A recent estimate suggests that we can potentially distinguish 2.3 million colors (1), and yet we achieve this by comparing the rates at which photons are absorbed in just three classes of retinal photopigment (Fig. 1 Lower). The photopigments consist of 11-cis-retinal bound to different “opsins,” which are members of the large family of G-protein-coupled receptors or heptahelicals. But our exquisite discrimination of hue requires that the three different opsins should be cleanly segregated into different cone cells in the retina. A new paper by Yanshu Wang et al. (2) bears on how such segregation may be maintained. The research group is led by Jeremy Nathans, whose now classic papers laid the basis of the molecular genetics of the cone pigments (3, 4).
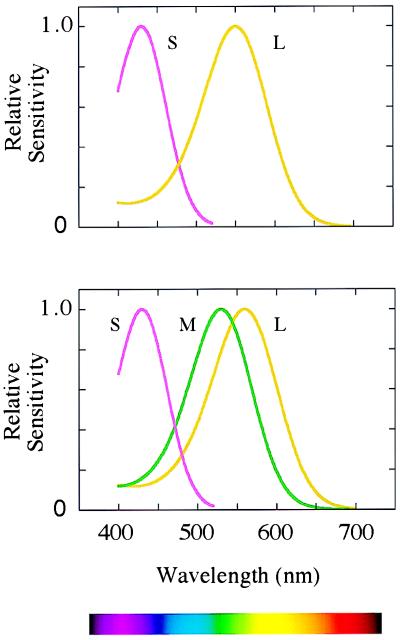
Figure 1.
(Lower) The sensitivities of the long-wave (L), middle-wave (M), and short-wave (S) photopigments found in the retinae of humans and of Old World primates. (Upper) The sensitivities of the pigments thought to have been present in ancestral mammals.
Old and New Subsystems of Color Vision.
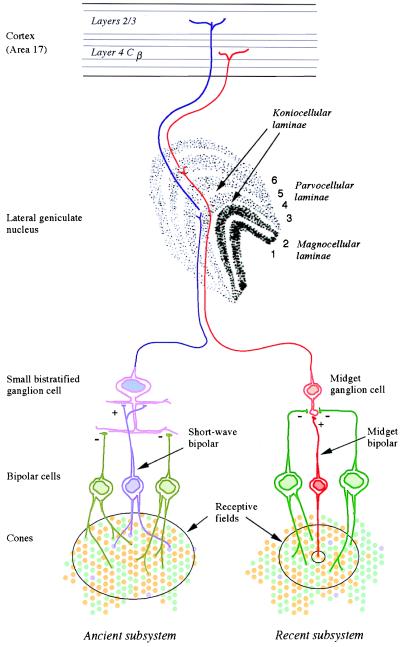
Our own trichromatic color vision depends on two subsystems, a phylogenetically recent one overlaid on a much more ancient one (5). Most mammals are dichromatic, having only two types of cone receptor in the retina (6). For the main business of vision—the detection of movement and form—they rely on a single class of cone, with its peak sensitivity at long wavelengths (500–570 nm). Among these cones lies a second sparser population of cones with peak sensitivity at short wavelengths (Fig. 1 Upper). A basic color vision is gained by comparing the rate of quantum catch in the short-wave cones with that in the long-wave cones. This ancient mammalian subsystem has its own morphological substrate in early stages of the visual system (Fig. 2): its signals are carried by the “blue cone” bipolar, by the small bistratified ganglion cell (7), and then by cells in koniocellular laminae 3 and 4 of the lateral geniculate nucleus (8), whence they pass directly to layers 2 and 3 of the striate cortex.
Figure 2.
The anatomical substrates of the two subsystems of human color vision. The phylogenetically ancient subsystem (Left) draws opposed inputs from the short-wave cones, on the one hand, and the long- and middle-wave cones on the other. Its signals are carried by small bistratified ganglion cells and by the koniocellular laminae of the lateral geniculate nucleus. The newer subsystem (Right) compares the signals of the recently differentiated long- and middle-wave cones. Its signals are thought to be carried by midget ganglion cells and the parvocellular laminae of the lateral geniculus.
It is with the second subsystem of color vision that Nathans and his colleagues are concerned. In the retinae of Old World primates, the single cone in the range 500 to 570 nm is replaced by the present long- and middle-wave cones (Fig. 1 Lower). The second subsystem depends on comparing the rates of quantum catch in the latter two types of cone, and its signals are thought to be carried by the midget bipolar cells, the midget ganglion cells, and the parvocellular laminae of the lateral geniculate nucleus, which project to layer 4Cβ of the striate cortex (Fig. 2). In our arboreal ancestors, the second subsystem may have evolved for detecting yellow fruit and other biological signals against a background of foliage: the spectral positions of the long- and middle-wave pigments are optimal for such tasks (9).
The critical stage in the evolution of the second subsystem is thought to have been the duplication of the X-chromosome gene that encoded the original long-wave pigment of mammalian cones (L in Fig. 1 Upper). The resulting genes then diverged, so as to encode the present long- and middle-wave photopigments (M and L in Fig. 1 Lower)—they may indeed have begun as variant alleles of a polymorphic gene before the duplication. The two genes remain juxtaposed on the q arm of the X-chromosome, and they are closely homologous in their sequences (3). It is not known exactly when the duplication occurred. Estimates have been made on the basis of the present nucleotide sequences of the exons, but these are unconvincing because the X-chromosome array of opsin genes appears to have often been subject to unequal crossing over, and it is suspicious that introns 2, 4, and 5 of the two genes are actually more homologous than are the exons (10, 11). But we can guess that the duplication occurred soon after the divergence of the Old and New World monkeys, perhaps 30 to 40 million years ago. For, in all the species so far studied, Old World monkeys exhibit long- and middle-wave pigments similar to those of humans (12), whereas most New World species appear to have a single polymorphic X-chromosome opsin locus (6, 13).
What Gives the Long- or Middle-Wave Cones Their Identities?
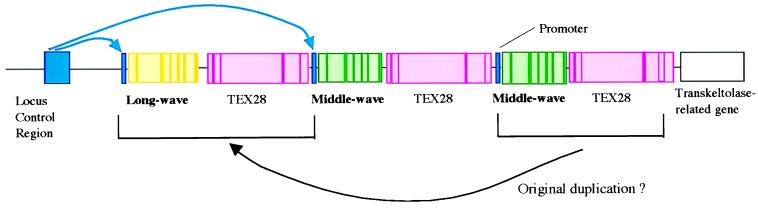
The distribution of the long- and middle-wave cones in primate and human retinae appears to be locally random (14, 15). Wang et al. ask what it is that makes a given cone long-wave or middle-wave. Their experiment is a test of a hypothesis put forward earlier by Nathans’ group (16). Upstream (3.5 kilobases) of the opsin gene array (Fig. 3) is a locus control region (LCR), without which none of the genes in the array are expressed (17). The hypothesis is that this stretch of DNA bends around to interact with a promoter region lying close to the first exon of one of the opsin genes. Only the favored gene is transcribed to RNA. These alternative couplings are indicated in blue in Fig. 3. Within a given cone cell, it is a stochastic matter whether the LCR favors the long-wave or the middle-wave gene, although—perhaps because it is physically closer—the first gene in the array may enjoy a statistical advantage over the second and the latter over all the others (18). The long-wave and middle-wave promoter regions, though closely homologous, are not identical (3), and Shaaban and Deeb, who have recently studied the expression of opsin genes in vitro in a retinoblastoma cell line (19), suggest that the middle-wave promoter is the more efficient, to compensate for its much greater distance from the LCR.
Figure 3.
The array of opsin genes on the q arm of the X-chromosome. Exons (the coding regions of the genes) are shown as narrow bars of saturated color, and introns are shown as less saturated. Typically there is only one copy of the gene for the long-wave pigment, but there may be more than one copy of the middle-wave gene. Interdigitated with these opsin genes are (truncated) copies of a gene “TEX28,” expressed in the testis (30), and from this we can infer the possible nature of the original duplication that gave rise to distinct long- and middle-wave genes. Upstream of the array is a LCR. The hypothesis of Wang and colleagues (2) is that the LCR interacts with the promoter region of just one of the opsin genes (alternative couplings are shown in blue), and this alone determines whether the cone is long-wave or middle-wave.
Wang and colleagues have now constructed an artificial piece of DNA that incorporates the locus control region and the promoter regions of the long-wave and middle-wave opsins (2). They replace the long-wave gene itself with a gene coding for one “reporter” molecule and replace the middle-wave gene with a gene coding for a second reporter. They introduce this DNA into mouse embryos to create transgenic animals, and later they examine retinal tissue histologically. The enzyme products of the two reporter genes can be revealed by different histochemical reactions. Conveniently, one of the gene products (β-galactosidase) is located in the cytoplasm of the cell, whereas the other (alkaline phosphatase) is located in the cell membrane. So, by light microscopy, it is relatively easy to tell whether one or both gene products is present in an individual cone. In retinal tissue from the transgenic mice, it turns out that many cones express only one or the other of the reporter genes, while some cones express neither or express both.
The finding that many cones express either the one reporter gene or the other is clearly in favor of the hypothesis being tested. More awkward to explain are those cones that express both genes. Wang et al. (2) suggest that from time to time the LCR becomes disconnected from its chosen promoter and is then free to make a fresh choice. This ability would have to be limited to the artificial constructs. The coupling in primate cones would have to be much more stable: electrophysiological recordings of single cones from macaque retina show that the cells fall tightly into two groups with standard deviations in the range 1.0–1.3 nm (20). There is little evidence of cones in transition between expressing one opsin and expressing the other.
The Evolution of the Second Subsystem.
Nathans’ hypothesis suggests that it would have been rather easy for the second subsystem of color vision to evolve once the duplication had occurred and once the genes differed at one of the codons that control the spectral sensitivity of the pigment (2). Random coupling of the LCR would automatically ensure that different photopigments were expressed in different cones.
But could this variation in cones be at once exploited by the visual system? Would not specialized neural machinery be required? Possibly not. The second subsystem may simply have been parasitic on an existing retinal pathway devoted to spatial analysis—the pathway formed by midget bipolars and midget ganglion cells (21). The midget ganglion cell draws a center input from just a single cone and draws a signal of opposite sign from surrounding cones (Fig. 2). Because they are found in dichromatic New World monkeys (22), such ganglion cells probably antedated trichromatic primate vision. Once the duplication had occurred and two slightly different opsin genes were present on the X-chromosome, a typical midget ganglion cell would automatically become chromatically opponent because its center input would have the spectral sensitivity of one opsin, and the antagonistic input would represent simply the average spectral sensitivity of the surrounding cones (13, 23). In turn, the individual midget ganglion cells would drive individual parvocellular units in the lateral geniculate nucleus, and the cerebral cortex itself seems above all designed to discover correlations in its inputs (24). So there could automatically emerge in the developing cortex some cells that respond primarily to long wavelengths and some that respond primarily to middle wavelengths. As Nathans and his coworkers themselves suggest (2), this central segregation of responses could be achieved through “Hebbian learning.” A Hebbian synapse is one that is strengthened if it is active when the postsynaptic cell is depolarized, i.e., when other excitatory synapses are concurrently active (24). Suppose that initially a given cortical cell, by chance, has more inputs from long-wave than from middle-wave units in the lateral geniculate. The Hebbian process will turn this small initial bias into strong selectivity for one class of inputs.
Wang et al. (2) discuss the possibility that even today the long- and middle-wave cones carry no distinguishing labels and that there is no special neural machinery for the second subsystem of color vision. There has been, they suggest, little time for special apparatus to evolve. On this argument, I should not myself place great weight: the whole apparatus of human speech and language is thought to have evolved within the last million years, whereas the duplication of the X-chromosome opsin gene took place 30 to 40 million years ago. One conceivable label might be the opsin itself, assuming that it is present throughout the cone membrane. The problem facing the retinal photoreceptors is faced, to a greater degree, by the many types of olfactory receptor neurons, which must make specific connections to the olfactory glomeruli. Their heptahelical receptor molecules are present in their axons as well as at the epithelial surface, and Singer, Shepherd, and Greer have identified amino acids in the second extracellular loop of these heptahelicals that are correlated with (and perhaps act as labels for) amino acids that lie within the binding pocket and are thought to determine specificity to a particular odorant (25). In the case of the long- and middle-wave opsins, a candidate for an extracellular label is site 298 in the amino acid sequence, a site that differentiates the two pigments and is encoded by a codon tightly linked to the exon 5 codons that determine spectral sensitivity. Both humans and chimpanzees differ at this site, but the hypothesis is weakened by the fact that gorillas and Old World monkeys have alanine at site 298 in both sequences (26).
Relative Numbers of Long- and Middle-Wave Cones.
On the basis of psychophysical experiments, there have been recurrent suggestions that long-wave cones are twice as common as middle-wave cones in the human fovea (27), and so psychophysicists will be interested by the fact that Wang and colleagues’ surrogate long-wave gene—the reporter gene closest to the LCR—was expressed more frequently than the surrogate middle-wave gene (2). However, we should not build too much on this aspect of their findings: first, the distances from the LCR to the first and to the second promoters are different in the artificial construct than in the natural case; second, even if the distances were the same in terms of total number of base pairs, the higher-order coiling of the DNA molecule may be different; third, the first intron of the opsin genes (which is not present in the artificial construct) may contain regulatory sequences that allow the expression of the gene to be enhanced or reduced—as is the case in other systems. Nor is it known yet whether the disproportion in expression would be maintained if the reporter genes were interchanged in position.
But Nathans and his group now have a powerful technique for examining the control of expression in the X-chromosome opsin gene array (2). By making artificial constructs in which the positions of the two promoters are reversed, or the same promoter is paired with each reporter, it will be possible to test whether sheer proximity to the LCR is the critical factor in favoring expression or whether, say, particular nucleotide differences between the two promoters are significant (19). And William Rushton (28), in the spirit of Gideon (29), would have wanted to see dew on the ground and not on the fleece: Is selective expression abolished if the artificial construct incorporates two LCRs, one for each promoter?
Footnotes
A commentary on this article begins on page 5251.
References
- 1.Pointer M R, Attridge G G. Color Res Appl. 1998;23:52–54. [Google Scholar]
- 2.Wang Y, Smallwood P M, Cowan M, Blesh D, Lawler A, Nathans J. Proc Natl Acad Sci USA. 1999;96:5251–5256. doi: 10.1073/pnas.96.9.5251. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Nathans J, Thomas D, Hogness D S. Science. 1986;232:193–202. doi: 10.1126/science.2937147. [DOI] [PubMed] [Google Scholar]
- 4.Nathans J, Piantanida T P, Eddy R L, Shows T B, Hogness D S. Science. 1986;232:203–210. doi: 10.1126/science.3485310. [DOI] [PubMed] [Google Scholar]
- 5.Mollon J D. J Exp Biol. 1989;146:21–38. doi: 10.1242/jeb.146.1.21. [DOI] [PubMed] [Google Scholar]
- 6.Jacobs G. Biol Rev. 1993;68:413–471. doi: 10.1111/j.1469-185x.1993.tb00738.x. [DOI] [PubMed] [Google Scholar]
- 7.Dacey D M, Lee B B. Nature (London) 1994;367:731–735. doi: 10.1038/367731a0. [DOI] [PubMed] [Google Scholar]
- 8.Martin P R, White A J R, Goodchild A K, Wilder H D, Sefton A E. Eur J Neurosci. 1997;9:1536–1541. doi: 10.1111/j.1460-9568.1997.tb01509.x. [DOI] [PubMed] [Google Scholar]
- 9.Regan B C, Julliot C, Simmen B, Viénot F, Charles-Dominique P, Mollon J D. Vision Res. 1998;38:3321–3327. doi: 10.1016/s0042-6989(97)00462-8. [DOI] [PubMed] [Google Scholar]
- 10.Shyue S-K, Li L, Chang B H-J, Li W-H. Mol Biol Evol. 1994;1:548–551. doi: 10.1093/oxfordjournals.molbev.a040134. [DOI] [PubMed] [Google Scholar]
- 11.Zhao Z, Hewett-Emmett D, Li W-H. J Mol Evol. 1998;46:494–496. doi: 10.1007/pl00013147. [DOI] [PubMed] [Google Scholar]
- 12.Bowmaker J K, Astell S, Hunt D M, Mollon J D. J Exp Biol. 1991;156:1–19. doi: 10.1242/jeb.156.1.1. [DOI] [PubMed] [Google Scholar]
- 13.Mollon J D, Bowmaker J K, Jacobs G H. Proc R Soc London Ser B. 1984;222:373–399. doi: 10.1098/rspb.1984.0071. [DOI] [PubMed] [Google Scholar]
- 14.Mollon J D, Bowmaker J K. Nature (London) 1992;360:677–679. doi: 10.1038/360677a0. [DOI] [PubMed] [Google Scholar]
- 15.Roorda A, Williams D R. Nature (London) 1999;397:520–522. doi: 10.1038/17383. [DOI] [PubMed] [Google Scholar]
- 16.Wang Y, Macke J P, Merbs S L, Klaunberg B, Bennett J, Zack D, Gearhart J, Nathans J. Neuron. 1992;9:429–440. doi: 10.1016/0896-6273(92)90181-c. [DOI] [PubMed] [Google Scholar]
- 17.Nathans J, Davenport I H, Maumenee R A, Lewis R A, Hejtmancik M, Litt E, Lovrien R, Weleber B, Bachynski B, Zwas F. Science. 1989;245:831–838. doi: 10.1126/science.2788922. [DOI] [PubMed] [Google Scholar]
- 18.Winderickx J, Battisti L, Motulsky A G, Deeb S S. Proc Natl Acad Sci USA. 1992;89:9710–9714. doi: 10.1073/pnas.89.20.9710. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Shaaban S A, Deeb S S. Invest Ophthalmol Visual Sci. 1998;39:885–896. [PubMed] [Google Scholar]
- 20.Baylor D A, Nunn B J, Schnapf J L. J Physiol. 1987;390:145–160. doi: 10.1113/jphysiol.1987.sp016691. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Mollon J D, Jordan G. Die Farbe. 1988;35/36:139–170. [Google Scholar]
- 22.Goodchild A K, Ghosh K K, Martin P R. J Comp Neurol. 1996;366:55–75. doi: 10.1002/(SICI)1096-9861(19960226)366:1<55::AID-CNE5>3.0.CO;2-J. [DOI] [PubMed] [Google Scholar]
- 23.Lennie P, Haake P W, Williams D R. In: Computational Models of Visual Processing. Landy M S, Movshon J A, editors. Cambridge, MA: MIT Press; 1991. pp. 71–82. [Google Scholar]
- 24.Linsker R. Annu Rev Neurosci. 1990;13:257–281. doi: 10.1146/annurev.ne.13.030190.001353. [DOI] [PubMed] [Google Scholar]
- 25.Singer M S, Shepherd G M, Greer C A. Nature (London) 1995;377:19–20. doi: 10.1038/377019b0. [DOI] [PubMed] [Google Scholar]
- 26.Dulai K S, Bowmaker J K, Mollon J D, Hunt D M. Vision Res. 1984;34:2483–2491. doi: 10.1016/0042-6989(94)90233-x. [DOI] [PubMed] [Google Scholar]
- 27.Vos J J, Walraven P L. Vision Res. 1971;11:799–818. doi: 10.1016/0042-6989(71)90003-4. [DOI] [PubMed] [Google Scholar]
- 28.Alpern M. In: Color Vision: Physiology and Psychophysics. Mollon J D, Sharpe L T, editors. London: Academic; 1983. pp. 590–596. [Google Scholar]
- 29.Judges 6:20–40.
- 30.Hanna M C, Platts J T, Kirkness E F. Genomics. 1997;43:384–386. doi: 10.1006/geno.1997.4830. [DOI] [PubMed] [Google Scholar]