Abstract
As part of the central core domain of the ribosome, helix 69 of 23S rRNA participates in an important intersubunit bridge and contacts several protein translation factors. Helix 69 is believed to play key roles in protein synthesis. Even though high-resolution crystal structures of the ribosome exist, the solution dynamics and roles of individual nucleotides in H69 are still not well defined. To better understand the influence of modified nucleotides, specifically pseudouridine, on the multiple conformational states of helix 69 in the context of 50S subunits and 70S ribosomes, chemical probing analyses were performed on wild-type and pseudouridine-deficient bacterial ribosomes. Local structural rearrangements of helix 69 upon ribosomal subunit association and interactions with its partner, helix 44 of 16S rRNA, are observed. The helix 69 conformational states are also magnesium dependent. The probing data presented in this study provide insight into the functional role of helix 69 dynamics and regulation of these conformational states by post-transcriptional pseudouridine modification.
Protein biosynthesis is carried out by the ribosome in all living organisms, and the overall mechanisms in this process are highly conserved throughout phylogeny. Bacterial ribosomes are composed of small (30S) and large (50S) subunits that associate into 70S ribosomes, with the catalytic core domain being composed of ribosomal RNA (rRNA).1–4 The rRNA plays a key role in decoding cognate tRNA-mRNA interactions,4, 5 catalyzing peptide-bond formation,6–8 and contributing to tRNA translocation.9, 10 Post-transcriptional nucleotide modifications, including pseudouridine (Ψ), are clustered within this functionally important domain of the ribosome.11–13
The subunit interactions required for 70S ribosome formation are supported by highly conserved intersubunit bridges.1, 14, 15 A key bridge, B2a, forms part of the decoding center and is composed of helix 69 (H69) of the 50S subunit and helix 44 (h44) of the 30S subunit.1 In ribosome crystal structures, residue A1912 of H69 projects into the minor groove of h44 and interacts with C1407 and G1494 of h44.3, 4 The positioning of A1912 seems to be supported by a reverse-Hoogsteen base pair with Ψ1917 (Figure 1a). Nucleotide A1919 has a minor groove interaction with U1406/U1495 of h44.3 This interaction is supported by another non-canonical Hoogsteen base pair between Ψ1911 and A1919 and a bridging 2′-OH of A1918 (Figure 1b). The B2a interaction plays an important role in maintaining subunit association as the 30S subunit rotates relative to the 50S subunit during translocation.16, 17 Results from mutation studies also reveal the biological significance of bridge B2a.18–20 Single-base substitutions of residues A1912 or A1919 reduce protein synthesis and peptidyl-transfer processivity, and lead to defects in 70S formation.19 Mutations at C1914, Ψ1915, A1916, and G1922 result in frameshifting and stop-codon readthrough.18, 20
Figure 1.
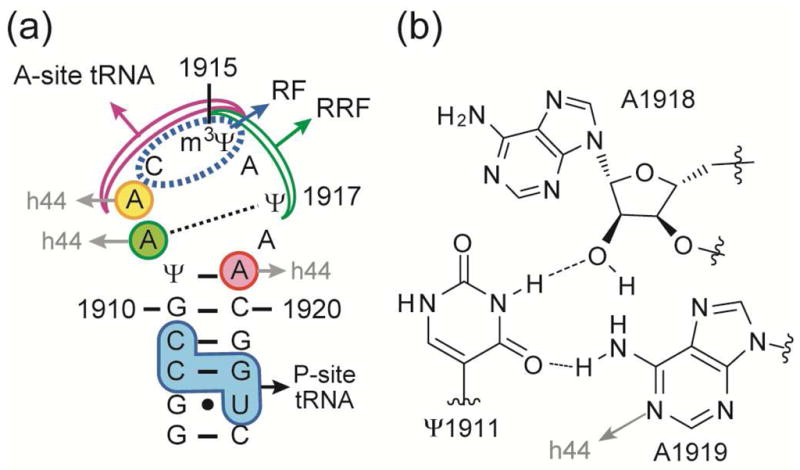
(a) The sequence and map of E. coli H69 RNA show key interactions with h44, tRNAs, and protein factors (RFs, RRF) (Ψ is pseudouridine and m3Ψ is 3-methylpseudouridine). (b) The Ψ1911-A1918-A1919 hydrogen-bonding network is observed in E. coli 70S ribosome structures.3
Helix 69 interacts with tRNAs,4, 21 release factors (RFs),22–24 and ribosome recycling factor (RRF)25 (summarized in Figure 1a). Contacts with A1913 are believed to play a role in A-site tRNA accommodation.26 The importance of proper H69-A-site-tRNA interactions is supported by the observation that an A1913U substitution suppresses mutant tRNA accommodation compared to the wild-type H69.27 The H69 stem (G1921–G1922) interacts with the D stem of the P-site tRNA,1 and G1922 mutants cause frameshifting and stop-codon readthrough.20 The tip of H69 (C1914) contacts the switch loop of RF1 during stop-codon recognition.22 Ribosomes lacking H69 also exhibit strong impairment in peptide release by RFs, although other translation steps are only moderately affected.24, 28 Direct interactions between RRF and H69 (Ψ1915–Ψ1917) are also observed.25 Overall, these interactions of H69 with protein factors and tRNAs appear to play key biological roles, which emphasizes the importance of structural dynamics and the ability of the RNA component to adopt multiple conformational states.
Recent dimethylsulfate (DMS) probing studies revealed that H69 within 50S subunits can exist in multiple conformational states that can be induced by changes in pH, Mg2+ concentration, and temperature, as well as being influenced by Ψ modifications.29 Several different conformational states of H69 are observed in crystal structures, in which the A1913 position forms a stacked (closed) conformation in 50S subunits and a flipped-out (open) conformation in 70S ribosomes.2, 3 The available biological and structural data suggest that these multiple conformational states of H69 play important roles in translation. Therefore, H69 has been proposed to be a suitable candidate for unique antibiotic targeting.29–31 A better understanding of the conformational states of H69 in the context of both 50S subunits and 70S ribosomes under solution conditions will help guide the design of new targeting agents. In this study, the H69 conformational states were analyzed by selective 2′-hydroxyl acylation and primer extension (SHAPE),32 and chemical probing with diethylpyrocarbonate (DEPC)33 and DMS.34 By comparing H69 conformational states in 70S ribosomes with those in 50S subunits, specific regions undergoing dynamic structural rearrangements upon subunit association were identified. In addition, similar studies on pseudouridine-deficient ribosomes have allowed the contributions of non-standard nucleotides to these rearrangements to be determined.
RESULTS AND DISCUSSION
Probing H69 Structural Rearrangements
Subunit association of the ribosome was expected to alter the accessibility of certain nucleotides towards chemical reagents. For example, the exposure of adenine residues of H69 under different conformational states in 70S ribosomes or 50S subunits can be revealed through DMS reactivity at the N1 position. To probe H69 conformational dynamics in 70S ribosomes, SHAPE analysis was employed. SHAPE reactivity is based on local nucleotide conformation-dependent 2′-hydroxyl nucleophilicity.32 If a base is involved in interactions such as Watson-Crick pairing or stacking, the corresponding nucleotide is constrained, which in turn reduces the 2′-OH nucleophilicity. On the other hand, if a base can move freely and is not involved in such interactions, the 2′-OH of this nucleotide is highly reactive towards the SHAPE reagent (N-methylisatonic anhydride, or NMIA).32 SHAPE analysis was therefore expected to provide useful information about the conformational states of H69 in 50S subunits and 70S ribosomes.
Changes in Conformational Flexibility of A1918 and A1913
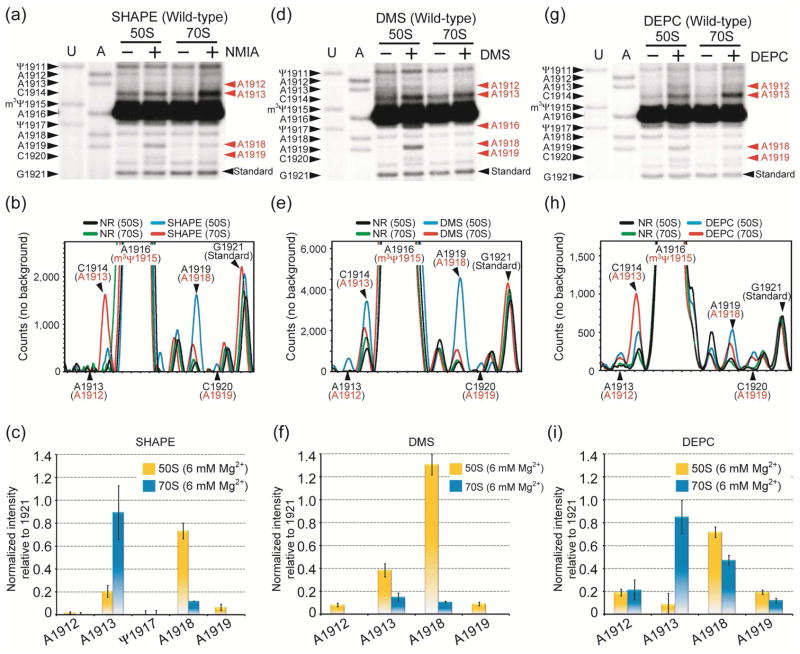
At 37 °C under physiological buffer conditions, SHAPE analysis reveals that the 2′-OH of A1918 is reactive (unconstrained) in 50S subunits, but much less reactive (seven-fold reduction; constrained) in 70S ribosomes (Figure 2a, b, c; these data were normalized to non-specific stop site 1921, which is located in a stem region). This probing pattern is consistent with X-ray crystal structures of 70S ribosomes, which show that the 2′-OH of A1918 is involved in a widened-Hoogsteen interaction with Ψ1911 and A1919 (Figure 1b).3 Reactivity of the A1918 2′-OH indicates that this site is flexible and accessible to NMIA in 50S subunits, but that ribosome assembly and bridge B2a formation confine its orientation. In contrast, the A1913 2′-OH shows greater than four-fold higher reactivity in 70S ribosomes relative to 50S subunits, indicating that A1913 is less constrained upon subunit (Figure 2c). Alternatively, A1913 might have a unique local conformation within the 70S ribosome that is ideal for increased SHAPE reactivity.32 The remaining H69 loop nucleotides (Ψ1911, A1912, C1914, m3Ψ1915, A1916, Ψ1917, A1919) are found to be constrained in both 50S and 70S ribosomes. These results suggest that H69 forms a well-ordered loop structure in 50S subunits and 70S ribosomes, but A1913 has increased flexibility while A1918 becomes more constrained upon subunit association. SHAPE analysis on isolated 70S ribosomes was also compared with reassociated 70S ribosomes (generated from 50S and 30S subunits) under the same reaction conditions, and no difference was observed (data not shown).
Figure 2.
Autoradiograms (top) and quantification (center, bottom) of SHAPE (a, b, c), DMS (d, e, f), and DEPC (g, h, i) probing analyses on wild-type 50S subunits and 70S ribosomes are shown. In panels a, d, and g, the E. coli H69 sequence corresponding to the dideoxy nucleotide stop sites (U and A reactions) is on the left side, and chemical modification sites, which cause a primer extension stop at the 3′ nucleotide, are in red on the right side. The normalized intensities for modified sites were calculated by subtracting non-specific primer extension stops (black and green traces, panels b, e, and h) from total intensity (blue and red, panels b, e, and h), and then normalizing to a uniform band (position 1921) (panels c, f, and i). The actual stop sites are indicated in black and the chemical modification sites are shown in red (panels b, e, and h).
Changes in Exposure of A1913 and A1918 in 70S Ribosomes
SHAPE analysis shows that A1913 gains flexibility upon subunit association. Nucleotide A1913 also has different conformations in crystal structures of 50S subunits and 70S ribosomes.2, 3 Therefore, DMS probing was employed to examine N1 accessibility of A1913 (Figure 2d, e). In this case, A1913 shows a two-fold difference in reactivity between 50S subunits and 70S ribosomes, with lower accessibility in 70S ribosomes (Figure 2f). Residue A1912 is weakly reactive in 50S subunits and unreactive in 70S ribosomes. In contrast, A1918 is strongly protected from DMS with a greater than ten-fold decrease in reactivity as a result of subunit association, consistent with previous probing data.35 This difference could result from a change in H69 structure or protection due to 30S subunit interactions.
To further explore the base accessibility in H69, DEPC was employed (Figure 2g, h). This reagent reacts with adenosine N7 and is also sensitive to variations in base-pairing or base-stacking interactions.33 In contrast to the DMS results, DEPC probing reveals that A1913 is a weakly reactive site in 50S subunits and eight-fold more reactive in 70S ribosomes (Figure 2i). This result suggests the release of A1913 from base-stacking interactions, most likely with A1912, upon subunit association. The results of DEPC probing at position 1918 also differ from those with DMS. Although the relative trend is similar, this residue shows less than a two-fold difference in reactivity between 50S subunits and 70S ribosomes. These differences are likely attributed to varying accessibilities of the N1 and N7 positions of A1913 and A1918. Indeed, in 70S ribosome high-resolution crystal structures, the N1 position of A1918 projects into the H69 loop; whereas, the N7 of A1918 faces the solvent-accessible surface.3
The Role of A1913 in H69
DEPC probing shows liberation of A1913 from a protected to an accessible state and SHAPE analysis shows its increased conformational flexibility in 70S ribosomes. In contrast, reduced DMS, DEPC, and NMIA reactivity of A1918 in 70S ribosomes compared to 50S subunits indicates protection of its N1, N7, and 2′-OH, respectively. These data suggest that formation of intersubunit bridge B2a results in confinement and/or protection of A1918, while in turn leading to conformational flexibility of A1913 on the opposite side of the H69 loop. Residue A1913 has been proposed to be important for cognate and near-cognate tRNA selection through direct contact with ribose 37 of the A-site tRNA anticodon stem loop.4, 36 A crystal structure of 70S ribosomes also shows that the A1913 base rotates around its C-N glycosidic bond upon A-site tRNA binding.36 Interestingly, the N7 of A1913 interacts with position 38 of cognate tRNAs through a Mg2+ ion; whereas, the Mg2+ ion bridges an interaction between A1913 of H69 and G1494 of h44 with near-cognate tRNA.36 Thus, it is possible that A1913 liberation from base-stacking interactions in the H69 loop and acquisition of local conformational elasticity upon 70S ribosome formation are necessary in order for the ribosome to have translational fidelity and to monitor incoming tRNAs.
The positioning of A1913 has also been proposed to be important for RF-mediated peptide release during translation termination.22 A crystal structure of the 70S ribosome-RF1 complex shows A1913 projecting into the minor groove of the A site of 16S rRNA.22 This interaction is believed to prevent extrusion of A1493 of h44 and promote RF binding to the ribosome.22 This model is supported by the observation that aminoglycosides, which induce A1492/A1493 base flipping in 16S rRNA,37 compete with RF binding.38 Interestingly, the RF- and A-site tRNA-binding regions of H69 are quite similar, although the A1913 positioning is quite different. Thus, the local nucleotide flexibility of residue A1913 and H69 structural rearrangements likely play an important role in sensing or regulating local interactions, in a manner similar to that of A1492/A1493 in 16S rRNA.
The Ψ1911-A1918-A1919 Network
DEPC probing shows slightly reduced reactivity at A1919 in 70S ribosomes when compared with 50S subunits (Figure 2g). Even though the band intensity is quite low (Figure 2i), this change is reproducible. In crystal structures of bacterial 70S ribosomes, a non-canonical Hoogsteen interaction between Ψ1911 and A1919 bridged by the 2′-OH of A1918 is observed (Figure 1b).3 However, SHAPE analysis reveals that the 2′-OH of A1918 is reactive in 50S subunits but unreactive in 70S ribosomes, and DMS probing shows that the N1 of A1918 is accessible in 50S subunits but protected in 70S ribosomes. These data suggest that A1918 does not participate in a Ψ1911-A1918-A1919 network in 50S subunits.
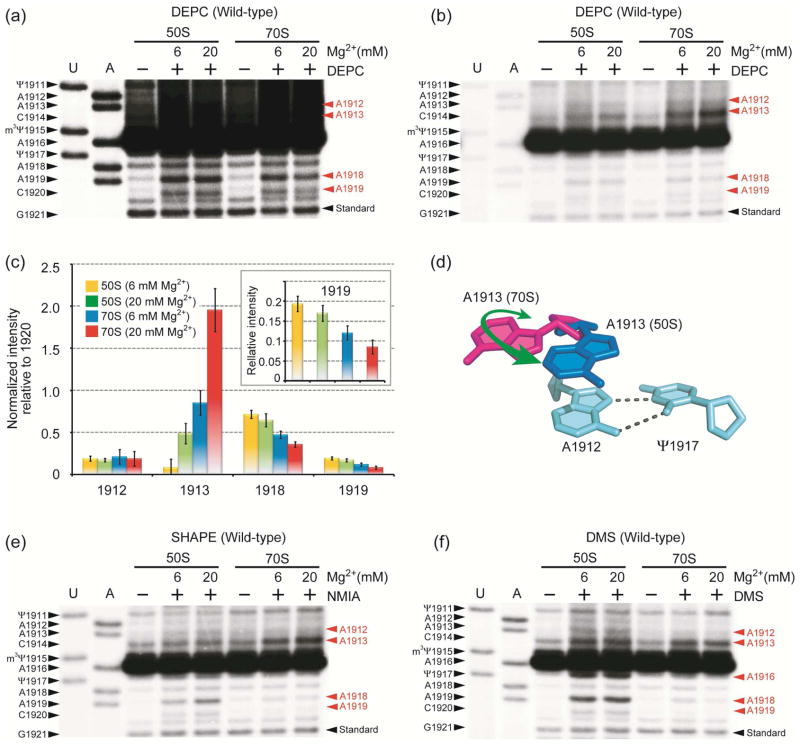
These probing results lead to the hypothesis that A1918 moves into the H69 loop upon subunit association and interacts with Ψ1911-A1919, leading to further protection of A1919 N7 from DEPC. Therefore, DEPC probing was carried out under higher Mg2+ concentration (20 mM) in order to stabilize the Ψ1911-A1918-A1919 interaction (Figure 3a–c). Further protection of A1919 N7 in 70S ribosomes is observed (Figure 3a, c). Sites in the neighboring helix 68 (H68) were used to confirm that DEPC reactivity is the same under these conditions (Supporting Information). In the case of A1918, its reactivity is further reduced (two-fold) in 70S ribosomes under the higher Mg2+ conditions, although not completely eliminated (Figure 3c).
Figure 3.
Autoradiograms show DEPC probing of H69 in wild-type 50S subunits and 70S ribosomes under low and high Mg2+ conditions with longer (a) and shorter (b) gel exposure times. Chemical modification sites are shown in red on the right side of the gel. (c) Quantification analysis for the DEPC probing is shown (inset: normalized band intensity at position 1919). (d) The proposed A1913 conformational change is illustrated. Autoradiograms for SHAPE (e) and DMS (f) analyses under 6 and 20 mM Mg2+ concentrations are shown.
Increased Exposure of A1913 at High Mg2+
In contrast to other residues in H69, A1913 exhibits elevated DEPC reactivity in 20 mM Mg2+ (Figure 3b, c). This result suggests that the Mg2+-stabilized ribosome conformation favors the flipped-out structure of A1913 (Figure 3d). SHAPE analysis (Figure 3e) supports this result, and increased reactivity of A1913 is observed under the higher Mg2+ conditions. In contract, the DMS-probing pattern for A1913 is not affected by changing the Mg2+ concentration (Figure 3f), suggesting solvent exposure of only one side of the base. The link between altered conformational dynamics with increased Mg2+ concentration and A1913 base flipping may play a role in factor-binding events. For example, binding of cognate tRNA or stop-codon recognition by RFs may cause reduced H69 conformational flexibility around A1918, but increased exposure of A1913. Moreover, it is known that elevated Mg2+ concentrations cause lower translational fidelity in vitro.39 This result would suggest that A1913 is able to interact with both cognate and near-cognate tRNAs in high Mg2+, perhaps due to improper positioning.
Role of Ψ Modifications in H69
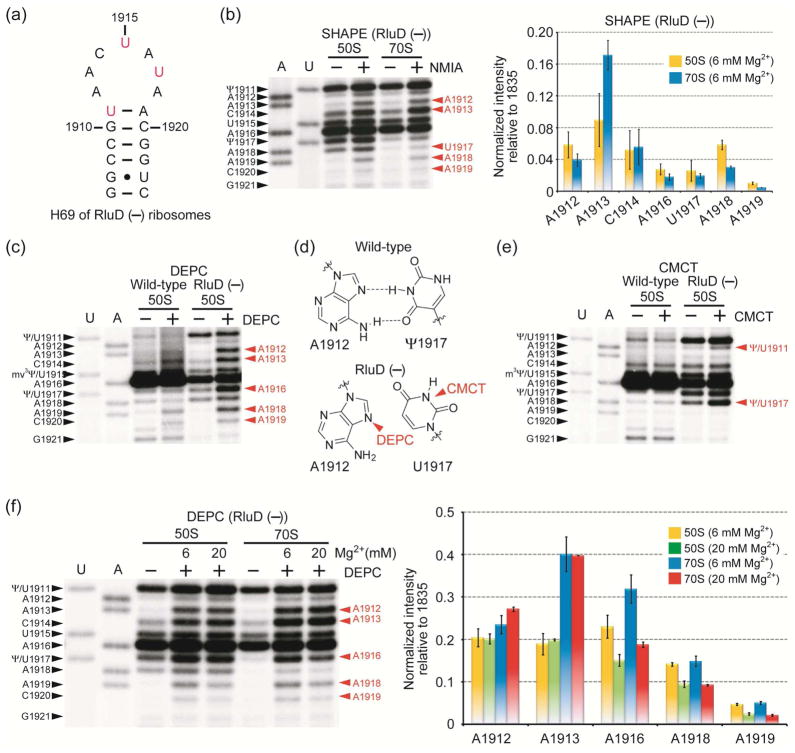
A distinct characteristic of H69 is the conserved modified nucleotides in its loop region, including Ψ at positions 1911 and 1917 and 3-methylpseudouridine (m3Ψ) at 1915.40, 41 Ribosomes lacking Ψ modifications in H69 exhibit slow growth rates in vivo and reduced subunit association in vitro.42 A mutant phenotype in a bacterial strain lacking H69 Ψ modifications (RluD(−)) can be rescued by mutations in RF2 protein at a site adjacent to H69-interacting residues.43 However, recent studies revealed that the RluD(−) phenotype is actually the consequence of mutant RF2 in the E. coli K-12 strains; whereas, E. coli B strains and Salmonella enterica containing fully active RF2 do not display the mutant phenotype, even upon deletion of the rluD gene encoding for H69 pseudouridine synthase.44 Although the role of Ψ residues in translation termination is questionable, H69 itself is still indispensable for efficient terminations by RFs24, 28 and its conformation is likely to be important.22 Previous model studies using small RNAs revealed that Ψs in H69 play an important role in maintaining the loop structure.45, 46 To better understand the role of Ψ modifications in H69 structure during subunit association, SHAPE analysis was also carried out on Ψ-deficient ribosomes (RluD(−)42) and compared with data on wild-type E. coli ribosomes (Figure 4a, unmodified H69).
Figure 4.
(a) The RluD(−) H69 RNA sequence is shown. (b) Autoradiogram (left) and quantification (right) of SHAPE reactivity on RluD(−) ribosomes are shown. Chemical modification sites are in red on the right side of the gel (m2G1835 was used for normalization). Comparisons of DEPC- (c) and CMCT-probing (e) patterns between wild-type and RluD(−) 50S subunits are shown. (d) DEPC and CMCT reactive sites in RluD(−) ribosomes are indicated. (f) Autoradiogram (left) and quantification (right) of DEPC reactivity of RluD(−) 50S and 70S ribosomes under low and high Mg2+ conditions are shown.
Changes in H69 Loop Flexibility
SHAPE analysis shows less than two-fold increased NMIA reactivity at A1913 in 70S RluD(−) ribosomes relative to 50S subunits and two-fold reduced reactivity of A1918 in 70S ribosomes relative to 50S subunits (compared to a four-fold increase at 1913 and seven-fold reduction at 1918 in wild-type ribosomes) (Figure 4b; in this case the data was normalized to m2G1835, a non-specific reverse transcriptase stop site (Supporting Information)). Furthermore, A1913 displays four-fold higher NMIA reactivity than A1918 in wild-type 50S subunits, but displays similar levels of reactivity in RluD(−) 50S subunits. The diminished reactivity at A1918 in RluD(−) compared to wild-type ribosomes implies a difference in helix 69 conformation. Substitution of U for Ψ at position 1911 could affect the 1911-1918-1919 network, resulting in reduced protection or increased flexibility of the A1918 2′-OH. This effect would, however, be indirect because the hydrogen-bonding capability of U on the Watson-Crick face is the same as Ψ (N3-H). A weakened 1911-1918-1919 interaction could in turn affect subunit association of RluD(−) ribosomes.
Nucleotides A1912, C1914, A1916, and U1917 are found to be less constrained in RluD(−) compared to wild-type 50S subunits (Figure 4b). The level of reactivity at C1914 on wild-type ribosomes is unclear, however, because this position is completely masked by a strong reverse transcription stop at m3Ψ1915. Nonetheless, the observation that many loop components are less constrained in RluD(−) ribosomes indicates a less organized H69 structure in comparison to wild-type H69, consistent with previous model studies.45, 46 Despite these differences, SHAPE analysis shows clear evidence for intersubunit bridge B7a3 formation in both wild-type and RluD(−) ribosomes, as indicated by decreased flexibility at position A1848 of the neighboring H68 upon subunit association (Supplementary Information).
Changes in H69 Nucleotide Accessibility
In support of the SHAPE data, DEPC probing on RluD(−) 50S subunits shows increased exposure of A1912 and A1916, as well as A1913 and A1918, relative to wild-type 50S subunits (Figure 4c). If A1912 interacts with U1917, either the N1 or N7 of A1912 are expected to be protected from DMS or DEPC, respectively; however, both sites in RluD(−) 50S subunits are highly reactive, suggesting a lack of such interaction in the unmodified RNA (Figure 4d).29 To further confirm the lack of an A1912-U1917 interaction, CMCT (1-cyclohexyl-(2-morpholinoethyl) carbodiimide metho-p-toluene sulfonate) was used to probe the pseudouridine/uridine residue. Although Ψ is known to react with CMCT to a lesser extent than U,47 a control experiment under denaturing conditions with the same CMCT concentration and reaction time revealed strong reactivity at Ψ1917 (Supporting Information). Under native RNA folding conditions, the N3 of Ψ1917 was protected from CMCT in wild-type ribosomes; whereas, the N3 of U1917 in RluD(−) 50S subunits showed high reactivity, suggesting greater accessibility than the modified RNA (Figure 4e). These results indicate the importance of Ψ1917 for reverse-Hoogsteen formation with A1912 (Figure 4d). These observations suggest that unmodified H69 forms a less structured loop conformation due to a lack of the 1912–1917 Hoogsteen interaction and base-stacking interactions, consistent with model studies.45, 46
Changes in Mg2+ Dependence
DEPC probing was also performed on RluD(−) ribosomes under varying Mg2+ conditions (Figure 4f). DEPC reactivity is increased three-fold and two-fold at A1913 and A1916, respectively, in 70S ribosomes relative to 50S subunits in 6 mM Mg2+. In contrast, the level of A1916 reactivity is similar in 50S subunits and 70S ribosomes under 20 mM Mg2+ conditions. The reactivity of A1912 is increased slightly (less than two-fold) in 70S ribosomes compared to 50S subunits. Residues A1918 and A1919 exhibit similar trends in relative reactivities in wild-type and Ψ-deficient ribosomes. The most noticeable difference between RluD(−) and wild-type ribosomes occurs at A1913. The elevated DEPC reactivity in 70S ribosomes with increased Mg2+ does not occur with Ψ-deficient ribosomes. These results suggest that in the absence of Ψs, H69 has an altered Mg2+-dependent conformation, which could alter the B2a interaction during subunit association. Furthermore, increased DEPC reactivity at A1916 in RluD(−) ribosomes could be interpreted as a loss in base-stacking interactions.46 Mutagenesis studies showed that deletions or additions at position 1916 (ΔA1916 and +AA1916) lead to diminished viability, and point or multiple mutations around positions 1914 to 1918 are lethal to bacteria.48 Combined with probing results, these data suggest that nucleotide positioning in H69, particularly at A1913 and A1916, is one of the key factors in regulating ribosome function. Mutations or Ψ deletions in H69 disrupt well-defined conformational states, and therefore could affect ribosome function, although the in vivo effects may be subtle.44
Conclusions
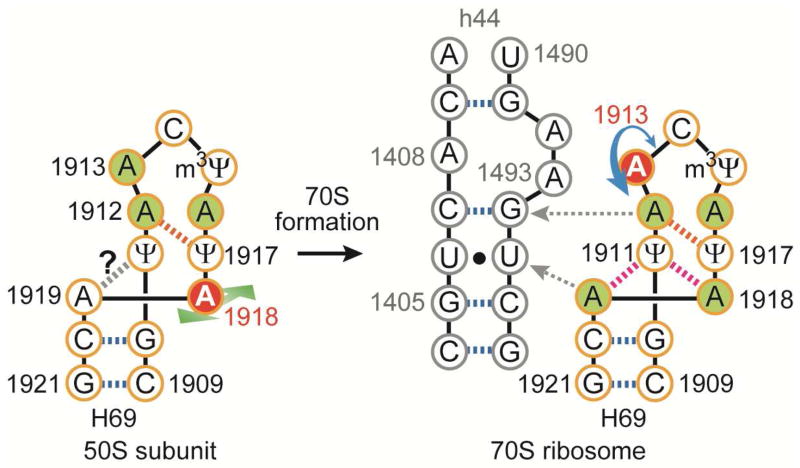
In this study, a detailed conformational analysis of the H69 region of 23S rRNA in 50S subunits and 70S ribosomes was carried out through a series of chemical-probing reactions. These solution data, together with previous crystallographic evidence, reveal conformational rearrangements of H69 that occur upon subunit association and are likely to play important roles in the translation mechanism. Residue A1913 is more exposed with greater flexibility in 70S ribosomes compared to 50S subunits (summarized in Figure 5). This movement of A1913 in 70S ribosomes might play a role in tRNA selection and peptide release by RFs. For example, A1913 could monitor the incoming aminoacyl tRNA through direct interactions with the anticodon stem loop.4 Residue A1913 also projects into the minor groove of h44 and base stacks with A1493, providing a structure that favors interactions with RF1.22 The fact that A1913 can accommodate multiple conformations in solution, as also observed in crystal structures, is perhaps not surprising given the important functional roles in translation. Furthermore, disruption or enhancement of such activity (e.g., high Mg2+ or loss of Ψs in H69) could cause abnormal ribosome activities, such as low translational fidelity39 or aberrant translation termination.43
Figure 5.
A schematic illustration shows the proposed H69 structural rearrangement at A1913 and A1918 upon subunit association and interactions with h44 of 16S rRNA (red circles = flexible nucleotides; green circles = protected nucleotides, as determined by SHAPE and DEPC; red dashed lines = non-canonical base-pair interactions). The probing data cannot confirm a hydrogen-bonding interaction between A1919 and Ψ1911.
Conformational adaptability at residues A1918 and A1919 may also play a role in forming the conformational states of H69 necessary for tRNA translocation and ratchet-like motions of the ribosome.17 Indeed, crystal structures having a hybrid tRNA state (P/E state) induced by RRF or RF3 show conformational compression of H69 and large movement of H69 toward the E site without disrupting the intersubunit bridge B2a.17, 49 An interchangeable network at the stem-loop junction of H69 involving A1918 and A1919, and also causing disruption of the terminal base pair (C1925-G1929) as observed in crystal structures, might play a role in ribosome energetics during translation.17
In conclusion, the results from chemical probing studies demonstrate that multiple positioning of specific nucleotides in H69 is possible, and that pseudouridine modifications do play a role in H69 dynamic behavior. Probing results provide a unique view of conformational complexity of H69 in solution, particularly with respect to subunit assembly, which likely plays a key role in the bacterial ribosome translation cycles. Developing a deeper understanding of rRNA structural changes and determination of the nucleotide accessibility during various functional stages such as ribosome assembly and disassembly will be useful for future antibiotic drug development with specific targeting of the individual RNA conformational states.
METHODS
Ribosome Isolation
Ribosomes were prepared as described elsewhere.50 Simply, E. coli MRE600 or RluD-deficient E. coli cells were grown to 0.5 OD600 and cell pellets were collected. Cell pellets were resuspended in lysis buffer (20 mM HEPES, pH 7.4 at 4 °C, 10 mM MgCl2, 100 mM NH4Cl, 4.6 mM 2-mercaptoethanol, and 0.5 mM EDTA) and lysed by passing twice through a French press at 12,000 psi. Lysate was centrifuged twice for 30 min at 11,000 rpm followed by ultracentrifugation for 4 h at 42,000 rpm. Crude ribosome pellets were gently resuspended in ribosome buffer (20 mM HEPES, pH 7.4 at 4 °C, 1 mM MgCl2, 200 mM NH4Cl, and 4.6 mM 2-mercaptoethanol for subunit isolation and 20 mM HEPES, pH 7.4 at 4 °C, 6 mM MgCl2, 100 mM NH4Cl, and 0.6 mM 2-mercaptoethanol for 70S ribosome isolation). The crude ribosome solution was layered on a 10–30% sucrose gradient containing 1 or 6 mM Mg2+. Ribosomes were separated by centrifugation at 19,000 rpm for 18 h, followed by elution from the bottom of the tube. The peaks of each ribosomal subunit and 70S ribosomes were observed at 260 nm and fractionated. Magnesium concentration of the pooled sucrose solution was adjusted to 10 mM and ribosomes were pelleted by ultracentrifugation for 24 h at 42,000 rpm for subunits and 24,000 rpm for 70S ribosomes. Purity of the subunits and 70S ribosomes was checked by both sucrose gradient and agarose gel electrophoresis of extracted rRNA. Isolated subunits and 70S ribosomes were quickly frozen and stored at −80 °C in stock buffer (20 mM HEPES, pH 7.3, 6 mM MgCl2, and 30 mM NH4Cl).
SHAPE Assay
SHAPE (2′-hydroxyl acylation analyzed by primer extension) was performed as described in the literature.32 Simply, ribosomes were reactivated in the presence of 150 mM KCl at 37 °C for 15 min, and buffer was exchanged with SHAPE buffer (50 mM HEPES, 150 mM KCl, 6 mM MgCl2, pH 8.0 at 37 °C) and the ribosome concentration was adjusted to 0.3 μM. Ribosomes in SHAPE buffer were incubated for over 15 min at 37 °C and the reaction was initiated by addition of 100 mM N-methylisatonic anhydride (NMIA) (Invitrogen) at a final concentration of 5 or 10 mM. The SHAPE reaction was done for 20 min at 37 °C and stopped by adding cold ethanol. Ribosomes were recovered by centrifugation and extracted with phenol-chloroform. The extracted rRNA was analyzed by standard primer extension assay with a 5′-32P-labeled DNA primer targeting positions 1929 to 1948 of 23S rRNA (5′-CGACAAGGAATTTCGCTACC-3′; 20mer DNA from Sigma-Genosys). Gel images were taken on a Typhoon 9200 (GE Healthcare). All probing experiments were performed three independent times and data analyses were completed with three experimental data sets to obtain standard errors. Band intensities were measured by using ImageQuant TL software (GE Healthcare). The background volume of a non-reacted control band was subtracted from the net band volume. Then, the relative intensity was calculated by dividing the corrected target band intensity by the standard band intensity.
DEPC, DMS, and CMCT Assays
DEPC (diethylpyrocarbonate), DMS (dimethylsulfate), and CMCT (1-cyclohexyl-(2-morpholinoethyl) carbodiimide metho-p-toluene sulfonate) probing reactions were carried out using a modified literature procedure.33, 34 Isolated ribosomes were reactivated in the presence of 100 mM NH4Cl at 37 °C for 15 min. Two μl of DEPC (Sigma-Aldrich) or 2 μl of 1:50 diluted DMS (Sigma-Aldrich) in cold ethanol were added to reactivated ribosomes in 40 μl of reaction buffer (80 mM HEPES, 100 mM NH4Cl, 6 mM MgCl2, pH 7.3 at 37°C), or an equal volume of CMCT solution (42 mg ml−1; Sigma-Aldrich) to reactivated ribosomes in 15 μl of CMCT reaction buffer (50 mM HEPES, 100 mM NH4Cl, 6 mM MgCl2, pH 8.1 at 37 °C). Samples were then incubated at 37 °C for 20 min (DEPC and DMS) or 10 min (CMCT) with gentle agitation in the case of DEPC probing. The reaction was terminated by adding 20 μl of stop buffer (3 M 2-mercaptoethanol, 1 M Tris-HCl, pH 7.5, 10 mM MgCl2) followed quickly by cold ethanol precipitation. Subsequent treatments were identical as the procedure described above for the SHAPE experiments.
Supplementary Material
Acknowledgments
We thank G. Garcia and A. Mankin for providing the E. coli strains, and P. Cunningham for assisting with ribosome isolations and helpful discussions. This project is supported by NIH (GM087596).
Footnotes
This material is available free of charge via the Internet at http://pubs.acs.org.
References
- 1.Yusupov MM, Yusupova GZ, Baucom A, Lieberman K, Earnest TN, Cate JH, Noller HF. Crystal structure of the ribosome at 5.5 Å resolution. Science. 2001;292:883–896. doi: 10.1126/science.1060089. [DOI] [PubMed] [Google Scholar]
- 2.Bashan A, Agmon I, Zarivach R, Schluenzen F, Harms J, Berisio R, Bartels H, Franceschi F, Auerbach T, Hansen HA, Kossoy E, Kessler M, Yonath A. Structural basis of the ribosomal machinery for peptide bond formation, translocation, and nascent chain progression. Mol Cell. 2003;11:91–102. doi: 10.1016/s1097-2765(03)00009-1. [DOI] [PubMed] [Google Scholar]
- 3.Schuwirth BS, Borovinskaya MA, Hau CW, Zhang W, Vila-Sanjurjo A, Holton JM, Cate JH. Structures of the bacterial ribosome at 3.5 Å resolution. Science. 2005;310:827–834. doi: 10.1126/science.1117230. [DOI] [PubMed] [Google Scholar]
- 4.Selmer M, Dunham CM, Murphy FV, 4th, Weixlbaumer A, Petry S, Kelley AC, Weir JR, Ramakrishnan V. Structure of the 70S ribosome complexed with mRNA and tRNA. Science. 2006;313:1935–1942. doi: 10.1126/science.1131127. [DOI] [PubMed] [Google Scholar]
- 5.Cochella L, Brunelle JL, Green R. Mutational analysis reveals two independent molecular requirements during transfer RNA selection on the ribosome. Nat Struct Mol Biol. 2007;14:30–36. doi: 10.1038/nsmb1183. [DOI] [PubMed] [Google Scholar]
- 6.Noller HF, Hoffarth V, Zimniak L. Unusual resistance of peptidyl transferase to protein extraction procedures. Science. 1992;256:1416–1419. doi: 10.1126/science.1604315. [DOI] [PubMed] [Google Scholar]
- 7.Khaitovich P, Mankin AS, Green R, Lancaster L, Noller HF. Characterization of functionally active subribosomal particles from Thermus aquaticus. Proc Natl Acad Sci USA. 1999;96:85–90. doi: 10.1073/pnas.96.1.85. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Nissen P, Hansen J, Ban N, Moore PB, Steitz TA. The structural basis of ribosome activity in peptide bond synthesis. Science. 2000;289:920–930. doi: 10.1126/science.289.5481.920. [DOI] [PubMed] [Google Scholar]
- 9.Moazed D, Noller HF. Interaction of tRNA with 23S rRNA in the ribosomal A, P, and E sites. Cell. 1989;57:585–597. doi: 10.1016/0092-8674(89)90128-1. [DOI] [PubMed] [Google Scholar]
- 10.Schmeing TM, Moore PB, Steitz TA. Structures of deacylated tRNA mimics bound to the E site of the large ribosomal subunit. RNA. 2003;9:1345–1352. doi: 10.1261/rna.5120503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Bakin A, Ofengand J. Four newly located pseudouridylate residues in Escherichia coli 23S ribosomal RNA are all at the peptidyltransferase center: analysis by the application of a new sequencing technique. Biochemistry. 1993;32:9754–9762. doi: 10.1021/bi00088a030. [DOI] [PubMed] [Google Scholar]
- 12.Decatur WA, Fournier MJ. rRNA modifications and ribosome function. Trends Biochem Sci. 2002;27:344–351. doi: 10.1016/s0968-0004(02)02109-6. [DOI] [PubMed] [Google Scholar]
- 13.Chow CS, Lamichhane TN, Mahto SK. Expanding the nucleotide repertoire of the ribosome with post-transcriptional modifications. ACS Chem Biol. 2007;2:610–619. doi: 10.1021/cb7001494. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Frank J, Verschoor A, Li Y, Zhu J, Lata RK, Radermacher M, Penczek P, Grassucci R, Agrawal RK, Srivastava S. A model of the translational apparatus based on a three-dimensional reconstruction of the Escherichia coli ribosome. Biochem Cell Biol. 1995;73:757–765. doi: 10.1139/o95-084. [DOI] [PubMed] [Google Scholar]
- 15.Gabashvili IS, Agrawal RK, Spahn CM, Grassucci RA, Svergun DI, Frank J, Penczek P. Solution structure of the E. coli 70S ribosome at 11.5 Å resolution. Cell. 2000;100:537–549. doi: 10.1016/s0092-8674(00)80690-x. [DOI] [PubMed] [Google Scholar]
- 16.Frank J, Agrawal RK. A ratchet-like inter-subunit reorganization of the ribosome during translocation. Nature. 2000;406:318–322. doi: 10.1038/35018597. [DOI] [PubMed] [Google Scholar]
- 17.Dunkle JA, Wang L, Feldman MB, Pulk A, Chen VB, Kapral GJ, Noeske J, Richardson JS, Blanchard SC, Cate JH. Structures of the bacterial ribosome in classical and hybrid states of tRNA binding. Science. 2011;332:981–984. doi: 10.1126/science.1202692. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.O’Connor M, Dahlberg AE. The involvement of two distinct regions of 23 S ribosomal RNA in tRNA selection. J Mol Biol. 1995;254:838–847. doi: 10.1006/jmbi.1995.0659. [DOI] [PubMed] [Google Scholar]
- 19.Kipper K, Hetényi C, Sild S, Remme J, Liiv A. Ribosomal intersubunit bridge B2a is involved in factor-dependent translation initiation and translational processivity. J Mol Biol. 2009;385:405–422. doi: 10.1016/j.jmb.2008.10.065. [DOI] [PubMed] [Google Scholar]
- 20.O’Connor M. Helix 69 in 23S rRNA modulates decoding by wild type and suppressor tRNAs. Mol Genet Genomics. 2009;282:371–380. doi: 10.1007/s00438-009-0470-6. [DOI] [PubMed] [Google Scholar]
- 21.Valle M, Zavialov A, Li W, Stagg SM, Sengupta J, Nielsen RC, Nissen P, Harvey SC, Ehrenberg M, Frank J. Incorporation of aminoacyl-tRNA into the ribosome as seen by cryo-electron microscopy. Nat Struct Biol. 2003;10:899–906. doi: 10.1038/nsb1003. [DOI] [PubMed] [Google Scholar]
- 22.Laurberg M, Asahara H, Korostelev A, Zhu J, Trakhanov S, Noller HF. Structural basis for translation termination on the 70S ribosome. Nature. 2008;454:852–857. doi: 10.1038/nature07115. [DOI] [PubMed] [Google Scholar]
- 23.Weixlbaumer A, Jin H, Neubauer C, Voorhees RM, Petry S, Kelley AC, Ramakrishnan V. Insights into translational termination from the structure of RF2 bound to the ribosome. Science. 2008;322:953–956. doi: 10.1126/science.1164840. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Korostelev A, Zhu J, Asahara H, Noller HF. Recognition of the amber UAG stop codon by release factor RF1. EMBO J. 2010;29:2577–2585. doi: 10.1038/emboj.2010.139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Agrawal RK, Sharma MR, Kiel MC, Hirokawa G, Booth TM, Spahn CM, Grassucci RA, Kaji A, Frank J. Visualization of ribosome-recycling factor on the Escherichia coli 70S ribosome: functional implications. Proc Natl Acad Sci USA. 2004;101:8900–8905. doi: 10.1073/pnas.0401904101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Ortiz-Meoz RF, Green R. Helix 69 is key for uniformity during substrate selection on the ribosome. J Biol Chem. 2011;286:25604–25610. doi: 10.1074/jbc.M111.256255. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Ortiz-Meoz RF, Green R. Functional elucidation of a key contact between tRNA and the large ribosomal subunit rRNA during decoding. RNA. 2010;16:2002–2013. doi: 10.1261/rna.2232710. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Ali IK, Lancaster L, Feinberg J, Joseph S, Noller HF. Deletion of a conserved, central ribosomal intersubunit RNA bridge. Mol Cell. 2006;23:865–874. doi: 10.1016/j.molcel.2006.08.011. [DOI] [PubMed] [Google Scholar]
- 29.Sakakibara Y, Chow CS. Probing conformational states of modified helix 69 in 50S ribosomes. J Am Chem Soc. 2011;133:8396–8399. doi: 10.1021/ja2005658. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Borovinskaya MA, Pai RD, Zhang W, Schuwirth BS, Holton JM, Hirokawa G, Kaji H, Kaji A, Cate JH. Structural basis for aminoglycoside inhibition of bacterial ribosome recycling. Nat Struct Mol Biol. 2007;14:727–732. doi: 10.1038/nsmb1271. [DOI] [PubMed] [Google Scholar]
- 31.Scheunemann AE, Graham WD, Vendeix FA, Agris PF. Binding of aminoglycoside antibiotics to helix 69 of 23S rRNA. Nucleic Acids Res. 2010;38:3094–3105. doi: 10.1093/nar/gkp1253. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Merino EJ, Wilkinson KA, Coughlan JL, Weeks KM. RNA structure analysis at single nucleotide resolution by selective 2′-hydroxyl acylation and primer extension (SHAPE) J Am Chem Soc. 2005;127:4223–4231. doi: 10.1021/ja043822v. [DOI] [PubMed] [Google Scholar]
- 33.Peattie DA, Gilbert W. Chemical probes for higher-order structure in RNA. Proc Natl Acad Sci USA. 1980;77:4679–4682. doi: 10.1073/pnas.77.8.4679. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Moazed D, Noller HF. Transfer RNA shields specific nucleotides in 16S ribosomal RNA from attack by chemical probes. Cell. 1986;47:985–994. doi: 10.1016/0092-8674(86)90813-5. [DOI] [PubMed] [Google Scholar]
- 35.Merryman C, Moazed D, Daubresse G, Noller HF. Nucleotides in 23S rRNA protected by the association of 30S and 50S ribosomal subunits. J Mol Biol. 1999;285:107–113. doi: 10.1006/jmbi.1998.2243. [DOI] [PubMed] [Google Scholar]
- 36.Jenner L, Demeshkina N, Yusupova G, Yusupov M. Structural rearrangements of the ribosome at the tRNA proofreading step. Nat Struct Mol Biol. 2010;17:1072–1078. doi: 10.1038/nsmb.1880. [DOI] [PubMed] [Google Scholar]
- 37.Ogle JM, Brodersen DE, Clemons WM, Jr, Tarry MJ, Carter AP, Ramakrishnan V. Recognition of cognate transfer RNA by the 30S ribosomal subunit. Science. 2001;292:897–902. doi: 10.1126/science.1060612. [DOI] [PubMed] [Google Scholar]
- 38.Youngman EM, He SL, Nikstad LJ, Green R. Stop codon recognition by release factors induces structural rearrangement of the ribosomal decoding center that is productive for peptide release. Mol Cell. 2007;28:533–543. doi: 10.1016/j.molcel.2007.09.015. [DOI] [PubMed] [Google Scholar]
- 39.Gromadski KB, Rodnina MV. Kinetic determinants of high-fidelity tRNA discrimination on the ribosome. Mol Cell. 2004;13:191–200. doi: 10.1016/s1097-2765(04)00005-x. [DOI] [PubMed] [Google Scholar]
- 40.Branlant C, Krol A, Machatt MA, Pouyet J, Ebel JP, Edwards K, Kossel H. Primary and secondary structures of Escherichia coli MRE 600 23S ribosomal RNA. Comparison with models of secondary structure for maize chloroplast 23S rRNA and for large portions of mouse and human 16S mitochondrial rRNAs. Nucleic Acids Res. 1981;9:4303–4324. doi: 10.1093/nar/9.17.4303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Kowalak JA, Bruenger E, Hashizume T, Peltier JM, Ofengand J, McCloskey JA. Structural characterization of U*-1915 in domain IV from Escherichia coli 23S ribosomal RNA as 3-methylpseudouridine. Nucleic Acids Res. 1996;24:688–693. doi: 10.1093/nar/24.4.688. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Raychaudhuri S, Conrad J, Hall BG, Ofengand J. A pseudouridine synthase required for the formation of two universally conserved pseudouridines in ribosomal RNA is essential for normal growth of Escherichia coli. RNA. 1998;4:1407–1417. doi: 10.1017/s1355838298981146. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Ejby M, Sørensen MA, Pedersen S. Pseudouridylation of helix 69 of 23S rRNA is necessary for an effective translation termination. Proc Natl Acad Sci USA. 2007;104:19410–19415. doi: 10.1073/pnas.0706558104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.O’Connor M, Gregory ST. Inactivation of the RluD pseudouridine synthase has minimal effects on growth and ribosome function in wild-type Escherichia coli and Salmonella enterica. J Bacteriol. 2011;193:154–162. doi: 10.1128/JB.00970-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Abeysirigunawardena SC, Chow CS. pH-Dependent structural changes of helix 69 from Escherichia coli 23S ribosomal RNA. RNA. 2008;14:782–792. doi: 10.1261/rna.779908. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Desaulniers JP, Chang YC, Aduri R, Abeysirigunawardena SC, SantaLucia J, Jr, Chow CS. Pseudouridines in rRNA helix 69 play a role in loop stacking interactions. Org Biomol Chem. 2008;6:3892–3895. doi: 10.1039/b812731j. [DOI] [PubMed] [Google Scholar]
- 47.Motorin Y, Muller S, Behm-Ansmant I, Branlant C. Identification of modified residues in RNAs by reverse transcription-based methods. Methods Enzymol. 2007;425:21–53. doi: 10.1016/S0076-6879(07)25002-5. [DOI] [PubMed] [Google Scholar]
- 48.Hirabayashi N, Sato NS, Suzuki T. Conserved loop sequence of helix 69 in Escherichia coli 23 S rRNA is involved in A-site tRNA binding and translational fidelity. J Biol Chem. 2006;281:17203–17211. doi: 10.1074/jbc.M511728200. [DOI] [PubMed] [Google Scholar]
- 49.Jin H, Kelley AC, Ramakrishnan V. Crystal structure of the hybrid state of ribosome in complex with the guanosine triphosphatase release factor 3. Proc Natl Acad Sci USA. 2011;108:15798–15803. doi: 10.1073/pnas.1112185108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Blaha G, Stelzl U, Spahn CM, Agrawal RK, Frank J, Nierhaus KH. Preparation of functional ribosomal complexes and effect of buffer conditions on tRNA positions observed by cryoelectron microscopy. Methods Enzymol. 2000;317:292–309. doi: 10.1016/s0076-6879(00)17021-1. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.