Abstract
Estrogen receptor (ER) β was discovered over a decade ago. The design of most studies on this receptor was based on knowledge of its predecessor, ERα. Although breast cancer (BCa) has been a main focus of ERβ research, its precise roles in breast carcinogenesis remain elusive. Data from in vitro models have not always matched those from observational or clinical studies. Several inherent factors may contribute to these discrepancies: a) several ERβ spliced variants are expressed at the protein level, and isoform-specific antibodies are unavailable for some variants; b) post-translational modifications of the receptor regulate receptor functions; c) the role of the receptor differs significantly depending on the type of ligands, cis-elements, and co-regulators that interact with the receptor; and d) the diversity of distribution of the receptor among intracellular organelles of BCa cells. This review addresses the gaps in knowledge in ERβ research as it pertains to BCa regarding the following questions: 1) is ERβ a tumor suppressor in BCa?; 2) do ERβ isoforms play differential roles in breast carcinogenesis?; 3) do nuclear signaling and extranuclear ERβ signaling differ in BCa?; 4) what are the consequences of post-translational modifications of ERβ in BCa?; 5) how do co-regulators and interacting proteins increase functional diversity of ERβ?; and 6) how do the types of ligand and regulatory cis-elements affect the action of ERβ in BCa? Insights gained from these key questions in ERβ research should help in prevention, diagnosis/prognosis, and treatment of BCa.
Keywords: ERbeta isoforms, tumor suppressor, post- translational modification, extranuclear localization, co-regulators, phytoestrogen
Introduction
Estrogen receptor beta (ERβ) is the second estrogen receptor (ER) identified in rat prostate and ovary in 1996 [1] and later in human testis in the same year [2], which is more than 30 years after discovery of the first ER (also referred to as ERα) [3]. Similar to the ERα, ERβ binds estradiol-17β (E2) with high affinity through its ligand-binding domain (LBD) but the two ERs share only moderate homology in their protein sequences (58% in human and 55% in rat) with the LBD of ERα [2]. Intriguingly, they have almost identical DNA-binding domains (DBDs, 96% homology in human and 95% in rat) capable of interacting with specific DNA elements (eg, estrogen-response element, ERE) and transactivating common and ER subtype-specific genes [4–7]. Significant information revolving around the involvement of ERβ in the development and progression of breast cancer (BCa) has emerged since its discovery [8], leading to new insights and directions in BCa research.
This review summarizes some of the major findings in this research area and highlights critical —missing pieces, with the hope that future investigations will fill these gaps. Throughout this review, we are aware that associations from clinical association data do not always agree with data obtained in vitro. No single cell model can truly duplicate the level of complexity found in tissues, which are subject to endocrine and paracrine influences from the surrounding macro- and micro-environment, respectively. We have chosen not to include tumor microenvironment in our review, as the topic is too broad. Instead, we have focused on six research areas we consider to be essential for improving our understanding of the function of ERβ in BCa: 1) ERβ as a possible tumor suppressor in BCa; 2) critical and distinct roles of ERβ isoforms in breast carcinogenesis; 3) differential roles of nuclear and extranuclear ERβ signaling in BCa; 4) the consequences of post-translational modifications of ERβ in BCa; 5) the increase in the diversity of the receptor’s function by co-regulators and interacting proteins of ERβ; and 6) the differential behaviors of ERβ elicited by different ligands in BCa. These topics were selected because a) of the need to clarify existing controversies; b) the emergence of new research that has not yet been extensively reviewed; and c) that are, on the basis of our knowledge, at the leading edge of ERβ research in BCa.
ERβ as a possible tumor suppressor in BCa
Immunohistochemical analyses have identified ERβ as the major form of ER in the normal breast that is localized in luminal epithelium, myoepithelium, and in fibroblasts and lymphocytes in the stroma [9]. Murphy et al. [10] have recently reviewed the literature on the expression of ERβ in ERα-positive and -negative BCa Approximately 58% of BCa express both ERs, 18% express only ERβ, and 14% express only ERα [10;11]. High levels of ERβ expression, regardless of ERα status, were found to associate with a better response to tamoxifen and a longer survival time [12–20]. In contrast, several studies of ERα-negative BCa demonstrated a positive correlation between high ERβ expression and poor prognostic phenotypes, such as elevated proliferation [21;22] and basal phenotype [23]. However, several studies reported opposite results, in which stronger ERβ immunopositivity predicted longer disease-free survival [14;24]. For example, Honma et al. showed that positivity for ERβ was associated with better survival in patients with ERBB2-positive or ERα-, PR- and ERBB2- negative (triple-negative) BCa and a better response to tamoxifen monotherapy [14]. Whether ERβ has multiple distinct roles in ERα-negative BCa needs further clarification. These contradictory results may reflect different therapeutic regimens among the patients or heterogeneity of the patient populations (postmenopausal vs mixed populations).
Several lines of evidence suggest that ERβ functions as a tumor suppressor in in vitro models. Ectopic expression of ERβ in ERα-positive BCa cells slowed down the mitogenic responses initiated by ERα [25–28], reduced cell motility and invasion [29;30], and inhibited tumor formation [26] and angiogenesis [31] in mouse xenografts. However, ERβ behaved differently in the ERα-negative MDA-MB-435 cells [32], whose identity as a true BCa cell line remains controversial [33]. The role of ERβ as a tumor suppressor is further supported by findings of several epidemiologic studies demonstrating a loss of ERβ in higher-grade vs lower-grade BCa tissue [27;34–38]. DNA methylation in the proximal promoter region of ERβ was identified as a potential cause of this gene silencing [39;40]. It has long been speculated that transcriptional silencing of ERβ is necessary for cancer progression, a phenomenon found not only in BCa but also in other hormone-sensitive cancers [41;42]. Overall, data from cancer-cell models and observational studies suggest that ERβ functions as a gatekeeper to inhibit tumor growth and progression.
Although ERβ seems to be a tumor suppressor in numerous cell models, its role in human breast carcinogenesis remains elusive. Presently, we know that E2 is a natural ligand for both ERα and ERβ. The proliferative effects caused by ERα are antagonized by the presence of ERβ signaling. ERβ, being the dominant ER should, in theory, be able to protect the normal mammary epithelial cells from any uncontrolled cell growth. But there is no consensus on how ERβ function should be studied in clinical BCa studies. Clearly, this remains as a gap in our understanding of the mechanisms of estrogen signaling and ERβ function in BCa. Association studies relying on immunodetection of nuclear ERβ in BCa specimens may be unable to define the native functions of ERβ in BCa, which may depend in large part on post-translational modifications of the receptor, the co-existence of ERβ with functionally unique ERβ isoforms, the involvement of extranuclear signaling, its differential modulations by interacting proteins and ligands, and hormone level (see below). Also, studies based on the measurement of ERβ transcripts with quantitative PCR in whole biopsy samples may not yield meaningful data because of the presence of a significant number of ERβ transcripts in the stroma, possibly masking expression in adjacent normal or malignant epithelial cells. Hence, future well-designed prospective population studies and/or large-scale clinical trials using specific ERβ agonists are needed to resolve these controversies.
Critical and distinct roles of ERβ isoforms in breast carcinogenesis
Immunohistochemical (IHC) analyses often are used to measure the expression of a protein in tissue sections. The success of this technique depends in large part on the specificity of an antibody against its target protein. Sometimes the alternative use of exons may result in the coexistence of multiple isoforms of the target protein in a tissue. Monoclonal antibodies that recognize only a common epitope or polyclonal antibodies that recognize multiple epitopes are necessary to differentiate the expression among various isoforms. ERβ is a prime example, whereby the use of a pan-ERβ antibody or isoform-specific antibodies for IHC studies may yield different results [43;44].
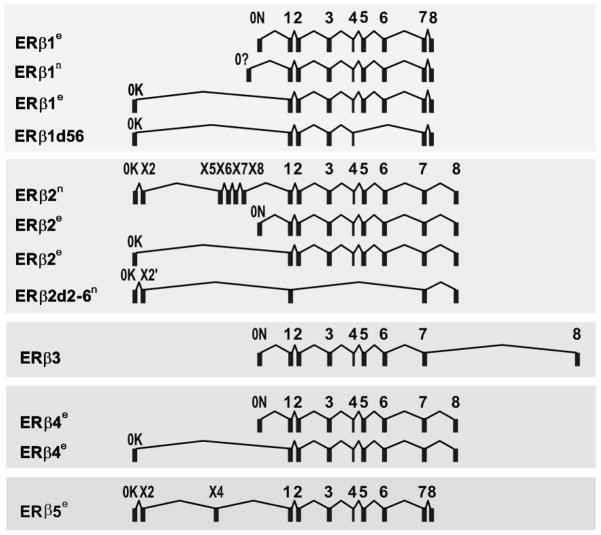
Early published data on human ERβ function/signaling were focused primarily on ERβ1, the originally cloned sequence [2]. Sequencing data suggested that multiple ERβ isoforms exist as a result of alternative splicing of the last coding exon (exon 8) (Figure 1) [45]. This is also supported by the availability of multiple ERβ isoform transcripts in the human genome project in the NCBI AceView database, as well as by our experimental data (Figure 1). With regard to nomenclature, the original ERβ is also called ERβ1. So far, four other ERβ isoforms (ERβ2, ERβ3, ERβ4, and ERβ5) have been identified. We and others have shown their existence as full-length transcripts, which have in common exon 1 through 7 plus one isoform-specific exon 8 (Figure 1) [46;47]. The molecular weights of ERβ1, 2, 4, and 5 have been determined as 59, 56, 54 and 53 kDa, respectively, according to protein sequence prediction programs, as well as ectopic protein-expression experiments [47]. Since all isoforms share exons 1 through 7, they all have the same AF1 domain, DBD, hinge domain, and LBD, leaving the AF2 domain (C-terminus) specific to each of the isoforms.
Figure 1.
Genomic arrangement of ERβ isoforms. Each ERβ transcript is composed of at least one 5′ non-coding exon (exon 0K, 0Xs, or 0N) and three to eight coding exons (exons 1–8). Light blue rectangular box represents an exon. Each full-length isoform (ERβ1, 2, 3, 4, 5) shares exons 1–7 plus an isoform-specific exon 8. Key: n=the transcripts published in NCBI database; e=transcripts discovered by sequencing experiments (unpublished).
An important question often asked in studying alternative-spliced variants is whether any of the endogenous proteins derived from the spliced variants are expressed in cells or tissues. Researchers tend to be skeptical about their existence because only transcripts, but not protein-products of the variants, are detectable in many cases. Studies carried out at the protein level depend on the availability of good antibodies, presenting a major obstacle in the study of protein isoforms such as ERβ because of the high sequence homology of the variants to their corresponding wild-type protein. Thus, the choice of antigenic region for raising antibody is limited. Fortunately, we [43] and others [12;44;48–53] have successfully raised antibodies to different ERβ isoforms. The specificity of a few isoform-specific antibodies has been validated [43;44], providing the needed tools for validating the clinical relevance of the expression of ERβ isoforms in BCa.
Shaaban and associates reviewed the role of ERβ isoforms in BCa in 2008 [54]. A few more studies have since been published. Up until 2011, at least 14 IHC studies on ERβ/ERβ1 in BCa have been published [55]. Seven studies found an association of ERβ1 expression with favorable outcomes [56–58] such as longer disease-free/overall survival [14;20], smaller tumor size, lymph-node negativity, lower histological grade [59], and responsiveness to tamoxifen [13]. Six studies did not find an association of ERβ1 expression with any clinical parameters [18;44;60–63]. Only one study showed an association of ERβ1 expression with increased cell proliferation in ERα-negative BCa [22]. Eleven of the fourteen studies also studied ERβ2. Five did not find a correlation of ERβ2 with any clinical outcome or survival [12;14;56;60;63]. Two investigations showed ERβ2 as a poor prognosticator [58;64], whereas two others found ERβ2 associated with better outcomes [59;61]. In one study, which also analyzed expression of progesterone receptor (PR), the presence of PR was found to associate with responsiveness to tamoxifen irrespective of ERβ2 level [65], but in a subset of PR-negative samples, ERβ2 expression was a predictor of resistance to tamoxifen [65]. In conclusion, with the use of ERβ-specific antibodies, it becomes clear that ERβ1 may be a predictor for good disease outcome; the role of other isoforms in BCa needs further studies using validated isoform-specific antibodies. Finally, only one study investigated ERβ1, 2, and 5 simultaneously and reported an association of expression of nuclear ERβ2 and cytosolic ERβ5 with BCa survival [44] (see Figure 2).
Figure 2.
Action of estrogen receptor β (ERβ) in breast cancer. Different ligands, such as estrogens, antiestrogens, and SERMs, activate non-genomic and genomic signaling of ERβ. In non-genomic signaling, ERβ interacts with membrane proteins (eg, G proteins, caveolin 1) and other interacting proteins (IPs) to activate kinase signaling pathways. In genomic signaling, liganded ERβ1 homodimerize or heterodimerize with ERβ isoforms or ERα and translocate from the cytoplasm to the nucleus. The homo- or heterodimer directly binds to estrogen response elements (EREs) or are tethered to other TFs (eg, AP1) in the promoter region or cis-regulatory sequences of target genes to facilitate gene transcription. Specific co-regulators (CoR) are believed to interact with ERβ to modulate gene transcription. ERβ may also translocate into mitochondria and interact with proteins involved in mitochrondrial ribosome synthesis and organization. ERβ has been found (or is expected to be) phosphorylated, ubiquitylated, or palmitoylated for gene transactivation, degradation, or membrane targeting, respectively. Expression of ERβ isoforms other than wild-type ERβ1 can be potential prognostic markers in breast cancer. For example, nuclear ERβ2 and ERβ5 were found to be associated with better patient survival; however, cytoplasmic ERβ2 was significantly correlated with worse outcome. Interactions of specific protein partners are believed to contribute to the functional roles of ERβ1 and its isoforms in breast cancer. Smiley and sad face represents good and bad prognosis in breast cancer, respectively.
Our present understanding of the molecular function of ERβ and its isoforms is still quite limited. ERβ1 was shown to form functional homodimers and heterodimers with other ERβ isoforms, as well as with ERα, and negates ERα-signaling [47;66;67]. ERβ2 was found to heterodimerize with ERα and to inhibit ERα-mediated estrogen action [68;69]. The major difference between ERβ1 and ERβ2 is that ERβ1 can counteract ERα signaling in two ways: by neutralizing the action of ERα via heterodimer formation and by directly triggering the anti-proliferative signal to counteract the pro-proliferative function of ERα. In contrast, ERβ2 can inhibit ERα signaling only through heterodimer formation, as this ER isoform was nonfunctional by itself [47;69]. So far, nothing has been published regarding the action of ERβ4 or ERβ5 on ERα signaling. With regard to ERβ signaling, ERβ isoforms 2, 4, and 5 can heterodimerize with ERβ1 and enhance ERβ1-induced transactivation in a ligand-dependent manner. Only ligands like E2 and bisphenol A, but not phytoestrogen, can initiate the dimer formation between ERβ1 and the other ERβ isoforms [47]. However, the functional role of each ERβ isoforms in BCa remains to be characterized.
Differential roles of nuclear and extranuclear ERβ signaling in BCa
Since the ERα is the single most powerful predictor of BCa prognosis, all patients with BCa are routinely scored for the amount and presence of ERα, but not for ERβ, in the nuclei of the normal or transformed epithelial cells. Clinical studies on the prognostic significance of ERβ in BCa have also focused on nuclear expression of ERβ [70;71]. Recent studies have noted, however, the existence of additional cellular ERβ pools in the cytoplasm, in the mitochondria, and at the plasma membrane of BCa cells (see Figure 2). One recent study found that nuclear ERβ1 and ERβ2 expression correlate with better overall survival and that nuclear ERβ2 correlates with better disease-free survival [44]. However, cytoplasmic ERβ2 expression alone, or in combination with nuclear ERβ2, predicted significantly worse overall survival. Patients with only cytoplasmic ERβ2 had a significantly poorer prognosis [44]. Cytoplasmic ERβ2 expression was also correlated with high-grade tumors, distant metastasis, recurrence, and death due to BCa. Nuclear ERβ2 also was strongly predictive of a twofold greater response to endocrine therapy. Thus, nuclear and cytoplasmic expressions of ERβ2 differentially affect outcome [58]. The distinct roles of ERβ isoforms at various cellular localizations have a clear prognostic significance. Here, we summarize what is known about ERβ in the nucleus, mitochondria, and plasma membrane in BCa cell models.
ERβ in the nucleus
Classical ERβ-mediated signaling involves binding of the ligand to the ERβ, resulting in translocation of ERβ to the nucleus, where it binds to DNA either directly to the classical ERE or indirectly to an NFκB-, AP1-, or Sp1-binding element via tethering with their respective transcription factors and recruits co-activators, thereby initiating downstream signaling cascades [72]. The ERβ1–5 isoforms retain the nuclear localization signal, and ERβ1, ERβ2, ERβ4, and ERβ5 are localized to the nucleus [43;70;71]. The question is whether the nuclear localization of the isoforms is really estrogen-dependent and whether ERβ1–5 isoforms are capable of activating transcription on their own when they are localized to the nucleus. In the yeast and HEK293 cell models, ERβ isoforms (ERβ2, 4, and 5) can form heterodimers with ERβ1, but not homodimers, and modulate gene expression in a ligand-dependent manner [47]. In ERα-expressing MCF7 cells, the constitutive expression of ERβ1 and ERβ2 diminished the ERE activity in these cells as compared with that of parental cells [65;73] as well as the expression of cathepsin D, a known target of ERα [74]. These findings indicate that both ERβ1 and ERβ2 inhibit ERα function, resulting in growth inhibition of ERα-positive BCa cells. These studies are especially relevant to the clinical situation, in which ERα is present initially and ERβ1 and its isoforms might repress ERα-targeted genes but have a different outcome in ERα-negative cancer.
ERβ in the mitochondria
Cytosolic ERβ was first demonstrated in MCF7 cells in experiments using pan-ERβ antiserum [75]. The observed cytoplasmic staining of ERβ was initially ignored as being either background staining or staining of inactive ERβ. At present, several lines of evidence support the presence of ERβ within the mitochondria and its association with mitochondrial proteins in BCa cells [76–79] (Figure 2). The mitochondrial ERβ in ERα-negative MCF-10F cells was involved in E2-induced expression of mitochondrial DNA (mtDNA)-encoded respiratory chain (MRC) proteins, cytochrome c oxidase subunits I and II, and NADPH dehydrogenase subunit 1. Using ERβ1 ectopic expression and tandem affinity purification followed by nano-LC-MS/MS, Nassa et al. identified the ERβ1 interactome in MCF7 cells, including the association of ERβ with several mitochondrial proteins [80]. Our laboratory performed yeast two hybrid–based interaction studies and also found the association of a number of mitochondrial proteins with the N-terminus of the ERβ (unpublished). The mitochondria are the energy powerhouse of a cell, and cellular processes such as cell proliferation, apoptosis, cell transformation, and tumorigenesis are closely related to MRC functions. Hence the physiological and pathological implications of ERβ-mediated mitochondrial effects in these cellular processes warrant further study. A putative mitochondrial targeting polypeptide signal (mtTP) has been identified in ERβ (between amino acids 220 and 270) and is present on all ERβ isoforms [81]. However, to date, no investigation has examined whether the mitochondrial association is an ERβ isoform – specific event.
To explore the role of ERβ in mitochondria, the Russo laboratory [81] treated the benign MCF10F cells with E2 to induce transformation. They found that ERβ shifted from a predominantly mitochondrial localization in normal and early transformed cells to a nuclear localization in association with the expression of progressive stages of cell transformation. A separate study found that ERs, especially ERβ, in mitochondria strongly prevented radiation-induced cell death in a BCa cell model [82]. Furthermore, when BCa cells were exposed to ultraviolet light in the presence of E2, PPT (an ERα agonist) or DPN (an ERβ agonist) [83], DPN was more potent than PPT in inhibiting cytochrome C release. The upregulation of manganese superoxide dismutase activity to quench reactive oxygen species, thereby preventing cell death signaling pathways, was proposed as the mediator of the ERβ action.
ERβ on the plasma membrane
Rapid signaling through plasma membrane ERβ is now believed to be the major venue of non-genomic action of the receptor [84–90]. Endogenous ERβ has been identified in the caveolae and cell membranes of endothelial, non-small cell lung tumor, and BCa cells [91;92]. Our yeast two-hybrid screen for ERβ partners has also uncovered a number of novel membrane-associated proteins (unpublished), suggesting that cell membrane ERβ per se or through tethering of other protein partners participates in rapid signaling. In this regard, a preponderance of evidence has emerged indicating that the membrane-associated ERα and ERβ can activate mitogen-activated protein kinase (MAPK) (both the extracellular signal- regulated kinase (ERK) and the c-jun kinase) pathways, and the cytoplasmic free calcium ([Ca2+]i) flux [91;93–95], pathways known to be involved in BCa cell functions. Recent works have defined motifs in the LBD of steroid receptors that are critical to membrane localization and function [96–98]. Mutation of these motifs prevents both receptor dimerization and signaling through ERK, PI3K, and cAMP. Loss of the former signals prevents the cell survival activity of E2 in breast and lung cancer cells.
In short, it is crucial to identify the key factors contributing to differential distribution of ERβ among the various subcellular compartments during the progression of breast carcinogenesis to enhance our understanding of the differential roles of nuclear versus non-nuclear ERβ.
The consequences of post-translational modifications of ERβ in BCa
Post-translational modification (PTM) refers to the covalent addition of functional groups to proteins; it includes phosphorylation, ubiquitylation, nitrosylation, palmitoylation, acetylation, sumolyation, glycosylation, and methylation [97;99–101]. These modifications allow proteins to respond to extracellular signals, intracellular stress, pharmacological agents, and morphogens at different developmental stages. Studies on human ERβ PTM are sparse. Here, we review PTM data on mouse ERβ and relevant information on human ERα, aiming to provide insights into the role of PTM in human ERβ function.
Phosphorylation is the most extensively studied PTM, in part because of its relative frequency and stability [99]. It is a reversible process at serine, threonine, and tyrosine residues. Phosphorylation may modify the function of the ERβ, and different phosphorylation sites may indicate different normal and pathological states of the receptor. A recent clinical study demonstrated an association of phosphorylation of ERβ at S105 with better survival in BCa, even in tamoxifen-resistant patients [102]. Similar studies on ERα have shown that various serine phosphorylation sites are valuable for the classification and prognosis of BCa [103–105]. Reviews summarizing the prognostic value of ERα PTM sites in BCa have been published [104;106], but since then the addition of new publications has been slow. In humans, stimulation of the p38 pathway enhanced the transcriptional function of ERβ (Figure 2) in endometrial adenocarcinoma Ishikawa cells [107] and BCa MCF-7 cells [108]. However, no single phosphorylation site on human ERβ has been identified de novo and functionally characterized. Using an unbiased mass spectrometry approach, our laboratory first identified three serines (S75, S87, and S105) as direct phosphorylation targets of ERK1/2 and p38 in the N-terminus of human ERβ. Functional analyses on the phosphorylation of ERβ at S105 demonstrated that this PTM inhibited migration and invasion in BCa cells [109].
Studies of the mouse ERβ have shed light on the functional role of phosphorylation in human ERβ [110;111]. For example, through prediction from mouse data, phosphorylation at S87 in the human ERβ was found to be a target of CXCL12/CXCR4 via activation of the ERK pathway in BCa cells [112]. Tremblay and co-workers extensively elucidated the function of mouse ERβ phosphorylation since they first cloned the gene in 1997 [111]. ERβ phosphorylation of S106 and S124 at the AF-1 domain of the mouse ERβ was ERK1/2-sensitive and associated with increased transactivation of the receptor [110]. Phosphorylation at these two sites was later shown to enhance recruitment of a steroid receptor co-activator 1 (SRC-1) and a co-activator CREB-binding protein (CBP) to the transcriptional complex [110;113] but ERβ phosphorylation at S255 was found to have an opposite effect [114]. Moreover, the same group reported that ERβ phosphorylation at S94 and S106 promoted degradation of the receptor through the ubiquitin-proteasome pathway [115]. A recent review provided a detailed summary of ERβ phosphorylation and its function in the mouse [116]. Although data on ERβ in mice has laid a foundation for human studies, not all information can be applied to human ERβ. The number of predicted kinase-specific motifs differs in humans and mice because of some major differences in the primary sequence, ie, some of the motifs are not preserved in humans, and the AF-1 domain of ERβ in humans is significantly shorter than that in mice. Emerging evidence indicates that phosphorylation of ERβ in mice may function differently from that in humans. For example, kinase p38 or ERβB2/ERβB3 activation repressed ERβ transactivation in mice [117], whereas p38 activation stimulated ERβ-mediated transcription in humans [107;108]. Therefore, further studies are necessary to understand the functions of each phosphorylation site and a combination of identified sites in human ERβ.
Ubiquitylation occurred at serine, threonine, and lysine residues of a protein [118]. Mono- and bi-ubiquitylation affects transcription, protein-protein interactions, and subcellular localization; and poly-ubiquitylation usually targets proteins for degradation through the 26S proteasome pathway. Human ERβ is degraded in an estrogen-dependent manner through ubiquitin/proteasome pathways in BCa cells in vitro, and the N-terminal 37-amino acid region is responsible in the recruitment of the ubiquitin ligase for ERβ degradation [119]. In addition, suppressor for Gal 1 (SUG1) interacts with and stimulates ubiquitin/proteasome-mediated degradation of human ERβ (and ERα), leading to reduced ER transactivation [120]. However, the exact sites of ubiquitylation have been difficult to identify owing to the instability of poly-ubiquitylated proteins. The exact ubiquitylation sites on ERβ remain to be revealed.
S-Nitrosylation is a liable and reversible reaction induced by nitric oxide on the cysteine residue of a protein. S-Nitrosylation of ERα at cysteine residues that coordinate Zn2+ within the two major DNA-binding Zn-finger domains inhibits the DNA binding of ERα at specific ERE [121]. Our laboratory, using mass spectrometry, recently identified three nitrosylation sites on human ERβ. In line with the findings for ERα, S-nitrosylation inhibited ERβ transactivation at ERE (unpublished).
A palmitoylation sequence was identified in the LBD of human ERβ based on sequence prediction, but the sites were not experimentally verified [98]. Palmitoylation has been shown to be essential for maintaining the juxtaposition of ERα with the plasma membrane, interacting with the membrane protein caveolin-1 (see Figure 2), and triggering non-genomic signaling pathways and cell proliferation [97]. Thus, it is reasonable to speculate that palmitoylation may function similarly to retain ERβ in the plasma membrane for rapid signaling, as the interaction between ERβ and caveolae has been shown to be crucial for the non-genomic action of ERβ [122].
Cell-surface and secreted proteins are usually modified by glycosylation. However, glycosylation with N-acetylglucosamine (O-GlcNAc) is more frequently detected in cytosolic and nuclear proteins on serine or threonine hydroxyl side chains [123]. S80 is a target for both phosphorylation and glycosylation in mouse ERβ, and these two modifications collaboratively modulate the degradation and activity of ERβ in the mouse [124]. Yet no glycosylation site has been identified on human ERβ. PTMs, including acetylation, sumolyation, and methylation, occur at lysine residues on steroid receptors [125;99;100]; however, no information on these modifications is currently available for ERβ.
Complex interplay among various types of PTMs on ERα exists: ERα S305 phosphorylation is reported to prevent K303 acetylation and to stimulate ERα activity [126]; and lysine sites (K266, K268, K299, K302, and K303) for acetylation are also common for sumoylation [127]. Until now, clinical studies on the de novo interaction between sumolyation and acetylation have been limited by the lack of antibodies specific to acetylated or sumoylated lysine; this area warrants further exploration. In conclusion, a greater effort should be made to investigate different ERβ PTMs and their inter-relationships because ERβ is expressed in 76% of BCa cases [11] and has been demonstrated to play important roles in cell functions in BCa and in its prognosis.
Increase in the diversity of the receptor’s function by co-regulators and interacting proteins of ERβ
ERβ transactivation requires co-regulators and other transcriptional machinery. Ligands such as E2 enhance the formation of heterodimers between ERs and increase the binding of co-activators to the receptors. SERMS such as tamoxifen conversely facilitate the binding of the co-repressors (see Figure 2). Acquisition of tamoxifen resistance has been shown to be associated with changes in the expression of co-regulators in the cell culture [128]. Co-regulators may play a role in directing which ERβ-regulating gene or gene set can be activated or repressed, thus further contributing to the functional diversity of the receptor. In this section, we focus on the literature on human ERβ co-regulators and their protein-binding partners.
The co-expression of ERs and co-activators correlates with different prognoses of BCa; however, such information remains limited [13;19;129–131]. Young and co-workers found an inverse correlation between steroid receptor co-activator 1 (SRC-1) and ERβ in BCa [13;19]. Whereas the expression of ERβ was associated with better prognosis and responsiveness to tamoxifen, SRC-1 expression had the reverse association [13;19]. However, in another study with only 25 specimens, ERβ expression was associated with SRC-1, transcription intermediary factor 2 (TIF2), and nuclear receptor co-repressor (NCoR) in malignant specimens [130]. Moreover, the expression of ERβ, protein 300 kDa/CREB-binding protein (p300/CBP) and amplified in breast cancer 1 (AIB1) (SRC3) were higher in invasive ductal carcinomas than in normal mammary tissue [130]. The expression of SRC3 within epithelial cells of BCa was positively associated with ERα but inversely associated with ERβ [131]. Although various co-regulators were shown to co-localize with ERβ in benign or malignant cells in BCa specimens, whether they are bona fide co-regulators of ERβ still needs to be determined experimentally. Cell and cell-free models have been used to analyze the ERβ co-regulatory activities of many co-regulators originally studied for their action in modifying ERα transcriptional activities. Both the type of ligand and cis-regulatory elements were important in determining the regulatory action of these co-regulators on ERβ; they often exert differential influences on the two ER subtypes [132].
Besides functioning as classical co-regulators that affect the transcriptional activities of ERβ, this nuclear receptor has also been shown to be influenced by its interaction with a variety of proteins with diverse functions. The better known ones now include inhibitor of differentiation protein (Id1) [133;134], insulin receptor substrate I (IRS-I)[135], and retinoblastoma protein 2 (pRb2/p130)[136]. With the advent of mass spectrometry, more than 300 proteins have been identified as putative ERβ1 binding partners [80]. Several previously proven ERβ1 co-regulators, such as SRC3 [132]; proline, glutamate, and leucine-rich protein 1 (PELP1) [137]; tripartite motif containing 24 (TRIM24) [137]; and mediator complex subunit 1 (MED1) [138] have been found. In addition, proteins involved in post-transcriptional modification of mRNA and actin filament-based processes were newly identified as ERβ protein partners [80]. ERβ was also shown to interact with proteins related to the regulation of apoptosis. They include a mitochondrial pro-apoptotic protein known as mitochondrial ribosomal protein S29 (MRPS29) [139], and Bcl-2-associated transcription factor 1 (BCLAF1), which is a transcriptional repressor localized to the nuclear enveloped and promotes apoptosis [140]. We recently identified BCL2-like 12 (proline rich)(BCL2L12) as a putative ERβ5 isoform-specific interacting protein that exerts anti- and pro-apoptotic functions in a cell-context manner (unpublished).
Looking ahead, the identification of ERβ-specific and ERβ isoform-specific co-regulators or protein-binding partners should help better define the functionality of ERβ in breast tissue, promising an improvement in BCa prognosis.
The differential behaviors of ERβ elicited by different ligands in BCa
E2 is the presumed ligand for ERβ. Will the diagnostic and prognostic value of ERβ vary with patients’ estrogen status? ERβ—ERβ1 in particular—was significantly associated with diminished biological aggressiveness in premenopausal women [141] but correlated with worst outcome in postmenopausal women [22]. Another study reported opposite results for postmenopausal women [14]. Other reports demonstrated no effects of menopausal status on the prognostic value of ERβ (or ERβ1) in BCa, with results independent of ERα expression or tamoxifen treatment [12;14;18;20;21;24;142]. Unfortunately, many of the early studies on ERβ did not determine the menopausal status of the patients, making data interpretation difficult. Better designed studies are needed to clarify this important issue. Tamoxifen has been one of the most common endocrine therapies for ERα-positive BCa for 30 years [143]. However, a notable proportion (~ 30–40%) of patients with BCa relapse within 5 years of post-treatment [144]. Although tamoxifen is believed to target ERα in ERα-positive BCa, this mixed agonist- antagonist can also transactivate ERβ, thus raising the question of whether the latter has prognostic value for tamoxifen responsiveness/resistance. In this regard, in patients with ERα-positive BCa, ERβ was an independent predictor of tamoxifen responsiveness [12]; thus higher levels of ERβ expression correlated with longer disease-free and overall survival (DFS and OS) following tamoxifen therapy [15]. In ERα-negative patients, ERβ expression was also found to associate with a longer duration of distant disease-free survival (DDFS) [24]. This association is further supported by the significant association of promoter hypermethylation of the ERβ promoter, which leads to ERβ silencing, with tamoxifen resistance in BCa [145]. These findings, taken together, indicate that ERβ expression is a good prognostic marker for tamoxifen responsiveness in both ER-positive and ER–negative BCa, although some studies had contrary findings [22].
Fulvestrant is used when patients with BCa patients experience a relapse following tamoxifen therapy. Fulvestrant has also been approved for the treatment of ERα-positive metastatic BCa in postmenopausal women [146]. However, most studies have established correlations only between fulvestrant treatment and ERα but not with ERβ. In a BCa cell model, fulvestrant had little or no effect on the antiproliferative action of ERβ [28], although it stabilized the ERβ protein but promoted the degradation of ERα [147].
The traditional view that ER binds to estrogen and transactivates at the classical vitellogenin ERE was formulated largely from studies on ERα [148]. More recent publications [149;150] now reveal that ERβ can transactivate via different cis-regulatory elements in a ligand-specific manner and thereby influence different cellular functions (Figure 2). Endoxifen, the most important metabolite of tamoxifen, was shown to exert its antiproliferative action through stabilization of ERβ and enhancement of the number of ERα/β heterodimers [151]. In global transcriptome analyses, E2- or SERMs-activated ERα or ERβ action showed few overlaps in gene-expression profiles [152]. ChIP-cloning and -sequencing approaches revealed more commonalities among tamoxifen- and raloxifene-regulated gene sets than those activated by E2 [153]. Consistent with the clinical observations (see above), ectopic expression of ERβ increased the sensitivity of ERα-positive BCa cells to tamoxifen [154], whereas siRNA-mediated knockdown of the receptor reduced the responsiveness of MCF-7 to estrogen and tamoxifen, supporting a role for ERβ as a tumor suppressor [155]. Noteworthy is the finding that antiestrogens (e.g. fulvestrant) and SERMs (e.g. raloxifene) inhibit ERβ transactivation at ERE but enhance the transcriptional activity at other cis-elements, such as AP1, Sp2, NFκB [111;156]. Apropos to the concept of cross-talk with non-ERE cis-elements is the finding that a number of AP1-regulated genes are regulated by ERβ1 through tethering on cJun/cFos complex at the AP1 site [149;157;158]. Using ChIP-seq approach, other studies have identified non-ERE ERβ binding sites, including molecules such as AP2, E2F, and Sp1 [159–161].
Evidence supporting a protective role of phytoestrogens against BCa is conflicting [162;163]. Whereas the increase in soy intake correlated with lower cancer risk in studies of Asian populations, no strong correlations between these two factors were observed in Western populations [164–166]. These findings suggest that lifelong or early-life exposure to dietary soy diet among Asians may be critical to conferring the protective effects of soy on the breast [167]. Soy consumption also influences the type of BCa developed in various populations, in that a diet high in soy products was positively correlated with decreased risk of HER2-negative/ERα-positive/PR-positive BCa in Asian countries [168;169].
Because phytoestrogens can act like agonists in a low-estrogen environment but antagonists in a high-estrogen environment [170;171], the actions of phytoestrogens may vary depending on menopausal status [172]. Moreover, the stronger binding affinity of phytoestrogens to ERβ compared with ERα [173;174] and the significant estrogenic potency of phytoestrogens on ERβ may contribute to their antiproliferative and inhibitory effects on tumor growth [175–177]. Global gene expression profiling studies showed that phytoestrogens have biphasic activity in BCa cells, depending on the relative levels of expression of the two ERs [178–180]. In the absence of ERβ, phytoestrogens induced the same transcriptome changes as E2 in ERα-positive T47D cells [178;179]. The upregulated genes included those involved in cell cycle, DNA replication, chromosome segregation, and inhibition of apoptosis. Ectopic expression of ERβ in T47D resulted in opposite responses to phytoestrogen stimulation, causing inhibition of cell growth and the induction of cell- cycle arrest and apoptosis [179;180]. Of interest, apigenin, a flavone found principally in camomile, chives, garlic, and parsley, also inhibits cell growth in MDA-MB-231 cells via ERβ signaling [181]. Furthermore, the growth inhibitory action of phytoestrogens may also be mediated via their actions in regulating the expression of the various ERβ isoforms. For example, genistein was found to induce the expression of ERβ1 and ERβ2 but not that of ERβ5 in T47D and BT20 BCa cells [182]. The ratio of the expression levels of these isoforms may be a key to determining whether an estrogen is pro- or antiproliferative. Finally, phytoestrogens can significantly affect the interaction between ERβ and co-regulators and thereby contribute to the tissue-dependent response [174]. Genistein preferentially promoted the binding of ERβ to SRC-1a (12,000-fold) and GRIP1 (33-fold) as compared with ERα [174].
Since breast carcinogenesis has been suggested to be related to estrogen-induced oxidative DNA damage [183;184], phytoestrogens may protect against tumorigenesis by reducing intracellular reactive oxygen species. Genistein, biochanin A, and resveratrol were shown to upregulate the expression of quinone reductase, a key enzyme in the maintenance of intracellular antioxidant capacity. The action of these phytochemicals was shown to be mediated preferentially through the transactivation of ERβ as opposed to that of ERα [185]. Biochanin A and resveratrol also significantly inhibited estrogen-induced oxidative DNA damage [186]. To conclude, the action of phytoestrogens in BCa may vary depending on the relative abundance of the two ER subtypes, the levels of E2, the utilization of cis-regulatory elements, and the expression levels of the various ERβ isoforms, hence making it difficult to predict their action in BCa prevention.
Finally, phytoestrogens may influence responses of patients with BCa to chemotherapy. Soy consumption was associated with a reduced risk of recurrence following tamoxifen treatment [187] or improved survival independent of tamoxifen use [188]. In BCa with both ERβ and HER2 expression, genistein was found to promote the growth inhibitory effect of trastuzumab, an anti-cancer drug targeting the HER2 receptor, in BT474 cells [189]. This finding suggests that ERβ-specific agonists potentiate the efficacy and enhance the potency of current BCa therapeutics.
Conclusion
ERβ is not simply a second ER. Its functions differ drastically from those of ERα and deviate more from those expected of a traditional nuclear receptor. It localizes in different cellular compartments and is susceptible to different PTMs. It is expressed in different variant forms, which interacts with multiple protein partners as well as ligands, utilizes canonical and non-canonical cis-elements, and heterodimerizes with ERα and its own isoforms, thereby creating a highly complex labyrinth of functions. In this review, we have summarized and discussed the existing literature in six key research areas of ERβ (see Figure 2). In our opinion, further investigations in these areas are essential to deepen our understanding of ERβ in BCa. Efforts should perhaps be focused on enhancing our understanding of the roles of ERβ isoforms and on devising ERβ-specific therapies that will help prevent or treat BCa.
We hope this review will stimulate additional studies in the following areas: 1) The function of ERβ isoforms in BCa. Do they respond to specific ligands? Do they regulate different gene sets and participate in physiological functions? Are SERMS or antiestrogens the ligands for the isoforms? 2) ERβ subcellular localization. How are ERβ isoforms targeted to different compartments and what are their physiological and pathophysiological roles? Does tamoxifen alter ERβ subcellular localization? 3) Determination of BCa relevant PTMs. What are the functional PTMs on human ERβ? Is there any cross-talk among PTMs? Is there a ligand-specific PTM signature? Is a PTM signature of a higher prognostic value than a single PTM? 4) Are there any ERβ interacting proteins that can dictate ERβ function in BCa? What is the mechanism? What are the relationships between these proteins with ERβ ligands? 5) Is E2 necessary to maintaining the basal function of ERβ in BCa? Is there any ERβ-specific ligand that can be used as a BCa therapeutic? Why are phytoestrogens preferential ligands for ERβ? Can phytoestrogens be used as chemopreventive drugs for BCa on the basis of ERβ status?
This review addresses the gaps in knowledge in ERβ research as it pertains to BCa regarding the following topics:
Tumor suppressive role of ERβ
ERβ isoforms, and its interacting proteins or co-regulators
Nuclear signaling and extranuclear ERβ signaling
Post-translational modifications of ERβ
Ligand and regulatory cis-elements of ERβ
Acknowledgments
We thank Nancy Voynow for her professional editing of this manuscript and thank Dr. Marian Miller for her valuable suggestions on the figures. This work was supported by grants from NIH (ES020988, ES019480, ES006096, CA15776, and CA112532 to SMH) and VA (BX000675 to SMH), and an internal funding source from the University of Cincinnati Medical Center.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
Reference List
- 1.Kuiper GG, Enmark E, Pelto-Huikko M, Nilsson S, Gustafsson JA. Cloning of a novel receptor expressed in rat prostate and ovary. Proc Natl Acad Sci U S A. 1996 Jun 11;93(12):5925–30. doi: 10.1073/pnas.93.12.5925. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Mosselman S, Polman J, Dijkema R. ER beta: identification and characterization of a novel human estrogen receptor. FEBS Lett. 1996 Aug 19;392(1):49–53. doi: 10.1016/0014-5793(96)00782-x. [DOI] [PubMed] [Google Scholar]
- 3.Jensen EV, Jacobsen HI. Basic guides to the mechanism of estrogen action. Rec Prog Horm Res. 1962;18:387–414. [Google Scholar]
- 4.Hall JM, McDonnell DP, Korach KS. Allosteric regulation of estrogen receptor structure, function, and coactivator recruitment by different estrogen response elements. Mol Endocrinol. 2002 Mar;16(3):469–86. doi: 10.1210/mend.16.3.0814. [DOI] [PubMed] [Google Scholar]
- 5.Hall JM, Korach KS. Analysis of the molecular mechanisms of human estrogen receptors alpha and beta reveals differential specificity in target promoter regulation by xenoestrogens. J Biol Chem. 2002 Nov 15;277(46):44455–61. doi: 10.1074/jbc.M200849200. [DOI] [PubMed] [Google Scholar]
- 6.Hyder SM, Chiappetta C, Stancel GM. Interaction of human estrogen receptors alpha and beta with the same naturally occurring estrogen response elements. Biochem Pharmacol. 1999 Mar 15;57(6):597–601. doi: 10.1016/s0006-2952(98)00355-4. [DOI] [PubMed] [Google Scholar]
- 7.Ramsey TL, Risinger KE, Jernigan SC, Mattingly KA, Klinge CM. Estrogen receptor beta isoforms exhibit differences in ligand-activated transcriptional activity in an estrogen response element sequence-dependent manner. Endocrinology. 2004 Jan;145(1):149–60. doi: 10.1210/en.2003-1043. [DOI] [PubMed] [Google Scholar]
- 8.Leygue E, Murphy LC. Comparative evaluation of ERalpha and ERbeta significance in breast cancer: state of the art. Expet Rev Endocrinol Metabol. 2011;6(3):333–43. doi: 10.1586/eem.11.27. [DOI] [PubMed] [Google Scholar]
- 9.Speirs V, Skliris GP, Burdall SE, Carder PJ. Distinct expression patterns of ER alpha and ER beta in normal human mammary gland. J Clin Pathol. 2002 May;55(5):371–4. doi: 10.1136/jcp.55.5.371. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Murphy L, Cherlet T, Lewis A, Banu Y, Watson P. New insights into estrogen receptor function in human breast cancer. Ann Med. 2003;35(8):614–31. doi: 10.1080/07853890310014579. [DOI] [PubMed] [Google Scholar]
- 11.Skliris GP, Leygue E, Watson PH, Murphy LC. Estrogen receptor alpha negative breast cancer patients: estrogen receptor beta as a therapeutic target. J Steroid Biochem Mol Biol. 2008 Mar;109(1–2):1–10. doi: 10.1016/j.jsbmb.2007.12.010. [DOI] [PubMed] [Google Scholar]
- 12.Esslimani-Sahla M, Simony-Lafontaine J, Kramar A, Lavaill R, Mollevi C, Warner M, et al. Estrogen receptor beta (ER beta) level but not its ER beta cx variant helps to predict tamoxifen resistance in breast cancer. Clin Cancer Res. 2004 Sep 1;10(17):5769–76. doi: 10.1158/1078-0432.CCR-04-0389. [DOI] [PubMed] [Google Scholar]
- 13.Fleming FJ, Hill AD, McDermott EW, O'Higgins NJ, Young LS. Differential recruitment of coregulator proteins steroid receptor coactivator-1 and silencing mediator for retinoid and thyroid receptors to the estrogen receptor-estrogen response element by beta-estradiol and 4-hydroxytamoxifen in human breast cancer. J Clin Endocrinol Metab. 2004 Jan;89(1):375–83. doi: 10.1210/jc.2003-031048. [DOI] [PubMed] [Google Scholar]
- 14.Honma N, Horii R, Iwase T, Saji S, Younes M, Takubo K, et al. Clinical importance of estrogen receptor-beta evaluation in breast cancer patients treated with adjuvant tamoxifen therapy. J Clin Oncol. 2008 Aug 1;26(22):3727–34. doi: 10.1200/JCO.2007.14.2968. [DOI] [PubMed] [Google Scholar]
- 15.Hopp TA, Weiss HL, Parra IS, Cui Y, Osborne CK, Fuqua SA. Low levels of estrogen receptor beta protein predict resistance to tamoxifen therapy in breast cancer. Clin Cancer Res. 2004 Nov 15;10(22):7490–9. doi: 10.1158/1078-0432.CCR-04-1114. [DOI] [PubMed] [Google Scholar]
- 16.Iwase H, Zhang Z, Omoto Y, Sugiura H, Yamashita H, Toyama T, et al. Clinical significance of the expression of estrogen receptors alpha and beta for endocrine therapy of breast cancer. Cancer Chemother Pharmacol. 2003 Jul;52( Suppl 1):S34–S38. doi: 10.1007/s00280-003-0592-1. [DOI] [PubMed] [Google Scholar]
- 17.Mann S, Laucirica R, Carlson N, Younes PS, Ali N, Younes A, et al. Estrogen receptor beta expression in invasive breast cancer. Hum Pathol. 2001 Jan;32(1):113–8. doi: 10.1053/hupa.2001.21506. [DOI] [PubMed] [Google Scholar]
- 18.Murphy LC, Leygue E, Niu Y, Snell L, Ho SM, Watson PH. Relationship of coregulator and oestrogen receptor isoform expression to de novo tamoxifen resistance in human breast cancer. Br J Cancer. 2002 Dec 2;87(12):1411–6. doi: 10.1038/sj.bjc.6600654. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Myers E, Fleming FJ, Crotty TB, Kelly G, McDermott EW, O'Higgins NJ, et al. Inverse relationship between ER-beta and SRC-1 predicts outcome in endocrine-resistant breast cancer. Br J Cancer. 2004 Nov 1;91(9):1687–93. doi: 10.1038/sj.bjc.6602156. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Nakopoulou L, Lazaris AC, Panayotopoulou EG, Giannopoulou I, Givalos N, Markaki S, et al. The favourable prognostic value of oestrogen receptor beta immunohistochemical expression in breast cancer. J Clin Pathol. 2004 May;57(5):523–8. doi: 10.1136/jcp.2003.008599. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Jensen EV, Cheng G, Palmieri C, Saji S, Makela S, Van Noorden S, et al. Estrogen receptors and proliferation markers in primary and recurrent breast cancer. Proc Natl Acad Sci U S A. 2001 Dec 18;98(26):15197–202. doi: 10.1073/pnas.211556298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.O'Neill PA, Davies MP, Shaaban AM, Innes H, Torevell A, Sibson DR, et al. Wild-type oestrogen receptor beta (ERbeta1) mRNA and protein expression in Tamoxifen-treated post-menopausal breast cancers. Br J Cancer. 2004 Nov 1;91(9):1694–702. doi: 10.1038/sj.bjc.6602183. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Skliris GP, Leygue E, Curtis-Snell L, Watson PH, Murphy LC. Expression of oestrogen receptor-beta in oestrogen receptor-alpha negative human breast tumours. Br J Cancer. 2006 Sep 4;95(5):616–26. doi: 10.1038/sj.bjc.6603295. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Gruvberger-Saal SK, Bendahl PO, Saal LH, Laakso M, Hegardt C, Eden P, et al. Estrogen receptor beta expression is associated with tamoxifen response in ERalpha-negative breast carcinoma. Clin Cancer Res. 2007 Apr 1;13(7):1987–94. doi: 10.1158/1078-0432.CCR-06-1823. [DOI] [PubMed] [Google Scholar]
- 25.Chang EC, Frasor J, Komm B, Katzenellenbogen BS. Impact of estrogen receptor beta on gene networks regulated by estrogen receptor alpha in breast cancer cells. Endocrinology. 2006 Oct;147(10):4831–42. doi: 10.1210/en.2006-0563. [DOI] [PubMed] [Google Scholar]
- 26.Paruthiyil S, Parmar H, Kerekatte V, Cunha GR, Firestone GL, Leitman DC. Estrogen receptor beta inhibits human breast cancer cell proliferation and tumor formation by causing a G2 cell cycle arrest. Cancer Res. 2004 Jan 1;64(1):423–8. doi: 10.1158/0008-5472.can-03-2446. [DOI] [PubMed] [Google Scholar]
- 27.Roger P, Sahla ME, Makela S, Gustafsson JA, Baldet P, Rochefort H. Decreased expression of estrogen receptor beta protein in proliferative preinvasive mammary tumors. Cancer Res. 2001 Mar 15;61(6):2537–41. [PubMed] [Google Scholar]
- 28.Strom A, Hartman J, Foster JS, Kietz S, Wimalasena J, Gustafsson JA. Estrogen receptor beta inhibits 17beta-estradiol-stimulated proliferation of the breast cancer cell line T47D. Proc Natl Acad Sci U S A. 2004 Feb 10;101(6):1566–71. doi: 10.1073/pnas.0308319100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Lazennec G, Bresson D, Lucas A, Chauveau C, Vignon F. ER beta inhibits proliferation and invasion of breast cancer cells. Endocrinology. 2001 Sep;142(9):4120–30. doi: 10.1210/endo.142.9.8395. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Platet N, Cunat S, Chalbos D, Rochefort H, Garcia M. Unliganded and liganded estrogen receptors protect against cancer invasion via different mechanisms. Mol Endocrinol. 2000 Jul;14(7):999–1009. doi: 10.1210/mend.14.7.0492. [DOI] [PubMed] [Google Scholar]
- 31.Hartman J, Lindberg K, Morani A, Inzunza J, Strom A, Gustafsson JA. Estrogen receptor beta inhibits angiogenesis and growth of T47D breast cancer xenografts. Cancer Res. 2006 Dec 1;66(23):11207–13. doi: 10.1158/0008-5472.CAN-06-0017. [DOI] [PubMed] [Google Scholar]
- 32.Hou YF, Yuan ST, Li HC, Wu J, Lu JS, Liu G, et al. ERbeta exerts multiple stimulative effects on human breast carcinoma cells. Oncogene. 2004 Jul 29;23(34):5799–806. doi: 10.1038/sj.onc.1207765. [DOI] [PubMed] [Google Scholar]
- 33.Chambers AF. MDA-MB-435 and M14 cell lines: identical but not M14 melanoma? Cancer Res. 2009 Jul 1;69(13):5292–3. doi: 10.1158/0008-5472.CAN-09-1528. [DOI] [PubMed] [Google Scholar]
- 34.Dotzlaw H, Leygue E, Watson PH, Murphy LC. Expression of estrogen receptor-beta in human breast tumors. J Clin Endocrinol Metab. 1997 Jul;82(7):2371–4. doi: 10.1210/jcem.82.7.4212. [DOI] [PubMed] [Google Scholar]
- 35.Iwao K, Miyoshi Y, Egawa C, Ikeda N, Tsukamoto F, Noguchi S. Quantitative analysis of estrogen receptor-alpha and -beta messenger RNA expression in breast carcinoma by real-time polymerase chain reaction. Cancer. 2000 Oct 15;89(8):1732–8. doi: 10.1002/1097-0142(20001015)89:8<1732::AID-CNCR13>3.0.CO;2-2. [DOI] [PubMed] [Google Scholar]
- 36.Leygue E, Dotzlaw H, Watson PH, Murphy LC. Altered estrogen receptor alpha and beta messenger RNA expression during human breast tumorigenesis. Cancer Res. 1998 Aug 1;58(15):3197–201. [PubMed] [Google Scholar]
- 37.Shaw JA, Udokang K, Mosquera JM, Chauhan H, Jones JL, Walker RA. Oestrogen receptors alpha and beta differ in normal human breast and breast carcinomas. J Pathol. 2002 Dec;198(4):450–7. doi: 10.1002/path.1230. [DOI] [PubMed] [Google Scholar]
- 38.Skliris GP, Munot K, Bell SM, Carder PJ, Lane S, Horgan K, et al. Reduced expression of oestrogen receptor beta in invasive breast cancer and its re-expression using DNA methyl transferase inhibitors in a cell line model. J Pathol. 2003 Oct;201(2):213–20. doi: 10.1002/path.1436. [DOI] [PubMed] [Google Scholar]
- 39.Zhao C, Lam EW, Sunters A, Enmark E, De Bella MT, Coombes RC, et al. Expression of estrogen receptor beta isoforms in normal breast epithelial cells and breast cancer: regulation by methylation. Oncogene. 2003 Oct 23;22(48):7600–6. doi: 10.1038/sj.onc.1207100. [DOI] [PubMed] [Google Scholar]
- 40.Rody A, Holtrich U, Solbach C, Kourtis K, von Minckwitz G, Engels K, et al. Methylation of estrogen receptor beta promoter correlates with loss of ER-beta expression in mammary carcinoma and is an early indication marker in premalignant lesions. Endocr Relat Cancer. 2005 Dec;12(4):903–16. doi: 10.1677/erc.1.01088. [DOI] [PubMed] [Google Scholar]
- 41.Zhu X, Leav I, Leung YK, Wu M, Liu Q, Gao Y, et al. Dynamic regulation of estrogen receptor-beta expression by DNA methylation during prostate cancer development and metastasis. Am J Pathol. 2004 Jun;164(6):2003–12. doi: 10.1016/s0002-9440(10)63760-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Yap OW, Bhat G, Liu L, Tollefsbol TO. Epigenetic modifications of the Estrogen receptor beta gene in epithelial ovarian cancer cells. Anticancer Res. 2009 Jan;29(1):139–44. [PMC free article] [PubMed] [Google Scholar]
- 43.Leung YK, Lam HM, Wu S, Song D, Levin L, Cheng L, et al. Estrogen receptor beta2 and beta5 are associated with poor prognosis in prostate cancer, and promote cancer cell migration and invasion. Endocr Relat Cancer. 2010 Sep;17(3):675–89. doi: 10.1677/ERC-09-0294. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Shaaban AM, Green AR, Karthik S, Alizadeh Y, Hughes TA, Harkins L, et al. Nuclear and cytoplasmic expression of ERbeta1, ERbeta2, and ERbeta5 identifies distinct prognostic outcome for breast cancer patients. Clin Cancer Res. 2008 Aug 15;14(16):5228–35. doi: 10.1158/1078-0432.CCR-07-4528. [DOI] [PubMed] [Google Scholar]
- 45.Moore JT, McKee DD, Slentz-Kesler K, Moore LB, Jones SA, Horne EL, et al. Cloning and characterization of human estrogen receptor beta isoforms. Biochem Biophys Res Commun. 1998 Jun 9;247(1):75–8. doi: 10.1006/bbrc.1998.8738. [DOI] [PubMed] [Google Scholar]
- 46.Poola I, Abraham J, Baldwin K, Saunders A, Bhatnagar R. Estrogen receptors beta4 and beta5 are full length functionally distinct ERbeta isoforms: cloning from human ovary and functional characterization. Endocrine. 2005 Aug;27(3):227–38. doi: 10.1385/ENDO:27:3:227. [DOI] [PubMed] [Google Scholar]
- 47.Leung YK, Mak P, Hassan S, Ho SM. Estrogen receptor (ER)-beta isoforms: A key to understanding ER–beta signaling. Proc Natl Acad Sci U S A. 2006 Aug 29;103(35):13162–7. doi: 10.1073/pnas.0605676103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Cammarata PR, Flynn J, Gottipati S, Chu S, Dimitrijevich S, Younes M, et al. Differential expression and comparative subcellular localization of estrogen receptor beta isoforms in virally transformed and normal cultured human lens epithelial cells. Exp Eye Res. 2005 Aug;81(2):165–75. doi: 10.1016/j.exer.2005.01.019. [DOI] [PubMed] [Google Scholar]
- 49.Chakravarty D, Srinivasan R, Ghosh S, Gopalan S, Rajwanshi A, Majumdar S. Estrogen receptor beta1 and the beta2/betacx isoforms in nonneoplastic endometrium and in endometrioid carcinoma. Int J Gynecol Cancer. 2007 Jul;17(4):905–13. doi: 10.1111/j.1525-1438.2006.00851.x. [DOI] [PubMed] [Google Scholar]
- 50.Chi A, Chen X, Chirala M, Younes M. Differential expression of estrogen receptor beta isoforms in human breast cancer tissue. Anticancer Res. 2003 Jan;23(1A):211–6. [PubMed] [Google Scholar]
- 51.Saunders PT, Millar MR, Macpherson S, Irvine DS, Groome NP, Evans LR, et al. ERbeta1 and the ERbeta2 splice variant (ERbetacx/beta2) are expressed in distinct cell populations in the adult human testis. J Clin Endocrinol Metab. 2002 Jun;87(6):2706–15. doi: 10.1210/jcem.87.6.8619. [DOI] [PubMed] [Google Scholar]
- 52.Scobie GA, Macpherson S, Millar MR, Groome NP, Romana PG, Saunders PT. Human oestrogen receptors: differential expression of ER alpha and beta and the identification of ER beta variants. Steroids. 2002 Nov;67(12):985–92. doi: 10.1016/s0039-128x(02)00047-8. [DOI] [PubMed] [Google Scholar]
- 53.Weitsman GE, Skliris G, Ung K, Peng B, Younes M, Watson PH, et al. Assessment of multiple different estrogen receptor-beta antibodies for their ability to immunoprecipitate under chromatin immunoprecipitation conditions. Breast Cancer Res Treat. 2006 Nov;100(1):23–31. doi: 10.1007/s10549-006-9229-5. [DOI] [PubMed] [Google Scholar]
- 54.Green CA, Peter MB, Speirs V, Shaaban AM. The potential role of ER beta isoforms in the clinical management of breast cancer. Histopathology. 2008 Oct;53(4):374–80. doi: 10.1111/j.1365-2559.2008.02968.x. [DOI] [PubMed] [Google Scholar]
- 55.Murphy LC, Leygue E. The role of estrogen receptor-beta in breast cancer. Semin Reprod Med. 2012 Jan;30(1):5–13. doi: 10.1055/s-0031-1299592. [DOI] [PubMed] [Google Scholar]
- 56.Honma N, Saji S, Kurabayashi R, Aida J, Arai T, Horii R, et al. Oestrogen receptor-beta1 but not oestrogen receptor-betacx is of prognostic value in apocrine carcinoma of the breast. APMIS. 2008 Oct;116(10):923–30. doi: 10.1111/j.1600-0463.2008.01122.x. [DOI] [PubMed] [Google Scholar]
- 57.Papadaki I, Mylona E, Giannopoulou I, Markaki S, Keramopoulos A, Nakopoulou L. PPARgamma expression in breast cancer: clinical value and correlation with ERbeta. Histopathology. 2005 Jan;46(1):37–42. doi: 10.1111/j.1365-2559.2005.02056.x. [DOI] [PubMed] [Google Scholar]
- 58.Yan M, Rayoo M, Takano EA, Fox SB. Nuclear and cytoplasmic expressions of ERbeta1 and ERbeta2 are predictive of response to therapy and alters prognosis in familial breast cancers. Breast Cancer Res Treat. 2011 Apr;126(2):395–405. doi: 10.1007/s10549-010-0941-9. [DOI] [PubMed] [Google Scholar]
- 59.Sugiura H, Toyama T, Hara Y, Zhang Z, Kobayashi S, Fujii Y, et al. Expression of estrogen receptor beta wild-type and its variant ERbetacx/beta2 is correlated with better prognosis in breast cancer. Jpn J Clin Oncol. 2007 Nov;37(11):820–8. doi: 10.1093/jjco/hym114. [DOI] [PubMed] [Google Scholar]
- 60.Miller WR, Anderson TJ, Dixon JM, Saunders PT. Oestrogen receptor beta and neoadjuvant therapy with tamoxifen: prediction of response and effects of treatment. Br J Cancer. 2006 May 8;94(9):1333–8. doi: 10.1038/sj.bjc.6603082. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Vinayagam R, Sibson DR, Holcombe C, Aachi V, Davies MP. Association of oestrogen receptor beta 2 (ER beta 2/ER beta cx) with outcome of adjuvant endocrine treatment for primary breast cancer--a retrospective study. BMC Cancer. 2007;7:131. doi: 10.1186/1471-2407-7-131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Wurster M, Ruoff A, Meisner C, Seeger H, Vogel U, Juhasz-Boss I, et al. Evaluation of ERalpha, PR and ERbeta isoforms in neoadjuvant treated breast cancer. Oncol Rep. 2010 Sep;24(3):653–9. [PubMed] [Google Scholar]
- 63.Yamashita H, Nishio M, Kobayashi S, Ando Y, Sugiura H, Zhang Z, et al. Phosphorylation of estrogen receptor alpha serine 167 is predictive of response to endocrine therapy and increases postrelapse survival in metastatic breast cancer. Breast Cancer Res. 2005;7(5):R753–R764. doi: 10.1186/bcr1285. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Esslimani-Sahla M, Kramar A, Simony-Lafontaine J, Warner M, Gustafsson JA, Rochefort H. Increased estrogen receptor betacx expression during mammary carcinogenesis. Clin Cancer Res. 2005 May 1;11(9):3170–4. doi: 10.1158/1078-0432.CCR-04-2298. [DOI] [PubMed] [Google Scholar]
- 65.Saji S, Omoto Y, Shimizu C, Warner M, Hayashi Y, Horiguchi S, et al. Expression of estrogen receptor (ER) (beta)cx protein in ER(alpha)-positive breast cancer: specific correlation with progesterone receptor. Cancer Res. 2002 Sep 1;62(17):4849–53. [PubMed] [Google Scholar]
- 66.Hall JM, McDonnell DP. The estrogen receptor beta-isoform (ERbeta) of the human estrogen receptor modulates ERalpha transcriptional activity and is a key regulator of the cellular response to estrogens and antiestrogens. Endocrinology. 1999 Dec;140(12):5566–78. doi: 10.1210/endo.140.12.7179. [DOI] [PubMed] [Google Scholar]
- 67.Peng B, Lu B, Leygue E, Murphy LC. Putative functional characteristics of human estrogen receptor-beta isoforms. J Mol Endocrinol. 2003 Feb;30(1):13–29. doi: 10.1677/jme.0.0300013. [DOI] [PubMed] [Google Scholar]
- 68.Zhao C, Matthews J, Tujague M, Wan J, Strom A, Toresson G, et al. Estrogen receptor beta2 negatively regulates the transactivation of estrogen receptor alpha in human breast cancer cells. Cancer Res. 2007 Apr 15;67(8):3955–62. doi: 10.1158/0008-5472.CAN-06-3505. [DOI] [PubMed] [Google Scholar]
- 69.Ogawa S, Inoue S, Watanabe T, Orimo A, Hosoi T, Ouchi Y, et al. Molecular cloning and characterization of human estrogen receptor betacx: a potential inhibitor of estrogen action in human. Nucleic Acids Res. 1998 Aug 1;26(15):3505–12. doi: 10.1093/nar/26.15.3505. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Omoto Y, Eguchi H, Yamamoto-Yamaguchi Y, Hayashi S. Estrogen receptor (ER) beta1 and ERbetacx/beta2 inhibit ERalpha function differently in breast cancer cell line MCF7. Oncogene. 2003 Aug 7;22(32):5011–20. doi: 10.1038/sj.onc.1206787. [DOI] [PubMed] [Google Scholar]
- 71.Skliris GP, Parkes AT, Limer JL, Burdall SE, Carder PJ, Speirs V. Evaluation of seven oestrogen receptor beta antibodies for immunohistochemistry, western blotting, and flow cytometry in human breast tissue. J Pathol. 2002 Jun;197(2):155–62. doi: 10.1002/path.1077. [DOI] [PubMed] [Google Scholar]
- 72.Heldring N, Pawson T, McDonnell D, Treuter E, Gustafsson JA, Pike AC. Structural insights into corepressor recognition by antagonist-bound estrogen receptors. J Biol Chem. 2007 Apr 6;282(14):10449–55. doi: 10.1074/jbc.M611424200. [DOI] [PubMed] [Google Scholar]
- 73.Hayashi SI, Eguchi H, Tanimoto K, Yoshida T, Omoto Y, Inoue A, et al. The expression and function of estrogen receptor alpha and beta in human breast cancer and its clinical application. Endocr Relat Cancer. 2003 Jun;10(2):193–202. doi: 10.1677/erc.0.0100193. [DOI] [PubMed] [Google Scholar]
- 74.Augereau P, Miralles F, Cavailles V, Gaudelet C, Parker M, Rochefort H. Characterization of the proximal estrogen-responsive element of human cathepsin D gene. Mol Endocrinol. 1994 Jun;8(6):693–703. doi: 10.1210/mend.8.6.7935485. [DOI] [PubMed] [Google Scholar]
- 75.Girdler F, Browell DA, Cunliffe WJ, Shenton BK, Hemming JD, Scorer P, et al. Use of the monoclonal antibody DAKO-ERbeta (8D5-1) to measure oestrogen receptor beta in breast cancer cells. Cytometry. 2001 Sep 1;45(1):65–72. doi: 10.1002/1097-0320(20010901)45:1<65::aid-cyto1145>3.0.co;2-8. [DOI] [PubMed] [Google Scholar]
- 76.Chen JQ, Yager JD. Estrogen's effects on mitochondrial gene expression: mechanisms and potential contributions to estrogen carcinogenesis. Ann N Y Acad Sci. 2004 Dec;1028:258–72. doi: 10.1196/annals.1322.030. [DOI] [PubMed] [Google Scholar]
- 77.Chen JQ, Eshete M, Alworth WL, Yager JD. Binding of MCF-7 cell mitochondrial proteins and recombinant human estrogen receptors alpha and beta to human mitochondrial DNA estrogen response elements. J Cell Biochem. 2004 Oct 1;93(2):358–73. doi: 10.1002/jcb.20178. [DOI] [PubMed] [Google Scholar]
- 78.Chen JQ, Delannoy M, Cooke C, Yager JD. Mitochondrial localization of ERalpha and ERbeta in human MCF7 cells. Am J Physiol Endocrinol Metab. 2004 Jun;286(6):E1011–E1022. doi: 10.1152/ajpendo.00508.2003. [DOI] [PubMed] [Google Scholar]
- 79.Clayton DA. Replication and transcription of vertebrate mitochondrial DNA. Annu Rev Cell Biol. 1991;7:453–78. doi: 10.1146/annurev.cb.07.110191.002321. [DOI] [PubMed] [Google Scholar]
- 80.Nassa G, Tarallo R, Ambrosino C, Bamundo A, Ferraro L, Paris O, et al. A large set of estrogen receptor beta-interacting proteins identified by tandem affinity purification in hormone-responsive human breast cancer cell nuclei. Proteomics. 2011 Jan;11(1):159–65. doi: 10.1002/pmic.201000344. [DOI] [PubMed] [Google Scholar]
- 81.Chen JQ, Russo PA, Cooke C, Russo IH, Russo J. ERbeta shifts from mitochondria to nucleus during estrogen-induced neoplastic transformation of human breast epithelial cells and is involved in estrogen-induced synthesis of mitochondrial respiratory chain proteins. Biochim Biophys Acta. 2007 Dec;1773(12):1732–46. doi: 10.1016/j.bbamcr.2007.05.008. [DOI] [PubMed] [Google Scholar]
- 82.Pedram A, Razandi M, Wallace DC, Levin ER. Functional estrogen receptors in the mitochondria of breast cancer cells. Mol Biol Cell. 2006 May;17(5):2125–37. doi: 10.1091/mbc.E05-11-1013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Harrington WR, Sheng S, Barnett DH, Petz LN, Katzenellenbogen JA, Katzenellenbogen BS. Activities of estrogen receptor alpha- and beta-selective ligands at diverse estrogen responsive gene sites mediating transactivation or transrepression. Mol Cell Endocrinol. 2003 Aug 29;206(1–2):13–22. doi: 10.1016/s0303-7207(03)00255-7. [DOI] [PubMed] [Google Scholar]
- 84.Hammes SR, Levin ER. Minireview: Recent Advances in Extranuclear Steroid Receptor Actions. Endocrinology. 2011 Oct 25; doi: 10.1210/en.2011-1470. [DOI] [PMC free article] [PubMed]
- 85.Levin ER. Cell localization, physiology, and nongenomic actions of estrogen receptors. J Appl Physiol. 2001 Oct;91(4):1860–7. doi: 10.1152/jappl.2001.91.4.1860. [DOI] [PubMed] [Google Scholar]
- 86.Levin ER. Plasma membrane estrogen receptors. Trends Endocrinol Metab. 2009 Dec;20(10):477–82. doi: 10.1016/j.tem.2009.06.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Levin ER. Minireview: Extranuclear steroid receptors: roles in modulation of cell functions. Mol Endocrinol. 2011 Mar;25(3):377–84. doi: 10.1210/me.2010-0284. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Mendelsohn ME, Karas RH. Rapid progress for non-nuclear estrogen receptor signaling. J Clin Invest. 2010 Jul 1;120(7):2277–9. doi: 10.1172/JCI43756. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Wu Q, Chambliss K, Umetani M, Mineo C, Shaul PW. Non-nuclear estrogen receptor signaling in the endothelium. J Biol Chem. 2011 Apr 29;286(17):14737–43. doi: 10.1074/jbc.R110.191791. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Zhao C, Dahlman-Wright K, Gustafsson JA. Estrogen signaling via estrogen receptor {beta} J Biol Chem. 2010 Dec 17;285(51):39575–9. doi: 10.1074/jbc.R110.180109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Levin ER, Pietras RJ. Estrogen receptors outside the nucleus in breast cancer. Breast Cancer Res Treat. 2008 Apr;108(3):351–61. doi: 10.1007/s10549-007-9618-4. [DOI] [PubMed] [Google Scholar]
- 92.Rai D, Frolova A, Frasor J, Carpenter AE, Katzenellenbogen BS. Distinctive actions of membrane-targeted versus nuclear localized estrogen receptors in breast cancer cells. Mol Endocrinol. 2005 Jun;19(6):1606–17. doi: 10.1210/me.2004-0468. [DOI] [PubMed] [Google Scholar]
- 93.Razandi M, Pedram A, Greene GL, Levin ER. Cell membrane and nuclear estrogen receptors (ERs) originate from a single transcript: studies of ERalpha and ERbeta expressed in Chinese hamster ovary cells. Mol Endocrinol. 1999 Feb;13(2):307–19. doi: 10.1210/mend.13.2.0239. [DOI] [PubMed] [Google Scholar]
- 94.Kelly MJ, Ronnekleiv OK. Membrane-initiated estrogen signaling in hypothalamic neurons. Mol Cell Endocrinol. 2008 Aug 13;290(1–2):14–23. doi: 10.1016/j.mce.2008.04.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Micevych PE, Mermelstein PG. Membrane estrogen receptors acting through metabotropic glutamate receptors: an emerging mechanism of estrogen action in brain. Mol Neurobiol. 2008 Aug;38(1):66–77. doi: 10.1007/s12035-008-8034-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Acconcia F, Bocedi A, Ascenzi P, Marino M. Does palmitoylation target estrogen receptors to plasma membrane caveolae? IUBMB Life. 2003 Jan;55(1):33–5. doi: 10.1080/1521654031000081256. [DOI] [PubMed] [Google Scholar]
- 97.Acconcia F, Ascenzi P, Bocedi A, Spisni E, Tomasi V, Trentalance A, et al. Palmitoylation-dependent estrogen receptor alpha membrane localization: regulation by 17beta-estradiol. Mol Biol Cell. 2005 Jan;16(1):231–7. doi: 10.1091/mbc.E04-07-0547. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Pedram A, Razandi M, Sainson RC, Kim JK, Hughes CC, Levin ER. A conserved mechanism for steroid receptor translocation to the plasma membrane. J Biol Chem. 2007 Aug 3;282(31):22278–88. doi: 10.1074/jbc.M611877200. [DOI] [PubMed] [Google Scholar]
- 99.Faus H, Haendler B. Post-translational modifications of steroid receptors. Biomed Pharmacother. 2006 Nov;60(9):520–8. doi: 10.1016/j.biopha.2006.07.082. [DOI] [PubMed] [Google Scholar]
- 100.Le RM, Poulard C, Cohen P, Sentis S, Renoir JM, Corbo L. Cracking the estrogen receptor's posttranslational code in breast tumors. Endocr Rev. 2011 Oct;32(5):597–622. doi: 10.1210/er.2010-0016. [DOI] [PubMed] [Google Scholar]
- 101.Thomas C, Gustafsson JA. The different roles of ER subtypes in cancer biology and therapy. Nat Rev Cancer. 2011 Aug;11(8):597–608. doi: 10.1038/nrc3093. [DOI] [PubMed] [Google Scholar]
- 102.Hamilton-Burke W, Coleman L, Cummings M, Green CA, Holliday DL, Horgan K, et al. Phosphorylation of estrogen receptor beta at serine 105 is associated with good prognosis in breast cancer. Am J Pathol. 2010 Sep;177(3):1079–86. doi: 10.2353/ajpath.2010.090886. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Murphy LC, Weitsman GE, Skliris GP, Teh EM, Li L, Peng B, et al. Potential role of estrogen receptor alpha (ERalpha) phosphorylated at Serine118 in human breast cancer in vivo. J Steroid Biochem Mol Biol. 2006 Dec;102(1–5):139–46. doi: 10.1016/j.jsbmb.2006.09.021. [DOI] [PubMed] [Google Scholar]
- 104.Murphy LC, Skliris GP, Rowan BG, Al–Dhaheri M, Williams C, Penner C, et al. The relevance of phosphorylated forms of estrogen receptor in human breast cancer in vivo. J Steroid Biochem Mol Biol. 2009 Mar;114(1–2):90–5. doi: 10.1016/j.jsbmb.2009.01.017. [DOI] [PubMed] [Google Scholar]
- 105.Skliris GP, Rowan BG, Al-Dhaheri M, Williams C, Troup S, Begic S, et al. Immunohistochemical validation of multiple phospho-specific epitopes for estrogen receptor alpha (ERalpha) in tissue microarrays of ERalpha positive human breast carcinomas. Breast Cancer Res Treat. 2009 Dec;118(3):443–53. doi: 10.1007/s10549-008-0267-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Murphy LC, Seekallu SV, Watson PH. Clinical significance of estrogen receptor phosphorylation. Endocr Relat Cancer. 2011 Feb;18(1):R1–14. doi: 10.1677/ERC-10-0070. [DOI] [PubMed] [Google Scholar]
- 107.Driggers PH, Segars JH, Rubino DM. The proto-oncoprotein Brx activates estrogen receptor beta by a p38 mitogen-activated protein kinase pathway. J Biol Chem. 2001 Dec 14;276(50):46792–7. doi: 10.1074/jbc.M106927200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Frigo DE, Basu A, Nierth-Simpson EN, Weldon CB, Dugan CM, Elliott S, et al. p38 mitogen-activated protein kinase stimulates estrogen-mediated transcription and proliferation through the phosphorylation and potentiation of the p160 coactivator glucocorticoid receptor-interacting protein 1. Mol Endocrinol. 2006 May;20(5):971–83. doi: 10.1210/me.2004-0075. [DOI] [PubMed] [Google Scholar]
- 109.Lam HM, Suresh Babu CV, Wang J, Yuan Y, Lam YW, Ho SM, et al. Phosphorylation of human estrogen receptor-beta at serine 105 inhibits breast cancer cell migration and invasion. Mol Cell Endocrinol. 2012 Feb 19; doi: 10.1016/j.mce.2012.02.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Tremblay A, Tremblay GB, Labrie F, Giguere V. Ligand-independent recruitment of SRC-1 to estrogen receptor beta through phosphorylation of activation function AF-1. Mol Cell. 1999 Apr;3(4):513–9. doi: 10.1016/s1097-2765(00)80479-7. [DOI] [PubMed] [Google Scholar]
- 111.Tremblay GB, Tremblay A, Copeland NG, Gilbert DJ, Jenkins NA, Labrie F, et al. Cloning, chromosomal localization, and functional analysis of the murine estrogen receptor beta. Mol Endocrinol. 1997 Mar;11(3):353–65. doi: 10.1210/mend.11.3.9902. [DOI] [PubMed] [Google Scholar]
- 112.Sauve K, Lepage J, Sanchez M, Heveker N, Tremblay A. Positive feedback activation of estrogen receptors by the CXCL12-CXCR4 pathway. Cancer Res. 2009 Jul 15;69(14):5793–800. doi: 10.1158/0008-5472.CAN-08-4924. [DOI] [PubMed] [Google Scholar]
- 113.Tremblay A, Giguere V. Contribution of steroid receptor coactivator-1 and CREB binding protein in ligand–independent activity of estrogen receptor beta. J Steroid Biochem Mol Biol. 2001 Apr;77(1):19–27. doi: 10.1016/s0960-0760(01)00031-0. [DOI] [PubMed] [Google Scholar]
- 114.Sanchez M, Sauve K, Picard N, Tremblay A. The hormonal response of estrogen receptor beta is decreased by the phosphatidylinositol 3-kinase/Akt pathway via a phosphorylation-dependent release of CREB-binding protein. J Biol Chem. 2007 Feb 16;282(7):4830–40. doi: 10.1074/jbc.M607908200. [DOI] [PubMed] [Google Scholar]
- 115.Picard N, Charbonneau C, Sanchez M, Licznar A, Busson M, Lazennec G, et al. Phosphorylation of activation function-1 regulates proteasome-dependent nuclear mobility and E6-associated protein ubiquitin ligase recruitment to the estrogen receptor beta. Mol Endocrinol. 2008 Feb;22(2):317–30. doi: 10.1210/me.2007-0281. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116.Sanchez M, Picard N, Sauve K, Tremblay A. Challenging estrogen receptor beta with phosphorylation. Trends Endocrinol Metab. 2010 Feb;21(2):104–10. doi: 10.1016/j.tem.2009.09.007. [DOI] [PubMed] [Google Scholar]
- 117.St-Laurent V, Sanchez M, Charbonneau C, Tremblay A. Selective hormone-dependent repression of estrogen receptor beta by a p38-activated ErbB2/ErbB3 pathway. J Steroid Biochem Mol Biol. 2005 Feb;94(1–3):23–37. doi: 10.1016/j.jsbmb.2005.02.001. [DOI] [PubMed] [Google Scholar]
- 118.Wang X, Herr RA, Chua WJ, Lybarger L, Wiertz EJ, Hansen TH. Ubiquitination of serine, threonine, or lysine residues on the cytoplasmic tail can induce ERAD of MHC–I by viral E3 ligase mK3. J Cell Biol. 2007 May 21;177(4):613–24. doi: 10.1083/jcb.200611063. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 119.Tateishi Y, Sonoo R, Sekiya Y, Sunahara N, Kawano M, Wayama M, et al. Turning off estrogen receptor beta-mediated transcription requires estrogen-dependent receptor proteolysis. Mol Cell Biol. 2006 Nov;26(21):7966–76. doi: 10.1128/MCB.00713-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 120.Masuyama H, Hiramatsu Y. Involvement of suppressor for Gal 1 in the ubiquitin/proteasome-mediated degradation of estrogen receptors. J Biol Chem. 2004 Mar 26;279(13):12020–6. doi: 10.1074/jbc.M312762200. [DOI] [PubMed] [Google Scholar]
- 121.Garban HJ, Marquez-Garban DC, Pietras RJ, Ignarro LJ. Rapid nitric oxide-mediated S-nitrosylation of estrogen receptor: regulation of estrogen-dependent gene transcription. Proc Natl Acad Sci U S A. 2005 Feb 15;102(7):2632–6. doi: 10.1073/pnas.0409854102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 122.Chambliss KL, Yuhanna IS, Anderson RG, Mendelsohn ME, Shaul PW. ERbeta has nongenomic action in caveolae. Mol Endocrinol. 2002 May;16(5):938–46. doi: 10.1210/mend.16.5.0827. [DOI] [PubMed] [Google Scholar]
- 123.Cheng X, Hart GW. Glycosylation of the murine estrogen receptor-alpha. J Steroid Biochem Mol Biol. 2000 Dec 15;75(2–3):147–58. doi: 10.1016/s0960-0760(00)00167-9. [DOI] [PubMed] [Google Scholar]
- 124.Cheng X, Hart GW. Alternative O-glycosylation/O-phosphorylation of serine-16 in murine estrogen receptor beta: post-translational regulation of turnover and transactivation activity. J Biol Chem. 2001 Mar 30;276(13):10570–5. doi: 10.1074/jbc.M010411200. [DOI] [PubMed] [Google Scholar]
- 125.Hochstrasser M. Origin and function of ubiquitin-like proteins. Nature. 2009 Mar 26;458(7237):422–9. doi: 10.1038/nature07958. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 126.Cui Y, Zhang M, Pestell R, Curran EM, Welshons WV, Fuqua SA. Phosphorylation of estrogen receptor alpha blocks its acetylation and regulates estrogen sensitivity. Cancer Res. 2004 Dec 15;64(24):9199–208. doi: 10.1158/0008-5472.CAN-04-2126. [DOI] [PubMed] [Google Scholar]
- 127.Sentis S, Le RM, Bianchin C, Rostan MC, Corbo L. Sumoylation of the estrogen receptor alpha hinge region regulates its transcriptional activity. Mol Endocrinol. 2005 Nov;19(11):2671–84. doi: 10.1210/me.2005-0042. [DOI] [PubMed] [Google Scholar]
- 128.Scott DJ, Parkes AT, Ponchel F, Cummings M, Poola I, Speirs V. Changes in expression of steroid receptors, their downstream target genes and their associated co-regulators during the sequential acquisition of tamoxifen resistance in vitro. Int J Oncol. 2007 Sep;31(3):557–65. doi: 10.3892/ijo.31.3.557. [DOI] [PubMed] [Google Scholar]
- 129.Girault I, Lerebours F, Amarir S, Tozlu S, Tubiana-Hulin M, Lidereau R, et al. Expression analysis of estrogen receptor alpha coregulators in breast carcinoma: evidence that NCOR1 expression is predictive of the response to tamoxifen. Clin Cancer Res. 2003 Apr;9(4):1259–66. [PubMed] [Google Scholar]
- 130.Hudelist G, Czerwenka K, Kubista E, Marton E, Pischinger K, Singer CF. Expression of sex steroid receptors and their co-factors in normal and malignant breast tissue: AIB1 is a carcinoma–specific co-activator. Breast Cancer Res Treat. 2003 Mar;78(2):193–204. doi: 10.1023/a:1022930710850. [DOI] [PubMed] [Google Scholar]
- 131.Mc IM, Fleming FJ, Buggy Y, Hill AD, Young LS. Tamoxifen-induced ER-alpha-SRC-3 interaction in HER2 positive human breast cancer; a possible mechanism for ER isoform specific recurrence. Endocr Relat Cancer. 2006 Dec;13(4):1135–45. doi: 10.1677/erc.1.01222. [DOI] [PubMed] [Google Scholar]
- 132.Klinge CM, Jernigan SC, Mattingly KA, Risinger KE, Zhang J. Estrogen response element-dependent regulation of transcriptional activation of estrogen receptors alpha and beta by coactivators and corepressors. J Mol Endocrinol. 2004 Oct;33(2):387–410. doi: 10.1677/jme.1.01541. [DOI] [PubMed] [Google Scholar]
- 133.Chen L, Qiu J, Yang C, Yang X, Chen X, Jiang J, et al. Identification of a novel estrogen receptor beta1 binding partner, inhibitor of differentiation-1, and role of ERbeta1 in human breast cancer cells. Cancer Lett. 2009 Jun 18;278(2):210–9. doi: 10.1016/j.canlet.2009.01.008. [DOI] [PubMed] [Google Scholar]
- 134.Sikder HA, Devlin MK, Dunlap S, Ryu B, Alani RM. Id proteins in cell growth and tumorigenesis. Cancer Cell. 2003 Jun;3(6):525–30. doi: 10.1016/s1535-6108(03)00141-7. [DOI] [PubMed] [Google Scholar]
- 135.Urbanska K, Pannizzo P, Lassak A, Gualco E, Surmacz E, Croul S, et al. Estrogen receptor beta-mediated nuclear interaction between IRS-1 and Rad51 inhibits homologous recombination directed DNA repair in medulloblastoma. J Cell Physiol. 2009 May;219(2):392–401. doi: 10.1002/jcp.21683. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 136.Macaluso M, Montanari M, Noto PB, Gregorio V, Surmacz E, Giordano A. Nuclear and cytoplasmic interaction of pRb2/p130 and ER-beta in MCF-7 breast cancer cells. Ann Oncol. 2006 Jun;17(Suppl 7):vii27–vii29. doi: 10.1093/annonc/mdl945. [DOI] [PubMed] [Google Scholar]
- 137.Thenot S, Bonnet S, Boulahtouf A, Margeat E, Royer CA, Borgna JL, et al. Effect of ligand and DNA binding on the interaction between human transcription intermediary factor 1alpha and estrogen receptors. Mol Endocrinol. 1999 Dec;13(12):2137–50. doi: 10.1210/mend.13.12.0387. [DOI] [PubMed] [Google Scholar]
- 138.Warnmark A, Almlof T, Leers J, Gustafsson JA, Treuter E. Differential recruitment of the mammalian mediator subunit TRAP220 by estrogen receptors ERalpha and ERbeta. J Biol Chem. 2001 Jun 29;276(26):23397–404. doi: 10.1074/jbc.M011651200. [DOI] [PubMed] [Google Scholar]
- 139.Kim HR, Chae HJ, Thomas M, Miyazaki T, Monosov A, Monosov E, et al. Mammalian dap3 is an essential gene required for mitochondrial homeostasis in vivo and contributing to the extrinsic pathway for apoptosis. FASEB J. 2007 Jan;21(1):188–96. doi: 10.1096/fj.06-6283com. [DOI] [PubMed] [Google Scholar]
- 140.Kasof GM, Goyal L, White E. Btf, a novel death-promoting transcriptional repressor that interacts with Bcl-2-related proteins. Mol Cell Biol. 1999 Jun;19(6):4390–404. doi: 10.1128/mcb.19.6.4390. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 141.Jarvinen TA, Pelto-Huikko M, Holli K, Isola J. Estrogen receptor beta is coexpressed with ERalpha and PR and associated with nodal status, grade, and proliferation rate in breast cancer. Am J Pathol. 2000 Jan;156(1):29–35. doi: 10.1016/s0002-9440(10)64702-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 142.Skliris GP, Carder PJ, Lansdown MR, Speirs V. Immunohistochemical detection of ERbeta in breast cancer: towards more detailed receptor profiling? Br J Cancer. 2001 Apr 20;84(8):1095–8. doi: 10.1054/bjoc.2001.1721. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 143.Osborne CK. Tamoxifen in the treatment of breast cancer. N Engl J Med. 1998 Nov 26;339(22):1609–18. doi: 10.1056/NEJM199811263392207. [DOI] [PubMed] [Google Scholar]
- 144.Ali S, Coombes RC. Endocrine-responsive breast cancer and strategies for combating resistance. Nat Rev Cancer. 2002 Feb;2(2):101–12. doi: 10.1038/nrc721. [DOI] [PubMed] [Google Scholar]
- 145.Chang HG, Kim SJ, Chung KW, Noh DY, Kwon Y, Lee ES, et al. Tamoxifen-resistant breast cancers show less frequent methylation of the estrogen receptor beta but not the estrogen receptor alpha gene. J Mol Med (Berl) 2005 Feb;83(2):132–9. doi: 10.1007/s00109-004-0596-2. [DOI] [PubMed] [Google Scholar]
- 146.Bross PF, Baird A, Chen G, Jee JM, Lostritto RT, Morse DE, et al. Fulvestrant in postmenopausal women with advanced breast cancer. Clin Cancer Res. 2003 Oct 1;9(12):4309–17. [PubMed] [Google Scholar]
- 147.Peekhaus NT, Chang T, Hayes EC, Wilkinson HA, Mitra SW, Schaeffer JM, et al. Distinct effects of the antiestrogen Faslodex on the stability of estrogen receptors-alpha and -beta in the breast cancer cell line MCF-7. J Mol Endocrinol. 2004 Jun;32(3):987–95. doi: 10.1677/jme.0.0320987. [DOI] [PubMed] [Google Scholar]
- 148.Klinge CM. Estrogen receptor interaction with estrogen response elements. Nucleic Acids Res. 2001 Jul 15;29(14):2905–19. doi: 10.1093/nar/29.14.2905. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 149.Ivanova MM, Luken KH, Zimmer AS, Lenzo FL, Smith RJ, Arteel MW, et al. Tamoxifen increases nuclear respiratory factor 1 transcription by activating estrogen receptor beta and AP-1 recruitment to adjacent promoter binding sites. FASEB J. 2011 Apr;25(4):1402–16. doi: 10.1096/fj.10-169029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 150.Leung YK, Ho SM. Estrogen receptor beta: switching to a new partner and escaping from estrogen. Sci Signal. 2011;4(168):e19. doi: 10.1126/scisignal.2001991. [DOI] [PubMed] [Google Scholar]
- 151.Wu X, Subramaniam M, Grygo SB, Sun Z, Negron V, Lingle WL, et al. Estrogen receptor-beta sensitizes breast cancer cells to the anti-estrogenic actions of endoxifen. Breast Cancer Res. 2011;13(2):R27. doi: 10.1186/bcr2844. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 152.Tee MK, Rogatsky I, Tzagarakis-Foster C, Cvoro A, An J, Christy RJ, et al. Estradiol and selective estrogen receptor modulators differentially regulate target genes with estrogen receptors alpha and beta. Mol Biol Cell. 2004 Mar;15(3):1262–72. doi: 10.1091/mbc.E03-06-0360. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 153.Levy N, Tatomer D, Herber CB, Zhao X, Tang H, Sargeant T, et al. Differential regulation of native estrogen receptor-regulatory elements by estradiol, tamoxifen, and raloxifene. Mol Endocrinol. 2008 Feb;22(2):287–303. doi: 10.1210/me.2007-0340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 154.Murphy LC, Peng B, Lewis A, Davie JR, Leygue E, Kemp A, et al. Inducible upregulation of oestrogen receptor-beta1 affects oestrogen and tamoxifen responsiveness in MCF7 human breast cancer cells. J Mol Endocrinol. 2005 Apr;34(2):553–66. doi: 10.1677/jme.1.01688. [DOI] [PubMed] [Google Scholar]
- 155.Treeck O, Lattrich C, Springwald A, Ortmann O. Estrogen receptor beta exerts growth-inhibitory effects on human mammary epithelial cells. Breast Cancer Res Treat. 2010 Apr;120(3):557–65. doi: 10.1007/s10549-009-0413-2. [DOI] [PubMed] [Google Scholar]
- 156.Paech K, Webb P, Kuiper GG, Nilsson S, Gustafsson J, Kushner PJ, et al. Differential ligand activation of estrogen receptors ERalpha and ERbeta at AP1 sites. Science. 1997 Sep 5;277(5331):1508–10. doi: 10.1126/science.277.5331.1508. [DOI] [PubMed] [Google Scholar]
- 157.Ramaswamy B, Majumder S, Roy S, Ghoshal K, Kutay H, Datta J, et al. Estrogen-mediated suppression of the gene encoding protein tyrosine phosphatase PTPRO in human breast cancer: mechanism and role in tamoxifen sensitivity. Mol Endocrinol. 2009 Feb;23(2):176–87. doi: 10.1210/me.2008-0211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 158.Sundberg S, Luurila OJ, Kohvakka A, Gordin A. The circadian heart rate but not blood pressure profile is influenced by the timing of beta-blocker administration in hypertensives. Eur J Clin Pharmacol. 1991;40(4):435–6. doi: 10.1007/BF00265862. [DOI] [PubMed] [Google Scholar]
- 159.Grober OM, Mutarelli M, Giurato G, Ravo M, Cicatiello L, De Filippo MR, et al. Global analysis of estrogen receptor beta binding to breast cancer cell genome reveals an extensive interplay with estrogen receptor alpha for target gene regulation. BMC Genomics. 2011;12:36. doi: 10.1186/1471-2164-12-36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 160.Louie MC, McClellan A, Siewit C, Kawabata L. Estrogen receptor regulates E2F1 expression to mediate tamoxifen resistance. Mol Cancer Res. 2010 Mar;8(3):343–52. doi: 10.1158/1541-7786.MCR-09-0395. [DOI] [PubMed] [Google Scholar]
- 161.Sheng S, Barnett DH, Katzenellenbogen BS. Differential estradiol and selective estrogen receptor modulator (SERM) regulation of Keratin 13 gene expression and its underlying mechanism in breast cancer cells. Mol Cell Endocrinol. 2008 Dec 16;296(1–2):1–9. doi: 10.1016/j.mce.2008.09.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 162.Martinez ME, Thomson CA, Smith-Warner SA. Soy and breast cancer: the controversy continues. J Natl Cancer Inst. 2006 Apr 5;98(7):430–1. doi: 10.1093/jnci/djj128. [DOI] [PubMed] [Google Scholar]
- 163.Rice S, Whitehead SA. Phytoestrogens and breast cancer--promoters or protectors? Endocr Relat Cancer. 2006 Dec;13(4):995–1015. doi: 10.1677/erc.1.01159. [DOI] [PubMed] [Google Scholar]
- 164.Dong JY, Qin LQ. Soy isoflavones consumption and risk of breast cancer incidence or recurrence: a meta-analysis of prospective studies. Breast Cancer Res Treat. 2011 Jan;125(2):315–23. doi: 10.1007/s10549-010-1270-8. [DOI] [PubMed] [Google Scholar]
- 165.Wu AH, Koh WP, Wang R, Lee HP, Yu MC. Soy intake and breast cancer risk in Singapore Chinese Health Study. Br J Cancer. 2008 Jul 8;99(1):196–200. doi: 10.1038/sj.bjc.6604448. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 166.Wu AH, Yu MC, Tseng CC, Pike MC. Epidemiology of soy exposures and breast cancer risk. Br J Cancer. 2008 Jan 15;98(1):9–14. doi: 10.1038/sj.bjc.6604145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 167.Hilakivi-Clarke L, Andrade JE, Helferich W. Is soy consumption good or bad for the breast? J Nutr. 2010 Dec;140(12):2326S–34S. doi: 10.3945/jn.110.124230. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 168.Dai Q, Shu XO, Jin F, Potter JD, Kushi LH, Teas J, et al. Population-based case-control study of soyfood intake and breast cancer risk in Shanghai. Br J Cancer. 2001 Aug 3;85(3):372–8. doi: 10.1054/bjoc.2001.1873. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 169.Suzuki T, Matsuo K, Tsunoda N, Hirose K, Hiraki A, Kawase T, et al. Effect of soybean on breast cancer according to receptor status: a case-control study in Japan. Int J Cancer. 2008 Oct 1;123(7):1674–80. doi: 10.1002/ijc.23644. [DOI] [PubMed] [Google Scholar]
- 170.Hwang CS, Kwak HS, Lim HJ, Lee SH, Kang YS, Choe TB, et al. Isoflavone metabolites and their in vitro dual functions: they can act as an estrogenic agonist or antagonist depending on the estrogen concentration. J Steroid Biochem Mol Biol. 2006 Nov;101(4–5):246–53. doi: 10.1016/j.jsbmb.2006.06.020. [DOI] [PubMed] [Google Scholar]
- 171.Morito K, Aomori T, Hirose T, Kinjo J, Hasegawa J, Ogawa S, et al. Interaction of phytoestrogens with estrogen receptors alpha and beta (II) Biol Pharm Bull. 2002 Jan;25(1):48–52. doi: 10.1248/bpb.25.48. [DOI] [PubMed] [Google Scholar]
- 172.Stubert J, Gerber B. Isoflavones - Mechanism of Action and Impact on Breast Cancer Risk. Breast Care (Basel) 2009;4(1):22–9. doi: 10.1159/000200980. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 173.Morito K, Hirose T, Kinjo J, Hirakawa T, Okawa M, Nohara T, et al. Interaction of phytoestrogens with estrogen receptors alpha and beta. Biol Pharm Bull. 2001 Apr;24(4):351–6. doi: 10.1248/bpb.24.351. [DOI] [PubMed] [Google Scholar]
- 174.Routledge EJ, White R, Parker MG, Sumpter JP. Differential effects of xenoestrogens on coactivator recruitment by estrogen receptor (ER) alpha and ERbeta. J Biol Chem. 2000 Nov 17;275(46):35986–93. doi: 10.1074/jbc.M006777200. [DOI] [PubMed] [Google Scholar]
- 175.Galluzzo P, Marino M. Nutritional flavonoids impact on nuclear and extranuclear estrogen receptor activities. Genes Nutr. 2006 Sep;1(3–4):161–76. doi: 10.1007/BF02829966. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 176.Kuiper GG, Carlsson B, Grandien K, Enmark E, Haggblad J, Nilsson S, et al. Comparison of the ligand binding specificity and transcript tissue distribution of estrogen receptors alpha and beta. Endocrinology. 1997 Mar;138(3):863–70. doi: 10.1210/endo.138.3.4979. [DOI] [PubMed] [Google Scholar]
- 177.Kuiper GG, Lemmen JG, Carlsson B, Corton JC, Safe SH, van der Saag PT, et al. Interaction of estrogenic chemicals and phytoestrogens with estrogen receptor beta. Endocrinology. 1998 Oct;139(10):4252–63. doi: 10.1210/endo.139.10.6216. [DOI] [PubMed] [Google Scholar]
- 178.Chang EC, Charn TH, Park SH, Helferich WG, Komm B, Katzenellenbogen JA, et al. Estrogen Receptors alpha and beta as determinants of gene expression: influence of ligand, dose, and chromatin binding. Mol Endocrinol. 2008 May;22(5):1032–43. doi: 10.1210/me.2007-0356. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 179.Dip R, Lenz S, Antignac JP, Le BB, Gmuender H, Naegeli H. Global gene expression profiles induced by phytoestrogens in human breast cancer cells. Endocr Relat Cancer. 2008 Mar;15(1):161–73. doi: 10.1677/ERC-07-0252. [DOI] [PubMed] [Google Scholar]
- 180.Sotoca AM, Gelpke MD, Boeren S, Strom A, Gustafsson JA, Murk AJ, et al. Quantitative proteomics and transcriptomics addressing the estrogen receptor subtype-mediated effects in T47D breast cancer cells exposed to the phytoestrogen genistein. Mol Cell Proteomics. 2011 Jan;10(1):M110. doi: 10.1074/mcp.M110.002170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 181.Mak P, Leung YK, Tang WY, Harwood C, Ho SM. Apigenin suppresses cancer cell growth through ERbeta. Neoplasia. 2006 Nov;8(11):896–904. doi: 10.1593/neo.06538. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 182.Cappelletti V, Miodini P, Di FG, Daidone MG. Modulation of estrogen receptor-beta isoforms by phytoestrogens in breast cancer cells. Int J Oncol. 2006 May;28(5):1185–91. [PubMed] [Google Scholar]
- 183.Bhat HK, Calaf G, Hei TK, Loya T, Vadgama JV. Critical role of oxidative stress in estrogen-induced carcinogenesis. Proc Natl Acad Sci U S A. 2003 Apr 1;100(7):3913–8. doi: 10.1073/pnas.0437929100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 184.Liehr JG. Is estradiol a genotoxic mutagenic carcinogen? Endocr Rev. 2000 Feb;21(1):40–54. doi: 10.1210/edrv.21.1.0386. [DOI] [PubMed] [Google Scholar]
- 185.Bianco NR, Perry G, Smith MA, Templeton DJ, Montano MM. Functional implications of antiestrogen induction of quinone reductase: inhibition of estrogen–induced deoxyribonucleic acid damage. Mol Endocrinol. 2003 Jul;17(7):1344–55. doi: 10.1210/me.2002-0382. [DOI] [PubMed] [Google Scholar]
- 186.Bianco NR, Chaplin LJ, Montano MM. Differential induction of quinone reductase by phytoestrogens and protection against oestrogen-induced DNA damage. Biochem J. 2005 Jan 1;385(Pt 1):279–87. doi: 10.1042/BJ20040959. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 187.Guha N, Kwan ML, Quesenberry CP, Jr, Weltzien EK, Castillo AL, Caan BJ. Soy isoflavones and risk of cancer recurrence in a cohort of breast cancer survivors: the Life After Cancer Epidemiology study. Breast Cancer Res Treat. 2009 Nov;118(2):395–405. doi: 10.1007/s10549-009-0321-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 188.Shu XO, Zheng Y, Cai H, Gu K, Chen Z, Zheng W, et al. Soy food intake and breast cancer survival. JAMA. 2009 Dec 9;302(22):2437–43. doi: 10.1001/jama.2009.1783. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 189.Lattrich C, Lubig J, Springwald A, Goerse R, Ortmann O, Treeck O. Additive effects of trastuzumab and genistein on human breast cancer cells. Anticancer Drugs. 2011 Mar;22(3):253–61. doi: 10.1097/CAD.0b013e3283427bb5. [DOI] [PubMed] [Google Scholar]