Abstract
PHACE syndrome (OMIM #606519) is a neurocutaneous syndrome of unknown etiology and pathogenesis. We report on an individual with PHACE syndrome with a complete deletion of SLC35B4 on 7q33. In order to further analyze this region, SLC35B4 was sequenced for 33 individuals with PHACE syndrome and one parental set. Common polymorphisms with a possible haplotype but no disease causing mutation were identified. Sixteen of 33 samples of the PHACE syndrome patients were also analyzed for copy number variations using high resolution oligo-Comparative Genomic Hybridization (CGH) microarray. A second individual in this cohort had a 26.5kb deletion approximately 80kb upstream of SLC35B4 with partial deletion of the AKR1B1 on 7q33. The deletions observed on 7q33 are not likely the singular cause of PHACE syndrome; however, it is possible that this region provides a genetic susceptibility to phenotypic expression with other confounding genetic or environmental factors.
Keywords: PHACE Syndrome, microarray, deletion, chromosome region 7q33, SLC35B4, AKR1B1
INTRODUCTION
Seven females with cervicofacial hemangiomas along with other anomalies were described by Pascual-Castroviejo in 1978 [Pascual-Castroviejo, 1978]. This association was further defined, and the acronym PHACE was proposed in 1996 by Frieden et al. [1996]. PHACE syndrome includes posterior fossa malformations, large infantile hemangioma of the head neck or face, arterial cerebrovascular anomalies, cardiac anomalies, and eye anomalies. Sternal clefting and/or supraumbilical raphe can also be present, and are then referred to as PHACES syndrome (OMIM # 606519) [Frieden et al., 1996] .
Large facial hemangiomas indicative of PHACE syndrome are often described as segmental; they comprise approximately 20% of hemangiomas identified in the head and neck region [Haggstrom et al.,2006]. In most patients, a diagnosis of PHACE syndrome includes a segmental hemangioma of > 5cm in diameter of the head, neck, or face and one extracutaneous anomaly. In 2009, leading experts established both major and minor criteria based on the particular organ system to better define PHACE syndrome [Metry et al., 2009a; Metry et al., 2009b]. The major criteria include: anomalies of the major cerebral arteries (dysplasia, arterial stenosis, and persistence of trigeminal artery), structural brain anomalies (Dandy-Walker complex, posterior fossa anomaly), cardiovascular anomalies (coarctation of the aorta, or aneurysm), ocular anomalies (posterior segment abnormality, retinal vascular or morning glory disc anomaly), and ventral or midline anomalies (sternal defects). Minor criteria include the persistence of embryonic arteries, midline anomalies, ventricular septal defects, cataracts, or hypopituitarism. A definition of PHACE syndrome based on these criteria would include a segmental hemangioma > 5cm in diameter of the head, neck, or face plus either one of the major criteria or 2 minor criteria.
To date there are more than 300 cases of PHACE syndrome reported in the literature [Haggstrom et al., 2010] but its etiology and pathogenesis remains unknown, Oligo-Comparative Genomic Hybridization (CGH) microarray used to evaluate a patient with PHACE syndrome identified a small novel deletion on 7q33. This region was subsequently analyzed in a larger cohort of PHACE syndrome patients and the results are reported here.
METHODS
Patients
DNA was obtained from five patients with PHACE syndrome through the Utah Clinical Genetics Research Program, Istanbul Medical Faculty, Istanbul University; and DNA from 28 additional individuals and two unaffected parents was obtained from the PHACE Syndrome International Clinical Registry and Genetic Repository. All affected subjects met diagnostic criteria for PHACE syndrome based on the diagnostic criteria by Metry et al. [2009a]. Diagnostic criteria include a large hemangioma on the face measuring 5 cm, plus at least one major criterion, such as a posterior fossa anomaly, coarctation of the aorta, developmental anomalies of the great cerebral arteries, developmental posterior segment eye anomalies and midline ventral defects, such as sternal cleft. Information was obtained from physical examinations and medical histories (e.g. results from echocardiograms, brain and vessel imaging, and ophthalmologic examinations).
Array CGH
A minimum of 500ng and up to 1μg of purified DNA was used on the microarray depending on the amount of sample available. Reference DNA from a pool of 10 healthy individuals was used for sex matched controls (Promega GmbH, Mannheim, Germany).
A total of 24 samples (22 propositi, 2 unaffected parents) out of 35 had a sufficient amount of DNA to perform the human CGH 3 × 720K whole genome tiling array (Roche Nimblegen, Inc., Madison, WI) DNA labeling (Nimblegen Dual-Color DNA Labeling Kit , Roche Nimblegen, Inc., Madison, WI), hybridization (Nimblegen Hybridization Kit, Roche Nimblegen, Inc., Madison, WI), and washing were performed according to the package insert. Data analysis was performed using Nimblescan (Roche Nimblegen, Inc., Madison, WI) and results were viewed using SignalMap (Roche Nimblegen, Inc., Madison, WI). Further data analysis was performed for all of the samples using the Nexus Copy Number software (BioDiscovery, Inc., El Segundo, CA). Deletions or duplications that were detected with 10 data points or more and had a log2 ratio greater than +0.3 were considered significant. Quality scores calculated using the Nexus software were used to determine if the sample data were valid. Samples with a quality score greater than 0.3 were excluded. In addition, sample data exhibiting greater than 1% variability in the genome were excluded. For purposes of this report, 7q33 was specifically analyzed.
Results from an additional archived 24 normal control samples were further analyzed for the 7q33 region exclusively. These archived normal control samples were processed using various CGH microarrays including 4 ×72K, 1 × 385K, and 1 × 2.1M microarrays.
This study was approved by the Institution Review Boards of the University of Utah, University of California San Francisco, Oregon Health & Science University and Children’s Hospital of Wisconsin and informed consent was obtained from all participants.
Sequencing
Whole genome amplification was performed on all of the samples for Sanger sequencing using the Illustra GenomiPhi DNA Amplification Kit (GE Healthcare Bio-Sciences Corp Piscataway, NJ). For cases in which there insufficient DNA to perform the microarray (<500ng), only Sanger sequencing was performed on the selected candidate gene (SLC35B4).
Based on the CGH microarray data from the index case, the candidate gene SLC35B4 was selected for sequencing. Sanger sequencing using big dye terminator was performed on the 10 coding exons for 35 samples including 33 probands, and one unaffected parental set. Primers were designed to sequence exonic regions including exon-intron boundaries. PCR was performed using approximately 50-75ng of genomic DNA. Forward and reverse primers were added at 10μM each with 12.5μL of High Fidelity PCR Mastermix (Roche, Mannheim, Germany). Bi-directional sequencing was performed on a 3730 ABI DNA Genetic Analyzer, using forward and reverse primers. Analysis was performed using Mutation Surveyor (SoftGenetics, LLC., State College, PA) and mutations have been named using Gene Bank reference sequence (NM_032826.4). Single nucleotide polymorphisms (SNP) and non-synonymous mutations identified were evaluated using Alamut (Interactive Biosoftware, France).
RESULTS
Index Case
Patient # 1 was born at 38 weeks gestation to a 21-year-old primiparous woman via cesarean due to abruptio placentae. A faint red birthmark involving the right side of the face in a segmental pattern was noted at birth. The infant was admitted to the neonatal intensive care unit. Magnetic resonance imaging (MRI) of the brain at age one week showed a dysplastic right superior vermis, an absent inferior vermis, a hypoplastic right dural venous sinus, and an abnormal enhancement of the anterior chamber of the right eye. Repeat MRI imaging at 1 month showed an increase in the size and number of proliferating hemangiomas within the right frontal scalp, right orbit and soft tissues of the face. Additional intracranial foci were identified in the right petrous apex, subarachnoid space, and pituitary gland. The circle of Willis was noted to be aberrant and there was a slight increase in the amount of flow-voids in the supracerebellar cistern. Computed tomography (CT) of the chest and abdomen showed multiple contrast-enhancing lesions within the chest wall, right hepatic lobe, and bowel wall. Ophthalmologic exam showed sclerocornea, a persistent papillary membrane, and a small superior optic nerve. An echocardiogram was normal. There was no obvious sternal cleft. The infant developed necrotizing enterocolitis with bowel perforation requiring bowel resection. At surgery multiple hemangiomas were visible throughout the small intestines. A second surgery was required in which the entire small intestine was determined to be non-viable. Support was withdrawn at approximately 2 months of age in a series by Drolet et al. [2011].
Molecular Results
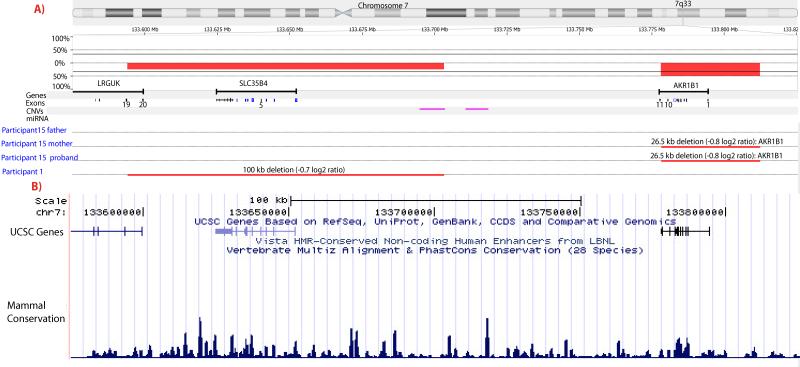
All probands met diagnostic criteria for PHACE syndrome [Metry et al., 2009a]. Using the Nimblegen 3 × 720K whole genome tiling array, a novel small deletion (-0.7 log2 ratio) of approximately 100kb in size located on chromosome 7q33 (chr7: 133,600,015-133,700,204, NCBI36/hg18) was identified in participant 1 (See Fig 1). This is a small region containing one gene, SLC35B4 (chr7: 133,624,630-133,652,343, NCBI36/hg18). Parental samples were not available to determine de novo or familial status of the deletion.
Figure 1.
A) Deletions identified in 7q33 using Nexus Copy Number™. Fifteen PHACE samples were analyzed using the Nexus software. Red lines indicate regions where deletions were identified. Purple lines demonstrate the known CNVs. A 100kb deletion (chr7: 133,600,015-133,700,204) was indentified in the index sample (Patient 1) including the entire SLC35B4 (chr7: 133,624,630-133,652,343) Another deletion of 26.5 kb was identified in Sample 15 including AKR1B1. The same deletion was also identified in the mother (Patient 15 mother) but not the father (Patient 15 father). Please note, Patients 2-14 are not shown because they did not show deletions or duplications in the region.
B) Image of conservation for the region (chr7: 133,575,000-133,825,500) from USCS Genome Bioinformatics (http://genome.ucsc.edu). This view contains conservation for 28 mammalian species including 17 placental mammal species and human. Observation of multiple species shows conservation for the region.
In the 33 propositi, in which sequencing was performed for one candidate gene (SLC35B4) based on the CGH microarray results, eleven probands showed no variation. In the other 22 propositi seven SNPs were identified in the introns and 1 synonymous SNP detected in the exon of SLC35B4 (rs447266, rs34664116, rs2075372, rs1832052, rs1421484, rs2241336, rs2505, rs2504). All SNPs were previously identified and common within the population. In all but two cases, these SNPs (7 intronic, 1 exonic) were found together in each sample, indicating a possible haplotype. As expected, Patient #1 with the SLC35B4 deletion did not show heterozygous SNPs .
Subsequently, 24 samples (22-proband, 1 parental set-father and mother of Patient #15) were run on the Nimblegen 3 × 720K whole genome tiling array. There was an insufficient DNA for 11 of 35 samples. Of 24 samples run on the array, six were eliminated because either the quality scores were above 0.3 or due to aberrant microarray results. A total of 16 samples from the propositi and unaffected parents were considered valid for analysis (Fig 1).
CGH array data from 24 randomly selected healthy control samples were analyzed exclusively for SLC35B4. There were no deletions or duplications identified in this region for the healthy controls.
Interestingly, Patient #15, had a 26.5kb deletion approximately 80kb upstream (chr7: 133,783,161-133,809,611, NCBI36/hg18) of SLC35B4. In this patient 16.3kb of the AKR1B1 (aldo-keto reductase Family 1, Member B1) is deleted (chr7:133,777,647-133,794,428, NCBI36/hg18). Participant #15 is a Caucasian female born full term to a primiparous 27-year-old woman. The birth weight was four kg. There were no reported pregnancy complications. The infant’s hemangioma involved the right side of the face and scalp with extension onto the neck and upper chest. A subglottic hemangioma was also present. The brain imaging was normal. An echocardiogram revealed an interrupted aortic arch and a ventricular septal defect. There were no eye anomalies or sternal defects. Additional complications included scoliosis and unilateral hearing loss. The identical deletion was also identified in her mother. The mother did not have vascular anomalies, hemangiomas or other reported medical conditions. She had a normal echocardiogram, but an angiogram has not been performed. When the mother’s CNV regions were compared with the participant’s, nine novel deletion/duplication regions were identified in the child’s sample. The percent variability in the genome was much higher for the child compared to the mother (0.13% versus 0.06%, respectively).
The other 14 individuals with PHACE syndrome had no significant changes in 7q33 on the CGH microarray. There were no other non-CNV regions in the whole genome containing genes and overlapping in at least two samples. Seven CNVs were present in at least 25% of the samples. All these CNV have been previously recognized as common in the normal population. Variability within the genome using our cut-off criteria for all samples ranged from 0.08% to 0.47% based on the total number of base pairs detected.
DISCUSSION
Out 16 individuals with PHACE syndrome in which CGH microarray results could be analyzed, two had small deletions on 7q33. This region contains SLC35B4 and AKR1B1. SLC35B4 (solute carrier Family 35, member B4) is a glycosyltransferase that transports sugar from the cytoplasm to the Golgi apparatus [Ashikov et al., 2005]. No disease causing mutations are reported in SLC35B4 and AKR1B1. However, repeat dinucleotide and single nucleotide polymorphisms in the AKR1B1 gene have been reported in association with diabetic macroangiopathy, diabetic retinopathy, and diabetic nephropathy, and cardiorenal complications [So et al., 2008; Thamotharampillai et al., 2006; Wolford et al., 2006; Xu et al., 2008]. None had a deletion of AKR1B1. In addition, we showed that 24 healthy controls did not have structural changes in this region and no CNVs have been reported in this deleted region. The deletions found in our two patients are novel changes.
The deletions in these two patients do not overlap; however, the breakpoints are 80 kb apart and located on the 5′ upstream region of SLC35B4. Regulatory proteins or transcription factors related to the regulation of SLC35B4 binding sites may have been affected. Studies have shown SLC35B4 expression in the fetal brain and heart, supporting it as a potential candidate gene [Fujita et al., 2011; Su et al., 2002; Su et al., 2004; Wu et al., 2009] (http://genome.ucsc.edu/, genome hg18.) We further investigated SLC35B4 and sequenced the exons and exon-intron boundaries for all individuals with PHACE syndrome in our cohort, but identified no mutations, suggesting that exonic sequence changes in SLC35B4 were not associated with PHACE syndrome in our cohort. It is unclear if the haplotype identified for some samples could be associated with PHACE syndrome.
The mother of Patient #15 has the identical deletion in AKR1B1 and as far as we know is clinically unaffected. When the mother’s CNV regions were compared with child’s, we identified nine novel deletion/duplication regions in the child’s sample. The differences in genetic background, including CNVs inherited by both parents and de novo deletions/duplications, could contribute to the presentation of PHACE syndrome via epistatic effects [Eichler et al.,2010]. However, we think it is unlikely that this small, inherited deletion is the causative genetic alteration in this patient. Although a single genetic etiology for PHACE syndrome has not been identified, it could be multifactorial and heterogeneous, contributing to the variable clinical presentation. Environmental factors or physical conditions such as pre-eclampsia and premature births may contribute to expression of the phenotype. The clinical variability of PHACE syndrome makes a diagnosis at times tenuous, but important for clinical care. So far, PHACE syndrome presents typically as simplex case without an obvious inheritance pattern. Although, the 7q33deletions are not likely the singular cause of PHACE syndrome it is possible that this region provides a genetic susceptibility in combination with other inherited or de novo CNVs [Girirajan and Eichler, 2010; Girirajan et al., 2010].
The presence of segmental hemangiomas is typical for PHACE syndrome. The segmental/unilateral findings in PHACE syndrome provide some support for a two-hit or multi-step model. We hypothesize that PHACE syndrome is multifactorial and that genetic predisposing factors in combination with other possible genetic and environmental influences result in the wide phenotypic spectrum. Molecular studies on DNA from different tissues for somatic events, including deletions of the 7q33, would be interesting. Our current cohort provides high resolution copy number change information with 720,000 probes throughout the whole genome in 16 individuals with PHACE syndrome. Although based on only two individuals, 7q33 is a potential candidate locus.
ACKNOWLEDGMENTS
We thank the families for their participation and Heather Hanson for help in sample collection. Support was provided by the Society of Pediatric Otolaryngology, Public Health Services research grant numbers #UL1-RR025764 and C06-RR11234 from the National Center for Research Resources, and the Children’s Health Research Center and Clinical Genetics Research Program at the University of Utah. Dr. Stevenson is supported by the Doris Duke Charitable Foundation Clinical Scientist Development Award, Thrasher Research Fund, and an NIH NINDS K23NS052500 award. Dr. Siegel was supported for this work with a Dermatology Foundation Career Development Award and Patient Directed Investigator Award, the Society of Pediatric Dermatology William L. Weston Grant and the Oregon Clinical and Translational Research Institute, grant number UL1 RR024140 01 from the National Center for Research Resources. Dr. Shieh is supported by NIH HNLBI. Dr. Drolet was involved in the enrollment and phenotyping of a subset of the subjects in the PHACE syndrome Clinical Registry and Genetic Repository. The content is solely the responsibility of the authors and does not necessarily represent the official views of the above granting agencies.
REFERENCES
- Ashikov A, Routier F, Fuhlrott J, Helmus Y, Wild M, Gerardy-Schahn R, Bakker H. The human solute carrier gene SLC35B4 encodes a bifunctional nucleotide sugar transporter with specificity for UDP-xylose and UDP-N-acetylglucosamine. J Biol Chem. 2005;280:27230–27235. doi: 10.1074/jbc.M504783200. [DOI] [PubMed] [Google Scholar]
- Drolet BA, Pope E, Juern AM, Sato T, Howell B, Puttgen KB, Lara-Corrales I, Gilliam A, Mancini A, Powell J, Siegel D, Metry D, Stevenson DA, Grimmer F, Frieden IJ. Gastointestinal bleeding in infantile hemangioma: A complication of segmental, rather than multifocal infantile hemangiomas. J Pediatrics. 2011 doi: 10.1016/j.jpeds.2011.12.026. [Accepted with minor revisions] [DOI] [PubMed] [Google Scholar]
- Eichler EE, Flint J, Gibson G, Kong A, Leal SM, Moore JH, Nadeau JH. Missing heritability and strategies for finding the underlying causes of complex disease. Nat Rev Genet. 2010;11:446–50. doi: 10.1038/nrg2809. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Frieden IJ, Reese V, Cohen D. PHACE syndrome. The association of posterior fossa brain malformations, hemangiomas, arterial anomalies, coarctation of the aorta and cardiac defects, and eye abnormalities. Arch Dermatol. 1996;132:307–311. doi: 10.1001/archderm.132.3.307. [DOI] [PubMed] [Google Scholar]
- Fujita PA, Rhead B, Zweig AS, Hinrichs AS, Karolchik D, Cline MS, Goldman M, Barber GP, Clawson H, Coelho A, Diekhans M, Dreszer TR, Giardine BM, Harte RA, Hillman-Jackson J, Hsu F, Kirkup V, Kuhn RM, Learned K, Li CH, Meyer LR, Pohl A, Raney BJ, Rosenbloom KR, Smith KE, Haussler D, Kent WJ. The UCSC Genome Browser database: update 2011. Nucleic Acids Res. 2011;39(Database issue):D876–882. doi: 10.1093/nar/gkq963. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Girirajan S, Eichler EE. Phenotypic variability and genetic susceptibility to genomic disorders. Hum Mol Genet. 2010;19:R176–187. doi: 10.1093/hmg/ddq366. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Girirajan S, Rosenfeld JA, Cooper GM, Antonacci F, Siswara P, Itsara A, Vives L, Walsh T, McCarthy SE, Baker C, Mefford HC, Kidd JM, Browning SR, Browning BL, Dickel DE, Levy DL, Ballif BC, Platky K, Farber DM, Gowans GC, Wetherbee JJ, Asamoah A, Weaver DD, Mark PR, Dickerson J, Garg BP, Ellingwood SA, Smith R, Banks VC, Smith W, McDonald MT, Hoo JJ, French BN, Hudson C, Johnson JP, Ozmore JR, Moeschler JB, Surti U, Escobar LF, El-Khechen D, Gorski JL, Kussmann J, Salbert B, Lacassie Y, Biser A, McDonald-McGinn DM, Zackai EH, Deardorff MA, Shaikh TH, Haan E, Friend KL, Fichera M, Romano C, Gecz J, DeLisi LE, Sebat J, King MC, Shaffer LG, Eichler EE. A recurrent 16p12.1 microdeletion supports a two-hit model for severe developmental delay. Nat Genet. 2010;42:203–209. doi: 10.1038/ng.534. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haggstrom AN, Drolet BA, Baselga E, Chamlin SL, Garzon MC, Horii KA, Lucky AW, Mancini AJ, Metry DW, Newell B, Nopper AJ, Frieden IJ. Prospective study of infantile hemangiomas: clinical characteristics predicting complications and treatment. Pediatrics. 2006;118:882–887. doi: 10.1542/peds.2006-0413. [DOI] [PubMed] [Google Scholar]
- Haggstrom AN, Garzon MC, Baselga E, Chamlin SL, Frieden IJ, Holland K, Maguiness S, Mancini AJ, McCuaig C, Metry DW, Morel K, Powell J, Perkins SM, Siegel D, Drolet BA. Risk for PHACE syndrome in infants with large facial hemangiomas. Pediatrics. 2010;126:e418–426. doi: 10.1542/peds.2009-3166. [DOI] [PubMed] [Google Scholar]
- Metry D, Heyer G, Hess C, Garzon M, Haggstrom A, Frommelt P, Adams D, Siegel D, Hall K, Powell J, Frieden I, Drolet B. Consensus Statement on Diagnostic Criteria for PHACE Syndrome. Pediatrics. 2009a;124:1447–1456. doi: 10.1542/peds.2009-0082. [DOI] [PubMed] [Google Scholar]
- Metry DW, Garzon MC, Drolet BA, Frommelt P, Haggstrom A, Hall J, Hess CP, Heyer GL, Siegel D, Baselga E, Katowitz W, Levy ML, Mancini A, Maronn ML, Phung T, Pope E, Sun G, Frieden IJ. PHACE syndrome: current knowledge, future directions. Pediatr Dermatol. 2009b;26:381–398. doi: 10.1111/j.1525-1470.2009.00944.x. [DOI] [PubMed] [Google Scholar]
- Pascual-Castroviejo I. Vascular and nonvascular intracranial malformation associated with external capillary hemangiomas. Neuroradiology. 1978;16:82–84. doi: 10.1007/BF00395211. [DOI] [PubMed] [Google Scholar]
- So WY, Wang Y, Ng MC, Yang X, Ma RC, Lam V, Kong AP, Tong PC, Chan JC. Aldose reductase genotypes and cardiorenal complications: an 8-year prospective analysis of 1,074 type 2 diabetic patients. Diabetes Care. 2008;31:2148–2153. doi: 10.2337/dc08-0712. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Su AI, Cooke MP, Ching KA, Hakak Y, Walker JR, Wiltshire T, Orth AP, Vega RG, Sapinoso LM, Moqrich A, Patapoutian A, Hampton GM, Schultz PG, Hogenesch JB. Large-scale analysis of the human and mouse transcriptomes. Proc Natl Acad Sci U S A. 2002;99:4465–4470. doi: 10.1073/pnas.012025199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Su AI, Wiltshire T, Batalov S, Lapp H, Ching KA, Block D, Zhang J, Soden R, Hayakawa M, Kreiman G, Cooke MP, Walker JR, Hogenesch JB. A gene atlas of the mouse and human protein-encoding transcriptomes. Proc Natl Acad Sci U S A. 2004;101:6062–6067. doi: 10.1073/pnas.0400782101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thamotharampillai K, Chan AK, Bennetts B, Craig ME, Cusumano J, Silink M, Oates PJ, Donaghue KC. Decline in neurophysiological function after 7 years in an adolescent diabetic cohort and the role of aldose reductase gene polymorphisms. Diabetes Care. 2006;29:2053–2057. doi: 10.2337/dc06-0678. [DOI] [PubMed] [Google Scholar]
- Wolford JK, Yeatts KA, Red Eagle AR, Nelson RG, Knowler WC, Hanson RL. Variants in the gene encoding aldose reductase (AKR1B1) and diabetic nephropathy in American Indians. Diabet Med. 2006;23:367–376. doi: 10.1111/j.1464-5491.2006.01834.x. [DOI] [PubMed] [Google Scholar]
- Wu C, Orozco C, Boyer J, Leglise M, Goodale J, Batalov S, Hodge CL, Haase J, Janes J, Huss JW, 3rd, Su AI. BioGPS: an extensible and customizable portal for querying and organizing gene annotation resources. Genome Biol. 2009;10:R130. doi: 10.1186/gb-2009-10-11-r130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu M, Chen X, Yan L, Cheng H, Chen W. Association between (AC)n dinucleotide repeat polymorphism at the 5′-end of the aldose reductase gene and diabetic nephropathy: a meta-analysis. J Mol Endocrinol. 2008;40:243–251. doi: 10.1677/JME-07-0152. [DOI] [PubMed] [Google Scholar]