Abstract
Pancreatic cancer is a disease caused by the accumulation of genetic alterations in specific genes. Elucidation of the human genome sequence, in conjunction with technical advances in the ability to perform whole exome sequencing, have provided new insight into the mutational spectra characteristic of this lethal tumour type. Most recently, exomic sequencing has been used to clarify the clonal evolution of pancreatic cancer as well as provide time estimates of pancreatic carcinogenesis, indicating that a long window of opportunity may exist for early detection of this disease while in the curative stage. Moving forward, these mutational analyses indicate potential targets for personalised diagnostic and therapeutic intervention as well as the optimal timing for intervention based on the natural history of pancreatic carcinogenesis and progression.
INTRODUCTION
Pancreatic ductal adenocarcinoma (hereafter referred to as pancreatic cancer) is among the deadliest of all solid malignancies. In both the USA and Europe, pancreatic cancer is the fourth leading cause of cancer deaths with a 5-year survival rate of only 4%.1,2 Extensive experimental and epidemiological data from the past two decades indicate that pancreatic cancer is a genetic disease. First, a variety of genetic mutations are recurrently identified in pancreatic cancers.3 Second, many of the identical mutations identified in pancreatic cancer can also be found in the precursor lesions that give rise to pancreatic cancer, often in the same pancreatectomy specimen.4–6 Third, pancreatic cancers are known to aggregate in some families, implying an inherited basis for some patients with this disease.7 Indeed, the genetic basis for a subset of high-risk individuals has been found.8–11 Fourth, genetically engineered mouse models that conditionally inactivate one or more genes known to play a role in pancreatic cancer can fully recapitulate the full spectrum of pancreatic carcinogenesis and metastasis as seen in humans.12–14
In light of the rapid accumulation of genetic data illustrating the genetic complexity of this disease, there is a need to summarise this information into its most practical essence that could be applied towards improving survival of patients with pancreatic cancer.15 In this basic science review the genetic basis of pancreatic cancer will be discussed as a preface to recent data outlining the clonal evolution of the pancreatic cancer genome that accompanies its progression and metastasis. These newfound discoveries provide a critical insight into the features of the pancreatic cancer genome that are most suited for early detection and therapeutic development.
PANCREATIC CARCINOGENESIS
Precursor lesions
A variety of precursor lesions have been described in the pancreas. These include pancreatic intraepithelial neoplasia (PanIN), intraductal papillary mucinous neoplasms (IPMNs) and mucinous cystadenomas (MCNs) (figures 1 and 2).16 PanIN is the term used to describe microscopic precursor lesions that arise in the small calibre (<5 mm) pancreatic ducts17 and that give rise to conventional ductal adenocarcinomas (figure 1). By contrast, IPMNs are macroscopically visible cystic neoplasms that arise in the mucin-producing main pancreatic duct or one of its branches (figure 2A). MCNs, the least common of these precursor lesions, are also macroscopically visible cystic neoplasms but they do not communicate with the pancreatic duct system (figure 2B). MCNs are characterised by a mucinous epithelial lining in association with an ovarian-like stroma (figure 2C).16 Although adeno-carcinoma may arise from any of the above precursor lesions, adenocarcinomas arising in association with PanINs are 13–100-fold more common in pancreaticoduodenectomy specimens than those arising from an IPMN or MCN.18 The reasons for this are multifactorial and beyond the scope of this review, but include a higher frequency of early diagnosis by non-invasive IPMNs and MCNs.19
Figure 1.
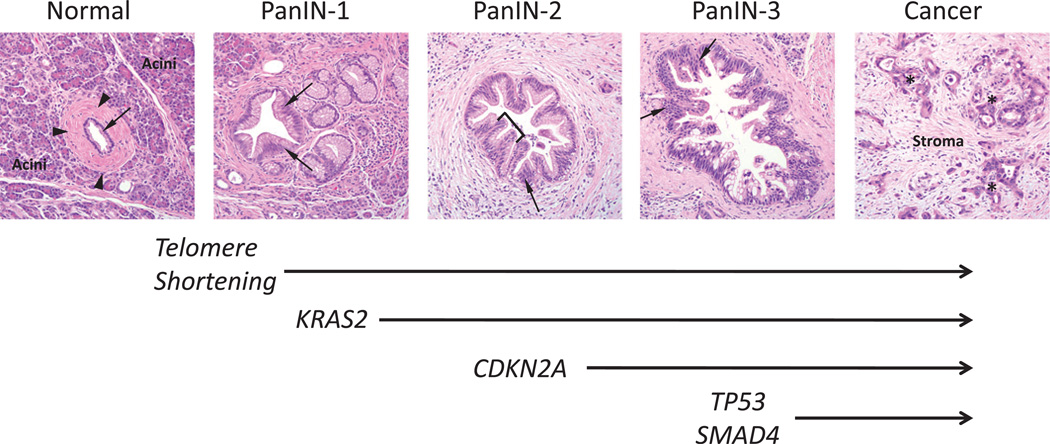
Morphological and genetic progression model of pancreatic carcinogenesis. Histological examples of a normal pancreatic duct, pancreatic intraepithelial neoplasia (PanIN) and pancreatic cancer are shown. Normal ducts are characterised by a low cuboidal epithelium (arrow) surrounded by a periductal fibrotic cuff (arrowheads). Ductal epithelium is relatively sparse compared with the surrounding acinar component. PanIN-1 lesions are differentiated from normal ductal epithelium by the presence of mucinous hyperplasia of the ductal cells (arrows) but without cytological atypia. By contrast, PanIN-2 lesions are notable for the presence of nuclear enlargement, atypia, crowding (arrow) and papillary infoldings of the epithelium (brackets). PanIN-3 lesions, synonymous to high-grade dysplasia/carcinoma in situ, show a complete loss of cell polarity (arrows) and marked cytological atypia in association with frequent mitotic figures and pseudopapillary growth of the neoplastic epithelium. PanIN-3 lesions may progress to invasive cancer, characterised by poorly formed neoplastic glands (asterisk) with an infiltrative growth pattern. Note the abundant desmoplastic stroma that is also a common feature of pancreatic cancer. Based on this progression model, the molecular alterations that accumulate during pancreatic carcinogenesis can be classified into early (telomere shortening and activating mutations in KRAS2), intermediate (inactivating mutations or epigenetic silencing of p16/CDKN2A) and late (inactivating mutations of TP53 and SMAD4) events. Mutations in additional genes may also occur during PanIN formation but are not illustrated in this example.
Figure 2.
Cystic precursors of pancreatic cancer. (A) Low power view of an intraductal papillary mucinous neoplasm. This precursor develops from ductal cells lining the main pancreatic duct (arrow) leading to cystic dilation and obstruction of the main pancreatic duct by the papillary neoplasm. (B) Low power view of a mucinous cystadenoma. Unlike IPMNs that arise within the main pancreatic duct, mucinous cystadenomas are not associated with the pancreatic duct system. Note the normal pancreatic tissue in the upper right of the image that is distinct from the cystic neoplasm. In this example the neoplasm also contains foci of borderline to high-grade dysplasia (indicated by arrow). (C) Higher power view of the cystic neoplasm shown in (B) to illustrate the presence of ovarian-like stroma that underlies the mucinous epithelium. IPMN, intraductal papillary mucinous neoplasm; MCN, mucinous cystadenoma.
Progression models
Accumulating evidence indicates that PanINs arise from a stem cell-like/progenitor cell population within the pancreas, either Hes1-positive centroacinar cells located at the junction of mature acini and ductal epithelium or mature acinar cells that transdifferentiate into ductal epithelium in response to cellular injury.20–22 The existence of a true duct epithelial stem cell has also been proposed.23 PanINs are represented by three stages characterised by increasing cytological atypia of the duct lining cells, called PanIN-1, PanIN-2 and PanIN-3 (figure 1).24 Whereas PanIN-1 is represented by mucinous differentiation of the ductal cells with minimal atypia, PanIN-3 corresponds to carcinoma in situ. Molecular analyses of PanINs from pancreatectomy specimens have indicated that increasing cytological atypia in ductal precursor lesions is highly associated with an accumulation of genetic alterations in specific genes, indicating that pancreatic carcinogenesis undergoes a genetic progression as described for other solid tumour types.17 Similar morphological progression models have been proposed for both IPMNs and MCNs, although the genetic features associated with these progression models is much less well characterised.25
Telomere length abnormalities
Telomeres are comprised of tandem repeats of the TTAGGG sequence that are progressively lost with each round of cell division.26 Telomere lengthening is mediated by telomerase, and critically short telomeres in normal cells activate a DNA damage response. The significance of this is because abnormally shortened telomeres lead to chromosome ends that become ‘sticky’, leading to chromosomal breakage–fusion–bridge cycles in dividing cells and consequent gene deletions and/or amplifications.27 Using an in situ hybridisation protocol to telomeric sequences, van Heek et al found that telomeres are abnormally shortened in all stages of PanIN compared with normal ductal epithelium, indicating that it is the earliest and most pervasive molecular abnormality identified in pancreatic carcinogenesis to date.28 Of interest, activation of telomerase expression is seen in the majority of pancreatic cancers, possibly as a protective mechanism against catastrophic DNA damage.29
PANCREATIC CANCER GENETICS BASED ON CANDIDATE GENE APPROACHES
Until recent years the genetic basis of pancreatic cancer has been elucidated using a candidate gene approach. Traditionally, this approach has relied on conventional dideoxy sequencing (‘Sanger sequencing’) using intronic primers that allow mutational analysis of the coding region of a gene and its associated exonic splice sites. Dideoxy sequencing has been highly sensitive at identifying tumour suppressor genes in pancreatic cancer when performed in association with loss of heterozygosity analyses using microsatellite markers spanning the allelic region of interest. This approach has identified the four genes most commonly ascribed to pancreatic cancer: KRAS2,CDKN2A,TP53 and SMAD4.
KRAS2
Activating mutations of the KRAS2 oncogene are the most common genetic abnormality known in pancreatic cancer, present in virtually all cases.30 KRAS2 encodes a member of the RAS family of guanosine triphosphate (GTP)-binding proteins that mediate a wide range of cellular functions including proliferation, cell survival and cytoskeletal remodelling. In normal cells, activated Kras2 that is bound to GTP is inactivated through guanosine triphosphatase-activating proteins that promote GTP hydrolysis and therefore attenuate Kras2 signalling. Activating mutations in KRAS2 impair this GTPase activity resulting in a Kras2 protein that is constitutively active, independent of extracellular or intracellular signal transduction.31 Mutations of the KRAS2 gene are one of the earliest genetic abnormalities observed in the progression model of pancreatic cancer, detectable as early as PanIN-1A lesions.4
p16/CDKN2A
The tumour suppressor gene CDKN2A (also known as p16 or INK4A) is the most commonly inactivated gene in pancreatic cancer.32 Loss of CDKN2A function is seen in approximately 90% of pancreatic cancers. This loss may occur by a variety of mechanisms such as homozygous deletion (40%), intragenic mutation with loss of the second allele (40%) or epigenetic silencing of gene expression by promoter methylation (10–15%).33 The protein encoded by this gene belongs to the cyclin-dependent kinase (CDK) inhibitor family and functions by inhibiting cell cycle progression through the G1–S checkpoint that is mediated by CDKs such as CDK4 and CDK6.34 Genetic inactivation of CDKN2A also occurs during pancreatic carcinogenesis and is seen as early as PanIN-2 lesions.4
TP53
Inactivation of the TP53 gene on chromosome 17p is present in approximately 50–75% of pancreatic cancers.35 Unlike CDKN2A that is inactivated by a variety of mechanisms, TP53 gene inactivation almost always occurs by an intragenic mutation combined with loss of the second allele.36 The p53 protein is a critical regulator of a variety of cellular functions including regulation of the G1–S cell cycle checkpoint, maintenance of G2–M arrest and the induction of apoptosis following cellular stress.37 Loss of p53 function allows cells to survive and divide despite the presence of damaged DNA, thus allowing the accumulation of additional genetic abnormalities and hence genetic instability.38 Mutations of TP53 are found as early as PanIN-3 stage lesions.39
SMAD4
Also known as DPC4, this gene is inactivated in approximately 55% of pancreatic cancers, either by homozygous deletion (30%) or by an intragenic mutation in association with loss of the second copy (25%).40 The Smad4 protein is a critical mediator of the transforming growth factor β (TGF-β) canonical signalling pathway that functions in cellular growth and differentiation.41 Activation of the TGF-β pathway occurs when the TGF-β protein binds to specific cell surface receptors, triggering an intracellular cascade that results in phosphorylation of the Smad transcription factors Smad 2/3. Smad2/3 then complexes with Smad4 and enters the nucleus where together they activate or repress gene transcription. Thus, loss of Smad4 in pancreatic cancer cells inhibits Smad-dependent TGF-β signalling, allowing an escape from TGF-β-induced growth inhibition.41 Similar to TP53, genetic inactivation of SMAD4 occurs at the stage of PanIN-3 lesions.5
Low-frequency targets and germline variants
A variety of genes are somatically inactivated at low frequency (<5%) in pancreatic cancer. These genes include the TGFBR1, TGFBR2 and ACVR1B receptors also within the TGF-β/activin signalling pathway42,43 and the protein kinase MKK4.44 Of interest, mutations in these genes do not confer the same negative impact on pancreatic cancer survival as does SMAD4, suggesting that alternative functions of the TGF-β pathway may be targeted by these mutations.45 MKK4, which encodes for a stress-activated protein kinase, is more often inactivated in pancreatic cancer metastases, suggesting that this gene may function as a metastasis suppressor.46
Subsets of low-frequency genetic targets represent germline variants associated with the familial aggregation of pancreatic cancer (reviewed by Hruban et al47). These variants include the serine/threonine kinase STK11/LKB148 and the DNA cross-linking repair genes BRCA2, FANCC and FANCG.6,49 Germline mutations in STK11/LKB1 are associated with the development of hamartomatous polyps in association with Peutz-Jeghers syndrome, and patients with this syndrome have a >100-fold increased risk of developing pancreatic cancer.47 However, STK11/LKB1 may be inactivated in sporadic pancreatic cancers as well.48 The main physiological function of STK11/LKB1 is in regulating cellular growth, metabolism and epithelial cell polarity.50 Inherited mutations in the DNA caretaker gene BRCA2 are perhaps the best characterised of the germline variants.6 In addition to an increased risk of developing cancers of the breast and ovary, BRCA2 mutations are associated with a 3.5–10-fold increased risk of developing pancreatic cancer. Brca2 functions in association with the Fanconi anaemia family of proteins to repair DNA cross-linking damage that normally occurs during cell replication, thus maintaining genomic fidelity while not directly influencing cell growth and proliferation.51 Following recognition of BRCA2 mutations in association with familial pancreatic cancer, germline mutations in the Fanconi anaemia family FANCC and FANCG genes have also been implicated in familial and young age of onset pancreatic cancer.49 PALB2 germline mutations, whose protein product interacts with Brca2, have also recently been identified in patients with familial pancreatic cancer.8
While CDKN2A is a known tumour suppressor gene in sporadic pancreatic cancers, germline mutations in CDKN2A cause familial atypical multiple mole melanoma syndrome (FAMMM syndrome).52 In addition to melanoma, patients with germline mutations in CDKN2A have a 9–47-fold increased risk of developing pancreatic cancer. Germline mutations in the serine protease PRSS1 and, to a lesser extent, the serine protease inhibitor SPINK1 cause autosomal dominant inherited chronic pancreatitis.53 Patients with familial pancreatitis have a lifetime risk of developing pancreatic cancer of as high as 40%.54
Recognition of the above germline variants in a patient with a family history of pancreatic cancer provides opportunities for therapeutic intervention. However, in most patients with a family history the genetic basis is unknown,55 prompting additional studies for identification of genes that confer an increased risk. To date, genome-wide association studies have identified pancreatic cancer susceptibility loci on chromosomes 9q34,56 13q22.1, 1q32.1 and 5p15.33.57 The locus on chromosome 9q34 corresponds to the ABO locus, whereas that of 5p15.33 corresponds to CLPTM1L-TERT which is also associated with multiple other cancer types. Linkage to chromosome 4q in a family with a unique form of autosomal dominant pancreatic cancer has also been reported.58 However, the PALLD gene implicated at this locus59 does not appear responsible for the majority of pancreatic cancer kindreds.60
INSIGHTS FROM EXOMIC SEQUENCING
While the candidate approach to pancreatic cancer gene discovery has yielded a variety of targets, until recently the extent to which additional genes throughout the genome are altered was unknown. However, a variety of technical advances have allowed for sequencing of the cancer exome (box 1), leading to greater insight into the mutational spectrum of human tumours including pancreatic cancer. For example, the publication of the human genome sequence in 2001,61 coupled with the development of high-throughput and automated methods for dideoxy sequencing analyses,62–64 have allowed for rapid analysis of genetic alterations in a variety of tumour types.65–71 Moreover, when sequencing analyses are performed using matched normal and tumour samples, these methods permit sensitive and specific identification of most types of coding genetic alterations in human cancer.
Box 1 Whole exome sequencing.
-
►
Whole exome sequencing is a type of high-throughput dideoxy sequencing analysis that specifically focuses on the coding portion of the genome.
-
►
Whole exome sequencing is different from whole genome sequencing in which both coding and non-coding regions of the genome are evaluated.
In 2008, such an approach was applied to the study of 24 pancreatic cancers with the goal of identifying the spectrum and extent of somatic mutations in pancreatic cancer and to identify the molecular pathways that were important for this tumour type.67 This effort focused on a set of 20 661 protein coding genes representing 99.6% of the known coding genome, the most curated set of genes at that time. Copy number chips to evaluate copy number gains or deletions were also used, as well as methods to interrogate the gene expression of each sample. To minimise sequencing artifacts, each sample was either xenograft-enriched or passaged in cell culture to remove contaminating normal cells that would otherwise mask the presence of these genetic alterations.
Among this set of genes 1562 somatic mutations were detected, most of which were base substitutions. A minority of small insertions and deletions, mutations at splice sites or the untranslated regions of these genes were also identified. Moreover, 1327 genes had at least one mutation and 148 had two or more mutations. Each mutation was also evaluated for their predicted effects on gene expression as the finding of a somatic mutation alone is not informative as to the potential consequences of that mutation for pancreatic cancer (box 2). For example, nonsense mutations that cause a stop codon prematurely end gene transcription, insertions or deletions of small numbers of bases may disrupt the amino acid sequence of the translated protein and splice site changes that interfere with mRNA processing; these types of mutations are frequently seen for the tumour suppressor genes CDKN2A, TP53 and SMAD4.72 By contrast, missense mutations that lead to a change in the amino acid at that site may or may not have effects on protein function. Thus, bioinformatics analyses were used to assess the predicted consequences of this class of mutations, indicating that only a minority was predicted to contribute to pancreatic cancer tumorigenesis (<20%).
Box 2 Types of genetic alterations in cancer.
-
►
Genetic alterations can be categorised into two types: driver genes and passenger genes.
-
►
Alterations in driver genes provide a selective advantage to the cancer cell in which they arise compared with other cells that do not contain these alterations.
-
►
Missense mutations that change protein function, nonsense mutations, frameshift mutations, gene deletions and gene amplifications are typical features of driver gene alterations.
-
►
Passenger mutations do not provide a selective advantage to the cancer cells in which they occur.
Gene deletions or amplifications were less common than base substitutions, but also provided insight into the pancreatic cancer genome. Atotal of 198 separate homozygous deletions were identified among the 24 pancreatic cancers. As expected, genes that were involved in a homozygous deletion again included the tumour suppressor genes CDKN2A, TP53 and SMAD4 as well as genes that had not previously been implicated in pancreatic cancer development. The number of deletions in a tumour was more variable than the number of somatic mutations, ranging between 2 and 20 per tumour. Moreover, 144 focal high copy amplifications were found in the 24 tumours, corresponding to genes such as KRAS and FOXA1. High copy number amplifications were the least common form of genetic alterations in pancreatic cancer compared with base substitutions or homozygous deletions.
Because the goal of this study was to identify genes likely to play a causal role in pancreatic cancer (‘driver genes’), mutated genes were categorised on the basis of two major criteria. First, only genes in which at least two genetic alterations were identified in the discovery screen were considered, at least one of which was potentially deleterious in nature based on its predicted effect on protein expression. Second, the mutation rate had to be >10 mutations per Mb. To determine those genes with mutations that fit this second criterion, passenger mutation rates for each gene were determined by taking into account the size of the gene, its nucleotide composition and other relevant features (details described by Jones et al67). However, because mathematical models used to calculate passenger mutation rates have inherent limitations,73,74 the estimated minimal, maximal and mid (average of minimal and maximal) passenger mutation rates were calculated. Based on the most lenient estimates of passenger mutation rate, this approach resulted in a list of 91 candidate cancer genes (CAN genes) (for details see supplementary table 7 in the paper by Jones et al67) that again included all genes previously known to play a significant role in pancreatic cancer through mutation or copy number change (KRAS, CDKN2A, TP53 and SMAD4), as well as numerous other genes of potential biological interest, many of which had not previously been identified as having a significant role in this tumour type (ARID1A). Of interest, some genes such as MLL3 were recently identified as mutated in pancreatic cancer in association with follow-up studies of breast and colon cancer CAN genes,75 suggesting a more universal role of MLL3 in carcinogenesis of multiple tissue types. Of note, most of the CAN genes were generally mutated at low frequency, consistent with the notion that the candidate approach to gene sequencing is unable to identify the majority of genetically altered genes in pancreatic cancer.76 Ultimately, additional validations and functional studies will be needed to confirm any role these predicted CAN genes play in pancreatic cancer.
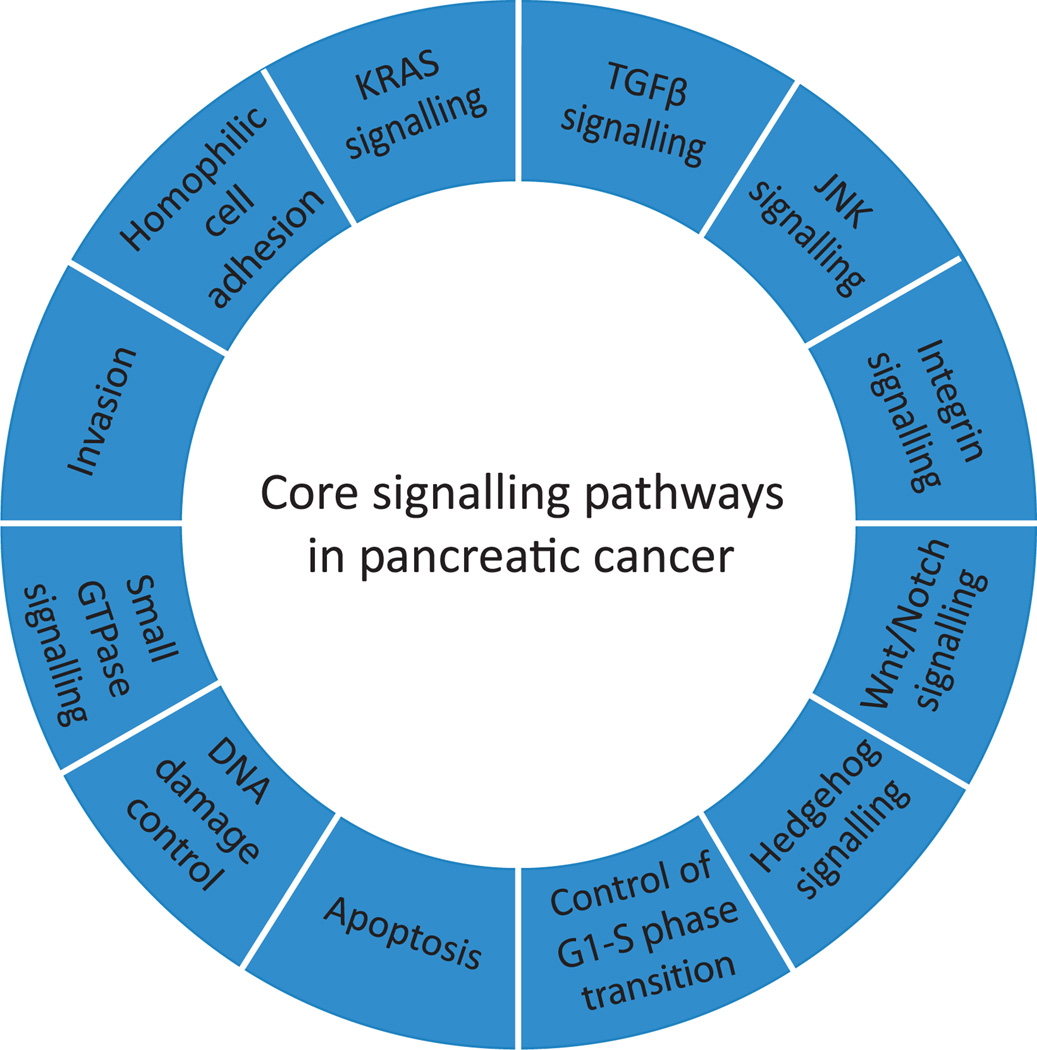
Cellular pathways and processes typically involve multiple proteins that function in a coordinated manner. The entire set of mutated genes were thus categorised into the cellular processes in which their protein products are involved, indicating that these genes corresponded to 69 gene sets that were genetically altered in the majority of the 24 cancers examined. For each gene set the statistical probability that a gene set contained a driver gene was also considered as well as whether the component genes were more likely to be affected by a genetic alteration than would be predicted by the passenger mutation rate for that set. Based on this approach, 31 of these 69 gene sets could be further distilled into 12 core signalling pathways and processes with a clear relevance to carcinogenesis (figure 3). Within each of these pathways the genes that were altered in any given carcinoma varied widely, yet only one gene member of each pathway was altered in a given carcinoma. This suggests that a small number of the core pathways—rather than large numbers of specific genes–provide the clue to understanding the biology of pancreatic cancer.67
Figure 3.
Core signalling pathways in pancreatic cancer. The 12 pathways and processes whose component genes were genetically altered in most pancreatic cancers based on whole exome sequencing are shown. Therapeutic targeting of one or more of these pathways, rather than specific gene alterations that occur within a pathway, provides a new paradigm for treatment of this disease. GTPase, guanosine triphosphatase; TGFβ, transforming growth factor β.
GENETIC EVOLUTION OF PANCREATIC CANCER
Until recently, a major limitation to studying pancreatic cancer progression has been the lack of tissue for study. This is because pancreatic cancer in its advanced stages is not a surgical disease77 and thus high quality tissues have not been available for study. This need has been addressed by our laboratory’s use of rapid autopsy protocols in which patients with end stage pancreatic cancer can consent premortem to an autopsy for the purpose of collecting high quality cancer tissues for research.78
Most patients with pancreatic cancer are diagnosed at a late stage.77 However, whether the poor prognosis of patients with pancreatic cancer compared with patients with other types of cancer is a result of late diagnosis or early dissemination of disease to distant organs was unknown, a distinction of critical importance towards improving survival of this disease. To address this important question, we relied on data generated from the pancreatic cancer genome project for seven patients for which one of their metastases was included in the genome survey. These data, together with the sample resources available through our autopsy resource, also provided the basis for characterising the clonal evolution of pancreatic cancer from its initiation within a normal cell until the time that it has disseminated to distant organs.
A comparison of the genetic features of these seven metastases with the 17 surgically resected (stages pT1–pT3) carcinomas also in the genome project indicated that they did not differ in their total number of genetic alterations, nor did they differ in the spectrum of mutations seen. Overall, these seven carcinomas contained an average of 61 mutations of which the vast majority were silent missense mutations (ie, did not change the amino acid at that site) that were not predicted to contribute to tumorigenesis. Thus, the genetic features of advanced stage pancreatic cancers do not differ from those seen in surgical resection specimens.
Comparative lesion sequencing
Comparative lesion sequencing is a method in which genetic alterations present in one cancer sample are analysed in additional geographically or temporally distinct samples from that same patient, permitting an evaluation of the clonal relatedness of different carcinoma samples within an individual.79,80 We performed comparative lesion sequencing for each of these seven patients by evaluating the presence or absence of each mutation in that patient’s index metastasis (ie, the metastasis sample sequenced in the genome project) in the matched primary carcinoma and in additional distinct metastases collected at rapid autopsy for each patient (figure 4). Using this approach, the mutations in each patient’s pancreatic cancer could be classified into two categories. The first category corresponded to mutations that were present in all samples analysed for each patient, called ‘founder’ mutations. Moreover, because they were present in both the primary carcinoma and the matched metastases, a logical assumption is that these mutations accumulated within the PanIN that ultimately gave rise to that pancreatic cancer and are thus present in the majority, if not all, of the cells of the tumour. Thus, founder mutations are genetic markers of the original parental clone of cells that formed that carcinoma. Consistent with this notion, founder mutations contained within the parental clones of each of the seven patients included all known driver mutations important for pancreatic cancer formation (KRAS2, CDKN2A, TP53 and SMAD4), as well as many CAN genes also identified by the pancreatic cancer genome project.67 The second category, called ‘progressor’ mutations, corresponded to those mutations that were present in a subset of the samples analysed for each patient. Because founder mutations were present in all samples analysed for each patient but progressor mutations were present in only a subset of those samples, it is likely that the progressor mutations occurred after founder mutations and thus represent subclonal evolution beyond the parental clone.
Figure 4.
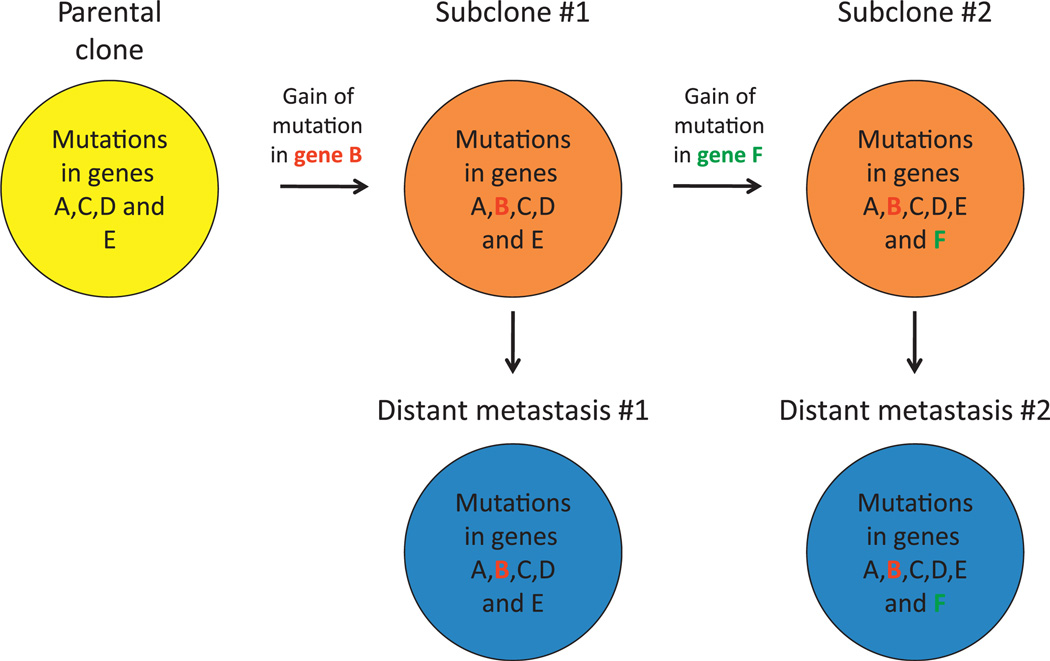
Comparative lesion sequencing. Comparative lesion sequencing (CSL) requires at least two geographically or temporally distinct samples of a patient’s cancer. These samples may be any number of synchronous metastases, the primary carcinoma and a subsequent metastatic recurrence, or even samples taken from different regions of a single primary carcinoma. In the first step of CSL, one sample (the ‘index’ lesion) is analysed by whole exome sequencing to identify all somatic mutations present in the coding fraction of the genome. In the second step of CSL, typically performed using standard sequencing methods, the presence or absence of all mutations found in the index lesion is assessed in additional samples for that patient. The ‘relatedness’ of the different samples to each other can then be derived based on the number of mutations shared. For example, mutations common to all samples are called founder mutations and reflect the genetic features of the clonal population (the ‘parental’ clone) that gave rise to all the samples analysed. By contrast, those mutations that are only present in a subset of the samples are called progressor mutations as they reflect clonal progression that occurred beyond formation of the parental clone. In the hypothetical example shown, whole exome sequencing is performed on one sample (the ‘index’ lesion shown in grey) that identified mutations in genes A–F. In the next step, two additional samples from this patient (shown in red) are analysed to determine if one or more of this set of six mutations were also present. A comparison of the findings indicates that mutations in genes A, C, D and E are common to all samples and thus represent founder mutations contained within the parental clone that gave birth to all three. By contrast, the mutations in genes B and F are only present in two of three samples and represent progressor mutations that occurred relatively later than the founder mutations in the clonal evolution of this carcinoma.
A characterisation of the specific genetic features of founder mutations versus progressor mutations allows an understanding of the genes specifically altered at the later stages of pancreatic cancer progression. Three major features of founder mutations were uncovered that distinguished them from progressor mutations. First, founder mutations were more numerous than progressor mutations, indicating that the majority of mutations present in a pancreatic cancer accumulated during PanIN progression and are contained within the parental clone. Second, founder mutations—and thus parental clones—contained the majority of deleterious mutations present in each carcinoma. Third, the vast majority of homozygous mutations—and thus allelic losses—also corresponded to founder mutations contained within the parental clone. Deleterious mutations and homozygous mutations are further characteristic of tumour suppressor genes in which once copy is lost in association with an inactivating mutation of the remaining gene copy. In total, these findings indicate that parental clones harbour the majority of genetic instability and deleterious mutations in a pancreatic cancer upon which additional mutations associated with clonal progression of pancreatic cancer are seen. This is also consistent with the findings of Campbell et al who showed, by massively parallel paired-end sequencing, that the majority of rearrangements within the pancreatic cancer genome occur early and before the development of metastatic disease.27
Mapping clonal evolution
While these patterns indicate the types of mutations seen in pancreatic cancer and their timing of accumulation, they do not reveal whether subclonal progression (indicated by progressor mutations) occurs within the primary carcinoma itself or after dissemination of cancer cells to distant sites. This can be addressed by evaluation of multiple geographically distinct regions of each patient’s primary carcinoma for each founder and progressor mutation. An example of a clonal progression model is shown in figure 5 and illustrates that, beyond the formation of the parental clone, many subclones are also present within the primary carcinoma. These subclones can be detected as samples of primary carcinoma that contained both the founder mutations and one or more progressor mutations. The genetic signature of different subclones within the primary carcinoma was also highly similar to the genetic signature of specific metastases in the same patient, indicating that subclones that develop within the primary carcinoma seed distant metastases. This does not rule out the possibility that metastases can also seed metastases, but does solidify the notion that metastatic subclones are pre-existent within the primary carcinoma.
Figure 5.
Clonal progression of pancreatic cancer. A representative example of the proposed clonal evolution of pancreatic cancer based on sequencing data of five different samples is shown. In this model, after development of the parental clone that seeded the infiltrating carcinoma (indicated in yellow), ongoing clonal evolution continues within the primary carcinoma leading to the development of a subclone characterised by the gain of a mutation in gene B. Additional clonal evolution beyond this subclone leads to the development of a second subclone characterised by the presence of a mutation in gene F. Subclones may then seed metastases to distant sites (indicated in blue) as reflected by the identical pattern of mutations in subclone 1 compared with distant metastasis 1, and in subclone 2 compared with distant metastasis 2.
Evolutionary timeline
The dataset is particularly powerful for estimations of the timing of clonal evolution of pancreatic cancer. Towards this goal, a computational model was created that relied on data generated from the genome project in conjunction with the published rates of cellular proliferation of normal and neoplastic pancreatic cancer cells,81 rates of passenger mutation per cell division79 and our categorisation of mutations into founders and progressors. The model relied on passenger mutations as they are unlikely to drive tumorigenesis, they accounted for the vast majority of mutations identified in each pancreatic cancer compared with the small number of driver mutations, they are accumulated independently in each cell lineage and they accumulate at a constant rate per cell division.82 Thus, the number of passenger mutations in the index metastasis sequenced in the genome project is proportional to the time taken to accumulate those mutations. Three critical times in the genetic evolution of pancreatic cancer for these seven patients were estimated (figure 6). The first time interval (T1) corresponded to the time taken from the initiating mutation in a normal ductal epithelial cell until the development of the parental clone as reflected by its unique number of founder mutations, the second time interval (T2) corresponded to the time taken for the development of the subclone within the primary carcinoma that seeded the index metastasis in that patient, and the third time interval (T3) corresponded to the subsequent time until the patient’s death. In essence, T1 can be related to PanIN formation until the infiltrating carcinoma first formed, T2 from that time until a metastatic subclone developed within the primary carcinoma and T3 as the time of metastatic dissemination of that subclone until the patient’s death. Based on this model, the conservative estimate of 11.7 years, 6.8 years and 2.7 years per interval, respectively, was reached, corresponding to an average of approximately 21 years from the initiating mutation until the patient’s death. Unfortunately, most patients with pancreatic cancer are diagnosed well towards the end of this time span,77 indicating that the overall poor prognosis is probably due to the diagnosis occurring far too late in the natural history of the disease.
Figure 6.
Estimates of time taken for the genetic progression of pancreatic cancer. Pancreatic carcinogenesis begins with an initiating mutation in a normal cell that confers a selective growth advantage. Successive waves of clonal expansion occur in association with the acquisition of additional mutations, corresponding to the progression model of pancreatic intraepithelial neoplasia (PanIN) and time T1. One founder cell within a PanIN lesion will seed the parental clone and hence initiate the infiltrating carcinoma. This is the beginning of time T2. Following additional waves of clonal expansion, the subclones that will give rise to one or more distant metastases will develop within the infiltrating carcinoma signifying the beginning of time T3. The estimated average time for each interval is also indicated and corresponds to a total of 21.2 years from tumour initiation until the patient’s death from metastatic disease. Unfortunately, most patients are not diagnosed until late in time T3 when the cells of these metastatic subclones have already escaped the pancreas and started to grow within distant organs.
SUMMARY AND IMPLICATIONS
The first and perhaps most significant repercussion of these recent data is their implication for the feasibility of screening to prevent pancreatic cancer deaths. This is important because the traditional view of pancreatic cancer is that it is an extremely fast-growing and aggressive malignancy.83 However, computational modelling indicates a large window of opportunity—at least a decade—for early detection of pancreatic cancer while in the curative stage, and several additional years until the development of metastatic subclones within the primary carcinoma.84 Thus, pancreatic cancer appears more similar to other tumour types in its growth rates and natural history than previously recognised.79 While this is promising news, it brings to light the sobering reality that the vast majority of patients are not diagnosed until this window has passed and metastatic dissemination has already occurred.77 This probably includes the majority of patients who also undergo surgical resection as most of those patients will also succumb to disease progression.85 The challenge for the future is thus to detect these tumours during the PanIN stage (interval T1) and before the development of metastatic ability within the primary carcinoma (early in time interval T2), and the protracted time interval now found for pancreatic cancer development and progression indicates that this is possible.
The development of technologies and biomarkers to detect pancreatic cancers during the curative stage will be a critical issue in coming years. However, most frequently mutated genes in pancreatic cancer that accumulate during PanIN progression, while providing insight into the biology of this disease, may not have value themselves as screening biomarkers, an issue of utmost importance as Ca19-9 alone lacks the sensitivity to detect pancreatic cancer in the majority of patients with the disease.86 One potential target for screening includes mutant KRAS2 DNA or protein that is shed into blood or stool.87,88 KRAS2 mutations are fairly specific to pancreatic, colorectal and lung cancers, and both colorectal and lung cancers can be excluded by currently available screening modalities.89,90 KRAS2 mutations are also an early event in pancreatic carcinogenesis,4 and thus a positive screening test may signify those patients who require additional evaluation by endoscopic ultrasound. Indeed, the technical ability to do this has already been demonstrated.88,91 However, while technically feasible, it is important to note that with identification of KRAS2 mutations runs the risk of overdiagnosis and overtreatment of pancreatic cancer precursors,47,92 particularly as surgical resection with its associated morbidities is the only known method of cure of this disease.18 Thus, prospective research aimed at biomarker identification and the optimal strategies for screening of this disease will continue to be needed.
Virtually all mutations that drive pancreatic cancer accumulate during intraductal progression (ie, PanINs). Thus, the genetic features present at the time of infiltrating cancer formation will dictate the behaviour of that carcinoma and will be present in all the cells of that patient’s disease. This concept is buttressed by data indicating that the genetic features of an infiltrating pancreatic carcinoma at diagnosis underlie its metastatic propensity.93 Thus, targeting specific cellular pathways appears a fruitful endeavour in the coming years because it is the pathways targeted by these genetic alterations, and not the specific genes, that may be most important.67 However, it remains to be seen whether different genes targeted within these pathways have equal effects on outcome or treatment efficacy. For example, genetic inactivation of SMAD4 is highly correlated with poor survival of pancreatic cancer after surgical resection whereas no such relationship was found when other members of this pathway are mutated.45
An understanding of the genetic features of pancreatic cancer is only useful if it can be used to improve the survival of patients with this disease.15 With this virtual explosion of genetic information about the pancreatic cancer genome, and its clonal evolution during the development of metastasis, comes the hope of newfound opportunities for screening and therapeutic development.
Acknowledgments
Funding CAI-D is supported by NIH/NCI grants CA140599, CA130938 and the V Foundation.
Footnotes
An additional table is published online only. To view this file please visit the journal online (http://gut.bmj.com).
There are no relevant financial conflicts to disclose.
Competing intrests None.
Provenance and peer review Commissioned; externally peer reviewed.
REFERENCES
- 1.Jemal A, Siegel R, Xu J, et al. Cancer statistics, 2010. CA Cancer J clin. 2010;60:277–300. doi: 10.3322/caac.20073. [DOI] [PubMed] [Google Scholar]
- 2.Malvezzi M, Arfe A, Bertuccio P, et al. European cancer mortality predictions for the year 2011. Ann Oncol. 2011;22:947–956. doi: 10.1093/annonc/mdq774. [DOI] [PubMed] [Google Scholar]
- 3.Maitra A, Hruban RH. Pancreatic cancer. Annu Rev Pathol. 2008;3:157–188. doi: 10.1146/annurev.pathmechdis.3.121806.154305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Moskaluk CA, Hruban RH, Kern SE. p16 and K-ras gene mutations in the intraductal precursors of human pancreatic adenocarcinoma. Cancer Res. 1997;57:2140–2143. [PubMed] [Google Scholar]
- 5.Wilentz RE, Iacobuzio-Donahue CA, Argani P, et al. Loss of expression of Dpc4 in pancreatic intraepithelial neoplasia: evidence that DPC4 inactivation occurs late in neoplastic progression. Cancer Res. 2000;60:2002–2006. [PubMed] [Google Scholar]
- 6.Goggins M, Hruban RH, Kern SE. BRCA2 is inactivated late in the development of pancreatic intraepithelial neoplasia: evidence and implications. Am J Pathol. 2000;156:1767–1771. doi: 10.1016/S0002-9440(10)65047-X. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Klein AP, Brune KA, Petersen GM, et al. Prospective risk of pancreatic cancer in familial pancreatic cancer kindreds. Cancer Res. 2004;64:2634–2638. doi: 10.1158/0008-5472.can-03-3823. [DOI] [PubMed] [Google Scholar]
- 8.Jones S, Hruban RH, Kamiyama M, et al. Exomic sequencing identifies PALB2 as a pancreatic cancer susceptibility gene. Science. 2009;324:217. doi: 10.1126/science.1171202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Hahn SA, Greenhalf B, Ellis I, et al. BRCA2 germline mutations in familial pancreatic carcinoma. J Natl Cancer Inst. 2003;95:214–221. doi: 10.1093/jnci/95.3.214. [DOI] [PubMed] [Google Scholar]
- 10.Rogers CD, Couch FJ, Brune K, et al. Genetics of the FANCA gene in familial pancreatic cancer. J Med Genet. 2004;41:e126. doi: 10.1136/jmg.2004.024851. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Rogers CD, van der Heijden MS, Brune K, et al. The genetics of FANCC and FANCG in familial pancreatic cancer. Cancer Biol Ther. 2004;3:167–169. doi: 10.4161/cbt.3.2.609. [DOI] [PubMed] [Google Scholar]
- 12.Bardeesy N, Aguirre AJ, Chu GC, et al. Both p16(Ink4a) and the p19(Arf)-p53 pathway constrain progression of pancreatic adenocarcinoma in the mouse. Proc Natl Acad Sci U S A. 2006;103:5947–5952. doi: 10.1073/pnas.0601273103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Hingorani SR, Wang L, Multani AS, et al. Trp53R172H and KrasG12D cooperate to promote chromosomal instability and widely metastatic pancreatic ductal adenocarcinoma in mice. Cancer Cell. 2005;7:469–483. doi: 10.1016/j.ccr.2005.04.023. [DOI] [PubMed] [Google Scholar]
- 14.Skoulidis F, Cassidy LD, Pisupati V, et al. Germline Brca2 heterozygosity promotes Kras(G12D)-driven carcinogenesis in a murine model of familial pancreatic cancer. Cancer Cell. 2010;18:499–509. doi: 10.1016/j.ccr.2010.10.015. [DOI] [PubMed] [Google Scholar]
- 15.McDermott U, Downing JR, Stratton MR. Genomics and the continuum of cancer care. N Engl J Med. 2011;364:340–350. doi: 10.1056/NEJMra0907178. [DOI] [PubMed] [Google Scholar]
- 16.Matthaei H, Schulick RD, Hruban RH, et al. Cystic precursors to invasive pancreatic cancer. Nat Rev Gastroenterol Hepatol. 2011;8:141–150. doi: 10.1038/nrgastro.2011.2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Hruban RH, Goggins M, Parsons J, et al. Progression model for pancreatic cancer. Clin Cancer Res. 2000;6:2969–2972. [PubMed] [Google Scholar]
- 18.Winter JM, Cameron JL, Campbell KA, et al. 1423 pancreaticoduodenectomies for pancreatic cancer: a single-institution experience. J Gastrointest Surg. 2006;10:1199–1210. doi: 10.1016/j.gassur.2006.08.018. discussion 210–11. [DOI] [PubMed] [Google Scholar]
- 19.Gaujoux S, Brennan MF, Gonen M, et al. Cystic lesions of the pancreas: changes in the presentation and management of 1,424 patients at a single institution over a 15-year time period. J Am Coll Surg. 2011;212:590–600. doi: 10.1016/j.jamcollsurg.2011.01.016. discussion 600–3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Miyamoto Y, Maitra A, Ghosh B, et al. Notch mediates TGF alpha-induced changes in epithelial differentiation during pancreatic tumorigenesis. Cancer Cell. 2003;3:565–576. doi: 10.1016/s1535-6108(03)00140-5. [DOI] [PubMed] [Google Scholar]
- 21.Stanger BZ, Stiles B, Lauwers GY, et al. Pten constrains centroacinar cell expansion and malignant transformation in the pancreas. Cancer Cell. 2005;8:185–195. doi: 10.1016/j.ccr.2005.07.015. [DOI] [PubMed] [Google Scholar]
- 22.Guerra C, Schuhmacher AJ, Canamero M, et al. Chronic pancreatitis is essential for induction of pancreatic ductal adenocarcinoma by K-Ras oncogenes in adult mice. Cancer Cell. 2007;11:291–302. doi: 10.1016/j.ccr.2007.01.012. [DOI] [PubMed] [Google Scholar]
- 23.Li C, Heidt DG, Dalerba P, et al. Identification of pancreatic cancer stem cells. Cancer Res. 2007;67:1030–1037. doi: 10.1158/0008-5472.CAN-06-2030. [DOI] [PubMed] [Google Scholar]
- 24.Hruban RH, Adsay NV, Albores-Saavedra J, et al. Pancreatic intraepithelial neoplasia: a new nomenclature and classification system for pancreatic duct lesions. Am J Surg Pathol. 2001;25:579–586. doi: 10.1097/00000478-200105000-00003. [DOI] [PubMed] [Google Scholar]
- 25.Furukawa T, Kloppel G, Volkan Adsay N, et al. Classification of types of intraductal papillary-mucinous neoplasm of the pancreas: a consensus study. Virchows Arch. 2005;447:794–799. doi: 10.1007/s00428-005-0039-7. [DOI] [PubMed] [Google Scholar]
- 26.Martinez P, Blasco MA. Telomeric and extra-telomeric roles for telomerase and the telomere-binding proteins. Nat Rev. 2011;11:161–176. doi: 10.1038/nrc3025. [DOI] [PubMed] [Google Scholar]
- 27.Campbell PJ, Yachida S, Mudie LJ, et al. The patterns and dynamics of genomic instability in metastatic pancreatic cancer. Nature. 2010;467:1109–1113. doi: 10.1038/nature09460. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.van Heek NT, Meeker AK, Kern SE, et al. Telomere shortening is nearly universal in pancreatic intraepithelial neoplasia. Am J Pathol. 2002;161:1541–1547. doi: 10.1016/S0002-9440(10)64432-X. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Seki K, Suda T, Aoyagi Y, et al. Diagnosis of pancreatic adenocarcinoma by detection of human telomerase reverse transcriptase messenger RNA in pancreatic juice with sample qualification. Clin Cancer Res. 2001;7:1976–1981. [PubMed] [Google Scholar]
- 30.Almoguera C, Shibata D, Forrester K, et al. Most human carcinomas of the exocrine pancreas contain mutant c-K-ras genes. Cell. 1988;53:549–554. doi: 10.1016/0092-8674(88)90571-5. [DOI] [PubMed] [Google Scholar]
- 31.Schubbert S, Shannon K, Bollag G. Hyperactive Ras in developmental disorders and cancer. Nat Rev. 2007;7:295–308. doi: 10.1038/nrc2109. [DOI] [PubMed] [Google Scholar]
- 32.Caldas C, Hahn SA, da Costa LT, et al. Frequent somatic mutations and homozygous deletions of the p16 (MTS1) gene in pancreatic adenocarcinoma. Nat Genet. 1994;8:27–32. doi: 10.1038/ng0994-27. [DOI] [PubMed] [Google Scholar]
- 33.Schutte M, Hruban RH, Geradts J, et al. Abrogation of the Rb/p16 tumor-suppressive pathway in virtually all pancreatic carcinomas. Cancer Res. 1997;57:3126–3130. [PubMed] [Google Scholar]
- 34.Kim WY, Sharpless NE. The regulation of INK4/ARF in cancer and aging. Cell. 2006;127:265–275. doi: 10.1016/j.cell.2006.10.003. [DOI] [PubMed] [Google Scholar]
- 35.Redston MS, Caldas C, Seymour AB, et al. p53 mutations in pancreatic carcinoma and evidence of common involvement of homocopolymer tracts in DNA microdeletions. Cancer Res. 1994;54:3025–3033. [PubMed] [Google Scholar]
- 36.Vogelstein B, Lane D, Levine AJ. Surfing the p53 network. Nature. 2000;408:307–310. doi: 10.1038/35042675. [DOI] [PubMed] [Google Scholar]
- 37.Raycroft L, Wu HY, Lozano G. Transcriptional activation by wild-type but not transforming mutants of the p53 anti-oncogene. Science. 1990;249:1049–1051. doi: 10.1126/science.2144364. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Goh AM, Coffill CR, Lane DP. The role of mutant p53 in human cancer. J Pathol. 2011;223:116–126. doi: 10.1002/path.2784. [DOI] [PubMed] [Google Scholar]
- 39.DiGiuseppe JA, Redston MS, Yeo CJ, et al. p53-independent expression of the cyclin-dependent kinase inhibitor p21 in pancreatic carcinoma. Am J Pathol. 1995;147:884–888. [PMC free article] [PubMed] [Google Scholar]
- 40.Hahn SA, Schutte M, Hoque AT, et al. DPC4, a candidate tumor suppressor gene at human chromosome 18q21.1. Science. 1996;271:350–353. doi: 10.1126/science.271.5247.350. [DOI] [PubMed] [Google Scholar]
- 41.Massague J. TGFbeta in Cancer. Cell. 2008;134:215–230. doi: 10.1016/j.cell.2008.07.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Goggins M, Shekher M, Turnacioglu K, et al. Genetic alterations of the transforming growth factor beta receptor genes in pancreatic and biliary adenocarcinomas. Cancer Res. 1998;58:5329–5332. [PubMed] [Google Scholar]
- 43.Su GH, Bansal R, Murphy KM, et al. ACVR1B (ALK4, activin receptor type 1B) gene mutations in pancreatic carcinoma. Proc Natl Acad Sci U S A. 2001;98:3254–3257. doi: 10.1073/pnas.051484398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Su GH, Hilgers W, Shekher MC, et al. Alterations in pancreatic, biliary, and breast carcinomas support MKK4 as a genetically targeted tumor suppressor gene. Cancer Res. 1998;58:2339–2342. [PubMed] [Google Scholar]
- 45.Blackford A, Serrano OK, Wolfgang CL, et al. SMAD4 gene mutations are associated with poor prognosis in pancreatic cancer. Clin Cancer Res. 2009;15:4674–4679. doi: 10.1158/1078-0432.CCR-09-0227. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Taylor JL, Szmulewitz RZ, Lotan T, et al. New paradigms for the function of JNKK1/MKK4 in controlling growth of disseminated cancer cells. Cancer Lett. 2008;272:12–22. doi: 10.1016/j.canlet.2008.05.012. [DOI] [PubMed] [Google Scholar]
- 47.Hruban RH, Canto MI, Goggins M, et al. Update on familial pancreatic cancer. Adv Surg. 2010;44:293–311. doi: 10.1016/j.yasu.2010.05.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Su GH, Hruban RH, Bansal RK, et al. Germline and somatic mutations of the STK11/LKB1 Peutz-Jeghers gene in pancreatic and biliary cancers. Am J Pathol. 1999;154:1835–1840. doi: 10.1016/S0002-9440(10)65440-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.van der Heijden MS, Yeo CJ, Hruban RH, et al. Fanconi anemia gene mutations in young-onset pancreatic cancer. Cancer Res. 2003;63:2585–2588. [PubMed] [Google Scholar]
- 50.Jansen M, Ten Klooster JP, Offerhaus GJ, et al. LKB1 and AMPK family signaling: the intimate link between cell polarity and energy metabolism. Physiol Rev. 2009;89:777–798. doi: 10.1152/physrev.00026.2008. [DOI] [PubMed] [Google Scholar]
- 51.Wang W. Emergence of a DNA-damage response network consisting of Fanconi anaemia and BRCA proteins. Nat Rev Genet. 2007;8:735–748. doi: 10.1038/nrg2159. [DOI] [PubMed] [Google Scholar]
- 52.Goldstein AM, Chan M, Harland M, et al. High-risk melanoma susceptibility genes and pancreatic cancer, neural system tumors, and uveal melanoma across GenoMEL. Cancer Res. 2006;66:9818–9828. doi: 10.1158/0008-5472.CAN-06-0494. [DOI] [PubMed] [Google Scholar]
- 53.Whitcomb DC, Gorry MC, Preston RA, et al. Hereditary pancreatitis is caused by a mutation in the cationic trypsinogen gene. Nat Genet. 1996;14:141–145. doi: 10.1038/ng1096-141. [DOI] [PubMed] [Google Scholar]
- 54.Lowenfels AB, Maisonneuve P, DiMagno EP, et al. Hereditary pancreatitis and the risk of pancreatic cancer. International Hereditary Pancreatitis Study Group. J Natl Cancer Inst. 1997;89:442–446. doi: 10.1093/jnci/89.6.442. [DOI] [PubMed] [Google Scholar]
- 55.Shi C, Hruban RH, Klein AP. Familial pancreatic cancer. Arch Pathol Lab Med. 2009;133:365–374. doi: 10.5858/133.3.365. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Amundadottir L, Kraft P, Stolzenberg-Solomon RZ, et al. Genome-wide association study identifies variants in the ABO locus associated with susceptibility to pancreatic cancer. Nat Genet. 2009;41:986–990. doi: 10.1038/ng.429. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Petersen GM, Amundadottir L, Fuchs CS, et al. A genome-wide association study identifies pancreatic cancer susceptibility loci on chromosomes 13q22.1, 1q32.1 and 5p15.33. Nat Genet. 2010;42:224–228. doi: 10.1038/ng.522. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Eberle MA, Pfutzer R, Pogue-Geile KL, et al. A new susceptibility locus for autosomal dominant pancreatic cancer maps to chromosome 4q32-34. Am J Hum Genet. 2002;70:1044–1048. doi: 10.1086/339692. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Pogue-Geile KL, Chen R, Bronner MP, et al. Palladin mutation causes familial pancreatic cancer and suggests a new cancer mechanism. PLoS Med. 2006;3:e516. doi: 10.1371/journal.pmed.0030516. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Klein AP, Borges M, Griffith M, et al. Absence of deleterious palladin mutations in patients with familial pancreatic cancer. Cancer Epidemiol Biomarkers Prev. 2009;18:1328–1330. doi: 10.1158/1055-9965.EPI-09-0056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Lander ES, Linton LM, Birren B, et al. Initial sequencing and analysis of the human genome. Nature. 2001;409:860–921. doi: 10.1038/35057062. [DOI] [PubMed] [Google Scholar]
- 62.Wang Z, Shen D, Parsons DW, et al. Mutational analysis of the tyrosine phosphatome in colorectal cancers. Science. 2004;304:1164–1166. doi: 10.1126/science.1096096. [DOI] [PubMed] [Google Scholar]
- 63.Parsons DW, Wang TL, Samuels Y, et al. Colorectal cancer: mutations in a signalling pathway. Nature. 2005;436:792. doi: 10.1038/436792a. [DOI] [PubMed] [Google Scholar]
- 64.Bardelli A, Parsons DW, Silliman N, et al. Mutational analysis of the tyrosine kinome in colorectal cancers. Science. 2003;300:949. doi: 10.1126/science.1082596. [DOI] [PubMed] [Google Scholar]
- 65.Sjoblom T, Jones S, Wood LD, et al. The consensus coding sequences of human breast and colorectal cancers. Science. 2006;314:268–274. doi: 10.1126/science.1133427. [DOI] [PubMed] [Google Scholar]
- 66.Stephens PJ, McBride DJ, Lin ML, et al. Complex landscapes of somatic rearrangement in human breast cancer genomes. Nature. 2009;462:1005–1010. doi: 10.1038/nature08645. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Jones S, Zhang X, Parsons DW, et al. Core signaling pathways in human pancreatic cancers revealed by global genomic analyses. Science. 2008;321:1801–1806. doi: 10.1126/science.1164368. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Parsons DW, Jones S, Zhang X, et al. An integrated genomic analysis of human glioblastoma multiforme. Science. 2008;321:1807–1812. doi: 10.1126/science.1164382. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Pleasance ED, Stephens PJ, O’Meara S, et al. A small-cell lung cancer genome with complex signatures of tobacco exposure. Nature. 2010;463:184–190. doi: 10.1038/nature08629. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Varela I, Tarpey P, Raine K, et al. Exome sequencing identifies frequent mutation of the SWI/SNF complex gene PBRM1 in renal carcinoma. Nature. 2011;469:539–542. doi: 10.1038/nature09639. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Jiao Y, Shi C, Edil BH, et al. DAXX/ATRX, MEN1, and mTOR pathway genes are frequently altered in pancreatic neuroendocrine tumors. Science. 2011;331:1199–1203. doi: 10.1126/science.1200609. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Rozenblum E, Schutte M, Goggins M, et al. Tumor-suppressive pathways in pancreatic carcinoma. Cancer Res. 1997;57:1731–1734. [PubMed] [Google Scholar]
- 73.Greenman C, Wooster R, Futreal PA, et al. Statistical analysis of pathogenicity of somatic mutations in cancer. Genetics. 2006;173:2187–2198. doi: 10.1534/genetics.105.044677. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Parmigiani G, Boca S, Lin J, et al. Design and analysis issues in genome-wide somatic mutation studies of cancer. Genomics. 2009;93:17–21. doi: 10.1016/j.ygeno.2008.07.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Balakrishnan A, Bleeker FE, Lamba S, et al. Novel somatic and germline mutations in cancer candidate genes in glioblastoma, melanoma, and pancreatic carcinoma. Cancer Res. 2007;67:3545–3550. doi: 10.1158/0008-5472.CAN-07-0065. [DOI] [PubMed] [Google Scholar]
- 76.Velculescu VE. Defining the blueprint of the cancer genome. Carcinogenesis. 2008;29:1087–1091. doi: 10.1093/carcin/bgn096. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Hidalgo M. Pancreatic cancer. N Engl J Med. 2010;362:1605–1617. doi: 10.1056/NEJMra0901557. [DOI] [PubMed] [Google Scholar]
- 78.Embuscado EE, Laheru D, Ricci F, et al. Immortalizing the complexity of cancer metastasis: genetic features of lethal metastatic pancreatic cancer obtained from rapid autopsy. Cancer Biol Ther. 2005;4:548–554. doi: 10.4161/cbt.4.5.1663. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Jones S, Chen WD, Parmigiani G, et al. Comparative lesion sequencing provides insights into tumor evolution. Proc Natl Acad Sci U S A. 2008;105:4283–4288. doi: 10.1073/pnas.0712345105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Navin N, Krasnitz A, Rodgers L, et al. Inferring tumor progression from genomic heterogeneity. Genome Res. 2010;20:68–80. doi: 10.1101/gr.099622.109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Klein WM, Hruban RH, Klein-Szanto AJ, et al. Direct correlation between proliferative activity and dysplasia in pancreatic intraepithelial neoplasia (PanIN): additional evidence for a recently proposed model of progression. Mod Pathol. 2002;15:441–447. doi: 10.1038/modpathol.3880544. [DOI] [PubMed] [Google Scholar]
- 82.Salk JJ, Horwitz MS. Passenger mutations as a marker of clonal cell lineages in emerging neoplasia. Semin Cancer Biol. 2010;20:294–303. doi: 10.1016/j.semcancer.2010.10.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Stathis A, Moore MJ. Advanced pancreatic carcinoma: current treatment and future challenges. Nat Rev Clin Oncol. 2010;7:163–172. doi: 10.1038/nrclinonc.2009.236. [DOI] [PubMed] [Google Scholar]
- 84.Yachida S, Jones S, Bozic I, et al. Distant metastasis occurs late during the genetic evolution of pancreatic cancer. Nature. 2010;467:1114–1117. doi: 10.1038/nature09515. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Amikura K, Kobari M, Matsuno S. The time of occurrence of liver metastasis in carcinoma of the pancreas. Int J Pancreatol. 1995;17:139–146. doi: 10.1007/BF02788531. [DOI] [PubMed] [Google Scholar]
- 86.Kim JE, Lee KT, Lee JK, et al. Clinical usefulness of carbohydrate antigen 19-9 as a screening test for pancreatic cancer in an asymptomatic population. J Gastroenterol Hepatol. 2004;19:182–186. doi: 10.1111/j.1440-1746.2004.03219.x. [DOI] [PubMed] [Google Scholar]
- 87.Caldas C, Hahn SA, Hruban RH, et al. Detection of K-ras mutations in the stool of patients with pancreatic adenocarcinoma and pancreatic ductal hyperplasia. Cancer Res. 1994;54:3568–3573. [PubMed] [Google Scholar]
- 88.Wang Q, Chaerkady R, Wu J, et al. Mutant proteins as cancer-specific biomarkers. Proc Natl Acad Sci U S A. 2011;108:2444–2449. doi: 10.1073/pnas.1019203108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Berg CD, Aberle DR. Computed tomography screening for lung cancer. JAMA. 2007;298:513–514. doi: 10.1001/jama.298.5.513-b. author reply 515–16. [DOI] [PubMed] [Google Scholar]
- 90.Philip AK, Lubner MG, Harms B. Computed tomographic colonography. Surg Clin North Am. 2011;91:127–139. doi: 10.1016/j.suc.2010.10.010. [DOI] [PubMed] [Google Scholar]
- 91.Shi C, Chandrasekaran A, Thuluvath PJ, et al. Ultrasensitive detection of KRAS2 mutations in bile and serum from patients with biliary tract carcinoma using LigAmp technology. J Mol Diagn. 2009;11:583–589. doi: 10.2353/jmoldx.2009.090061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Greenhalf W, Grocock C, Harcus M, et al. Screening of high-risk families for pancreatic cancer. Pancreatology. 2009;9:215–222. doi: 10.1159/000210262. [DOI] [PubMed] [Google Scholar]
- 93.Iacobuzio-Donahue CA, Klimstra DS, Adsay NV, et al. Dpc-4 protein is expressed in virtually all human intraductal papillary mucinous neoplasms of the pancreas: comparison with conventional ductal adenocarcinomas. Am J Pathol. 2000;157:755–761. doi: 10.1016/S0002-9440(10)64589-0. [DOI] [PMC free article] [PubMed] [Google Scholar]