gene 2 mutants 37, the basis of our screen. We note that CID 2743870 [D] has been identified as an active compound in many screens listed on PubChem and may inhibit many cellular components.
Identification of Potent Inhibitors of AddAB and RecBCD Enzymes
By direct nuclease assays in the presence of ~20 compounds structurally related to each of the two initial compounds CID 1045135 [A] and CID 2295461 [B], we found more potent inhibitors. In our view, the most interesting are CID 697851 [B] (most active against AddAB) and CID 1517823 [A] (most active against RecBCD); their structures are in Fig. 2. CID 697851 [B] inhibits AddAB nuclease with an IC50 of ~15 μM, but like its parent compound CID 2295461 [B] it does not detectably inhibit AddAB unwinding activity under the conditions used (Figs. 5, S2, and S5; AK, unpublished data).
Like its parent compound CID 1045135 [A], compound CID 1517823 [A] inhibited all of the activities of RecBCD tested, both with purified enzyme and with cell-based recombination assays (Figs. 3, 4, 5, 6, S5, and S7; Tables 1 and 2). IC50 values were ~3 μM for nuclease and Chi-cutting with purified enzyme (AK, unpublished data) and ~0.3 μM for Hfr recombination and ~5 μM for phage λ recombination promoted by RecBCD; it also significantly reduced Chi hotspot activity in λ crosses (Table 1). It only marginally inhibits recombination by the E. coli RecE and RecF pathways, which do not employ RecBCD (Table 2). In the T4 gene 2 mutant screen, it only slightly inhibits E. coli growth in the absence of phage but strongly inhibits in the presence of phage (Fig. S3). These observations indicate that CID 1517823 [A] specifically inhibits RecBCD in cells. We expect this compound would have been detected in our initial screen of 326,100 compounds had it been in that library. Thus, compound CID 1517823 [A] has the properties that we expected for a specific inhibitor of RecBCD enzyme and is a strong validation of our screen.
Unexpected properties of RecBCD inhibitors CID 1045135 and 1517823
We were surprised that the IC50 values of these compounds were ~10 times lower in the intracellular assays for Hfr recombination than in assays with purified enzyme. This result may reflect differences in the enzyme’s environment during the assays, or it may reflect some activity of RecBCD not yet assayed, such as the loading of RecA protein after action at Chi 38, that is even more sensitive to inhibition than the nuclease, DNA unwinding, and Chi-cutting. In any case, this result encourages the possibility that these or related compounds may be effective antibacterial drugs, as discussed below.
The biphasic dose-response curve for inhibition of DNA unwinding and Chi-cutting by CID 1045135 [A] suggests a complex interaction between this compound and RecBCD enzyme. At ~50 μM compound, unwinding and Chi-cutting were partially inhibited, but at ~500 μM unwinding appeared to be stimulated and DNA was cut at novel positions (Fig. 4). Since RecBCD has two helicases and a nuclease involved in these activities, the compound may have differential effects on two or more primary activities. For example, one helicase may be simply inhibited at low concentrations, and the other stimulated or altered at high concentrations. The cutting of DNA at novel positions is reminiscent of the behavior of RecBCD enzyme from two mutants altered in the RecB helicase domain, recB2732 (Y803H) and recB2734 (V804E). These mutant enzymes cut not at Chi but at a position that depends on the length of the DNA substrate 39. We hypothesize that the RecB nuclease cuts wherever it is on the DNA when the faster helicase, RecD, reaches the end of the substrate, and that the recB mutations sensitize the enzyme to a signal from Chi through RecC to stop RecD when RecBCD encounters Chi. CID 1045135 [A] may similarly sensitize the enzyme to signaling between RecD and RecB. In any case, further studies of the effects of CID 1045135 [A] and related compounds, such as CID 1517823 [A], may help elucidate how this complex enzyme works.
Searching for more effective inhibitors related to CID 1045135 [A], we tested CID 4831 [A], which has the pyrimidopyridone part but not the benzene ring part of CID 1045135 [A] (Fig. 2). CID 4831 [A] did not detectably inhibit the nuclease activity of purified AddAB or RecBCD enzyme (Fig. S5), but it strongly inhibited Hfr and phage λ recombination (Figs. 6 and S7). In the T4 gene 2 mutant assay it inhibited E. coli growth with or without addition of phage (Fig. S3). These results indicate that CID 4831 [A] inhibits some cellular component other than RecBCD, such as DNA gyrase, which is inhibited by CID 4831 [A] (pipemidic acid) 31, 32 and is required for cell growth 40 and for recombination 30. These facts suggest, in turn, that the benzene ring part of CID 1045135 [A] is critical for inhibition of RecBCD. Indeed, CID 1517823 [A], the m-trifluoromethyl isomer of CID 1045135 [A], is more potent than the o-trifluoromethyl parent compound in assays of both RecBCD and AddAB nuclease (Fig. 5). The results of this limited SAR survey suggests that potent inhibitors of both RecBCD and AddAB nuclease can be identified, an outcome important for preparing broad-spectrum antibiotics (see below).
Mode of Action of Inhibitors
Our primary screens were based on inhibition of the nuclease activity of AddAB or RecBCD, but because the nuclease activity depends on the helicase and ATPase activities, the inhibitors could act on any of these activities. None of the inhibitors significantly affected the AddAB helicase activity under the conditions used (Fig. S2). Although further experiments, including ATPase assays, are required to test more rigorously an effect on AddAB helicase, the inhibitors of AddAB nuclease activity, such as CID 2295461 [B] and its derivatives (Figs. 3, 5, and S5), may directly inhibit the nuclease. For RecBCD, the most potent inhibitors of the nuclease activity also inhibited the unwinding activity (Table 1), suggesting that these inhibitors (CID 1045135 [A], CID 1517823 [A], CID 2295461 [B], and CID 697851 [B]) may act primarily on the helicase or ATPase. In contrast, three compounds (CID 771978 [C], CID 5341736 [C], and CID 3577588 [C]) appear to inhibit the helicase but not the nuclease activity of RecBCD. This result may reflect the 200-fold higher ATP concentration used for the helicase assay than for the nuclease assay. Determining the primary target of the inhibitors will require more investigation, including the isolation of mutant enzymes resistant to the inhibitors. The requirement for RecBCD for growth of certain E. coli mutants, such as ligA, polA, and rnh 41, 42 may enable isolation of resistant mutants and thereby determine, with the aid of the crystal structure of RecBCD 43, the site of action of the inhibitors.
Potential for Identification of Novel Antibacterial Drugs
Inhibitors of AddAB and RecBCD are potentially useful antibacterial drugs for two reasons. First, these enzymes are required for repair of DNA damage inflicted upon bacteria by their host cells upon infection. Salmonella recB mutants are unable to kill a mouse 44, 45, and H. pylori addAB mutants less effectively colonize the mouse stomach than wild type 10. Second, RecBCD, and perhaps AddAB, is required for the induction of the SOS response to DNA damage, which includes the induction of mutagenic polymerases responsible for most induced mutations 46. Inhibition of RecBCD should thus lessen the evolution of bacteria resistant to the inhibitor, an important goal in current antibacterial drug therapy. The AddAB and RecBCD class of enzymes is found in ~90% of all bacterial species whose genomes have been sequenced and reported 47. Thus, their inhibitors may be effective broad-spectrum antibiotics. CID 1045135 [A] and its derivatives may be especially effective, since they contain the pyrimidopyridone moiety that inhibits DNA gyrase and leads to dsDNA breaks whose repair requires RecBCD enzyme 45. A derivative that inhibits both gyrase and RecBCD would thus lead to both DNA damage and the failure to repair it. Such a compound might be a single-molecule combination drug more effective than two drugs. The screen described here may be useful in finding such a drug.
METHODS
Bacterial and Phage Strains
The E. coli strains used are listed in Table S3, and phage λ strains in Table S4. Phage T4 wild type and gene 2 amN51 mutant are from our collection 37. A derivative bearing three adjacent nonsense mutations is described below. Stocks of T4 phage were grown in strain V67, which lacks RecBCD and does not suppress the nonsense mutation(s) so that the phage particles contain DNA not protected by gene 2 protein.
Plasmids and Oligonucleotides
Plasmids are listed in Table S5, and oligonucleotides in Table S6.
Bacterial Growth Media
Luria-Bertani (LB) broth contains 1.0% Tryptone (Difco), 0.5% yeast extract (Difco), and 0.5% NaCl. (All percentages in this paragraph are w/v.) LB agar is LB broth with 1.5% agar (Difco). Cation-adjusted Mueller-Hinton broth was purchased from Becton-Dickinson. TB contains 1.0% Tryptone and 0.5% NaCl; for phage λ crosses, 0.1% maltose was added. BBL top and bottom agar contain TB with 0.75% and 1% agar, respectively. BBL-YE is BBL bottom agar supplemented with 0.2% yeast extract. For growth of strains with plasmids, media were supplemented with ampicillin (100 μg/ml) or chloramphenicol (15 μg/ml). Cultures were grown at 37 °C.
Compound Libraries and Additional Compounds
For 1536-well screening, the Molecular Libraries Small Molecule Repository (MLSMR) library was provided by the NIH’s Roadmap: Molecular Libraries Initiative. Details regarding compound selection for this library can be found online at http://mli.nih.gov/mli/compound-repository/mlsmr-compounds/. The MLSMR library is a highly diversified collection of small molecules, with more that 50% of compounds having molecular weights between 350 and 410 g/mol. These compounds comprise both synthetic and natural products, from either commercial or academic sources, that can be grouped into three categories: (1) specialty sets of known bioactive compounds, such as drugs and toxins, (2) focused libraries aimed at specific target classes, and (3) diversity sets covering a large area of the chemical space. Powders of compounds in Table 1 were obtained from ChemBridge, except for Chemical Identifier (CID) 693996, 4831, and 1517823 (Vitas-M lab), 2473636 and 2345741 (Enamine), 2743870 (Maybridge), 3577588 (ChemDiv), and 4189453, 4426551, and 43945278 (Life Chemicals). An additional library of ~18,400 compounds from Life Chemicals (Burlington, Canada) was a generous gift of Kineta, Inc. (Seattle, WA).
RecBCD and AddAB Enzyme Assays
Nuclease assays measured the formation of TCA-soluble radioactive material from phage T7 [3H] DNA (2 μg/ml; 6 μM nucleotides) substrate in a 20 min incubation at 37 °C 48. AddAB assays were in 50 mM Tris-HCl (pH 8.5), 10 mM MgCl2, polyvinylpyrrolidone (1 mg/ml), 1 mM DTT, and 50 μM ATP. RecBCD assays used the same condition but with 25 μM ATP. Compounds were diluted in DMSO and added to enzyme in assay buffer on ice; final DMSO concentration was 5.0% (v/v) in each assay. DNA substrate was added, and after <5 min the reactions were started by transferring the samples to 37 °C. Reactions were stopped by addition of calf thymus DNA to 0.2 mg/ml and TCA to 5% (w/v). After 10 min on ice, the mixtures were centrifuged for 5 min at 16,100 × g, and the soluble radioactive material determined in a scintillation counter.
Helicase assays measured the formation of ss DNA from 5′ [32P] pBR322 χ+F (or χo control) DNA (0.1 nM molecules) linearized by digestion with HindIII enzyme. AddAB assays were in 25 mM Tris-acetate (pH 7.5), 2.0 mM Mg(OAc)2, 5 mM ATP, 1.5 μM SSB and used 1 nM enzyme. RecBCD assays used the same condition but with 0.15 nM enzyme and without SSB. Compounds were added to the reaction mixture containing all the reagents except ATP; final DMSO concentration was 2.5% (v/v) for each assay. (All other percentages in this paragraph are w/v.) Reactions were started by addition of ATP and were at 37 °C for 1 min (RecBCD) or 2 min (AddAB). Reactions were stopped by addition of 1/3 vol of stop buffer (2.5% SDS, 100 mM EDTA, 0.125% bromophenol blue, 0.125 % xylene cyanol FF, and 10% Ficoll), and the products subjected to electrophoresis in a 1.25% agarose gel at 5 V/cm for 2.5 h in TAE buffer (40 mM Tris base, 20 mM acetic acid, 1 mM EDTA). Gels were dried under vacuum, and the products detected by autoradiography or analyzed with a Typhoon Trio PhosphorImager (GE Lifesciences) and ImageQuant TL software (Amersham). With RecBCD, this assay also detects cutting of DNA at the Chi site χ+F, which produces a 1.46 kb fragment 49.
Genetic Assays
E. coli Hfr recombination, phage λ recombination, and Chi hotspot activity assays were conducted as described 37.
To test for inhibition of Hfr recombination, recipient cells were grown in LB to an optical density (OD) of 0.25 at 650 nm. Compound was added and incubation continued until the OD reached 0.5 (typically 30 to 45 min later). An aliquot was mixed with donor strain V1306 (Hfr PO44) in the ratio of one donor cell per ten recipient cells. After 8 min, the mixture was diluted 1:50 into LB with compound. After 20 additional min, cells were vortexed to separate mating pairs, diluted, and plated to select recombinants. Viability of the cells was not significantly affected by compounds during this 1.5 h incubation.
To test for inhibition of phage λ recombination and Chi hotspot activity, E. coli strain V66 (recBCD+)was grown as above except in TB plus 0.1% (w/v) maltose. Cells were infected with λ phages at an MOI of 5 each; cross 1 was λ 1081 × λ 1082, and cross 2 was λ 1083 × λ 1084. After 15 min, cells were diluted 1:100 into TB with compound at the same concentration, incubated at 37 °C for 90 min, and treated with chloroform. Phage were titered on strain 594 (sup+) for J+ R+ recombinants and on strain C600 (supE44) for total phage. Chi hotspot activity was measured as √(T1/C1)/(T2/C2), where T1/C1 is the ratio of turbid (c+) to clear (cI857) J+ R+ recombinants in cross 1 and T2/C2 is the same for cross 2 34.
Construction of T4 gene 2 am149 Triple Nonsense Mutant Phage
Phage T4 gene 2, including 851 bp 5′ and 842 bp 3′ of the ORF, was amplified from a lysate of phage T4 gene 2 amN51 in a DNA polymerase chain reaction using oligonucleotides OL2636 and OL2637, Platinum Taq Polymerase (Invitrogen), and the manufacturer’s suggested conditions. The product was purified on a QIAquick column (Qiagen), digested with HindIII (New England Biolabs), and ligated into HindIII-cleaved pBR322 to yield plasmid pSA520. The sequence of gene 2 in this plasmid was that of wild type except for 5′ TAG 3′ at codon 247 (5′ TGG 3′ in wild type). Two additional nonsense mutations were introduced into this gene at codons 248 (5′ GCG 3′ → 5′ TAG 3′) and 249 (5′ AAC 3′→ 5′ TAA 3′) using a QuikChange reaction (Statagene-Agilent Technologies) and oligonucleotides OL2652 and OL2653 to yield plasmid pSA524. Strain V67 (recB21) transformed with this plasmid was grown in TB; ~1 × 106 cells were embedded in BBL top agar on an LB agar plate, and ~1 × 105 T4 wild-type phage spotted on this lawn. After overnight incubation at 37 °C, phage were harvested, diluted, and plated on strain V67. About 100 small plaques, from a total of about 600 plaques, were transferred with toothpicks to a lawn of V67 and to a lawn of strain V66 (recB+). Phage that grew on V67 but not on V66, about 10% of the total tested, were plaque-purified, grown in V67, and confirmed to contain the expected triple non-sense mutations. This complex mutation is designated gene 2 am149.
Compound Screen in 96-well Format
A fresh overnight culture of strain V66 in LB was diluted 1:100 into LB broth with 0.1% (v/v) DMSO and grown with aeration at 37o to an OD650 of 0.05. Each well of a 96-well plate (Costar, Corning Inc.) was prepared by adding 10 μl of compound in 20% (v/v) DMSO or the appropriate control (LB with 20% (v/v) DMSO) and then 100 μl of the bacterial culture (containing about 2.5 × 106 cells) or uninoculated medium was added. The OD650 of each well was read on a VERSAmax microplate reader (Molecular Devices), and the plates were incubated without shaking at 37 °C. After about 1 h the OD650 of the cultures was approximately 0.1, and 10 μl of a phage suspension containing 5 × 104 phage or 2.5 μg of chloramphenicol was added to the appropriate wells. The plates were incubated for approximately 20 h and the OD650 determined.
Compound Screen in 1536-well Format
These assays, including assay statistics, are summarized in Table S2. Detailed assay protocols and data generated for the high-throughput screen (HTS) and probe development are found at PubChem AID 449731: pubchem.ncbi.nlm.nih.gov/assay/assay.cgi?aid=449731.
AddAB (Strain V3065) HTS assay
All reagents were purchased from Sigma unless noted otherwise below. Prior to assay, strainsV3065 (addAB+) and V3069 (vector control) were grown at 37 °C to an OD600 of 0.05 (~2.5 × 107 cfu/mL). Three μL of assay buffer containing glycerol (0.1% v/v) and ampicillin (100 μg/mL) (Fisher) in Cation-adjusted Mueller Hinton Broth (Becton-Dickinson) were added to each well of a 1,536-well clear-bottom plate (Aurora, Nexus Biosystems). Sixty nL of test compound (12 μM final concentration), ciprofloxacin (0.95 μg/mL final concentration, as a control for complete inhibition,), or DMSO alone (1.2% v/v final concentration) were added to the appropriate wells; compounds and ciprofloxacin were in DMSO. One μL of strain V3065 (addAB+) or strain V3069 (vector control) was dispensed into the appropriate wells, and plates were incubated for 60 min at 37 °C. One μL of phage T4 gene 2 am149 mutant was dispensed to the appropriate wells at a multiplicity of infection (MOI) of 0.02. Plates were centrifuged, and after 18 h of incubation at 37 °C the absorbance, as OD600, was read on an Envision microplate reader (PerkinElmer). All data were normalized to that of the positive control (wells containing strain V3065, ciprofloxacin, and phage) and negative control (wells containing strain V3065, DMSO, and phage). This protocol was used for primary, secondary, and titration screening assays (PubChem AID 435030, 488942, and 492957, respectively). The hit-cutoff used to qualify active compounds in the primary assay was calculated as the average percentage activity of all compounds tested plus three times their standard deviation 50. The secondary assay used the same conditions as the primary screening assay, except that plates were assessed in triplicate and results for each compound were reported as the average percentage activity of the three measurements, plus or minus the associated standard deviation. For titration experiments, assay protocols were identical to those described above, except that compounds were prepared in 10 point, 1:3 serial dilutions starting at a nominal test concentration of 120 μM, and assessed in triplicate 51.
Bacterial Viability HTS assay
This assay was identical to the AddAB (strain V3065) screening assay, except that no phage was added to the wells. All data were normalized to that of the positive control (wells containing strain V3065 and ciprofloxacin) and negative control (wells containing strain V3065 and DMSO). This protocol was used for primary, secondary, and titration screening assays (PubChem AID 449728, 488956, and 492958, respectively)
RecBCD (Strain V66) HTS assay
This assay was identical to the primary AddAB screening assay except that strains V66 (recBCD+) and V67 (recB21) replaced strains V3065 and V3069, respectively. All data were normalized to that of the positive control (wells containing strain V66, ciprofloxacin, and phage) and negative control (wells containing strain V66, DMSO, and phage). This protocol was used for secondary and titration screening assays (PubChem AID 488955, and 492957, respectively).
Supplementary Material
Acknowledgments
We are especially grateful to Julian Simon and Antonio Bedalov (Fred Hutchinson Cancer Research Center; FHCRC) for discussions of inhibitor screens and, with Gemma Park, for aiding our preliminary screens for RecBCD and AddAB inhibitors, which were supported by grants from the FHCRC Synergy Fund and the Bill and Melinda Gates Foundation Grand Challenges Explorations. We thank Pierre Baillargeon and Lina DeLuca (Lead Identification Division, Scripps Florida) for compound management, Peter Chase (Lead Identification, Scripps Florida) for assistance in screening activities, Nina Salama (FHCRC) for discussions of H. pylori, Julian Simon for comments on the manuscript, Micah Ferrel (Seattle Structural Genomics Center for Infectious Disease) for purified H. pylori AddAB enzyme, Andrew Taylor (FHCRC) for purified E. coli RecBCD enzyme, and Kineta, Inc. for a compound library from Life Chemicals. This work was supported, in part, by grants from the FHCRC Synergy Fund (GRS), Bill and Melinda Gates Foundation Grand Challenges Explorations grant 53290 (GRS), and National Institutes of Health grants R01 GM031693-S1 (GRS), R03 AI083736 (GRS), and U54 MH084512 (PH).
Footnotes
ASSOCIATED CONTENT
Supporting information. This material is available free of charge via the Internet at http://pubs.acs.org.
References
- 1.Tse YC, Kirkegaard K, Wang JC. Covalent bonds between protein and DNA. Formation of phosphotyrosine linkage between certain DNA topoisomerases and DNA. J Biol Chem. 1980;255:5560–5. [PubMed] [Google Scholar]
- 2.Gellert M, Mizuuchi K, O’Dea MH, Itoh T, Tomizawa J. Nalidixic acid resistance: A second genetic character involved in DNA gyrase activity. Proc Natl Acad Sci USA. 1977;74:4772–4776. doi: 10.1073/pnas.74.11.4772. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Smith GR. Homologous recombination near and far from DNA breaks: Alternative roles and contrasting views. Annu Rev Genet. 2001;35:243–274. doi: 10.1146/annurev.genet.35.102401.090509. [DOI] [PubMed] [Google Scholar]
- 4.Dillingham MS, Kowalczykowski SC. RecBCD enzyme and the repair of double-stranded DNA breaks. Microbiol Mol Biol Rev. 2008;72:642–71. doi: 10.1128/MMBR.00020-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Smith GR. How RecBCD enzyme and Chi promote DNA break repair and recombination– A molecular biologist’s view. Microbiol Mol Biol Rev. doi: 10.1128/MMBR.05026-11. in press. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Ponticelli AS, Schultz DW, Taylor AF, Smith GR. Chi-dependent DNA strand cleavage by RecBC enzyme. Cell. 1985;41:145–151. doi: 10.1016/0092-8674(85)90069-8. [DOI] [PubMed] [Google Scholar]
- 7.Touzain F, Petit MA, Schbath S, El Karoui M. DNA motifs that sculpt the bacterial chromosome. Nat Rev Microbiol. 2011;9:15–26. doi: 10.1038/nrmicro2477. [DOI] [PubMed] [Google Scholar]
- 8.Goldmark PJ, Linn S. Purification and properties of the RecBC DNase of Escherichia coli K-12. J Biol Chem. 1972;247:1849–1860. [PubMed] [Google Scholar]
- 9.Buchmeier NA, Libby SJ, Xu Y, Loewen PC, Switala J, Guiney DG, Fang FC. DNA repair is more important than catalase for Salmonella virulence in mice. J Clin Invest. 1995;95:1047–53. doi: 10.1172/JCI117750. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Amundsen SK, Fero J, Hansen LM, Cromie GA, Solnick JV, Smith GR, Salama NR. Helicobacter pylori AddAB helicase-nuclease and RecA promote recombination-related DNA repair and survival during stomach colonization. Molec Microb. 2008;69:994–1007. doi: 10.1111/j.1365-2958.2008.06336.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Taylor AF. RecBCD enzyme of Escherichia coli. In: Kucherlapati R, Smith GR, editors. Genetic Recombination. American Society for Microbiology; Washington, DC: 1988. pp. 231–263. [Google Scholar]
- 12.Dziegielewska B, Beerman TA, Bianco PR. Inhibition of RecBCD enzyme by antineoplastic DNA alkylating agents. J Mol Biol. 2006;361:898–919. doi: 10.1016/j.jmb.2006.06.068. [DOI] [PubMed] [Google Scholar]
- 13.Kooistra J, Haijema BJ, Hesseling-Meinders A, Venema G. A conserved helicase motif of the AddA subunit of the Bacillus subtilis ATP-dependent nuclease (AddAB) is essential for DNA repair and recombination. Molec Microb. 1997;23:137–49. doi: 10.1046/j.1365-2958.1997.1991570.x. [DOI] [PubMed] [Google Scholar]
- 14.Taylor AF, Smith GR. Unwinding and rewinding of DNA by the RecBC enzyme. Cell. 1980;22:447–457. doi: 10.1016/0092-8674(80)90355-4. [DOI] [PubMed] [Google Scholar]
- 15.Rosamond J, Telander KM, Linn S. Modulation of the RecBC enzyme of Escherichia coli K12 by Ca2+ J Biol Chem. 1979;254:8646–8652. [PubMed] [Google Scholar]
- 16.Karu AE, Sakaki Y, Echols H, Linn S. The gamma protein specified by bacteriophage lambda. Structure and inhibitory activity for the RecBC enzyme of Escherichia coli. J Biol Chem. 1975;250:7377–7387. [PubMed] [Google Scholar]
- 17.Murphy KC. The lambda Gam protein inhibits RecBCD binding to dsDNA ends. J Mol Biol. 2007;371:19–24. doi: 10.1016/j.jmb.2007.05.085. [DOI] [PubMed] [Google Scholar]
- 18.Court R, Cook N, Saikrishnan K, Wigley D. The crystal structure of lambda-Gam protein suggests a model for RecBCD inhibition. J Mol Biol. 2007;371:25–33. doi: 10.1016/j.jmb.2007.05.037. [DOI] [PubMed] [Google Scholar]
- 19.Murphy KC. Lambda Gam protein inhibits the helicase and chi-stimulated recombination activities of Escherichia coli RecBCD enzyme. J Bacteriol. 1991;173:5808–21. doi: 10.1128/jb.173.18.5808-5821.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Oliver DB, Goldberg EB. Protection of parental T4 DNA from a restriction exonuclease by the product of gene 2. J Mol Biol. 1977;116:877–881. doi: 10.1016/0022-2836(77)90276-5. [DOI] [PubMed] [Google Scholar]
- 21.Yu M, Souaya J, Julin DA. Identification of the nuclease active site in the multifunctional RecBCD enzyme by creation of a chimeric enzyme. J Mol Biol. 1998;283:797–808. doi: 10.1006/jmbi.1998.2127. [DOI] [PubMed] [Google Scholar]
- 22.Hsieh S, Julin DA. Alteration by site-directed mutagenesis of the conserved lysine residue in the consensus ATP-binding sequence of the RecB protein of Escherichia coli. Nucleic Acids Res. 1992;20:5647–5653. doi: 10.1093/nar/20.21.5647. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Amundsen SK, Taylor AF, Chaudhury AM, Smith GR. recD: The gene for an essential third subunit of exonuclease V. Proc Natl Acad Sci USA. 1986;83:5558–5562. doi: 10.1073/pnas.83.15.5558. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Masterson C, Boehmer PE, McDonald F, Chaudhuri S, Hickson ID, Emmerson PT. Reconstitution of the activities of the RecBCD holoenzyme of Escherichia coli from the purified subunits. J Biol Chem. 1992;267:13564–13572. [PubMed] [Google Scholar]
- 25.Amundsen SK, Fero J, Salama NR, Smith GR. Dual nuclease and helicase activities of Helicobacter pylori AddAB are required for DNA repair, recombination, and mouse infectivity. J Biol Chem. 2009;284:16759–66. doi: 10.1074/jbc.M109.005587. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Haijema BJ, Meima R, Kooistra J, Venema G. Effects of lysine-to-glycine mutations in the ATP-binding consensus sequences in the AddA and AddB subunits on the Bacillus subtilis AddAB enzyme activities. J Bacteriol. 1996;178:5130–7. doi: 10.1128/jb.178.17.5130-5137.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Sinha KM, Unciuleac MC, Glickman MS, Shuman S. AdnAB: a new DSB-resecting motor-nuclease from mycobacteria. Genes Devt. 2009;23:1423–37. doi: 10.1101/gad.1805709. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Miller ES, Kutter E, Mosig G, Arisaka F, Kunisawa T, Ruger W. Bacteriophage T4 genome. Microbiol Molec Biol Rev. 2003;67:86–156. doi: 10.1128/MMBR.67.1.86-156.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Wang GR, Vianelli A, Goldberg EB. Bacteriophage T4 self-assembly: in vitro reconstitution of recombinant gp2 into infectious phage. J Bacteriol. 2000;182:672–9. doi: 10.1128/jb.182.3.672-679.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Ennis DG, Amundsen SK, Smith GR. Genetic functions promoting homologous recombination in Escherichia coli: A study of inversions in phage λ. Genetics. 1987;115:11–24. doi: 10.1093/genetics/115.1.11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Zweerink MM, Edison A. Inhibition of Micrococcus luteus DNA gyrase by norfloxacin and 10 other quinolone carboxylic acids. Antimicrob Agents Chemother. 1986;29:598–601. doi: 10.1128/aac.29.4.598. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Shen LL, Pernet AG. Mechanism of inhibition of DNA gyrase by analogues of nalidixic acid: the target of the drugs is DNA. Proc Natl Acad Sci USA. 1985;82:307–11. doi: 10.1073/pnas.82.2.307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Clark AJ. Recombination deficient mutants of E. coli and other bacteria. Annu Rev Genet. 1973;7:67–86. doi: 10.1146/annurev.ge.07.120173.000435. [DOI] [PubMed] [Google Scholar]
- 34.Stahl FW, Stahl MM. Recombination pathway specificity of Chi. Genetics. 1977;86:715–725. doi: 10.1093/genetics/86.4.715. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Aiello D, Barnes MH, Biswas EE, Biswas SB, Gu S, Williams JD, Bowlin TL, Moir DT. Discovery, characterization and comparison of inhibitors of Bacillus anthracis and Staphylococcus aureus replicative DNA helicases. Bioorg Med Chem. 2009;17:4466–76. doi: 10.1016/j.bmc.2009.05.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Taylor AF, Smith GR. RecBCD enzyme is a DNA helicase with fast and slow motors of opposite polarity. Nature. 2003;423:889–893. doi: 10.1038/nature01674. [DOI] [PubMed] [Google Scholar]
- 37.Schultz DW, Taylor AF, Smith GR. Escherichia coli RecBC pseudorevertants lacking Chi recombinational hotspot activity. J Bacteriol. 1983;155:664–680. doi: 10.1128/jb.155.2.664-680.1983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Anderson DG, Kowalczykowski SC. The translocating RecBCD enzyme stimulates recombination by directing RecA protein onto ssDNA in a χ regulated manner. Cell. 1997;90:77–86. doi: 10.1016/s0092-8674(00)80315-3. [DOI] [PubMed] [Google Scholar]
- 39.Amundsen SK, Taylor AF, Reddy M, Smith GR. Intersubunit signaling in RecBCD enzyme, a complex protein machine regulated by Chi hot spots. Genes Dev. 2007;21:3296–307. doi: 10.1101/gad.1605807. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Gottesman MM, Hicks ML, Gellert M. Genetics and function of DNA ligase in Escherichia coli. J Mol Biol. 1974;77:531–547. doi: 10.1016/0022-2836(73)90221-0. [DOI] [PubMed] [Google Scholar]
- 41.Kushner SR. In vivo studies of temperature-sensitive recB and recC mutants. J Bacteriol. 1974;120:1213–1218. doi: 10.1128/jb.120.3.1213-1218.1974. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Kogoma T, Hong X, Cadwell GW, Barnard KG, Asai T. Requirement of homologous recombination functions for viability of the Escherichia coli cell that lacks RNase HI and exonuclease V activities. Biochimie. 1993;5:89–99. doi: 10.1016/0300-9084(93)90029-r. [DOI] [PubMed] [Google Scholar]
- 43.Singleton MR, Dillingham MS, Gaudier M, Kowalczykowski SC, Wigley DB. Crystal structure of RecBCD enzyme reveals a machine for processing DNA breaks. Nature. 2004;432:187–93. doi: 10.1038/nature02988. [DOI] [PubMed] [Google Scholar]
- 44.Buchmeier NA, Lipps CJ, So MY, Heffron F. Recombination-deficient mutants of Salmonella typhimurium are avirulent and sensitive to the oxidative burst of macrophages. Mol Microbiol. 1993;7:933–6. doi: 10.1111/j.1365-2958.1993.tb01184.x. [DOI] [PubMed] [Google Scholar]
- 45.Cirz RT, Chin JK, Andes DR, de Crecy-Lagard V, Craig WA, Romesberg FE. Inhibition of mutation and combating the evolution of antibiotic resistance. PLoS Biol. 2005;3:e176. doi: 10.1371/journal.pbio.0030176. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.McPartland A, Green L, Echols H. Control of recA gene RNA in E. coli: regulatory and signal genes. Cell. 1980;20:731–737. doi: 10.1016/0092-8674(80)90319-0. [DOI] [PubMed] [Google Scholar]
- 47.Cromie GA. Phylogenetic ubiquity and shuffling of the bacterial RecBCD and AddAB recombination complexes. J Bacteriol. 2009;191:5076–84. doi: 10.1128/JB.00254-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Eichler DC, Lehman IR. On the role of ATP in phosphodiester bond hydrolysis catalyzed by the RecBC deoxyribonuclease of Escherichia coli. J Biol Chem. 1977;252:499–503. [PubMed] [Google Scholar]
- 49.Smith GR, Kunes SM, Schultz DW, Taylor A, Triman KL. Structure of Chi hotspots of generalized recombination. Cell. 1981;24:429–436. doi: 10.1016/0092-8674(81)90333-0. [DOI] [PubMed] [Google Scholar]
- 50.Hodder P, Cassaday J, Peltier R, Berry K, Inglese J, Feuston B, Culberson C, Bleicher L, Cosford ND, Bayly C, Suto C, Varney M, Strulovici B. Identification of metabotropic glutamate receptor antagonists using an automated high-throughput screening system. Anal Biochem. 2003;313:246–54. doi: 10.1016/s0003-2697(02)00608-5. [DOI] [PubMed] [Google Scholar]
- 51.Madoux F, Li X, Chase P, Zastrow G, Cameron MD, Conkright JJ, Griffin PR, Thacher S, Hodder P. Potent, selective and cell penetrant inhibitors of SF-1 by functional ultra-high-throughput screening. Mol Pharmacol. 2008;73:1776–84. doi: 10.1124/mol.108.045963. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Lloyd RG, Buckman C. Identification and genetic analysis of {IsbcC} mutations in commonly used recBC sbcB strains of Escherichia coli K12. J Bacteriol. 1985;164:836–844. doi: 10.1128/jb.164.2.836-844.1985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Horii Z-I, Clark AJ. Genetic analysis of the RecF pathway to genetic recombination in Escherichia coli K12: Isolation and characterization of mutants. J Mol Biol. 1973;80:327–344. doi: 10.1016/0022-2836(73)90176-9. [DOI] [PubMed] [Google Scholar]
- 54.Lloyd RG, Buckman C, Benson FE. Genetic analysis of conjugational recombination in Escherichia coli K12 strains deficient in RecBCD enzyme. J Gen Microbiol. 1987;133:2531–2538. doi: 10.1099/00221287-133-9-2531. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.