Abstract
Opioid receptors (ORs) mediate the actions of endogenous and exogenous opioids for many essential physiological processes including regulation of pain, respiratory drive, mood, and, in the case of κ-opioid receptors (KOR), dysphoria and psychotomimesis. Here we report the crystal structure of the human KOR (hKOR) in complex with the selective antagonist JDTic, arranged in parallel-dimers, at 2.9 angstrom resolution. The structure reveals important features of the ligand binding pocket that contribute to JDTic’s high affinity and subtype-selectivity for hKOR. Modeling of other important KOR-selective ligands, including the morphinan-derived antagonists nor-BNI and GNTI, and the diterpene agonist salvinorin A analog RB-64, reveals both common and distinct features for binding these diverse chemotypes. Analysis of site-directed mutagenesis and ligand structure-activity relationships confirms the interactions observed in the crystal structure, thereby providing a molecular explanation for hKOR subtype-selectivity along with insight essential for the design of hKOR compounds with new pharmacological properties.
The four opioid receptors (ORs),μ, δ, κ (MOR, DOR, KOR, and the nociceptin/orphanin FQ peptide receptor) belong to the class A (Rhodopsin-like) γ subfamily of G protein-coupled receptors (GPCRs)1 with a common seven-transmembrane (7TM) helical architecture and are coupled predominantly to heterotrimeric Gi/Go proteins; their activation by endogenous or exogenous ligands are linked to a number of neuropsychiatric sequelae including analgesia, sedation, depression, dysphoria, and euphoria2. The three closely related subtypes, MOR, DOR and KOR, share ~70% sequence identity in their 7TM domains, with more variations in the extracellular loops (ECLs) and very little similarity in their N and C termini2. The majority of endogenous opioid peptides have a defined preference to specific subtypes, for example, endorphins act via DORs and MORs, whereas dynorphins preferentially activate KORs. However, most exogenous and synthetic opioid ligands interact promiscuously (see Ki Database; http://pdsp.med.unc.edu/pdsp.php), likely due to the high degree of similarity of opioid-binding pockets. While decades of focused medicinal chemistry efforts have yielded reasonably selective ligands for all four ORs (see Ki Database), substantial interest continues for the development of subtype-selective agonists and antagonists.
Recent breakthroughs in elucidating high resolution structures of GPCRs in complex with small molecule3–7 and peptide8 ligands are providing details of their function9, leading to numerous rational ligand discovery studies10,11. However, while most of these structures belong to the α subfamily of class A GPCRs1, the highly diverse peptide-binding γ subfamily is represented only by the CXCR4 chemokine receptor8; additional structural coverage is needed to elucidate the repertoire of features12 that define the pharmacological profile of the subfamily. KOR, identified based on studies with the κ-type prototypic agonist ketocyclazocine13, represents an attractive target for structure determination. Several KOR-selective partial agonists and antagonists have been developed as potential antidepressants, anxiolytics, and anti-addiction medications14, whereas a widely abused, naturally-occurring hallucinogen Salvinorin A (SalA) was also found to be a highly selective KOR agonist15. Although many KOR agonists and antagonists have not demonstrated desirable pharmacological properties, lacking specificity or displaying frank psychotomimetic actions in humans14,16, some have shown to be viable drug candidates. A KOR ligand in advanced stages of clinical development, JDTic, ((3R)-7-hydroxy-N-[(1S)-1-(((3R,4R)-4-(3-hydroxyphenyl)-3,4-dimethyl-1-piperidinyl)methyl)-2-methylpropyl]-1,2,3,4-tetrahydro-3-isoquinoline-carboxamide), was originally designed as a novel selective KOR antagonist17 that blocks the κ-agonist U50,488-induced antinociception, while not antagonizing μ-agonist induced analgesia18. JDTic also displays robust activity in rodent models of depression, anxiety, stress-induced cocaine relapse, and nicotine withdrawal18,19. Here, we report the crystal structure of a human KOR (hKOR) construct (hKOR-T4L) in complex with JDTic at 2.9 Å resolution. The results provide structural insights into the atomic details of molecular recognition and subtype-selectivity of KOR and related ORs, and should catalyze the structure-based design of advanced hKOR agonists and antagonists with improved pharmacological profiles and enhanced therapeutic efficacies.
Overall architecture of hKOR
Structural studies were carried out using an engineered hKOR construct (see Methods and Supplementary Fig. 1) and crystallized in cholesterol-doped monoolein lipidic cubic mesophase (see Methods). The construct used displayed pharmacological behavior similar to that of a native receptor expressed in HEK 293-T cells (Supplementary Tables 2 and 3). Data collection and refinement statistics are shown in Supplementary Table 1.
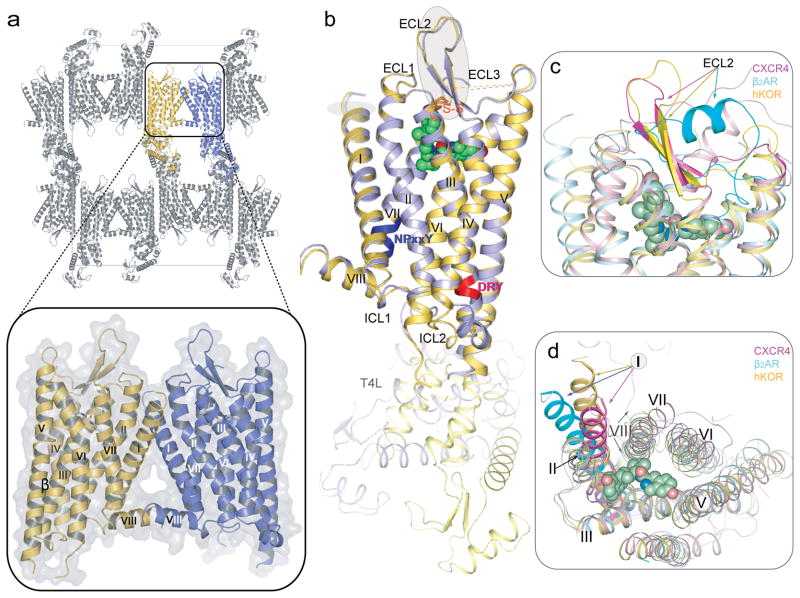
The structure of hKOR-JDTic was determined at 2.9 Å in the P 212121 space group. The asymmetric unit (ASU) consists of a two receptors forming a parallel dimer (Fig. 1a). The dimer interface with ~1100 Å2 buried surface area is formed through contacts between helices I, II and VIII (Fig. 1a, insert). Previously, parallel receptor dimers have been identified in crystal structures of activated rhodopsin (involving helices I, II and VIII)20, β2 adrenergic receptor (β2AR; cholesterol mediated)3 and CXCR4 (involving helices IV, V and VI)8. Consistent with these crystallographic data, recent biochemical studies have suggested the existence of two dimerization interfaces: along helices IV and V - sensitive to receptor activation, and along helix I – insensitive to the state of activation21. While the orientations of the two T4-lysozyme (T4L) copies in the receptor monomers in one ASU differ by ~ 60° rotation, both copies of the receptor are highly similar (Fig. 1b) and will be treated identically except where otherwise noted. The remaining residues of the N- and C-termini in the hKOR/JDTic structure are disordered as in other class A GPCRs.
Figure 1. Crystal packing and overview of the hKOR structure in complex with JDTic, and comparison with the inactive CXCR4 and β2AR structures.
(a) hKOR-T4L crystal packing. The parallel-dimer in one ASU is highlighted by insert. (b) Overall architecture of hKOR-T4L in complex with JDTic. A molecule (yellow) and B molecule (blue) in one ASU are aligned through the receptor part. The DRY and NPxxY motifs are highlighted in red and blue, respectively. JDTic is shown in a green sphere representation and the disulfide bonds are colored orange. (c) Side and (d) extracellular views of a structural alignment of hKOR (yellow); CXCR4 (PDB ID: 3ODU; magenta) and β2AR (PDB ID: 2RH1; cyan). The graphics were created by PyMOL.
The main fold of the hKOR consists of a canonical 7TM bundle of α helices followed by an intracellular helix VIII that runs parallel to the membrane. (Fig. 1a, b), resembling previously solved GPCR structures3–8. Structural comparison with other GPCRs suggests that hKOR has striking similarities in the ECL region with CXCR4, another peptide binding receptor in the γ subfamily. In the 7TM region, however, the hKOR structure is closer to aminergic receptors belonging to the α subfamily (RMSDCα ~2.3 Å for β2AR, ~1.9 Å for Dopamine D3 receptor (D3R), and ~2.7Å for CXCR4). The structure reveals distinctive features of hKOR, including: (i) conformation of the extracellular end of helix I that deviates from the position observed in CXCR4, where the tip of helix I is pulled towards the TM bundle by a disulfide bond between the N-terminus and ECL3. (ii) ECL2, the largest extracellular loop of hKOR, forms a β-hairpin similar to that observed in CXCR4, despite the low sequence similarity in this domain between the two receptors. Conservation of this feature between these peptide receptors suggests that the β-hairpin could be a common motif in the ECL2 of other γ subfamily receptors, where interactions between ECL2 and their endogenous peptide ligands are deemed important for ligand recognition and selectivity22. (iii) Unlike other solved non-rhodopsin class A GPCRs which have more than one disulfide bond, hKOR has only one formed between Cys1313.25 (superscripts indicate residue numbering using the Ballesteros-Weinstein nomenclature23) and Cys210, bridging ECL2 to the end of helix III. These two cysteines are conserved in all ORs and this disulfide bond is the canonical one shared by all other solved class A GPCRs. (iv) intracellular loop 2 (ICL2) adopts slightly different structures in the two hKOR molecules in the ASU, involving a two-turn α helix in molecule B, and only a one-turn α helix in molecule A (Supplementary Fig. 2), possibly reflecting the conformational plasticity of this region5. (v) ECL3 of hKOR is disordered. Of the approximately eleven residues in this loop (residues 300–310), six residues in molecule A and three in molecule B do not have interpretable electron density.
A common feature of the class A GPCRs is the presence of a conserved sequence motif Asp/Glu3.49-Arg3.50-Tyr3.51 (D/ERY) located at the cytoplasmic end of helix III. A salt bridge interaction between Arg3.50 and Asp/Glu6.30 from the cytoplasmic end of helix VI constitutes an “ionic lock”, which is thought to stabilize the inactive conformation of rhodopsin and other rhodopsin-like class A GPCRs5,24, while its absence can enhance constitutive activity6,23. Although hKOR lacks either of the acidic residues Asp/Glu at position 6.30, Arg1563.50 forms a hydrogen bond to another helix VI residue, Thr2736.34 (Supplementary Fig. 3a) in this inactive hKOR structure, thereby conceivably stabilizing the inactive receptor conformation. The NPxxY motif located at the cytoplasmic side of helix VII, which is composed of Asn3267.49, Pro3277.50, Ile3287.51, Leu3297.52 and Tyr3307.53 in hKOR, is another highly conserved functional motif that is proposed to act as one of the molecular switches responsible for class A GPCR activation25,26. Comparison of hKOR with inactive β2AR and A2A adenosine receptor (A2AAR) structures (Supplementary Fig. 3b) reveals a similar conformation of this motif in these receptors, thereby supporting the hypothesis that the observed hKOR-JDTic complex structure corresponds to the inactive state. To further establish that JDTic stabilizes an inactive conformation, we evaluated its ability to modulate Gi-mediated and β-arrestin-mediated signaling in transfected HEK293-T cells. We found that JDTic was devoid of agonist activity at both canonical and non-canonical pathways and completely blocked the effects of the prototypic agonist U69593 (Supplementary Fig. 4).
hKOR ligand binding pocket
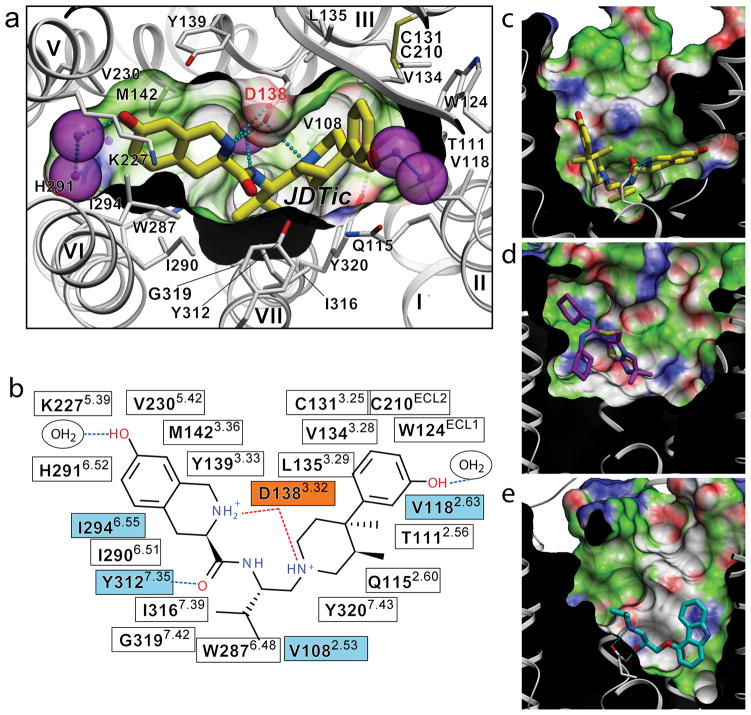
The hKOR ligand binding pocket displays a unique combination of key characteristics both shared with and distinct from those in the chemokine and aminergic receptor families. While the hKOR binding pocket is comparatively large and partially capped by the ECL2 β-hairpin, as in CXCR4, it is also much narrower and deeper than in CXCR4 (Fig. 2c, d and Supplementary Fig. 5). In addition to a different set of side chains lining the pocket, the shape differences result from an approximately 4.5 Å inward shift of the extracellular tip of helix VI in hKOR as compared to CXCR4. The electron density clearly shows the position of the JDTic ligand (Supplementary Fig. 6), which reaches deep into the pocket to form ionic interactions with the Asp1383.32 side chain (Fig. 2a). The Asp3.32 residue is conserved in all aminergic GPCRs, thereby playing a critical role in the selectivity of aminergic receptors toward protonated amine-containing ligands. Asp3.32 is conserved in all ORs and for which modeling and mutagenesis studies27 suggest an essential role in anchoring positively charged hKOR ligands.
Figure 2. Binding of the high affinity selective antagonist JDTic in the hKOR crystal structure.
(a) Conformation of the binding pocket with JDTic shown by sticks with yellow carbons. The protein is displayed in cartoon representation looking down from the extracellular side, with the 22 contact residues within 4.5 Å from the ligand shown by white sticks. The pocket surface is shown as a semitransparent surface colored according to binding properties (green: hydrophobic, blue: H-bond donor, red: H-bond acceptor). Salt bridges and hydrogen bonds are shown as dotted lines. Structured water molecules are shown as large magenta spheres. (b) Diagram of ligand interactions in the binding pocket side chains at 4.5 Å cutoff. Salt bridges are shown in red and direct hydrogen bonds in blue dashed lines. Ballesteros-Weinstein numbering is shown as superscript. Residues that vary among MOR, DOR and KOR subtypes are highlighted in cyan, and residue Asp1383.32 inferred in hKOR ligand binding by mutagenesis data, is highlighted orange. Side views of the sliced binding pocket in (c) hKOR-JDTic, (d) CXCR4-IT1t, and (e) β2AR-carazolol complexes. The pocket surfaces are colored as in panel A, the protein interior is black and the extracellular space is white. Ligands are shown as capped sticks with carbons colored yellow (JDTic), magenta (IT1t) and cyan (carazolol). Asp3.32 side chains in hKOR-JDTic and β2AR-carazolol complexes are shown by thin sticks with grey carbons. The graphics were prepared using ICM molecular modeling package (Molsoft LLC).
Structural basis of JDTic selectivity
JDTic, developed as a derivative of the trans-(3R,4R)-dimethyl-4-(3-hydroxyphenyl) piperidine scaffold17, has exceptionally high affinity (Ki = 0.32 nM), potency (Ki = 0.02 nM in GTPγS assays)17,28, long duration of action and a more than 1000-fold selectivity for hKOR as compared to other OR subtypes18. Extensive structure–activity relationship (SAR) analyses performed on JDTic analogues have yielded important insights into key determinants of JDTic activity28–30, although reliable identification of the interaction mode(s) and contact residues of these ligands has not been feasible without a receptor crystal structure.
The crystal structure of hKOR–JDTic shows a tight fit of the ligand in the bottom of the binding cleft (Fig. 2a), forming ionic, polar, and extensive hydrophobic interactions with the receptor (Fig. 2b). The protonated amines in both piperidine and isoquinoline moieties of the ligand form salt bridges to the Asp1383.32 side chain (3.0 and 3.1 Å N–O distances, respectively). The piperidine amine is a part of the original trans-(3R,4R)-dimethyl-4-(3-hydroxyphenyl)piperidine scaffold, and is essential for binding31. SAR studies of JDTic analogues show that the isoquinoline nitrogen can be replaced by carbon, oxygen or sulfur atoms with only ~10- to 50-fold reduction in affinity30. Similar to the observed JDTic conformation in the hKOR-JDTic complex, a V-shaped conformation was found in the crystal structure of JDTic by itself and which showed its amino groups coordinating a water molecule (Supplementary Fig. 7a). While several rotatable bonds within the JDTic molecule allow for the sampling of different conformations (see Supplementary Fig. 7b) and facilitate the ligand passage through the narrow binding pocket entrance, the anchoring-type interaction of two amino groups with Asp1383.32 likely fixes the ligand in this characteristic V-shape.
SAR studies have also underscored the importance of the distal hydroxyl groups on both the piperidine and isoquinoline moieties of JDTic, the removal of which did result in about a 100-fold reduction of affinity. A much smaller effect was observed upon methylation of these hydroxyls or their replacement by other polar groups28. These SAR results suggest the importance of water-mediated interactions between these two hydroxyl groups and the receptor. Indeed, while the crystal structure does not show direct hydrogen bonding with the receptor for both hydroxyl groups, there is clear electron density for several structured water molecules that mediate their polar interactions (Supplementary Fig. 6).
The structure provides important clues for understanding the structural basis for the exceptional subtype selectivity of JDTic. Among many extensive contacts, JDTic interacts with four residues of the binding pocket that differ in other closely related ORs, which are thought to contribute to the subtype selectivity of JDTic and other KOR-selective ligands32 (human MOR (hMOR) and human DOR (hDOR) amino acids shown in parentheses, respectively): Val1082.53 (Ala and Ala), Val1182.63 (Asn and Lys), Ile2946.55 (Val and Val), and Tyr3127.35 (Trp and Leu) (Fig. 2b and Supplementary Fig. 8). Analysis of JDTic binding into hKOR-based hMOR and hDOR homology models, as well as JDTic SAR results17,28,30 (Supplementary Fig. 9), suggest that all described residues can contribute to the JDTic selectivity profile. Thus, changes in the Val1182.63 side chain, where larger hydrophilic residues, Asn2.63 and Lys2.63 are found in hMOR and hDOR, respectively, are likely to introduce unfavorable contacts with JDTic. Additionally, changing Tyr3127.35 to the Trp7.35 and Leu7.35 residues found in hMOR and hDOR, respectively, are likely to result in the loss of an important polar interaction with the JDTic amide. The remaining two hydrophobic side chains replacements, Val to Ala at position 2.53 and Ile to Val at position 6.55, may cause a reduction of the hydrophobic contact between JDTic and the receptor.
The isopropyl group from JDTic reaches deep in the orthosteric pocket to form a hydrophobic interaction with a conserved Trp2876.48 side chain (aka the “rotamer toggle switch”), possibly playing a critical role in the pharmacological properties of this ligand. Trp6.48 is thought to be a key part of the activation mechanism in many class A GPCRs including rhodopsin26 and A2AAR25, and similar hydrophobic contacts have been implicated in blocking activation-related conformational changes in the dark state visual rhodopsin by 11-cis retinal, and by inverse agonists in the A2AAR and D3R.
Binding of KOR-selective morphinans
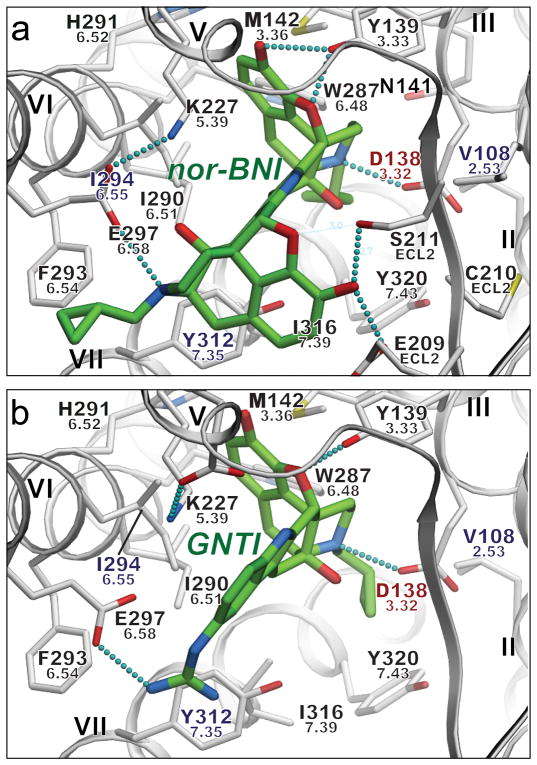
Prior mutagenesis and modeling studies suggested that many small molecule opioid ligands can interact with KOR, as well as with MOR and DOR, by forming a salt bridge with the highly conserved Asp3.32 (ref 33,34). This is consistent with our mutagenesis studies (Supplementary Table 3) and flexible docking35 of a series of morphine analogues, including selective KOR antagonists nor-BNI and GNTI (Fig. 3 and Supplementary Fig. 10). To assess the compatibility of these bulky and rigid ligands with the observed hKOR protein backbone conformation, we performed global energy optimizations of nor-BNI and GNTI in the binding cavity of hKOR, keeping side chains of the binding pocket fully flexible. Multiple independent runs consistently resulted in low energy conformations with essentially identical poses and receptor contacts for the common naltrexone moieties of both nor-BNI and GNTI (RMSD = 0.85 Å). In addition to a highly complementary van der Waals interface, both compounds formed an amino group salt bridge to the Asp1383.32 side chain and a hydrogen bond to the Tyr1393.33 side chain, both of which are important anchoring points for binding of morphine-based ligand, as supported by previous mutagenesis studies34.
Figure 3. Putative interaction modes of morphine-based high affinity hKOR selective antagonists nor-BNI (a) and GNTI (b).
Ligands are depicted as capped sticks with green carbons, and contact side chains of the receptor within 4 Å from the ligand are shown with grey carbons. Key hydrogen bonds and salt bridges are indicated with small cyan spheres and residues unique to KOR are labeled in blue. Residue Asp1383.32, which also shows critical impact on GNTI and nor-BNI binding in mutagenesis studies, is highlighted red. Ballesteros-Weinstein residue numbers are shown under the hKOR residue numbers. The graphics were prepared using ICM molecular modeling package (Molsoft LLC).
Moreover, unlike JDTic, both nor-BNI and GNTI compounds have a second basic moiety located more than 10 Å away from the first amino group (the other morphine in nor-BNI and the guanidine moiety in GNTI). In the predicted models of hKOR- nor-BNI/GNTI complexes, these additional amino groups of both ligands form a salt bridge with Glu2976.58 located at the entrance to the ligand binding pocket, which was previously characterized as a residue critical for subtype selectivity of hKOR-selective morphinan derivatives36. This interaction is also supported by our mutagenesis results (Supplementary Table 3), where a Glu297Ala mutation induced a significant drop in both nor-BNI and GNTI binding, but did not affect JDTic binding. Hydrophobic interactions at the KOR-specific residue Ile294 were also found for both nor-BNI and GNTI; consistent with our mutagenesis results (Supplementary Table 3) and suggesting that Ile294 may also be important for developing KOR subtype selective morphinan derivatives. Additional polar interactions with hKOR-specific residues, Glu209 and Ser211 in ECL2, are found for nor-BNI, which may further enhance hKOR-selectivity of this bulky ligand. Another side chain of the pocket, His2916.52, which is involved in the highly conserved aromatic cluster around Trp6.48 and thought to play a critical role in the receptor activation process37, forms hydrophobic contacts with JDTic, nor-BNI and GNTI. His2916.52 can be mutated to another aromatic residue, phenylalanine, without disrupting binding of these antagonists (Supplementary Table 3). The non-conservative His2916.52Lys mutation, however, totally abolished binding of all tested ligands, likely because of the disruption of the aromatic cluster induced by the lysine side chain. Interestingly, the cyclopropyl moiety of both nor-BNI and GNTI in these binding poses has the same position as the isopropyl moiety of JDTic, making hydrophobic contact with the conserved residue Trp2876.48. This cyclopropyl moiety is generally implicated in conversion of opioid agonists into antagonists (e.g. agonist oxymorphone into antagonist naltrexone), and this effect may be partially explained by a direct interaction with the Trp2876.48 side chain.
Overall, these structure-based docking results support the ‘message-address’ model38 in applications to morphine-based ligands nor-BNI and GNTI36, which points to Glu2976.58 as a key side chain that controls hKOR selectivity by anchoring the ‘address’ moieties of these compounds. The crystal structure of hKOR-JDTic complex (Fig. 2 and 3), however demonstrates that even in lieu of “address” interaction with Glu2976.58 more than 1000-fold subtype-selectivity to hKOR can be achieved for JDTic and some of its derivatives. Importantly, then, the ‘message-address’ hypothesis does not uniformly apply to all hKOR-selective antagonists.
Binding of Salvinorins
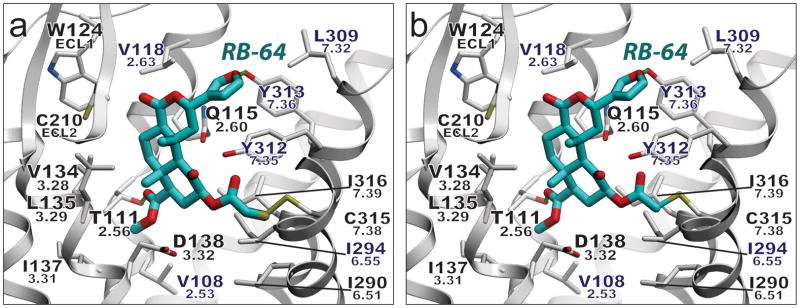
Salvinorin A (SalA), a naturally occurring diterpene from the widely abused hallucinogenic plant Salvia divinorum, represents an exceedingly potent (EC50 = 1 nM) and selective KOR agonist (>1000-fold)15. SalA is unique compared to other KOR ligands in that it lacks a charged or polar nitrogen atom to anchor it in the binding pocket. Extensive site-directed mutagenesis, SCAM (substituted cysteine-accessibility mutagenesis) and SAR studies on SalA and its analogues have been performed, indicating (among others) that the 2-acetoxy moiety interacts with Cys3157.38 (ref 39). Possible modes of interaction between the cysteine-reactive and ultra-potent agonist and SalA analog 22-thiocyanatosalvinorin A (RB-64; Ki = 0.59 nM; EC50 = 0.077 nM)39, and the hKOR structure were thus evaluated. Exposure of hKOR to RB-64 produces irreversibly-bound, wash-resistant adducts that are tethered to Cys3157.38 (ref 39). As the thiocyanate group contains two electrophilic centers, two distinct adducts may be formed, increasing the mass by either 463 or 431 amu. Docking studies using GOLD40 predict that the salvinorin 2-position can access Cys3157.38 while maintaining many of the interactions implicated by site-directed mutagenesis for SalA, providing a possible mechanism for the formation of the KOR–RB-64 adduct (Fig. 4, Supplementary Tables 4–5 and Figs. 11–12). Additionally, the docking results serve as a model of the initial recognition process of SalA-related agonists of the hKOR in an inactive state, although additional studies will be needed to fully elucidate the nature of the SalA-induced activation mechanism.
Figure 4. Model of covalently-bound RB-64.
Putative binding mode of (a) the RB-64 +463 amu and (b) the RB-64 +431 amu adduct. Residues within 4 Å of the ligand are displayed. Rendering: ligand, capped sticks/cyan carbons; hKOR side chains, capped sticks; hydrogen bonds, small green spheres; hKOR-unique residues labeled in blue. Ballesteros-Weinstein residue numbers are shown under the hKOR residue numbers. The graphics were prepared using ICM molecular modeling package (Molsoft LLC).
Conclusions
The JDTic-hKOR crystal structure has uncovered a combination of key features shared with chemokine and aminergic GPCRs along with unique structural details characteristic of the opioid subfamily. The hKOR was crystallized as a parallel-dimer with contacts involving helices I, II and VIII. While the existence of GPCR dimers in vivo and their physiological relevance remain highly debatable, several distinct potential dimer interfaces are starting to emerge from crystallographic and biochemical studies. Such multiple dimerization interfaces may serve to support different functional pathways, as well as to promote oligomeric assembly of GPCRs. Analysis of the ligand-receptor interactions has revealed important molecular details responsible for the exceptionally high affinity and subtype selectivity of JDTic–a small molecule antagonist with a broad therapeutic potential. The elucidation of a large binding cavity with a multitude of potential anchoring points begins to explain both the extreme structural diversity of hKOR drugs and differences in their receptor interaction modes, as supported by differential effects of various site-directed mutations on the binding properties of chemically diverse prototypic ligands. The structure clearly provides a long anticipated molecular framework for understanding opioid drug action, and thereby affords valuable new opportunities for a structure-based discovery of new drugs with ideal pharmacological properties.
Methods Summary
hKOR-T4L was expressed in Spodoptera frugiperda (Sf9) cells. Ligand-binding and functional assays were performed as described in Methods. Sf9 cells were solubilized using 1% (w/v) n-dodecyl-β-D-maltopyranoside (DDM) and 0.2% (w/v) cholesteryl hemisuccinate (CHS), and purified by immobilized metal ion affinity chromatography (IMAC), followed by reverse IMAC after cleaving N-terminal FLAG-10xHis tags by His-tagged Tobacco Etch Virus (TEV) protease. The purified protein was mixed with monoolein and cholesterol in a ratio of 40%:54%:6% (w/w) to form lipidic cubic phase (LCP) from which the receptor was crystallized. Crystals were grown at 20 °C in 45 nl protein-laden LCP boluses overlaid by 800 nl of precipitant solutions as described in Methods. Crystals were harvested from the LCP matrix and flash frozen in liquid nitrogen. X-ray diffraction data were collected on the 23ID-B/D beamline (GM/CA CAT) at the Advanced Photon Source, Argonne, IL using a 10 μm minibeam at wavelength of 1.0330 Å. Data collection, processing, structure solution and refinement are described in Methods. Modeling of JDTic analogues and hKOR-selective morphine derivatives nor-BNI and GNTI was performed using ICM-Pro; SYBYL-X 1.3 and GOLD Suite 5.1 were used to model RB-64 complexes, as described in Methods.
Full Methods and any associated references are available in the online version of the paper at www.nature.com/nature.
Supplementary Material
Acknowledgments
This work was supported by PSI:Biology grant U54 GM094618 (R.C.S.) for biological studies and structure production, NIH Roadmap grant P50 GM073197 (R.C.S.) for technology development and R01 DA017624 (B.L.R., E.V., R.B.M., P.D.M.), R01 DA027170 (B.L.R.), the NIMH Psychoactive Drug Screening Program Contract (B.L.R., X.-P.H.), the Michael Hooker Distinguished Chair of Pharmacology (B.L.R.), and the NIH grant R01 DA009045 (F.I.C.). D.W. is supported by a Boehringer Ingelheim Fonds Ph.D. Fellowship. The JDTic X-ray structure was conducted by Laboratory for the Structure of Matter, Naval Research Laboratory, Washington, DC. We thank J. Velasquez for help on molecular biology; T. Trinh, K. Allin and M. Chu for help on baculovirus expression; V. Setola for help with functional activity assays; J. Evans for help acquiring compounds; the National Institute of Drug Abuse Drug Supply Program for supplying JDTic and other opioid ligands used in these studies; K. Kadyshevskaya for assistance with figure preparation; E. Abola for assistance with manuscript preparation; A. Walker for assistance with manuscript preparation; J. Smith, R. Fishetti, and N. Sanishvili for assistance in development and use of the minibeam and beamtime at GM/CA-CAT beamline 23-ID at the Advanced Photon Source, which is supported by National Cancer Institute grant Y1-CO-1020 and National Institute of General Medical Sciences grant Y1-GM-1104.
Footnotes
Supplementary Information is linked to the online version of the paper at www.nature.com/nature.
Author Contributions H.W. assisted protein expression, optimized the constructs, purified and crystallized the receptor in LCP, optimized crystallization conditions, grew crystals for data collection, collected the data and processed diffraction data, and prepared the manuscript. D.W. assisted with protein expression, purified the receptor, performed thermal stability assay and assisted with preparing the manuscript. V.K. performed nor-BNI/GNTI-receptor docking and prepared the manuscripts. M.M. assisted with protein expression, purified the receptor, tested the JDTic compound, and performed the thermal stability assay. G.W.H. solved and refined the structure and assisted with preparing the manuscript. E.V. created the initial tagged hKOR constructs and E.V. and X.-P.H. performed the ligand-binding and site-directed mutagenesis studies. W.L. assisted with construct optimization and crystallization in LCP. A.A.T. refined the structure and assisted with preparing the manuscript. F.I.C. and S.W.M. provided JDTic crystal structure, performed conformational studies of JDTic, and assisted with preparing the manuscript. R.B.W. and P.D.M. performed RB-64-receptor docking and prepared the manuscript. V.C. assisted with the crystallization in LCP, processed diffraction data, refined the structure and prepared the manuscript. B.L.R. suggested the JDTic compound for structural studies, supervised the pharmacology and mutagenesis studies and prepared the manuscript. R.C.S. was responsible for the overall project strategy and management and led the manuscript preparation and writing.
Author Information The coordinates and the structure factors have been deposited in the Protein Data Bank under the accession code (4DJH). Reprints and permissions information is available at www.nature.com/reprints. R.C.S. is a founder and BOD member of Receptos, a GPCR drug discovery company. Readers are welcome to comment on the online version of this article at www.nature.com/nature.
Literature Cited
- 1.Fredriksson R, Lagerstrom MC, Lundin LG, Schioth HB. The G-protein-coupled receptors in the human genome form five main families. Phylogenetic analysis, paralogon groups, and fingerprints. Mol Pharmacol. 2003;63:1256–1272. doi: 10.1124/mol.63.6.1256. [DOI] [PubMed] [Google Scholar]
- 2.Waldhoer M, Bartlett SE, Whistler JL. Opioid receptors. Annu Rev Biochem. 2004;73:953–990. doi: 10.1146/annurev.biochem.73.011303.073940. [DOI] [PubMed] [Google Scholar]
- 3.Cherezov V, et al. High-resolution crystal structure of an engineered human beta2-adrenergic G protein-coupled receptor. Science. 2007;318:1258–1265. doi: 10.1126/science.1150577. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Jaakola VP, et al. The 2.6 angstrom crystal structure of a human A2A adenosine receptor bound to an antagonist. Science. 2008;322:1211–1217. doi: 10.1126/science.1164772. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Chien EY, et al. Structure of the human dopamine D3 receptor in complex with a D2/D3 selective antagonist. Science. 2010;330:1091–1095. doi: 10.1126/science.1197410. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Warne T, et al. Structure of a beta1-adrenergic G-protein-coupled receptor. Nature. 2008;454:486–491. doi: 10.1038/nature07101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Shimamura T, et al. Structure of the human histamine H1 receptor complex with doxepin. Nature. 2011;475:65–70. doi: 10.1038/nature10236. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Wu B, et al. Structures of the CXCR4 chemokine GPCR with small-molecule and cyclic peptide antagonists. Science. 2010;330:1066–1071. doi: 10.1126/science.1194396. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Rasmussen SG, et al. Crystal structure of the beta2 adrenergic receptor-Gs protein complex. Nature. 2011;477:549–555. doi: 10.1038/nature10361. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Katritch V, Cherezov V, Stevens RC. Diversity and modularity of G protein-coupled receptor structures. Trends Pharmacol Sci. 2011 doi: 10.1016/j.tips.2011.09.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Congreve M, Langmead CJ, Mason JS, Marshall FH. Progress in structure based drug design for G protein-coupled receptors. J Med Chem. 2011;54:4283–4311. doi: 10.1021/jm200371q. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Kufareva I, Rueda M, Katritch V, Stevens RC, Abagyan R. Status of GPCR modeling and docking as reflected by community-wide GPCR Dock 2010 assessment. Structure. 2011;19:1108–1126. doi: 10.1016/j.str.2011.05.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Martin WR, Eades CG, Thompson JA, Huppler RE, Gilbert PE. The effects of morphine- and nalorphine- like drugs in the nondependent and morphine-dependent chronic spinal dog. J Pharmacol Exp Ther. 1976;197:517–532. [PubMed] [Google Scholar]
- 14.Carlezon WA, Jr, Beguin C, Knoll AT, Cohen BM. Kappa-opioid ligands in the study and treatment of mood disorders. Pharmacol Ther. 2009;123:334–343. doi: 10.1016/j.pharmthera.2009.05.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Roth BL, et al. Salvinorin A: a potent naturally occurring nonnitrogenous kappa opioid selective agonist. Proc Natl Acad Sci U S A. 2002;99:11934–11939. doi: 10.1073/pnas.182234399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Walsh SL, Strain EC, Abreu ME, Bigelow GE. Enadoline, a selective kappa opioid agonist: comparison with butorphanol and hydromorphone in humans. Psychopharmacology (Berl) 2001;157:151–162. doi: 10.1007/s002130100788. [DOI] [PubMed] [Google Scholar]
- 17.Thomas JB, et al. Identification of the first trans-(3R,4R)- dimethyl-4-(3-hydroxyphenyl)piperidine derivative to possess highly potent and selective opioid kappa receptor antagonist activity. J Med Chem. 2001;44:2687–2690. doi: 10.1021/jm015521r. [DOI] [PubMed] [Google Scholar]
- 18.Carroll I, et al. Pharmacological properties of JDTic: a novel kappa-opioid receptor antagonist. Eur J Pharmacol. 2004;501:111–119. doi: 10.1016/j.ejphar.2004.08.028. [DOI] [PubMed] [Google Scholar]
- 19.Jackson KJ, Carroll FI, Negus SS, Damaj MI. Effect of the selective kappa-opioid receptor antagonist JDTic on nicotine antinociception, reward, and withdrawal in the mouse. Psychopharmacology (Berl) 2010;210:285–294. doi: 10.1007/s00213-010-1803-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Salom D, et al. Crystal structure of a photoactivated deprotonated intermediate of rhodopsin. Proc Natl Acad Sci U S A. 2006;103:16123–16128. doi: 10.1073/pnas.0608022103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Mancia F, Assur Z, Herman AG, Siegel R, Hendrickson WA. Ligand sensitivity in dimeric associations of the serotonin 5HT2c receptor. EMBO Rep. 2008;9:363–369. doi: 10.1038/embor.2008.27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Wang JB, Johnson PS, Wu JM, Wang WF, Uhl GR. Human kappa opiate receptor second extracellular loop elevates dynorphin’s affinity for human mu/kappa chimeras. J Biol Chem. 1994;269:25966–25969. [PubMed] [Google Scholar]
- 23.Ballesteros JA, Weinstein H. Integrated methods for the construction of three-dimensional models and computational probing of structure-function relations in G protein-coupled receptors. Methods Neurosci. 1995;25:366–428. [Google Scholar]
- 24.Palczewski K, et al. Crystal structure of rhodopsin: A G protein-coupled receptor. Science. 2000;289:739–745. doi: 10.1126/science.289.5480.739. [DOI] [PubMed] [Google Scholar]
- 25.Xu F, et al. Structure of an Agonist-Bound Human A2A Adenosine Receptor. Science. 2011;332:322–327. doi: 10.1126/science.1202793. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Standfuss J, et al. The structural basis of agonist-induced activation in constitutively active rhodopsin. Nature. 2011;471:656–660. doi: 10.1038/nature09795. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Subramanian G, Paterlini MG, Larson DL, Portoghese PS, Ferguson DM. Conformational analysis and automated receptor docking of selective arylacetamide-based kappa-opioid agonists. J Med Chem. 1998;41:4777–4789. doi: 10.1021/jm9803166. [DOI] [PubMed] [Google Scholar]
- 28.Cai TB, et al. Synthesis and in vitro opioid receptor functional antagonism of analogues of the selective kappa opioid receptor antagonist (3R)-7-hydroxy-N-((1S)-1-{[(3R,4R)-4-(3-hydroxyphenyl)-3,4-dimethyl-1-pipe ridinyl]methyl}-2-methylpropyl)-1,2,3,4-tetrahydro-3-isoquinolinecarboxami de (JDTic) J Med Chem. 2008;51:1849–1860. doi: 10.1021/jm701344b. [DOI] [PubMed] [Google Scholar]
- 29.Thomas JB, et al. Importance of phenolic address groups in opioid kappa receptor selective antagonists. J Med Chem. 2004;47:1070–1073. doi: 10.1021/jm030467v. [DOI] [PubMed] [Google Scholar]
- 30.Runyon SP, et al. Analogues of (3R)-7-hydroxy-N-[(1S)-1-{[(3R,4R)-4-(3-hydroxyphenyl)-3,4-dimethyl-1-pipe ridinyl]methyl}-2-methylpropyl)-1,2,3,4-tetrahydro-3-isoquinolinecarboxami de (JDTic). Synthesis and in vitro and in vivo opioid receptor antagonist activity. J Med Chem. 2010;53:5290–5301. doi: 10.1021/jm1004978. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Zimmerman DM, Nickander R, Horng JS, Wong DT. New structural concepts for narcotic antagonists defined in a 4-phenylpiperidine series. Nature. 1978;275:332–334. doi: 10.1038/275332a0. [DOI] [PubMed] [Google Scholar]
- 32.Vortherms TA, Mosier PD, Westkaemper RB, Roth BL. Differential helical orientations among related G protein-coupled receptors provide a novel mechanism for selectivity. Studies with salvinorin A and the kappa-opioid receptor. J Biol Chem. 2007;282:3146–3156. doi: 10.1074/jbc.M609264200. [DOI] [PubMed] [Google Scholar]
- 33.Surratt CK, et al. -mu opiate receptor. Charged transmembrane domain amino acids are critical for agonist recognition and intrinsic activity. J Biol Chem. 1994;269:20548–20553. [PubMed] [Google Scholar]
- 34.Befort K, et al. The conserved aspartate residue in the third putative transmembrane domain of the delta-opioid receptor is not the anionic counterpart for cationic opiate binding but is a constituent of the receptor binding site. Mol Pharmacol. 1996;49:216–223. [PubMed] [Google Scholar]
- 35.Totrov M, Abagyan R. Flexible protein-ligand docking by global energy optimization in internal coordinates. Proteins Suppl. 1997;1:215–220. doi: 10.1002/(sici)1097-0134(1997)1+<215::aid-prot29>3.3.co;2-i. [DOI] [PubMed] [Google Scholar]
- 36.Metzger TG, Paterlini MG, Portoghese PS, Ferguson DM. Application of the message-address concept to the docking of naltrexone and selective naltrexone-derived opioid antagonists into opioid receptor models. Neurochem Res. 1996;21:1287–1294. doi: 10.1007/BF02532369. [DOI] [PubMed] [Google Scholar]
- 37.Chen S, et al. Mutation of a single TMVI residue, Phe(282), in the beta(2)-adrenergic receptor results in structurally distinct activated receptor conformations. Biochemistry. 2002;41:6045–6053. doi: 10.1021/bi012189c. [DOI] [PubMed] [Google Scholar]
- 38.Chavkin C, Goldstein A. Specific receptor for the opioid peptide dynorphin: structure--activity relationships. Proc Natl Acad Sci U S A. 1981;78:6543–6547. doi: 10.1073/pnas.78.10.6543. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Yan F, et al. Structure-based design, synthesis, and biochemical and pharmacological characterization of novel salvinorin A analogues as active state probes of the kappa-opioid receptor. Biochemistry. 2009;48:6898–6908. doi: 10.1021/bi900605n. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Verdonk ML, Cole JC, Hartshorn M, Murray CW, Taylor R. Improved Protein-Ligand Docking Using GOLD. Proteins. 2003;52:609–623. doi: 10.1002/prot.10465. [DOI] [PubMed] [Google Scholar]
- 41.Rosenbaum DM, et al. GPCR engineering yields high-resolution structural insights into beta2-adrenergic receptor function. Science. 2007;318:1266–1273. doi: 10.1126/science.1150609. [DOI] [PubMed] [Google Scholar]
- 42.Caffrey M, Cherezov V. Crystallizing membrane proteins using lipidic mesophases. Nat Protoc. 2009;4:706–731. doi: 10.1038/nprot.2009.31. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Cherezov V, Peddi A, Muthusubramaniam L, Zheng YF, Caffrey M. A robotic system for crystallizing membrane and soluble proteins in lipidic mesophases. Acta Crystallogr D Biol Crystallogr. 2004;60:1795–1807. doi: 10.1107/S0907444904019109. [DOI] [PubMed] [Google Scholar]
- 44.Cherezov V, et al. Rastering strategy for screening and centring of microcrystal samples of human membrane proteins with a sub-10 microm size X-ray synchrotron beam. J R Soc Interface. 2009;6 (Suppl 5):S587–597. doi: 10.1098/rsif.2009.0142.focus. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Otwinowski Z, Minor W. Processing of X-ray diffraction data collected in oscillation mode. Method Enzymol. 1997;276:307–326. doi: 10.1016/S0076-6879(97)76066-X. [DOI] [PubMed] [Google Scholar]
- 46.McCoy AJ, et al. Phaser crystallographic software. J Appl Crystallogr. 2007;40:658–674. doi: 10.1107/S0021889807021206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Murshudov GN, Vagin AA, Dodson EJ. Refinement of macromolecular structures by the maximum-likelihood method. Acta Crystallogr D Biol Crystallogr. 1997;53:240–255. doi: 10.1107/S0907444996012255. [DOI] [PubMed] [Google Scholar]
- 48.BUSTER. 2.8.0. Global Phasing Ltd; Cambridge, U.K: 2009. [Google Scholar]
- 49.Emsley P, Lohkamp B, Scott WG, Cowtan K. Features and development of Coot. Acta Crystallogr D Biol Crystallogr. 2010;66:486–501. doi: 10.1107/S0907444910007493. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.