Abstract
Developing methods that provide adequate vascular perfusion is an important step toward engineering large functional tissues. Meanwhile, an imaging modality to assess the three-dimensional (3-D) structures and functions of the vascular channels is lacking for thick matrices (>2~3mm). Herein, we report on an original approach to construct and image 3-D dynamically perfused vascular structures in thick hydrogel scaffolds. In this work, we integrated a robotic 3-D cell-printing technology with a mesoscopic fluorescence molecular tomography imaging system, and demonstrated the capability of the platform to construct perfused collagen scaffolds with endothelial lining and to image both the fluid flow and fluorescent-labeled living endothelial cells at high-frame rates, with high sensitivity and accuracy. These results establish the potential of integrating both 3-D cell-printing and fluorescence mesoscopic imaging for functional and molecular studies in complex tissue engineered tissues.
Keywords: Endothelial cells, Collagen scaffold, Perfused vascular construct, 3-D cell culture, 3-D cell printing, Mesoscopic fluorescence molecular tomography
1. Introduction
Despite tremendous progresses in the field of tissue engineering [1], there are still significant challenges in creating thick tissue constructs. Especially, there is a critical need to develop methods that provide adequate vascular perfusion for engineering large functional tissues [2-4]. Thus, creating a dynamically perfused vasculature within 3-D matrix represents an important step toward this goal, and this realistic physiologically relevant system allows us to study the growth and maturation process of the tissue engineered thick construct.
However, an imaging modality to assess the 3-D structures and functions of the vascular channels is lacking for thicker matrices (>2~3mm) [5]. Confocal and two-photon microscopes are useful tools for this research, but they can only image tissues with a depth of less than a few hundred microns. The configuration of 3-D construct in the perfusion chamber makes it even more challenging to access the sample for imaging. Another difficulty arises from the scaffold materials, which can be translucent or opaque depending on the matrix biomaterial and its density, and can become non-transparent as more matrices are produced with tissue growth. This results in optically high-scattering thick tissue samples. Typically, investigations of engineered tissues mostly rely on histological sections. However, time-consuming procedures and sample fixations make this approach not suitable for high-throughput applications and/or obtaining dynamic information of living samples [5]. Thus, it is urgent to develop imaging techniques that can image well beyond the penetration limits of conventional microscopy (several hundred microns) to evaluate in real-time the maturation process and functions of thick tissues up to a few millimeters.
To fulfill this unmet imaging need, we have developed a fluorescence mesoscopic imaging technique based on laminar optical tomography (LOT) [6] principles. LOT is a non-contact laser-scanning imaging technique, which harnesses scattered light to probe both absorbing and fluorescent contrast in living tissues. LOT is able to obtain depth-resolved 3-D quantitative images to depths of several millimeters with high sensitivity. Similar to fluorescence molecular tomography (FMT) [7], LOT achieves sensitivity to depth-resolved absorption and fluorescence by exploiting scattering photons emerging from the tissue. In FMT, due to lager source-detector (S-D) separation, more uncertainty from scattering light propagation though the tissue limits the resolution to macroscopic scale, which makes it difficult to visualize small-diameter engineered vasculature. LOT is able to achieve higher resolution than FMT by shortening S-D separation, thus enabling quantitative and non-invasive 3-D imaging for assessing distributions of fluorescence biomarkers in thick tissues. Conversely to optical coherence tomography (OCT), LOT is sensitive to fluorescence signal whereas OCT is sensitive to index of refraction mismatch [8]. Conversely to photoacoustic tomography (PAT), LOT allows imaging of gene reporters.
In this study, we integrated a robotic cell printing system with a mesoscopic fluorescence molecular tomography (MFMT) imaging system for tissue engineering applications. Herein, we report on the 3-D cell printing technology (Supplementary Fig. S1) based on solid free-form fabrication (SFF) technology [9-15] and the methods (Fig. 1) we developed to construct a dynamically perfused vascular channel containing luminal endothelial cell (EC) lining within hydrogel biomaterials. We also report on the MFMT system performances and ability to image both fluid flow and fluorescent-labeled living endothelial cells at high-frame rates in a perfused collagen scaffold.
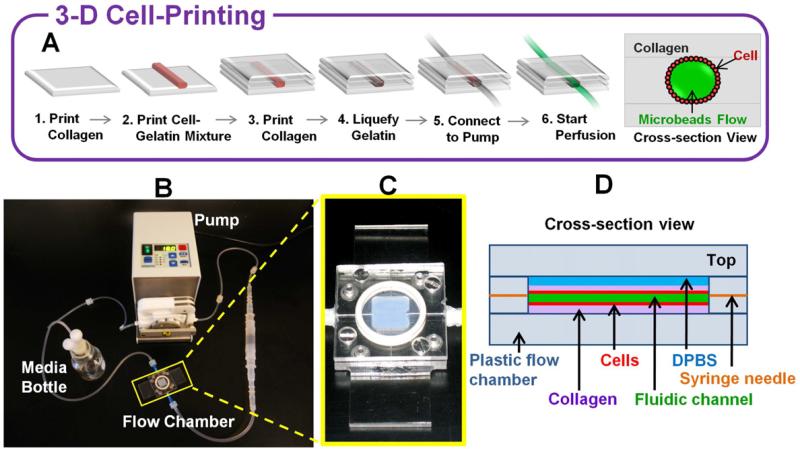
Fig. 1.
3-D vascular channel construct in perfusion system. (A) Schematics of channel construction procedure and cross-section view. (B) The actual perfusion pump system. Flow chamber is marked by a yellow rectangle. (C) Specially designed flow chamber. (D) Schematic diagram of cross –section view of the printed vascular channel construct in the flow chamber.
2. Materials and Methods
2.1 Scaffold materials and gelation
Collagen, rat tail, type I (BD Bioscience) was used as a scaffold material in this study. The collagen precursor from stock was diluted to 6-9 mg/ml with 1X Dulbecco's phosphate buffered saline (DPBS). The collagen precursor, which is acidic in stock, becomes collagen gel in neutral pH. To induce gelation of the collagen, we used 0.8M sodium bicarbonate (NaHCO3) solutions. The solutions were gently applied to collagen precursor as fine misty via nebulization. Gelatin from porcine skin (Sigma-Aldrich) was used as a sacrificial material for perfusion sample preparation and also used for fine patterning. Right before printing, 20% gelatin solution dissolved in DPBS was prepared and mixed with cell suspension to obtain desired concentration.
2.2 Cell culture and labeling
Human umbilical vein endothelial cells (HUVECs) were cultured at 37°C in 5% CO2 in EGM®-2 Endothelial Cell Growth Medium-2 (Lonza). Culture media was changed every other day. HUVECs were routinely passaged onto tissue culture flask and were discarded after passage number 10. Right before printing, cells were harvested using 0.25% Trypsin-EDTA (Mediatech). After harvesting, cell suspensions were kept on ice until they were mixed with gelatin for printing. For MFMT imaging, HUVECs were labeled with 5μM cell tracker (1,1'-Dioctadecyl-3,3,3',3'-Tetramethylindodicarbocyanine-5,5'-Disulfonic Acid (DiIC18(5)-DS), Invitrogen) before being harvested. Fluorescence excitation and emission wavelength of the cell tracker were 650nm and 670nm, respectively.
2.3 3-D cell-printing system
3-D cell printing system (Supplementary Fig. S1) consists of eight independently-controlled electromechanical valve dispensers mounted onto a 3-axis, high-precision x-y-z robot stage (bidirectional reproducibility of 5 μm) to enable printing of multiple cell types and scaffold materials simultaneously. The dispenser (non-contact type, minimum 25nl droplet size) is capable of dispensing various cell types with high cell viability and is compatible with most hydrogels used in tissue engineering. The volume of the droplet can be adjusted by digitally controlled pressure and pulse duration. The duration of the valve opening/closing is provided by the conventional 5V Transistor-Transistor Logic pulse (100~750μs in duration) to the control driver, and normal operation allows continuous dispensing with an actuation frequency of 1kHz, which renders the high throughput capability in dispensing. The repeatability of the mechanical gating is excellent (variability < ±1%). To facilitate printing sophisticated 3-D tissue constructs, we developed a user-friendly software interface to control the printing sequence. The software interface is compatible with multi-modal image sources such as commercially available computer-aided design (CAD) programs, radiological images (from computer tomography or magnetic resonance imaging), and common image formats. A MATLAB based graphic-user-interface (GUI) was built to assist in converting slice-by-slice 3-D structural information to the appropriate robot-control codes. The sampled printing coordinates are routed to the path planner algorithm (either through vector or in coordinate-by-coordinate fashion), which prescribes the printing sequence.
2.4 3-D printing optimization
Printing pressure and dispenser valve opening time were decided considering viscosity of materials and droplet size. Lowest pressure, which showed stable dispensing without clogging, was defined for collagen precursors and cell-gelatin mixture. Collagen precursor was more viscose than cell suspension, thus high pressure was applied for collagen printing. There were no significant difference in viscosity and droplet size with regards to various cell types and cell suspension density. 6.0-7.0 psi of pressure and 750μs of valve opening time were used for collagen printing. 4.0-6.0 psi of pressure and 750μs of valve opening time were used for cell printing. Spacing (distance between printed droplets) and cell suspension density were also chosen considering viscosity of the materials and droplet size. 500μm spacing was used for collagen and 300-400μm spacing for cell-gelatin mixture.
2.5 Construction of perfused vascular channel
3-D vascular channel was constructed using collagen, gelatin and HUVECs (Fig. 1A). Hydrogel and cells were printed on a specially designed flow chamber (Fig. 1B-D). 10% Gelatin was prepared as solution and used as a sacrificial material to generate fluidic channel [16, 17]. Before printing was started, nebulized NaHCO3 vapor was applied on the bottom surface of the flow chamber to quickly polymerize first collagen layer and increase adhesion to the bottom surface. After printing of one collagen layer, nebulized NaHCO3 vapor was applied onto the printed collagen layer for gelation. Nebulization guaranteed gentle and uniform gelation without destroying 3-D pattern. It also acted as an adherent between the bottom gelated layer and newly printed layer. After nebulization, one minute was allowed for the gelation of the collagen layer, which provided a firm base for next layer. After 3~4 layers of collagen were printed at the bottom, HUVECs mixed with gelatin were printed as one straight line on top of the collagen surface. The printed cell-gelatin mixture became solidified within 1~2 minutes when it was exposed to room temperature. The flow chamber was kept in 4°C for 20 minutes to complete gelatin solidification. After this, collagen layers were printed on top of it until collagen layers filled the whole chamber. The time required for construction of whole structure was about 40 minutes. The flow chamber was sealed and placed in 37°C for one hour to liquefy the gelatin and to allow the cells to be attached to the surface of the channel. During the incubation, we flipped the flow chamber every 10-15 minutes to seed cells uniformly throughout the channel. Following this step, the flow chamber was connected to dispensing pump (Ismatec) as shown in Fig. 1B and gentle media flow was applied to wash out the gelatin and to obtain a perfused vascular channel. Flow rate was gradually increased until the average shear stress in the channel reaches 10dyn/cm2.
2.6 MFMT system
In this work, we developed an ultra-fast multi-wavelength mesoscopic fluorescence imaging platform, to visualize tissue-engineered vascular structures consisting of living ECs lining the fluidic lumen within a collagen scaffold. Our MFMT platform is a 2-color imaging system that can acquire excitation and emission data at two wavelengths (658nm and 488nm) with frame rates of 7.5Hz (<150ms/frame) over a 5.5×8mm area. MFMT serially injects light, and detects emerging light at multiple discrete positions on a tissue surface. In a typical case, the system acquires 15,360 source positions with 7 detectors measurements for each source position (107,520 total measurements). The optical chain of our MFMT system is provided in Supplementary Fig. S2 and is similar to the one developed by Ouakli et al [18] for in vivo functional imaging. Two laser diodes at 488nm (L488P060MLD, Thorlabs) and 658nm (L658P040, Thorlabs) are used to illuminate the sample. The power of each laser diode can be easily tuned by a laser diode controller (LTC100-NL-SP, Thorlabs). The back-scattered signal is collected by the objective lens (LB1471, Thorlabs) and passes a second polarizer to remove the specular reflections and fluorescence bandpass filters (FF01-496/LP-25 for blue laser diode, LP02-664RS-25 for red laser diode, Semrock). The back-scattered fluorescence signal is then imaged on a 4×8 avalanche photodiode detector (APD) array (S8550, Hamamatsu). 7 elements of the APD are employed to capture the fluorescence at seven different radial distances away from the illumination point, which is recorded by 16-bit data acquisition card (PCI-6143, National Instruments). Note that the first element is used for alignment with the optical axis of the system rather than data acquisition due to the high dynamical range. The whole system is mounted on a vertical translation stage and linear translation stage (M-432, Newport) to adjust the focusing depth on the sample. In this configuration, the physical distances between the sampling points on the specimen are 0.6mm at minimum and 4mm at maximum.
2.7 Monte Carlo optical reconstruction algorithms
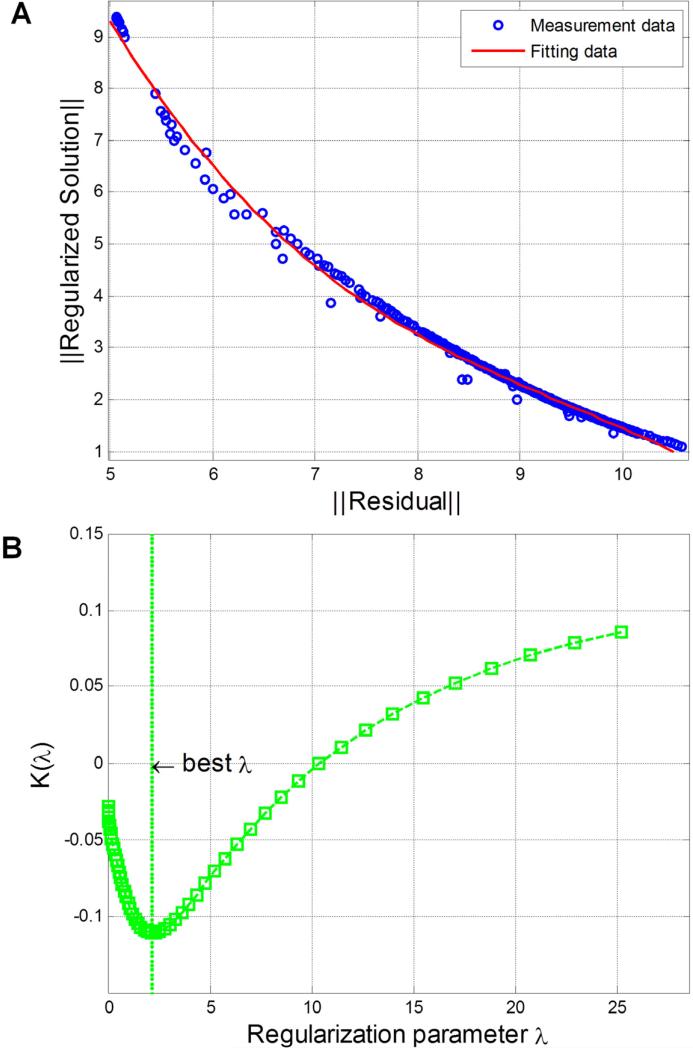
To interpret the measured MFMT data, we employed a Monte Carlo (MC) based light propagation model to reconstruct 3-D visualization of the depth-resolved absorption and the fluorescence contrast. We employed a fast mesoscopic reconstruction algorithm based on a Graphical Processor Unit (GPU) implementation to produce 3-D images of the volumes imaged with our MFMT system[19]. The algorithm is based on a MC code to create Jacobians with the adjoint method for computational efficiency [20]. The MFMT image reconstruction process can be expressed using the following equation: (ATA+λD)x′=ATb, where b is obtained from the fluorescence measurement subtracting background; x′ is reconstructed fluorescence distribution; λ is a regularization depth-dependent parameter. A is the whole Jacobian that is computed based on a MC forward model for accuracy. Thanks to the symmetry of the imaging space, A is populated for all the positions of the raster scanned beam based on a single set of sensitivity matrices, which are computed for one optode set (1 source and 7 detectors). The MC forward model is computed with 106 photons, which is performed on a GPU (GTX260) under less than 5 minutes for the typical 107,520 data measurements. λD is depth-dependent regularization parameter which is employed to mitigate the ill-posedness of the reflectance geometry. D is a diagonal matrix [21] whose diagonal elements are the square-root of the corresponding diagonal elements of ATA. The optimized scaling factor (λ) applied to experimental reconstructions is chosen by L-curve method and its curvature function k(λ) (Fig. 2) [22]. The conjugate gradient method is applied to solve this linear system. The optical properties of the collagen matrices are estimated non-concurrently on our time-resolved preclinical imager [23] and were used as a priori information for the reconstruction processes. In this study, voxel size of all the reconstructions is 0.125mm.
Fig. 2.
Optimal regularization parameter chosen from L-curve analysis. (A) L-curve from experimental reconstructions is used to balance the size of residual (2-norm of Ax′ – b) and size of regularized solution (2-norm of x′). A is the whole Jacobian matrix computed based on a MC forward model; x′ is reconstructed fluorescence distribution; b is obtained from the fluorescence measurement. (B) Curvature of L-curve [22] with respect to regularization parameters is used to choose the best regularization parameter.
2.8 Micro-CT imaging
We non-concurrently imaged the samples with a micro-CT (Scanco Medical Viva CT40) to validate the spatial accuracy of the MFMT reconstructions. For micro-CT imaging of collagen based phantom and vascular channel construct (Fig. 4C), a piece of cotton was slightly moistened with DPBS and put in the micro-CT holder covered with sealing film to prevent collagen scaffold from drying up during long acquisition time (~40 minutes). The collagen phantom with glass capillaries was visualized with a field of view (FOV) of 32.8 mm2 at 16μm voxel size. Air was pumped into the open lumen of vascular channel construct to improve contrast for visualization by micro-CT with a FOV of 25.6 mm2 at 25μm voxel size (Fig. 7A). Acquisition time was approximately 20 minutes.
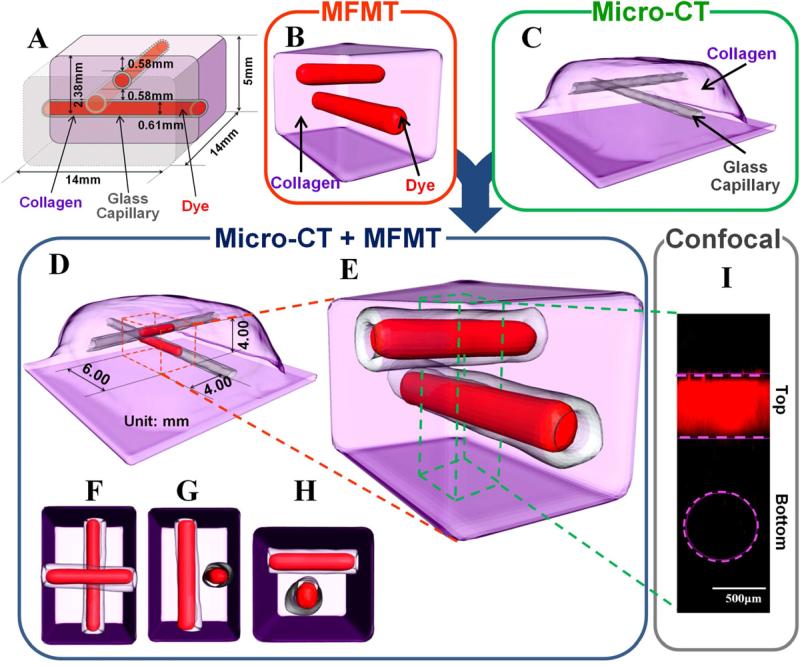
Fig. 4.
In-vitro validation of vascular structure mimicking in a collagen scaffold. (A) Schematic diagram of phantom design shows the locations of two capillaries filled with fluorescence dye in the collagen scaffold. (B) 3-D Surface based on threshold segmentation (T=25%) of reconstructed fluorescent capillaries by MFMT. (C) Localizations of glass walls of two capillaries in the collagen scaffold from micro CT scanner. (D) and (E) Merged 3-D image of (B) and (C). The red dashed cuboid shows the reconstructed volume (4mm×6mm×4mm). The green dashed cuboid shows the rendering volume by confocal microscope (0.89mm×0.89mm×3mm). (F-H) show top view, front view, and right side view of (E). (I) 3-D maximum intensity projection of capillaries obtained by confocal microscope (objective 10x, LSM 510Meta; Zeiss). The pink dashed lines shows the top capillary, and the pink dashed circle corresponds to the position of the deeper inclusion as located by micro-CT.
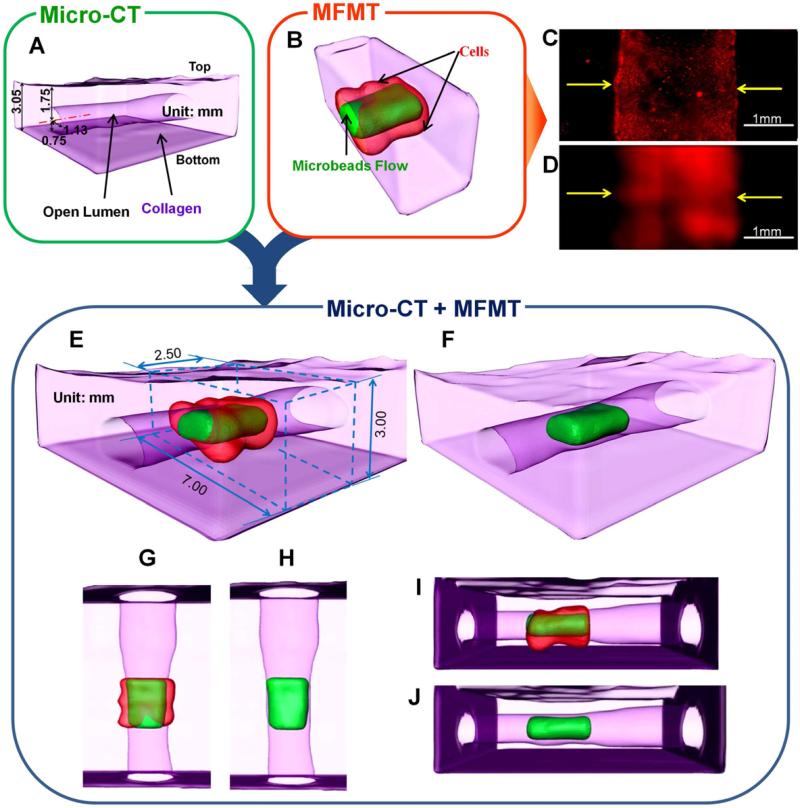
Fig. 7.
Imaging of 3-D perfused vascular channel construct in thick collagen scaffold. (A) Localizations and dimensions of the open lumen from a micro CT scanner. (B) 3-D surface rendering based on threshold segmentation (T=35%) from MFMT image reconstruction of Open lumen (green) and luminal endothelial lining (red). (C) Fluorescence image of luminal endothelial lining in the printed vascular channel by a wide-field microscope (objective 4x). (D) 2-D maximum intensity Z projection of MFMT reconstruction of luminal endothelial lining. Scale: 1mm. (E) Merged 3-D image of (A) and (B). The reconstructed volume is marked by the blue dashed cuboid (2.5mm×7mm×3mm). (F) Merged 3-D image of reconstructed open lumen and (A). (G-J) are top view and right side view of (E) and (F), respectively.
2.9 Image post-processing
Amira 5 software (Visage Imaging) was used for 3-D visualization of collagen based phantom and vascular channel construct (Fig. 4 and Fig. 7). The micro-CT images of both samples were resampled to the same voxel size (0.125mm) as MFMT reconstruction. The corresponding polygonal surface models were generated after threshold segmentation for areas of interest of micro-CT reconstruction images and MFMT reconstruction images.
2.10 Confocal imaging
The confocal imaging of collagen phantom (Fig. 4I) was acquired by an inverted objective (10x/0.25 N.A., LSM 510Meta; Zeiss). The inverted collagen based phantom was placed in the glass bottom dish. There was a small gap between the cover glass and the surface of collagen filled with DPBS to avoid drying up during the laser scanning acquisition. The excitation wavelength was 633 nm, and the fluorescence was detected between 670 and 720 nm.
2.11 Wild-field imaging
A wide-field microscope (Eclipse Ti, Nikon) was used for time-lapse phase contrast imaging (Fig. 5) and fluorescence imaging (Fig. 6B). For fluorescence imaging, HUVECs labeled with cell tracker (DiIC18 (5)-DS) was visualized in red color using G-2E/C filter (λex=540 nm, λem=620 nm). Green fluorescent beads (0.2μm, 1.5 ×105 particles/ml, λex=480nm, λem=520nm, Bangs Laboratories, Inc.) were injected to visualize the flow. B-2E/C filter (λex=480 nm, λem=535 nm) was used for beads flow imaging.
Fig. 5.
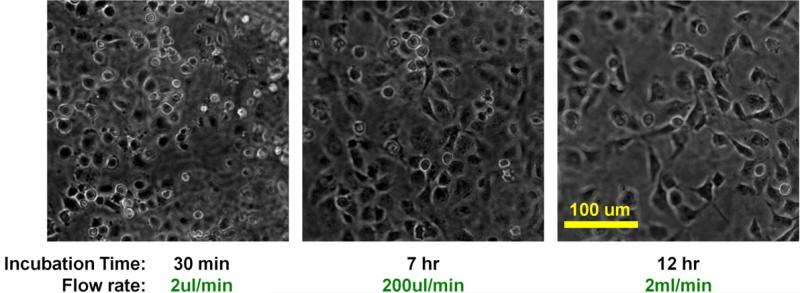
Time-lapse phase contrast images of HUVECs under fluidic condition by a wide-field microscope (objective 10x). Perfusion started with gentle flow (2μl/min). Cells were round at this point. Flow rate was gradually increased until it reached 20ml/min. After 7 hours of perfusion, flow rate was increased to 200μl/min and HUVECs began to spread out. After 12 hours of perfusion, flow rate was increased to 2ml/min and maintained thereafter.
Fig. 6.
(A) Schematic diagram of the printed vascular channel construct. (B) Fluorescence image of the printed vascular channel construct by a wide-field microscope (objective 4x). HUVECs were visualized in red color, beads flow in green color.
3. Results and Discussions
3.1 3-D cell printing optimization
Collagen precursor in 9mg/ml concentration restricted migration of ECs into the scaffold under the flow condition, keeping the tubular shape as its pattern was being printed. Among various types and concentration of gelatin, 10% gelatin from porcine skin showed good cell viability (Supplementary Fig. S3), appropriate viscosity at 37°C, and adequate rigidity after solidification. HUVECs mixed in 10% gelatin were printed on prepared collagen layer with various cell density and spacing (distance between droplets). The viability was high (>95%) regardless of cell density. However, we choose 1~2×107cells/ml for appropriate seeding and faster vasculature formation. High cell density is required because we use <10μL of cell-gelatin mixture for channel patterning. 300-400μm of spacing was chosen to achieve high viability, pattern accuracy and desired channel thickness.
3.2 Depth-dependent validation of the MFMT system in vitro
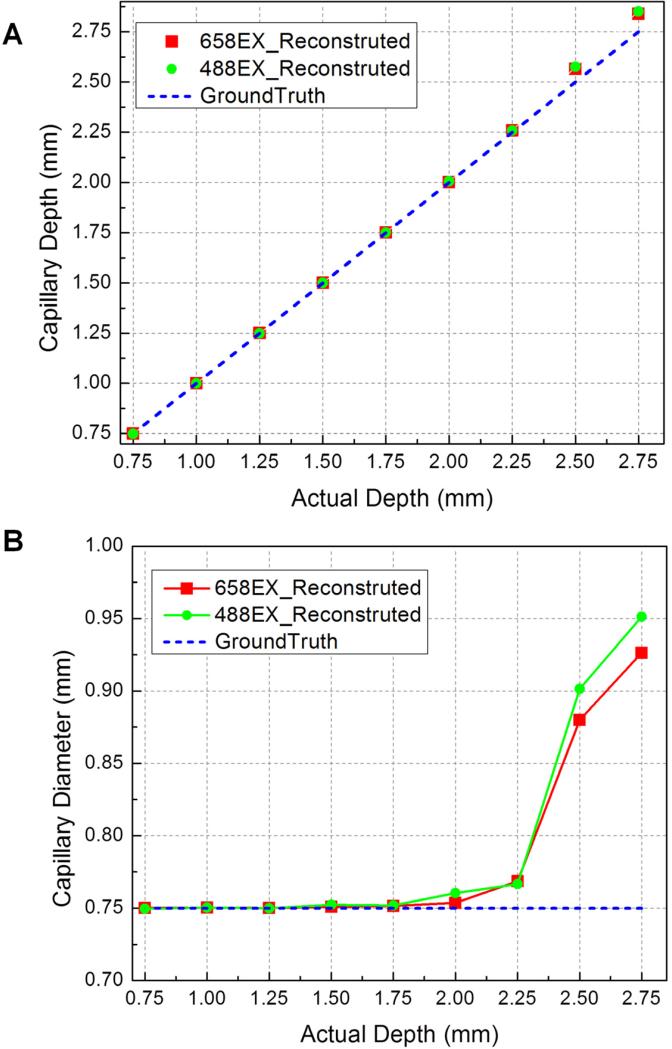
The imaging performance of the MFMT system was thoroughly tested in phantom experiments. A capillary (douter=1mm, dinner=0.75mm) containing 5 μM of NIR-664-iodoacetamide (λex=664nm, λem=689nm; Sigma-Aldrich) and Rhodamine 110 (λex=498nm, λem=520nm; Sigma-Aldrich), respectively, was placed in an intralipid-india ink solution mimicking in vivo muscle optical properties (μa=0.008mm-1; μs'=0.5mm-1). A linear translation stage was used to translate the fluorescence capillary at multiple positions along the vertical direction up to 2.75mm at 0.25mm each step. The MC image reconstruction algorithm was used to retrieve the 3-D fluorescence image. The reconstructed depth and diameter of capillary were compared at different excitation wavelengths. The reconstructed depth has good agreement with the actual depth (Fig. 3A). When depth was increasing, the diameter became lager and lager due to higher scattering, but the error remained smaller than 27%. The system is able to image fluorescent structure up to 3mm deep with a resolution of ~200μm.
Fig. 3.
Fluorescence inclusions at different depths can be resolved by MFMT system. (A) Reconstructed depths recovery at 488nm excitation and 658nm excitation in comparison to actual depths. (B) Comparison of reconstructed diameters at 488nm excitation and 658nm excitation with actual diameters in different depths.
3.3 MFMT imaging of complex structure in collagen-based phantom and validation
The capacity of the MFMT system to image complex structure has been cross-validated with micro-CT and confocal microscopy. An example of such cross-validation is provided below. Two sealed glass capillaries (douter=1mm, dinner=0.61mm) containing 5μM of NIR-664-iodoacetamide in methanol were positioned in collagen (9mg/ml) scaffold at two different depths in an orthogonal fashion (Fig. 4A). Immediately following MFMT imaging, the phantom was transported in DPBS and kept in a freezing box (0-4°C) for micro-CT and confocal imaging, sequentially. Micro-CT system was used to obtain a precise template of the phantom (Fig. 4C), which provided the localization of the capillaries due to the contrast imparted by the glass (micro-CT cannot image fluorescence contrasts). Confocal microscope was used to obtain Z stacks of the overlap volume of two capillaries from the surface of collagen scaffold to the bottom of the deeper capillary for creating 3-D maximum intensity projections (Fig. 4I). As shown in Fig. 4I, the confocal microscope was able to resolve the capillary closest to the surface but was unable to image the capillary embedded deeper. By comparison, both capillaries reconstructed by MFMT system presented accurate location, configuration and size (Fig. 4B, Fig. 4D-H). The two capillaries were distinctively reconstructed even in the area where they overlapped, which demonstrates the ability of MFMT to image vascular-like fluorescence patterns in thick scaffold with high-resolution and accuracy.
3.4 A 3-D dynamically perfused vascular channel construct within collagen scaffold
Time-lapse phase images were taken at 30 minutes, 7 hours, and 12 hours after the media perfusion began (Fig. 5, Supplementary Video S1). Flow rate was gradually increased from 2μl/min to 2ml/min. We observed some cell loss during liquefaction and early medium perfusion due to weak cell attachment in the beginning. When perfusion started, non-attached cells were washed away with gradually increasing flow. Live ECs close to the wall were uniformly attached to the luminal surface of the fluidic channel, and they started to migrate and spread along the lumen. However, we were not able to obtain clear phase images after Day 1 as HUVECs remodeled the scaffold collagen, which became more opaque. As shown in Fig. 6, green fluorescence microspheres in the perfusion medium flowed through the open lumen of the printed structures with ECs seeded around the channel. The microspheres were not able to diffuse through the collagen gel so it clearly indicated the lumen where the flow occurred (Fig. 6, Supplementary Video S2). Long-term culture under perfusion (up to 2-3 weeks) will be performed in the future to investigate the functional properties of the vascular channel.
It is a current challenge to create very dense capillary network with open lumen and physiological perfusion at the same time. The developed platform is able to create vascular channel of 500μm at a distance of 700μm between multiple channels, which is still far from the density of the capillary network seen in vivo. However, this developed technology provides a path to support the perfusion and cell growth and remodeling within the thick hydrogel scaffold, and capillaries can be induced from the engineered vessel via angiogenesis process by introducing the supporting mural cells in the matrix. Currently, we are investigating this process between two printed vascular channels to determine conditions that can result in a perfused capillary network, which will enable generation of denser capillary network in the future.
3.5 MFMT imaging of 3-D perfused vascular channel construct collagen scaffold and validation
The 2-color 3-D MFMT images clearly identify the open lumen and ECs located around the lumen (Fig. 7B). To validate the locations and dimensions of open lumen, the vascular construct was obtained by the micro-CT system (Fig. 7A). The mechanical impact of the printing droplet on top of the gelatin layer can also be seen clearly as the wrinkled lumen surface. The reconstructed open lumen and the endothelial lining also confirm this observation. The 2-D maximum intensity Z projection form MFMT 3-D reconstruction of luminal endothelial lining (Fig. 7D) also matched the fluorescence image (Fig. 7C) obtained by a conventional wide-field fluorescence microscope, which does not have intrinsic 3-D imaging capability due to insufficient out-of-focus suppression that obscures the in-focus signals. It is also important to note that our MFMT is able to acquire depth-resolved images of the vascular structure within the perfusion chamber (behind a 2.5mmm-thick plastic cover), and the whole imaging acquisition process takes only 1 minute. Hence, the technique is well suited for dynamical and longitudinal studies of cell behavior within scaffolds in bioreactors without disturbing their environment.
This in vitro fluorescence imaging of vascular construct can give structural and functional information with depth embedded up to 3mm, which can help to more clearly understand the mechanisms of vascularization in large tissue-engineered constructs. Since the in vitro vascular model was created using only cells and extracellular matrix, we can also use MFMT to study the tissue development process, structures and functionalities of the synthesized structure with regard to matrix properties and culture parameter, or the biological interactions between ECs and surrounding cells.
In addition to the potential application of this integrated platform, we can image the distribution of different fluorophores in this in vitro vascular model to study angiogenesis process by introducing the supporting mural cells in the matrix, where capillaries will occur around the perfused vascular channel and fluid flow can achieve capillary lumen though the vascular channel, which closely mimics the in vivo angiogenesis. The high concentration of collagen used in the study might restrict the capillary formation from the perfused vascular channel. Recently, we have implemented this method to construct vascular channel using different concentrations of collagen (from 3mg/ml to 9mg/ml, data not shown). It is suggested that cell migration will be very slow in high concentration collagen gels while the process will be faster in low concentration gel. It will be interesting to study these under long term (2~3 weeks) perfused condition. Ongoing studies in the lab will investigate the functional properties of the vascular channel and capillary formation at different gel density. Although much research remains to be done, it appears that this technique has a great potential to bridge the gap between vascularization and tissue regeneration, and thus facilitate clinical applications of tissue-engineered constructs. The distribution and intensity of fluorescence may provide an insight into the sequence of events that leads to angiogenesis process and may help elucidate vascularization mechanisms.
The unique advantages of the 3-D cell-printing and fluorescence mesoscopic imaging system will make this integrated platform a new and important tool in the field of tissue engineering. It will allow us to construct and study tissue structures that are previously unachievable with other technologies. For example, it can be used to construct complex 3-D tissue structures containing multiple cell types with vasculature, as well as to study molecular expression and the dynamic interactions of cellular/tissue processes surrounding the vascular perfusion in engineered thick tissues. Further development of this integrated platform promises to bring significant value to functional tissue engineering.
4. Conclusion
In this study, we developed an integrated platform by combining the 3-D cell printing technique with MFMT system. Our results demonstrated the capability of cell printing technology to create in vitro vascular tissues with perfusion, and to allow rapid, non-contact imaging of live tissue up to a few millimeters in depth. The 3-D cell printing system is able to dispense live cells, soluble factors, and phase-changing hydrogel in desired pattern maintaining high cell viability. ECs were incorporated to from the inner lining of the vascular construct, since ECs can regulate permeability and form lumens as well as remodel of vascular networks, allow tissue growth and regeneration for our future study. We also developed an MFMT system to investigate the function and structure of vascular construct. The two-color MFMT system is able to acquire quantitative 3-D absorption and fluorescence contrasts of thick tissues (up to 3mm) with high frame rates of 7.5Hz over an adaptable large FOV (5.5×8mm at maximum). Meanwhile, a fast MC method based algorithm was also developed to reconstruct 3-D fluorescence distribution of imaged volume.
Supplementary Material
Acknowledgments
We thank N.Ouakli, E.Guevara, and F. Lesage (Ecole Polytechnique Montreal, Canada) for help with integrating MFMT system and funding support from National Institutes of Health R21HL102773.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
Author Contributions
G.D. and X.I. conceived and initiated project. S-S.Y. and V.K.L. designed the 3-D bio-fabrication technology. X.I. and L.Z. developed the MFMT system. L.Z. and V.K.L. designed and performed experiments, analyzed data and wrote the manuscript. G.D. and X.I. supervised the project.
Competing Financial Interests
The authors declare no competing financial interests.
Appendix
Supplementary Data
Supplementary data associated with this article can be found in the online version.
References
- 1.Khademhosseini A, Vacanti JP, Langer R. Progress in tissue engineering. Sci Am. 2009;300:64–71. doi: 10.1038/scientificamerican0509-64. [DOI] [PubMed] [Google Scholar]
- 2.Griffith LG, Swartz MA. Capturing complex 3D tissue physiology in vitro. Nat Rev Mol Cell Biol. 2006;7:211–24. doi: 10.1038/nrm1858. [DOI] [PubMed] [Google Scholar]
- 3.Jain RK, Au P, Tam J, Duda DG, Fukumura D. Engineering vascularized tissue. Nat Biotechnol. 2005;23:821–3. doi: 10.1038/nbt0705-821. [DOI] [PubMed] [Google Scholar]
- 4.Lovett M, Lee K, Edwards A, Kaplan DL. Vascularization strategies for tissue engineering. Tissue Eng Part B Rev. 2009;15:353–70. doi: 10.1089/ten.teb.2009.0085. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Ntziachristos V. Going deeper than microscopy: the optical imaging frontier in biology. Nat Methods. 2010;7:603–14. doi: 10.1038/nmeth.1483. [DOI] [PubMed] [Google Scholar]
- 6.Hillman EM, Burgess SA. Sub-millimeter resolution 3D optical imaging of living tissue using laminar optical tomography. Laser Photon Rev. 2009;3:159–79. doi: 10.1002/lpor.200810031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Ntziachristos V. Fluorescence molecular imaging. Annu Rev Biomed Eng. 2006;8:1–33. doi: 10.1146/annurev.bioeng.8.061505.095831. [DOI] [PubMed] [Google Scholar]
- 8.Huang D, Swanson EA, Lin CP, Schuman JS, Stinson WG, Chang W, et al. Optical coherence tomography. Science. 1991;254:1178–81. doi: 10.1126/science.1957169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Sun W, Darling A, Starly B, Nam J. Computer-aided tissue engineering: overview, scope and challenges. Biotechnology and Applied Biochemistry. 2004;39:29–47. doi: 10.1042/BA20030108. [DOI] [PubMed] [Google Scholar]
- 10.Borenstein JT, Weinberg EJ, Orrick BK, Sundback C, Kaazempur-Mofrad MR, Vacanti JP. Microfabrication of three-dimensional engineered scaffolds. Tissue Eng. 2007;13:1837–44. doi: 10.1089/ten.2006.0156. [DOI] [PubMed] [Google Scholar]
- 11.Norotte C, Marga FS, Niklason LE, Forgacs G. Scaffold-free vascular tissue engineering using bioprinting. Biomaterials. 2009;30:5910–7. doi: 10.1016/j.biomaterials.2009.06.034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Mironov V, Trusk T, Kasyanov V, Little S, Swaja R, Markwald R. Biofabrication: a 21st century manufacturing paradigm. Biofabrication. 2009;1:022001. doi: 10.1088/1758-5082/1/2/022001. [DOI] [PubMed] [Google Scholar]
- 13.Mironov V, Boland T, Trusk T, Forgacs G, Markwald RR. Organ printing: computer-aided jet-based 3D tissue engineering. Trends Biotechnol. 2003;21:157–61. doi: 10.1016/S0167-7799(03)00033-7. [DOI] [PubMed] [Google Scholar]
- 14.Lee W, Debasitis JC, Lee VK, Lee J-H, Fischer K, Edminster K, et al. Multi-layered culture of human skin fibroblasts and keratinocytes through three-dimensional freeform fabrication. Biomaterials. 2009;30:1587–95. doi: 10.1016/j.biomaterials.2008.12.009. [DOI] [PubMed] [Google Scholar]
- 15.Lee W, Lee V, Polio S, Keegan P, Lee J-H, Fischer K, et al. On-demand three-dimensional freeform fabrication of multi-layered hydrogel scaffold with fluidic channels. Biotechnol Bioeng. 2010;105:1178–86. doi: 10.1002/bit.22613. [DOI] [PubMed] [Google Scholar]
- 16.Golden AP, Tien J. Fabrication of microfluidic hydrogels using molded gelatin as a sacrificial element. Lab Chip. 2007;7:720–5. doi: 10.1039/b618409j. [DOI] [PubMed] [Google Scholar]
- 17.Boland T, Xu T, Damon B, Cui X. Application of inkjet printing to tissue engineering. Biotechnol J. 2006;1:910–7. doi: 10.1002/biot.200600081. [DOI] [PubMed] [Google Scholar]
- 18.Ouakli N, Guevara E, Dubeau S, Beaumont E, Lesage F. Laminar optical tomography of the hemodynamic response in the lumbar spinal cord of rats. Opt Express. 2010;18:10068–77. doi: 10.1364/OE.18.010068. [DOI] [PubMed] [Google Scholar]
- 19.Fang QQ, Boas DA. Monte Carlo Simulation of Photon Migration in 3D Turbid Media Accelerated by Graphics Processing Units. Optics Express. 2009;17:20178–90. doi: 10.1364/OE.17.020178. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Chen J, Intes X. Comparison of Monte Carlo methods for fluorescence molecular tomography computational efficiency. Medical Physics. 2011;38(10) doi: 10.1118/1.3641827. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Endoh R, Fujii M, Nakayama K. Depth-adaptive regularized reconstruction for reflection diffuse optical tomography. Optical review. 2008;15:51–6. [Google Scholar]
- 22.Intes X, Ripoll J, Chen Y, Nioka S, Yodh AG, Chance B. In vivo continuous-wave optical breast imaging enhanced with Indocyanine Green. Med Phys. 2003;30:1039–47. doi: 10.1118/1.1573791. [DOI] [PubMed] [Google Scholar]
- 23.Venugopal V, Chen J, Intes X. Development of an optical imaging platform for functional imaging of small animals using wide-field excitation. Biomed Opt Express. 2010;1:143–56. doi: 10.1364/BOE.1.000143. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.