SUMMARY
The molecular determinants of spleen organogenesis and the etiology of isolated congenital asplenia (ICA), a life-threatening human condition, are unknown. We previously reported that Pbx1 deficiency causes organ growth defects including asplenia. Here, we show that mice with splenic mesenchyme-specific Pbx1 inactivation exhibit hyposplenia. Moreover, the loss of Pbx causes down-regulation of Nkx2-5 and derepression of p15Ink4b in spleen mesenchymal progenitors, perturbing the cell cycle. Removal of p15Ink4b in Pbx1 spleen-specific mutants partially rescues spleen growth. By whole-exome sequencing of a multiplex kindred with ICA, we identify a heterozygous missense mutation (P236H) in NKX2-5 showing reduced transactivation in vitro. This study establishes that a Pbx/Nkx2-5/p15 regulatory module is essential for spleen development.
Keywords: Spleen, Organ growth, Human Isolated Congenital Asplenia, Cell cycle, Pbx, p15Ink4b, Nkx2-5
INTRODUCTION
The vertebrate spleen is a secondary lymphoid organ and red blood cell repository (Brendolan et al., 2007). It plays important roles in host defense, via the maturation of B cells and the phagocytosis of circulating microbes by macrophages. Consistent with this critical immunological function, congenital or acquired asplenia is life-threatening, due to invasive bacterial infections. Spleen morphogenesis is achieved during development through interactions between mesenchymal and invading endothelial and hematopoietic cells. Genetically engineered mouse models have led to the discovery of genes, mostly encoding transcription factors, which are required for the temporal and spatial coordination of cell-fate specification, cell proliferation, and differentiation during spleen development. Transcription factor-encoding genes Nkx2-3, Nkx3-2, Pbx1, Sox11, Wt1, and Tcf21 are expressed within the spleen primordium, and mice deficient for these proteins exhibit spleen agenesis or hypoplasia, with other organogenesis defects (Brendolan et al., 2005; Herzer et al., 1999; Lettice et al., 1999; Lu et al., 2000; Pabst et al., 1999; Sock et al., 2004). Additionally, the homeobox gene Nkx2-5 marks progenitor cells of the spleen anlage, albeit its role in spleen development has not been elucidated since Nkx2-5 mutants die in utero before spleen specification (Lyons et al., 1995). In contrast, Tlx1-null mice exhibit isolated asplenia without other abnormalities (Kanzler and Dear, 2001; Roberts et al., 1994), mimicking human isolated congenital asplenia (ICA, OMIM#271400; Mahlaoui et al., 2011). Overall, only a few genes are known to control spleen development in mice, by hitherto elusive mechanisms and unknown interactions.
Human congenital asplenia can result from laterality defects, i.e. failure to establish left–right (L-R) axis specification, as in heterotaxy (Mahlaoui et al., 2011; Zhu et al., 2006), including Ivemark syndrome with congenital anomalies of the heart or great vessels (OMIM#208530). While >80 genes have been implicated in L-R axis specification in model organisms, only about 20 (including LEFTYA, CRYPTIC, and NKX2-5) have been associated with human heterotaxy (Zhu et al., 2006). Congenital asplenia can also be isolated and independent of laterality defects (OMIM #271400). Interestingly, necropsy reveals hypoplastic spleens in some patients (Mahlaoui et al., 2011). Since its first description, only 70 ICA patients (29 sporadic and 41 familial) have been reported in the literature, mostly in childhood. We reported the only clinical series of ICA patients (Mahlaoui et al., 2011). However, ICA is probably under-diagnosed, as it often manifests with rapidly lethal infections that prevent diagnosis unless necropsy is performed. Most known children with ICA die of fulminant bacterial disease – invasive pneumococcal infection in particular. Unlike heterotaxy, there is no known genetic etiology for ICA, and mutations in TLX1, which cause isolated asplenia in the mouse, have not been reported in human ICA (Brendolan et al., 2007; unpublished data). Our previous studies using a mouse strain with global loss of Pbx1 (Pbx1−/−), which encodes a TALE homeodomain transcription factor (Moens and Selleri, 2006), revealed general organ hypoplasia and asplenia in Pbx1−/− (Brendolan et al., 2005; Kim et al., 2002). Accordingly, we set out to decipher the mechanisms by which Pbx1 controls spleen morphogenesis and growth by generating mice with conditional Pbx1 inactivation in splenic mesenchymal progenitors. By this approach, we tested the hypothesis that a Pbx-dependent spleen regulatory network may be disrupted in ICA patients.
RESULTS
Spleen hypoplasia results from Pbx1 inactivation in spleen mesenchymal progenitors
In the early mouse embryo, only mesenchyme and endothelium form the spleen anlage, until hematopoietic cells invade at E13.5 (Brendolan et al., 2007). Given the prime role of Pbx1 in spleen organogenesis, we created a conditional allele for spleen mesenchymal Pbx1 inactivation (Pbx1flox/flox; Figure S1), to prevent in utero lethality and non cell-autonomous effects of Pbx1 loss in non-splenic tissues. Ubiquitous Cre-mediated Pbx1 inactivation with a β-actin Cre strain (Lewandoski and Martin, 1997) recapitulated Pbx1−/− phenotypes (Figure S2A–F).
We reasoned that crossing the Pbx1 conditional strain to a line in which Cre expression is driven by endogenous Nkx2-5 cis-regulatory elements (Stanley et al., 2002), would yield abnormal spleen growth, given findings that: 1) Nkx2-5 marks splenic progenitors in Xenopus (Patterson et al., 2000) and mouse (Burn et al., 2008; Hecksher-Sørensen et al., 2004); 2) Pbx1 expression precedes Nkx2-5 in lateral plate mesoderm (LPM; Figure S2G; Capellini et al., 2006), dorsal mesentery (DM; Figure S2H), and spleno-pancreatic mesenchyme, including the adjacent splanchnic mesodermal plate (Smp; Figures 1F and S2I), which give rise to the spleen anlage; 3) Pbx1 controls Nkx2-5 expression (Brendolan et al., 2005); and 4) Pbx1 is required for cell proliferation in most embryonic organs, including spleen (Brendolan et al., 2005). We inferred that splenic Pbx1 inactivation would occur after onset of Nkx2-5 Cre expression, enabling Pbx1 to fulfill its role as a spleen specification determinant in this strain. Thus, this model allows the study of spleen morphogenesis and expansion, independent of specification.
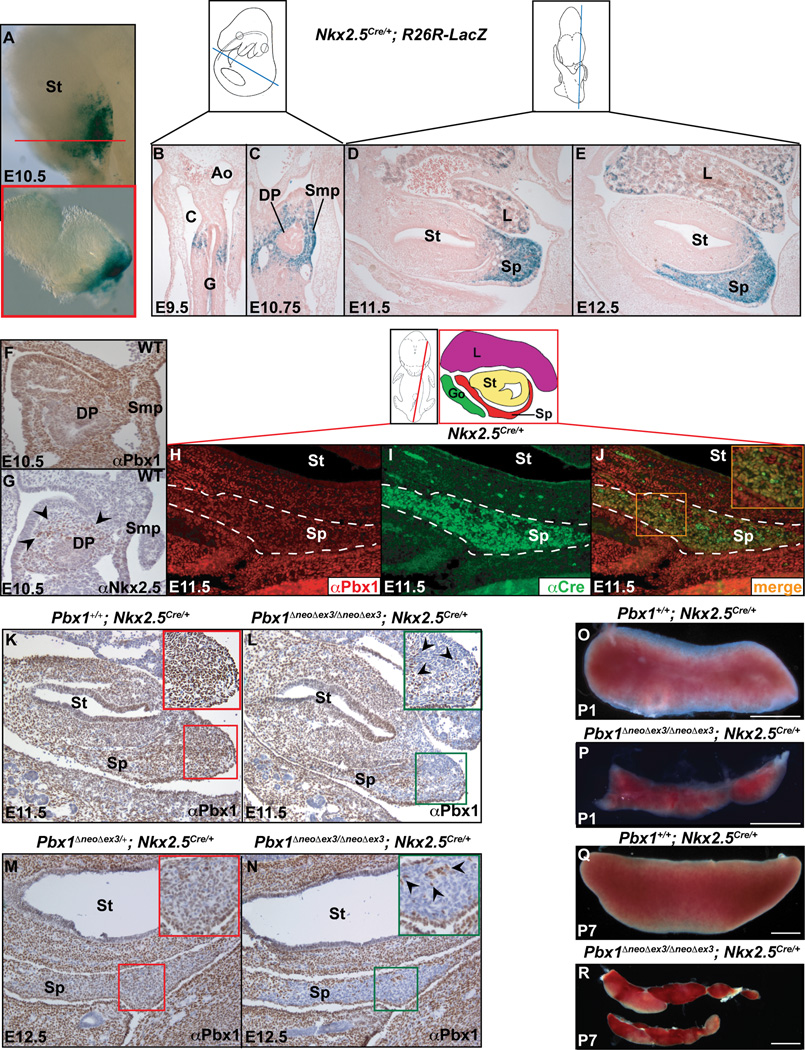
Figure 1. Pbx1 inactivation in Nkx2-5-positive mesenchyme causes spleen hypoplasia.
(A–E) Whole mount (A), transverse (A inset, red box corresponds to plane of section indicated by red line; B–C), and sagittal (D–E) sections of Nkx2-5Cre/+;R26R-LacZ embryos from E9.5 to E12.5, stained by β galactosidase. (F) IHC for Pbx1b and (G) Nkx2-5 (black arrowheads) in E10.5 WT transverse sections. (H–J) IF on E11.5 Nkx2-5Cre/+sagittal sections. Co-localization (orange, J inset) of Pbx1 (red), which is widespread in the spleen, and Cre (green) in spleen mesenchyme (outlined by white dashes). (K–N) IHC with α-Pbx1b on E11.5 and 12.5 sagittal sections from control (K,M) and Pbx1 conditional mutant (L,N) embryos, showing Cre-mediated Pbx1 loss in all but a few positive cells (black arrowheads; insets). (O–R) P1 and P7 control (O,Q) and homozygous mutant spleens (P,R). Ao, aorta; C, coelomic cavity; DP, dorsal pancreas; G, gut; Go, gonad; L, liver; Smp, splanchnic mesodermal plate; Sp, spleen; St, stomach; WT, Wildtype. See also Figures S1 and S2.
Using R26RLacZ mice (Figure 1A–E; Soriano, 1999), we revealed that Nkx2-5 Cre is first detectable at E9.5 on both sides of the visceral mesoderm (Figure 1B), where spleen progenitors will arise. By E10.75, Cre marked more of the spleno-pancreatic mesenchyme (Figure 1C), and was confined to the splenic anlage to the left of the stomach by E11.5–12.5. The prospective spleen capsule was devoid of Cre (Figure 1D,E). We also detected Nkx2-5 Cre in other domains, including a minority of liver cells, as reported (Figure 1D,E; Stanley et al., 2002). At E10.5, while Pbx1 was widespread in spleno-pancreatic mesenchyme and Smp, Nkx2-5 was detected in only a few mesenchymal cells (Figure 1F,G). By E11.5, Pbx1 and Nkx2-5 Cre co-localized in most mesenchymal cells of the anlage (Figure 1H–J). Thus, the Nkx2-5 Cre line appeared suitable for assessing Pbx1 roles in organ growth, after splenic fate specification.
We demonstrated efficient Cre-mediated Pbx1 loss in spleen mesenchyme of Pbx1ΔneoΔex3/ΔneoΔex3; Nkx2-5Cre/+ mice (hereafter Pbx1/Nkx2-5 Cre; Figure 1K–N and insets). Pbx1 loss mirrored Nkx2-5 Cre activity at E11.5 and E12.5 (compare Figure 1L with Figure 1D) and only 3–5% of mesenchymal cells in Pbx1/Nkx2-5 Cre spleens retained Pbx1 (insets in Figure 1L,N). The prospective spleen capsule, which does not express Nkx2-5 (Brendolan et al., 2005), retained Pbx1 (Figure 1L,N) and cells associated with splenic small vessels, which do not arise from Nkx2-5-positive mesenchyme, also showed low Pbx1 levels, as in postnatal day 3 (P3) mutant spleens (Figure S2P–S). Thus, Pbx1 loss was permanent (Figure S2S). All mutant mice (with neo [Pbx1Δex3/Δex3;Nkx2-5Cre/+] or without neo [Pbx1ΔneoΔex3/ΔneoΔex3;Nkx2-5Cre/+]) formed hypoplastic and fragmented spleens (Figure 1P,R) with full penetrance. Since Wt1 also marks spleen mesenchyme (Brendolan et al., 2005; Hecksher-Sørensen et al., 2004), we inactivated Pbx1 using the Wt1 Cre line (Wilm et al., 2005), which resulted in similarly hypoplastic spleens (Figure S1D,E), confirming that Pbx1 controls splenic growth.
Spleen hypoplasia, resulting from a Tlx1 (Hox11)-independent proliferation defect, is exacerbated by Pbx1/Pbx2 compound loss
Loss of even one allele of Pbx2, which co-localizes with its family member Pbx1 in the majority of spleen mesenchymal progenitors (Figure S2K), on a Pbx1/Nkx2-5 Cre background, exacerbated spleen hypoplasia and fragmentation (Figure S6A–F). Thus, Pbx1/2 exhibit overlapping functions in spleen morphogenesis and growth, as in skeletal development (Capellini et al., 2006). We uncovered a significant decrease of mitotic mesenchymal cells in the anlagen of Pbx1/Nkx2-5 Cre embryos versus controls at different gestational days (Figure S5A,B; quantifications in Figure 5I), while apoptosis was unaffected (Figure S2T,U). We previously reported reduced mesenchymal proliferation in Pbx1−/− spleens, as in deficiency of Tlx1 (known as Hox11). While Tlx1 was undetectable in Pbx1−/− splenic anlage (Brendolan et al., 2005), we found normal Tlx1 expression in E14.5 Pbx1Δex3/Δex3;Pbx2+/−;Nkx2-5Cre/+ anlagen versus controls (Figure S5C,D). Moreover, we observed that Tlx1-positive spleen progenitors are similarly present in WT and Pbx1 mutant embryos at early stages of spleen development (E11.5; Figure S5E,F). Since Tlx1 is a splenic fate marker (Kanzler and Dear, 2001), required for cell cycle progression (Kawabe et al., 1997), and Tlx1 loss-of-function (LOF) mice exhibit only asplenia (Roberts et al., 1994), our findings confirmed that splenic specification is unperturbed in this model, and that the hyposplenia is not due to inadequate specification of spleen progenitors. Instead, expansion of these progenitors was perturbed. Despite splenic hypoplasia, colonization of E14.5 mutant anlagen by erythroid (Vannucchi et al., 2000) and endothelial (Baldwin et al., 1994) progenitors appeared grossly normal (Figure S2X,Y). Since erythroid colonization commences only around E14.5 (Sasaki and Matsumura, 1988), the proliferation defect of Pbx mutant spleen anlagen (Figure S5A,B) precedes this process. In addition, in the embryonic red pulp, lymphocytes constitute only approximately 2% of hematopoietic cells during development (Sasaki and Matsumura, 1988). Therefore, even if hematopoietic cells are deficient in Pbx mutants, they constitute a minority of the total population of the normal splenic anlage at these gestational stages. These data, together with reduced proliferation in Pbx-deficient mesenchymal cultures (below), underscore that spleen hypoplasia in this model resulted from a Tlx1-independent defect in mesenchymal proliferation, highlighting the primary, cell-autonomous role of Pbx in promoting splenic progenitor expansion.
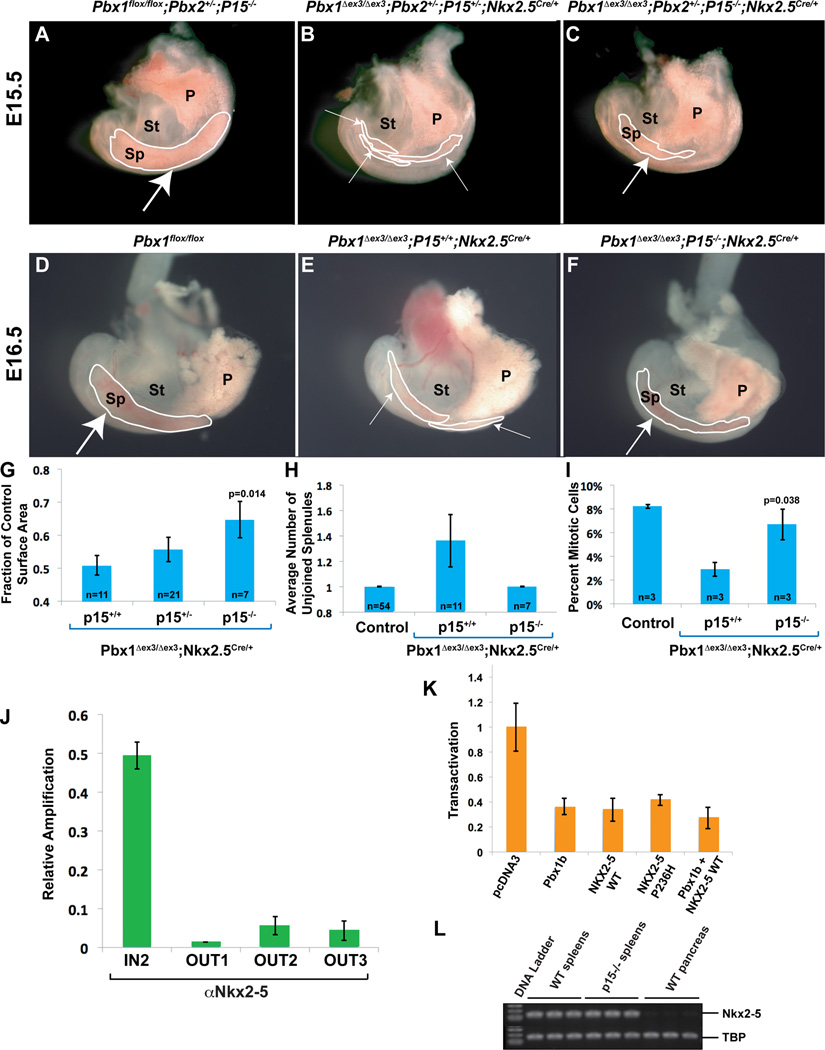
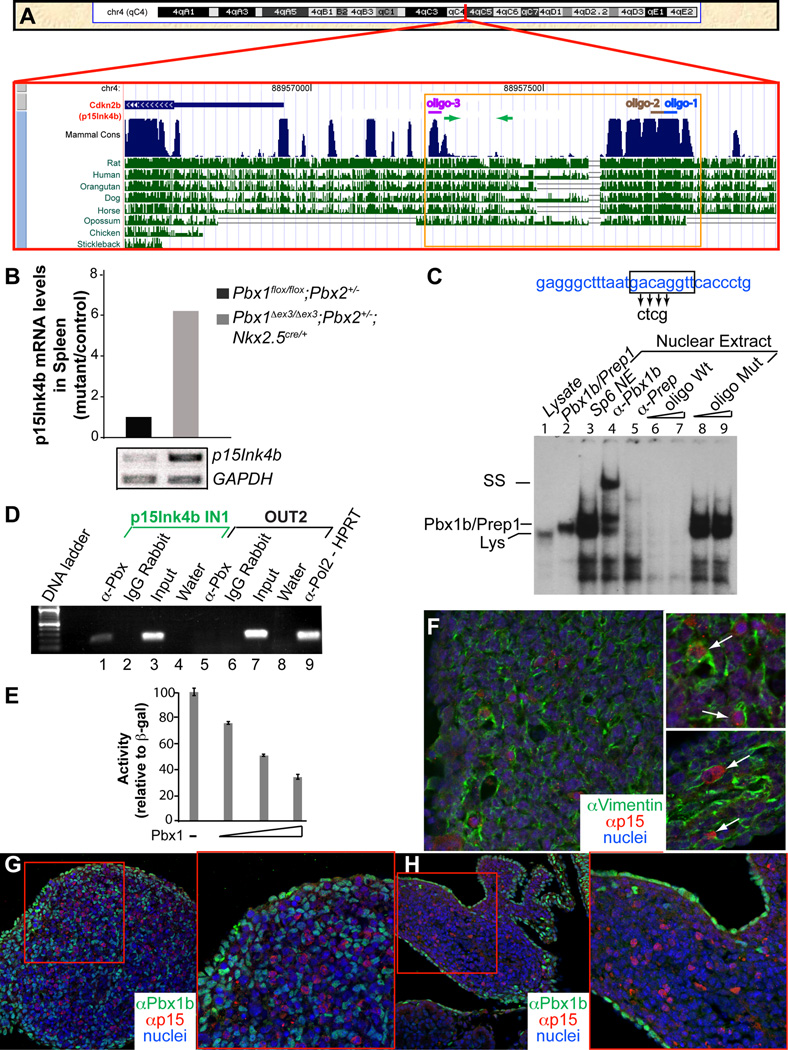
Figure 5. Genetic ablation of p15Ink4b, which is bound by Nkx2-5 in its cis-regulatory elements, partially rescues the spleen phenotypes.
(A–F) Partial rescue of spleen size and morphogenesis in E15.5 (A–C) and E16.5 (D–F) Pbx1Δex3/Δex3;Nkx2-5Cre/+ mutants on a p15-deficient background. Embryonic spleens (outlined in white) in which Pbx1 has been deleted in the mesenchyme either alone (E,F) or with Pbx2 (B,C), show a significant increase in spleen size and rescued morphology on a p15-deficient background (medium white arrows; C,F), versus littermates which retain p15 (thin white arrows; B,E). Control spleens (thick white arrows; A,D) for comparison. (G–I) Summary of genetic rescue data at E16.5. (G) Spleen surface area measurements (Experimental Procedures) normalized to controls reveal that p15-null Pbx1 mutant spleens are significantly larger than p15+/+ mutant spleens (p=0.014). (H) All rescued embryos develop one single spleen anlage. (I) Proliferation rate in p15−/− mutant spleen mesenchyme is restored to near WT levels, significantly higher than in p15+/+ Pbx1 mutants (p=0.038). Number of samples (n) indicated on corresponding bar of each graph. P, pancreas; Sp, spleen; St, stomach. (J) ChIP shows Nkx2-5 binding to the p15 promoter. Primers located within the p15 promoter (p15Ink4b IN2), but not non-specific primers (OUT1, OUT2, and OUT3), amplify a specific product from αNkx2-5-immunoprecipitated chromatin from SPCL2 cells, as assessed by qPCR and normalized to amplification of input chromatin. (K) Transient transfection of NIH 3T3 cells with 800ng of Pbx1b, NKX2-5 WT or NKX2-5 P236H, or co-transfection of Pbx1b and NKX2-5 in combination (500ng each), causes a reduction in p15 promoter activity. Transactivation normalized to empty expression vector (pcDNA3). Data are mean ± SEM of one out of three independent experiments performed in triplicate. (L) Semi-qRT-PCR on E16.5 mouse splenic RNA does not reveal perturbations of Nkx2-5 in p15−/− mutants versus controls. E16.5 WT pancreas RNA used as negative control for Nkx2-5 expression. All data are mean ± SEM. TBP, TATA-binding protein. See also Figure S6.
Pbx directly maintains Nkx2-5 splenic mesenchymal expression, which is essential for spleen growth
Like Tlx1, Nkx2-5 is detected in E10.5 spleno-pancreatic mesenchyme and in spleen progenitors thereafter (Figure 1A–E,G; Brendolan et al., 2005; Hecksher-Sørensen et al., 2004). It also has roles in heart progenitor proliferation (Prall et al., 2007). Pbx1 is required for Nkx2-5 initiation, as Pbx1−/− embryonic spleens lack Nkx2-5 (Brendolan et al., 2005). In contrast, Nkx2-5 Cre-mediated Pbx1 inactivation at E10.5, after Nkx2-5 expression onset, enabled assessment of the continued requirement of Pbx1 for Nkx2-5 maintenance during splenic growth. Nkx2-5 was detected in a minority of mutant spleen cells (Figure 2B), compared to controls in which more than 90% of spleen mesenchymal cells exhibit Nkx2-5 protein (Figure 2A). Additionally, Nkx2-5 mRNA was significantly reduced in mutant versus control spleens (Figure 2C). Though Nkx2-5 is an early spleen mesenchymal marker, its roles in spleen organogenesis are unknown, due to early in utero lethality of null mutants (Lyons et al., 2005). Different Nkx2-5 partial loss of function (LOF) alleles (Experimental Procedures), including Nkx2-5+/− mice (Lyons et al., 2005), Nkx2-5Cre/GFP hypomorphic embryos (Prall et al., 2007), and Nkx2-5Y-A:IRESLacZ/+-wildtype-chimeras, a dominant-negative model of Nkx2-5 deficiency conferred by mutation of a conserved tyrosine-rich domain (Elliott et al., 2006), showed hyposplenia (Figures 2D–G and 6D). These mouse models demonstrated that Nkx2-5 is critical for spleen growth, as reduced Nkx2-5 dosage and mutations in different parts of the protein yield spleen hypoplasia. Chromatin immunoprecipitation (ChIP) on the Nkx2-5 spleen-stomach enhancer (Figure 2H; Reecy et al., 1999), which contains two predicted binding sites for Pbx-Hox and one for Pbx-Prep (Figure S3), using SPCL2 cells with an α-Pbx antibody (Ab), showed marked amplification of a region-specific PCR product, but not of an outside control DNA fragment (Figure 2I). Luciferase (Luc) reporter assays conducted by transfecting a pGL3 reporter (containing the Nkx2-5 spleen enhancer upstream of Luc) into HEK293T cells revealed a consistent two-fold transactivation relative to empty pGL3 vector. Critically, this transactivation was abolished when using a Nkx2-5 spleen enhancer reporter in which the Pbx-Hox and Pbx-Prep binding sites were mutated (Figure 2J). These findings established the requirement of Nkx2-5 for spleen development and of Pbx1/2 for maintenance of its expression in spleen progenitors in vivo, via binding to its spleen enhancer.
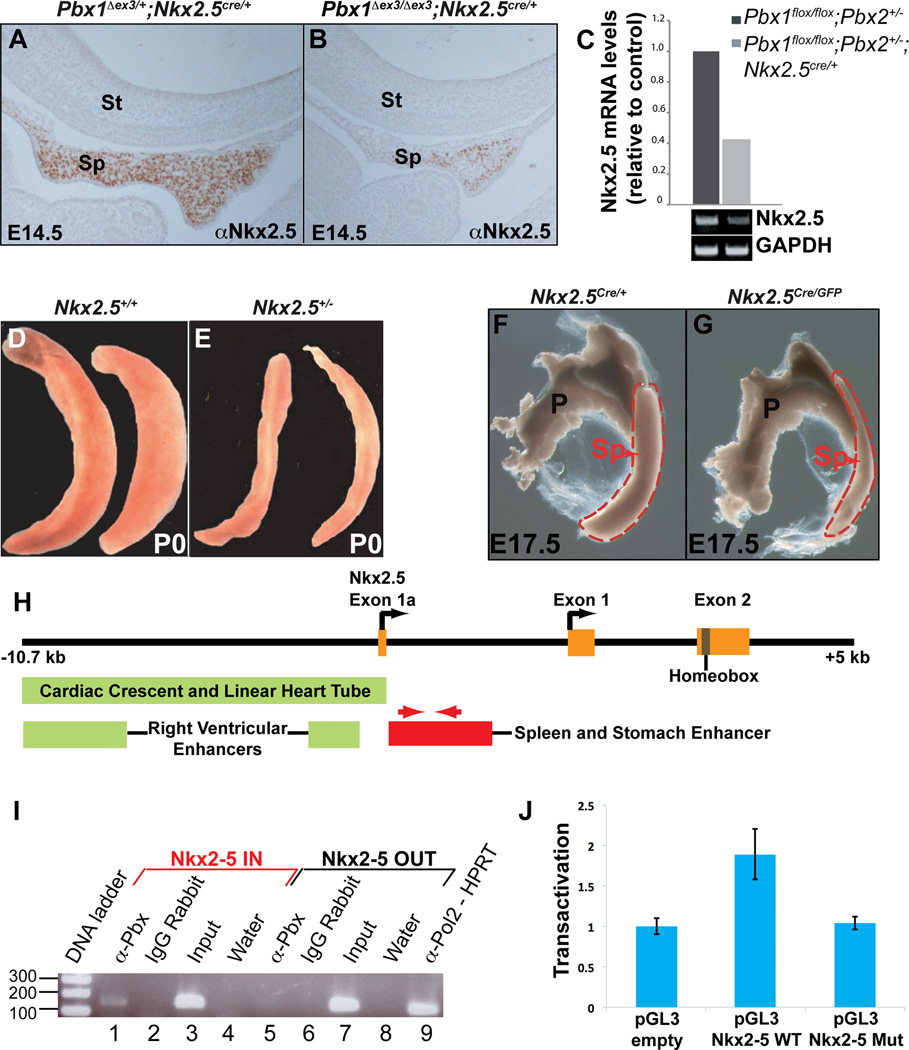
Figure 2. Pbx1 ablation in spleen mesenchyme causes down-regulation of Nkx2-5, essential for spleen growth.
(A,B) IHC (E14.5 sagittal sections) shows few Nkx2-5-positive cells in Pbx1 conditional mutants (B) versus widespread Nkx2-5 in controls (A). (C) qRT-PCR reveals a significant reduction of Nkx2-5 mRNA in Pbx1 mutant versus control spleen mesenchyme. (D–G) Reduced Nkx2-5 levels in Nkx2-5+/− (E), and Nkx2-5Cre/GFP (G) mice result in smaller spleens versus controls (D,F: P0 and E17.5). (H) Diagram of the murine Nkx2-5 locus. Enhancers directing expression in heart (green boxes) and spleen-stomach (red box) indicated (Figure S3). (I) Pbx proteins bind to the Nkx2-5 spleen-stomach enhancer, as assessed by ChIP. Primers located within the enhancer (H, red arrows), but not non-specific primers (lane 5), amplify a specific band from α-Pbx immunoprecipitated chromatin from SPCL2 cells (lane 1; Experimental Procedures). (J) A Luc reporter containing WT Nkx2-5 spleen-stomach enhancer (pGL3 Nkx2-5 WT) is transactivated approximately two-fold when transiently transfected into HEK293T cells versus a control Luc reporter (pGL3-empty). This transactivation is abolished when three predicted Pbx binding sites within the enhancer have been mutated (pGL3 Nkx2-5 Mut). Data are mean ± SEM of three independent experiments performed in triplicate. P, pancreas; Sp, spleen; St, stomach. See also Figure S3.
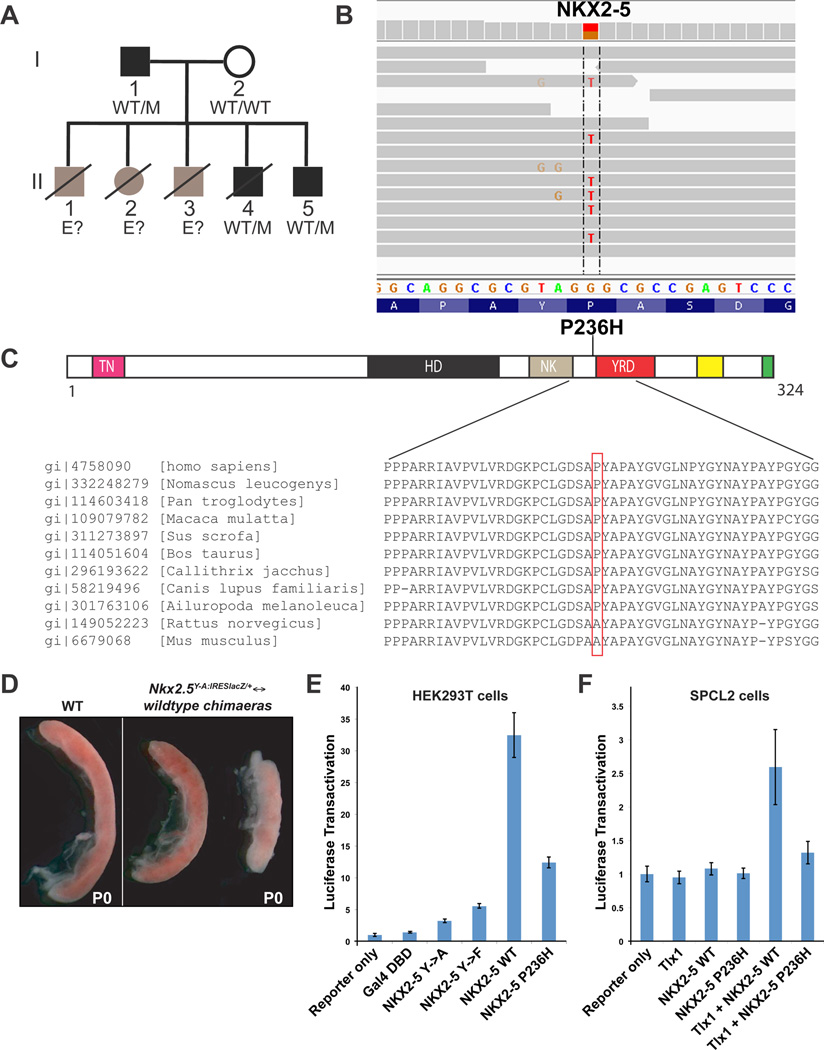
Figure 6. Characterization of an inherited NKX2-5 mutation in ICA patients.
(A) Pedigree of the family. Black symbols indicate confirmed ICA cases. Gray symbols indicate siblings who died early in childhood with clinical symptoms similar to those of II.4. Haplotypes of NKX2-5 are indicated: M stands for c.707C>A, and WT for the wild-type allele. E? indicates unknown haplotype. (B) Illumina sequencing reads displayed for patient I.1. Reads overlapping the mutation in exon 2 of NKX2-5 (bp position g.172,659,827 – g.172,659,852, hg19) reveal the heterozygous substitution of C to A (reads are reversed). (C) The P236H mutation is plotted on a schematic diagram of the NKX2-5 protein (drawn to scale) showing conserved and/or functional domains (homeodomain: HD; tinman/Nkx2-5 domain: TN; NK2 specific domain: NK; tyrosine rich domain: YRD). Two other conserved domains, the NKX2-5 box and the GIRAW motif, are indicated by yellow and green boxes, respectively. Evolutionary conservation of the NKX2-5 region containing amino acid P236 is represented below. (D) Depicted are spleens from Nkx2-5Y-A:IRESlacZ/+↔wildtype chimaeric P0 mouse pups (Prall et al., 2007). Chimaeric spleens (right panel), with reduced Nkx2-5 function, are smaller and dysmorphic, compared to WT (left panel). (E) An expression vector encoding a construct consisting of the Gal4 DNA-binding domain fused to the C-terminus of WT human NKX2-5 (NKX2-5 WT) causes a greater than 30-fold increase in transactivation when transiently co-transfected into HEK293T cells with a UAS-Luc reporter. This transactivation is reduced by more than 50% when a fusion construct bearing the P236H mutation (NKX2-5 P236H) is used. Constructs expressing the Gal4-DBD alone, or fusion constructs in which all Tyrosines in the YRD have been substituted with Alanines or Phenylalanines (NKX2-5 Y->A and NKX2-5 Y->F, respectively; Elliott et al., 2006), used as negative controls. Data are mean ± SEM of two independent experiments performed in triplicate. (F) Human NKX2-5 WT expression vector transiently co-transfected into SPCL2 cells with Tlx1 expression vector and a Luc reporter containing the Nkx2-5 spleen-stomach enhancer (pGL3-Nkx2-5-Luc) increased transactivation nearly three-fold relative to reporter alone. Co-transfection of Tlx1 with mutated NKX2-5 expression construct bearing the P236H mutation (NKX2-5 P236H) did not transactivate the Luc reporter, nor did NKX2-5 WT, NKX2-5 P236H, or Tlx1 transfected singly. Data are mean ± SEM of four independent experiments performed in triplicate. See also Figure S7 and Tables S2–S5.
Pbx1 directly represses the cell cycle inhibitor p15Ink4b in spleen expansion
In RT-PCR arrays (Experimental Procedures), only the CDK inhibitor p15 (known as Cdkn2b; Figure 3A) was significantly perturbed in Pbx mutant spleens among cell cycle regulators (Miller et al., 2007). It was up-regulated more than 6-fold in E13.5 and E14.5 mutant spleens (E13.5 in Figure 3B) (Table S1), but was not significantly up-regulated in Pbx1/Nkx2-5 Cre mutant hearts or pancreata (Figure S4D,E). While p15 often acts downstream of TGF-β signaling (Reynisdottir and Massague, 1997), we did not observe dysregulation of other TGF-β-associated genes in mutant spleens (Table S1).
Figure 3. Binding of Pbx1 to the p15Ink4b promoter is associated with p15Ink4b repression.
(A) p15 locus on mouse chromosome 4. Region upstream of p15 (red box) with evolutionarily conserved sequences (blue peaks). Oligonucleotides for EMSA assays: colored lines. Primers for ChIP: green arrows. Sequence of the p15 promoter (orange box) shown in Figure S4A. (B) qRT-PCR of total RNA reveals significant upregulation of p15 in mutant embryonic spleens versus controls. GAPDH mRNA, internal control. (C) EMSA with radiolabeled oligo-1 (blue line; panel A) containing a Pbx-Prep/Meis binding site (black box). Spleen mesenchymal cell nuclear extracts probed with oligo-1, and treated with α-Pbx Ab (lane 4), resulted in a supershifted band, indicating Pbx binding. Binding specificity confirmed by competition from excess unlabeled WT (lanes 6–7), but not mutated oligo (lanes 8–9). (D) ChIP shows Pbx binding to the p15 promoter. Primers within the promoter (p15Ink4b IN1; panel A; green arrows), but not non-specific primers (OUT2; lane 5) amplify a distinct band from α-Pbx immunoprecipitated chromatin (lane 1). (E) Transient transfection of NIH 3T3 cells with Pbx1 causes a dose-dependent reduction in p15 promoter activity. Data are mean ± SEM of one out of four independent experiments performed in triplicate. (F) IF on E13.5 WT transverse sections. p15 (red) co-localizes in cells (white arrows, insets) exhibiting high levels of the mesenchymal marker vimentin (green). (G–H) IF on sections from E16.5 WT (G) and Pbx1Δex3/Δex3;Nkx2-5Cre/+ (H) spleens. Insets correspond to areas within red boxes. p15 (red) is not present in Pbx1b-positive cells (green). See also Figure S4 and Table S1.
The p15 cis-regulatory elements (Figure 3A; Staller et al., 2001), conserved among vertebrates, bear three Pbx-Prep/Meis binding sites (Figure S4A). Electrophoretic mobility shift assays (EMSA) on SP6 nuclear extracts with oligonucleotides (oligos 1–3, Figure 3a) containing one of the binding sites, and an α-Pbx1b Ab, identified a supershifted band (Figures 3C and S4B,C; Berthelsen et al., 1998), indicating binding of a Pbx1b-Prep/Meis complex. ChIP assays on SPCL2 cells using primers within the promoter region bearing Pbx-Prep/Meis binding sites (Figure 3A, green arrows) showed amplification of a specific band from α-Pbx-immunoprecipitated DNA (Figures 3D, lane 1 and S4F), demonstrating Pbx binding to the p15 promoter in the spleen anlage. Co-transfection of increasing concentrations of Pbx1b expression vector with a Luc reporter containing p15 regulatory sequences in NIH-3T3 cells (Figure 3E) revealed a dose-dependent repression of Luc activity. In contrast, a Luc reporter controlled by a ubiquitous promoter showed no decrease in activity when co-transfected with Pbx1b. These findings demonstrated that Pbx binding represses p15 transcription in the spleen anlage, in contrast to human hepatocellular carcinoma cell lines, in which p15 was cooperatively activated by Pbx1/Meis (Bjerke et al., 2011).
In WT spleen anlagen, p15 co-localized with the mesenchymal marker vimentin (Figure 3F and insets), indicating that the proliferation defect is intrinsic to mesenchyme, and not to colonizing cells. p15 was not detected in Pbx1-positive cells in E16.5 (Figure 3G,H) WT and mutant spleen anlagen. Cells negative for both proteins were likely non-mesenchymal (hematopoietic and endothelial) cells colonizing the spleen by E16.5. The mutual exclusion of p15 and Pbx1 was consistent with direct p15 repression by Pbx1 in the spleen anlage.
Loss of Pbx1/2 in cultured spleen mesenchymal cells reduces proliferation and increases p15 levels
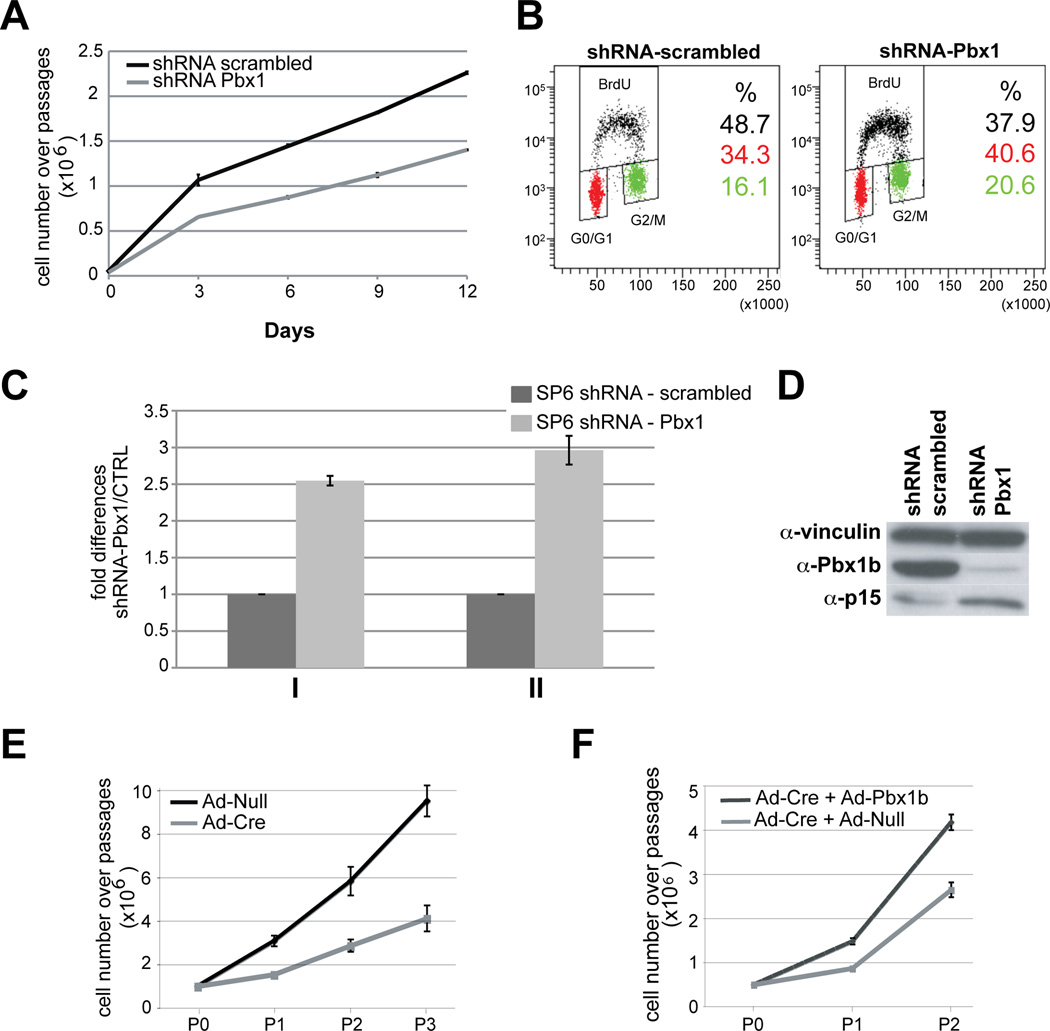
Spleen mesenchymal cultures (Experimental Procedures) in which Pbx1, Tlx1 and Nkx2-5 co-localize, reproduce the embryonic splenic mesenchymal environment in a controlled system in vitro (Figure S5G–L). Pbx1 loss (by infection with lentiviral shRNA-Pbx1; Experimental Procedures) in SP6 cells significantly reduced cell proliferation versus controls (shRNA-scrambled; Figure 4A). Asynchronous cells infected by shRNA-Pbx1 showed reduced BrdU incorporation and accumulated in G0/G1 and G2/M as compared to controls (Figure 4B). Moreover, loss of Pbx1/2 increased p15 mRNA and protein levels in SP6 cells (Figure 4C,D). Pbx1 loss in SpM cells infected with adenovirus-Cre (Ad-Cre) also significantly reduced cell proliferation versus controls infected by Ad-Null (Figure 4E). Restoration of Pbx1b expression via Ad-Pbx1b infection in Pbx1/2 mutant cells ameliorated the proliferation defect versus Pbx1/2 mutant cells infected with Ad-Null (Figure 4F). These results established cell-autonomous requirements for Pbx1/2 in spleen mesenchymal cell proliferation in vivo and in vitro. We do not exclude that hematopoietic or endothelial cells may also be affected by Pbx loss in the splenic mesenchyme, but all the data described above indicate that they are not primarily responsible for the observed organ hypoplasia.
Figure 4. Loss of Pbx1/2 in cultured spleen mesenchymal cells reduces proliferation and increases p15Ink4b levels.
(A) SP6 cells after infection with a lentivirus containing short hairpin RNAs specific for Pbx1 (shRNA Pbx1, gray line), exhibit significantly reduced cell number over four passages, versus cells infected with a control lentivirus carrying a scrambled shRNA sequence (shRNA scrambled, black line). (B) Asynchronous SP6 cells infected with shRNA Pbx1 show reduced BrdU incorporation versus scrambled shRNA, and accumulate in G0/G1 and G2/M. Loss of Pbx1 reduces percentage of cells in S phase of the cell cycle (p=0.003), and increases percentage of cells in G0/ G1 (p=0.004) and G2/ M (p=0.032). (C) Asynchronous (I) and serum-released (II) growing SP6 cells infected by shRNA Pbx1 exhibit increased p15 mRNA versus shRNA scrambled. (D) Western blot shows reduced Pbx1b and increased p15 in shRNA Pbx1-infected SP6 cells versus control. α-Vinculin, loading control. (E) Pbx1 inactivation in SpM cells by infection with a Cre-expressing adenovirus (Ad-Cre, gray line) significantly reduces cell number over three passages (P), versus SpM cells infected by control adenovirus (Ad-Null, black line). Data are mean ± SEM of one representative experiment out of a total of three performed in triplicate. (F) Reintroduction of Pbx1b by an adenovirus expressing Pbx1b (Ad-Pbx1b), performed four days after Ad-Cre-mediated Pbx1 inactivation, produces a significant increase in cell number (black line) after two passages (P), versus SpM cells infected by Ad-Null (gray line). Data are mean ± SEM of one representative experiment out of a total of two performed in triplicate. See also Figure S5.
Genetic ablation of p15Ink4b, which is bound by Nkx2-5 in its cis-regulatory elements, partially rescues the spleen phenotypes
p15−/− mice are fertile (Latres et al., 2000), and we observed that their spleens are indistinguishable from WT during development. E15.5–E17.5 spleens from Pbx1Δex3/Δex3;p15−/−;Nkx2-5Cre/+ embryos appeared morphologically larger and closer to WT size, versus Pbx1Δex3/Δex3;p15+/+;Nkx2-5Cre/+ littermates (Figures 5A–F and 6G–I). Thus, splenic expansion was partially rescued on a p15 null background. In each spleen, we quantified: 1) size; 2) morphology (number of discrete anlagen); and 3) mesenchymal proliferation. Surface area measurement was chosen as opposed to volumetric analysis, due to the fragility and fragmentation of the minute, thin mutant spleens (Experimental Procedures). Defects in mutant spleen volume or weight, as well as the degree of rescue, would likely be more striking. We observed a significant (p<0.02) increase of about 30% in the size of rescued spleens versus mutants (Figure 5G). Also, rescued spleens, lacking p15, always formed one single, compact anlage, as in controls, while Pbx1 mutants on a p15+/+ background formed multiple, distinct splenules (Figures 5H and S6F). We confirmed that Pbx1 inactivation in spleen mesenchyme significantly reduced proliferation, while loss of p15 function in Pbx1/Nkx2-5 Cre mutants significantly (p<0.05) increased proliferation to near WT levels (Figure 5I). In sum, absence of p15 in Pbx1;Nkx2-5 Cre mutants resulted in: 1) significant rescue of spleen size; 2) complete rescue of spleen fragmentation; and 3) significant rescue of mesenchymal proliferation. Together, these findings demonstrated that repression of p15 by Pbx is required for organ morphogenesis and growth in vivo.
ChIP revealed binding of Nkx2-5 to the p15 promoter (Figures 5J and S4F), which is also bound by Pbx, suggesting control of p15 also by Nkx2-5. Supporting this hypothesis, transfection experiments revealed that transient expression of NKX2-5 in NIH 3T3 cells represses transcription of a Luc reporter controlled by p15 regulatory elements, similarly to transfection of Pbx1b (Figure 5K). RT-PCR analysis on p15−/− spleens did not show substantial perturbation of Nkx2-5 mRNA levels relative to WT (Figure 5L), indicating likely unidirectional control of p15 by Nkx2-5, and highlighting the central role of Nkx2-5 within this spleen regulatory module.
Heterozygous missense NKX2-5 mutation in a human kindred with ICA
We set out to identify genetic etiologies of human ICA by whole-exome sequencing (WES; Alcais et al., 2010; Bolze et al., 2010; Byun et al., 2010). We investigated an African kindred (Family E; Mahlaoui et al., 2011) with 3 ascertained cases (I.1, II.4 and II.5) of ICA (Figure 6A; case report in Experimental Procedures). We hypothesized that ICA segregated as fully penetrant, autosomal dominant (AD), Mendelian trait in this family. The number of reads and the exome coverage metrics (Table S2) show inferior quality of the assembly for the exome of patient II.4 compared to the 2 other patients (42% of target bases covered at 10X compared to 72% and 71%), probably due to suboptimal quality of II.4 genomic DNA (gDNA), which was extracted from necropsy samples. Therefore, we selected candidate variations present in all three patients (I.1, II.4 and II.5), or present in I.1 and II.5 and not covered by WES in II.4 (Table S3). After filtering out known polymorphisms (Experimental Procedures), we identified only 32 variants that could underlie ICA in this family (Tables S3 and S4).
Of the 32 candidate variants identified, a substitution in NKX2-5 was the only one affecting a gene involved in mouse spleen development (Table S4). This variant is a missense heterozygous c.707C>A in protein-coding exon 2 of NKX2-5 (NM _004387.3, MIM600584) that changes Proline at amino acid position 236 to Histidine, p.P236H (P236H hereafter; Figure 6B,C). Sanger sequencing of peripheral blood gDNA and cDNA from EBV-transformed B cells (EBV-B) of ICA patients confirmed that the P236H variant segregated with ICA in all family members examined. All patients, but no healthy relatives, were heterozygous for P236H (Figures 6A and S7A). The variant was not found in 1,052 additional healthy individuals from the Centre d’Etude du Polymorphisme Humain and Human Genome Diversity panels, nor in the 1,197 samples sequenced by the 1,000 Genomes project, which together include 327 individuals of Sub-Saharan African origin (Table S5). These results suggest that P236H is a rare, potentially ICA-causing variant rather than an irrelevant polymorphism. Lastly, Proline at position 236 is evolutionarily conserved, although an Alanine is present at this position in Mus musculus (Figure 6C). No species in which Nkx2-5 has been sequenced bears a Histidine at this position. We also examined copy number variants (CNV) throughout the II.5 genome (Supplementary Experimental Procedures). We did not observe any CNV larger than 50 kb that was not present in the DGV database (http://projects.tcag.ca/variation/) or our own database of 150 samples. Overall, these genetic data suggest that P236H is associated with ICA in this multiplex kindred.
Biological characterization of the human NKX2-5 mutant allele
Because POLYphen II (Adzhubei et al., 2010) predicted that the P236H mutation is benign and residue 236 is outside the NKX2-5 homeodomain, we hypothesized that the mutation may not impair the production of the protein or its DNA binding. Western blot and EMSA confirmed that NKX2-5 P236H is produced and binds to DNA similarly to WT protein (Figure S7). We hypothesized that the mutation may disrupt transactivation by NKX2-5, probably through interaction with spleen-specific cofactors. P236 lies immediately adjacent to the first of nine Tyrosines that define a conserved Tyrosine-rich domain (YRD; residues 237–275; Figure 6C), which our previous work established as a critical domain for the in vivo function of Nkx2-5, as well as its transcriptional activity in a heterologous context (Elliott et al., 2006). Mouse chimeras composed partly of mutant cells in which Tyrosines in the Nkx2-5 YRD have been replaced by Alanines (Elliott et al., 2006), develop hypoplastic spleens (Figure 6D). Thus, we posited that the P236H mutation could potentially impact YRD function and the interaction of NKX2-5 with splenic cofactors.
To test the effects of P236H on the transactivation potential of Nkx2-5, we employed a heterologous system previously used to study Nkx2-5 function in mouse heart development (Elliott et al., 2006). The human NKX2-5 C-terminal domain, containing either WT or mutant sequence, was fused to the Gal4 DNA-binding domain (DBD) and co-transfected into HEK293T cells with a Luc reporter (pGL4.31) downstream of UAS elements. While the WT construct increased Luc activity about 30-fold versus control, the construct bearing P236H induced approximately three-fold lower activity (Figure 6E). Mouse constructs bearing mutations of critical YRD domain Tyrosines showed low Luc activity in this assay. Additionally, since Nkx2-5 is subject to autoregulation in heart development (Prall et al., 2007), we transfected SPCL2 cells with the murine Nkx2-5 spleen enhancer Luc reporter (Figure S3) and expression vectors for WT or P236H NKX2-5, with or without Tlx1, which is critical in mouse spleen development (Kanzler and Dear, 2001; Roberts et al., 1994). NKX2-5 WT co-transfected with Tlx1 activated Nkx2-5 Luc approximately 2.5-fold above control, whereas this transactivation was abolished when using NKX2-5 P236H (Figure 6F). The same Nkx2-5/Tlx1 co-transfection experiment performed with HEK293T cells did not activate the Nkx2-5 Luc reporter, highlighting that this co-operative transactivation of Nkx2-5 is spleen-specific. These results demonstrated that P236H impairs transactivation by NKX2-5, both in a heterologous transactivation assay and in the context of spleen mesenchymal progenitors. Given the findings in our mouse models, in which reduced Nkx2-5 gene dosage or YRD function yield hyposplenia, and the observed in vitro functional deficiency of the NKX2-5 P236H rare allele identified in an ICA kindred, we conclude that Nkx2-5 plays a central role in the development and growth of the mammalian spleen.
DISCUSSION
Pbx1 is a prime regulator of the organogenesis of the spleen, a vital but understudied organ. Here, we identified regulatory pathways that control spleen growth in development. Pbx1−/− mice exhibit numerous developmental defects (Capellini et al., 2006; Ferretti et al., 2011; Kim et al., 2002; Selleri et al., 2001), notably organ hypoplasia with diminished cell proliferation and asplenia (Brendolan et al., 2005). Our mouse model with spleen-specific Pbx1 loss afforded the dissection of Pbx roles in spleen organogenesis distinct from those in specification. Pbx1/Nkx2-5 Cre mice form spleen anlagen with defects in morphogenesis and growth, and exhibit multiple, unjoined splenules, indicating that Pbx loss affects the growth and concomitantly the fusion of splenic progenitor condensations. This strain therefore provides a model also for human polysplenia, a condition that occurs when independent mesodermal condensations fail to fuse, as normally occurs in human development, to form one cohesive spleen (Moore and Persaud, 2007; Porembka et al., 2008). The growth defects observed in this mouse model, in which Tlx1 expression is initiated and maintained, suggest that spleen fate determination occurs normally, prior to Pbx1 inactivation, and that the proliferation defect is Tlx1-independent. Pbx1 loss in Pbx1/Nkx2-5 Cre spleen anlage down-regulates Nkx2-5, for which we uncover roles in spleen growth. The central role of Nkx2-5 in spleen organogenesis is demonstrated here by hyposplenia in mouse models that have either reduced Nkx2-5 dosage or mutations affecting Nkx2-5 function. Transactivation of Nkx2-5 is one likely mechanism whereby Pbx1 governs spleen expansion (Figure 7D). Interestingly, Nkx2-5 promotes cardiac progenitor cell proliferation (Prall et al., 2007).
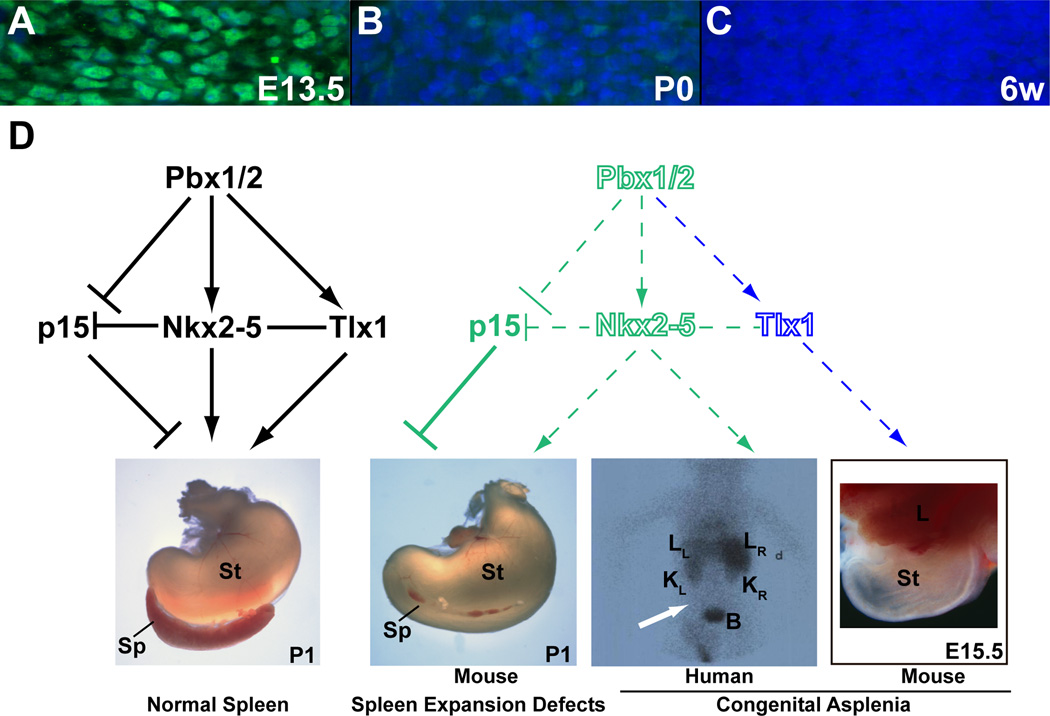
Figure 7. Pbx1 is present in the early spleen anlage where it directs a regulatory module that converges on Nkx2-5, controlling organ growth and morphogenesis.
(A–C) Present at high levels in E13.5 spleen mesenchyme (A), Pbx1 (green) is markedly reduced by P0 (B), and almost completely absent from 6 week-old mouse spleens (C). (D) During normal development, Pbx1/2 promote spleen expansion via multiple pathways (black lettering and lines; left panel): Pbx genes transactivate Nkx2-5 (and Tlx1, as reported; Brendolan et al., 2005) in the spleen mesenchyme, and repress the CDK-inhibitor p15. Moreover, like Pbx, Nkx2-5 can also bind to the regulatory elements of p15 and repress its transcription. Additionally, Nkx2-5 and Tlx1 likely act cooperatively (solid line) to transactivate target genes in spleen mesenchyme. The pivotal role of Nkx2-5 within the Pbx-directed regulatory module is highlighted by its central position within the depicted network. Disruption in any component of this module (illustrated by hollow fonts and dashed lines; right panel) can result in hyposplenia or asplenia in mice and humans. In particular, complete asplenia results from global loss of Pbx1 or Tlx1 in the mouse (Brendolan et al., 2005; Roberts et al., 1994), and is associated with mutation of NKX2-5 in ICA patients (scintigraphy image of patient II.4: white arrow indicates absence of uptake of 99mTc-labeled autologous red cells in the left abdomen below the liver, where the spleen should be located). Also, both spleen-specific Pbx1/2 inactivation and reduced Nkx2-5 dosage in mouse models yield hyposplenia and perturbed spleen morphogenesis. Green lettering and lines indicate findings described in this study; blue lettering and lines indicate previously reported hierarchical genetic control (Brendolan et al., 2005; Roberts et al., 1994). B, bladder; d, dextrum (right side); KL, left kidney; KR, right kidney; LL, left lobe of liver; LR, right lobe of liver; Sp, spleen; St, stomach.
Pbx controls spleen growth also by transcriptional repression of the CDK inhibitor p15Ink4b. p15−/− mice develop normally, though they acquire lymphoproliferative disorders as adults, and compound loss of p15 with p18Ink4c results in splenomegaly (Latres et al., 2000). While it has been reported that Pbx1 binds and transactivates p15 in human hepatic cancer cell lines (Bjerke et al., 2011), our mouse models demonstrate a repressive transcriptional control of p15 by Pbx in vivo in the spleen anlage. This repression is validated by the partial rescue of spleen growth and morphogenesis when Pbx1 is ablated from spleen mesenchyme in the absence of p15. Nkx2-5 also binds the p15 promoter, linking the two Pbx-regulated pathways, and partly accounting for spleen growth defects in models of Nkx2-5 deficiency. Indeed, both Pbx1b and NKX2-5, either alone or in combination, transcriptionally repress p15 in NIH 3T3 cells (Figure 7). Interestingly, the NKX2-5 P236H mutant protein also represses p15 transcription. This finding suggests that this particular mutation may perturb interaction of Nkx2-5 with spleen-specific cofactors, and/or it may disrupt the regulation of target genes other than p15 (such as Nkx2-5 itself; see Figure 6E,F). Additionally, Nkx2-5 may control other cell-cycle regulators, as suggested by the incomplete rescue of spleen size by p15 loss, which further underscores the central role of Nkx2-5 within this regulatory module.
To date, no existing mouse models with hyposplenia or asplenia have contributed to elucidating the genetic etiology of ICA, a life-threatening asplenic condition without other abnormalities, underreported and often undetected at birth (Mahlaoui et al., 2011). Here, we delineate candidate genes and regulatory modules that govern mammalian spleen organogenesis. Pbx target genes identified in the mouse spleen anlage in this study guided the analysis of WES data obtained from a human kindred with ICA. This approach led to the identification of a missense mutation in NKX2-5, which is a central component within the Pbx-directed module in the mouse. This variant, not found as a polymorphism in human populations, segregates with ICA in all family members examined. P236H diminished transactivation by Nkx2-5 in cultured spleen cells, which occurs only in the presence of the splenic cofactor Tlx1, suggesting that spleen-specific interactions are disrupted by this mutation. Interestingly, all known congenital heart disease (CHD) patients with previously identified NKX2-5 mutations do not display asplenia (Harvey et al., 2002), suggesting that these mutations, unlike P236H, preferentially compromise heart development. Although associated with ICA in vivo and deleterious in vitro, it is probable but not certain that the P236H allele is the key factor underlying ICA in this kindred. The generation of knock-in mutant mouse lines or the identification of NKX2-5 mutations in other ICA kindreds would provide further evidence. Meanwhile, this study paves the way for analyses of ICA patients using WES and other genome-wide approaches, and underscores the effectiveness of evaluating human genetic research in conjunction with analyses in mouse models.
EXPERIMENTAL PROCEDURES
Mice
Pbx1 conditional mice were engineered by homologous recombination in E14 129/Ola ES cells (Figure S1; Hooper et al., 1987). Previously described mice (Pbx1−/−, Pbx2−/−, Nkx2-5 Cre, Wt1 Cre, β-actin Cre, Nkx2-5+/−, Nkx2-5Y-A:IRESlacZ, Nkx2-5Cre/GFP, R26R-LacZ, and p15Ink4b) were used and genotyped as described (Biben et al., 2000; Capellini et al., 2006; Latres et al., 2000; Lewandoski and Martin, 1997; Lyons et al., 1995; Prall et al., 2007; Selleri et al., 2001; Soriano, 1999; Stanley et al., 2002; Wilm et al., 2005). Oligonucleotide primers for PCR genotyping in Supplementary Experimental Procedures.
Assessment of β-galactosidase activity, histology, immunohistochemistry (IHC), immunofluorescence (IF), whole-mount and section in situ hybridization (ISH), and TUNEL assays
Protocols as described (Brendolan et al., 2005; Selleri et al., 2001). Antibodies in Supporting Materials. Single-stranded sense and antisense riboprobes specific for Tlx1 (Brendolan et al., 2005) and Pbx1 (Capellini et al., 2006) were used for ISH.
RT-Profiler array screening and qRT-PCR
Total RNA and cDNA were generated from pools of 8–10 E13.5 and E14.5 Pbx1Δex3/Δex3;Pbx2−/−;Nkx2-5Cre/+ and Pbx1flox/flox;Pbx2−/− spleens with commercial kits (QIAGEN; SuperArray Bioscience). Expression of 168 genes was assessed by RT-Profiler PCR Array (SuperArray Bioscience). Differentially-expressed candidate genes were validated by qRT-PCR using SYBR Green (SuperArray Bioscience). For semi-qRT-PCR, cDNA prepared as above.
Chromatin immunoprecipitation
ChIP was performed as described (Brendolan et al., 2005; Capellini et al., 2006). Abs and PCR primers listed in Supplementary Experimental Procedures.
In vitro transcriptional assays
Typically, 800 ng of reporter plasmid was transiently transfected into NIH 3T3 cells, Pbx1−/− MEFs, SPCL2 cells, or HEK293T using Lipofectamine 2000 (Invitrogen), or FuGENE HD Transfection Reagent (Roche). Reporter plasmids were: p15Luc −1040/+70 (Li et al., 1995); pGL3-Luc containing 1 kb of the murine WT Nkx2-5 spleen-stomach enhancer (Figure S3) or the enhancer in which the Pbx-Hox and Pbx-Prep binding sites were mutated; 200–1000 ng of expression constructs containing cDNA of human WT or P236H mutant NKX2-5; or of a pcDNA3 construct containing the Pbx1b cDNA (Berthelsen et al., 1998); and 50 ng of control plasmid (pCMV-β-gal or Renilla luciferase). For GAL4-UAS assays, 800 ng of the luciferase reporter vector pGL4.31 (Promega) was co-transfected into HEK293T cells with a pCMV construct expressing the Gal4 DNA-binding domain fused to the YRD of human NKX2-5 (Elliott et al., 2006), either WT or bearing the P236H mutation.
Electrophoretic mobility shift assays
EMSA as described (Brendolan et al., 2005) using nuclear extracts from embryonic spleen mesenchymal cells, HEK293T cells, or in vitro translated proteins. Oligonucleotides and Abs in Supplementary Experimental Procedures.
Derivation of spleen stromal cell lines
Cell suspensions from embryonic spleens were expanded for 10–15 passages, according to the NIH 3T3 protocol (Todaro and Green, 1963). Two immortal lines (SP2 and SP6) were obtained from E16.5 Pbx1+/+;Pbx2−/−; one line (SpM) from E17–18 Pbx1flox/flox;Pbx2−/−; and one line (SPCL2) from E16.5 WT C57Bl/6 spleens. Immortalized lines were used for growth curves, FACS analysis, or ChIP.
Analysis of genetic rescue
Fixed, unstained E15.5, E16.5, and E17.5 spleens, stomach, and pancreas were positioned flat and photographed using QCapture software (QImaging) and spleen surface areas were measured with ImageJ (http://rsbweb.nih.gov/ij). Within each E16.5 litter, the sizes of Pbx1 mutant and rescued (lacking p15) spleens were normalized to the average WT spleen size within that litter. Sections from E16.5 WT, Pbx1 mutant, and rescued spleens were immunostained with α-pH3 Ab, and mitotic cells counted using the StereoInvestigator software (MBF Bioscience). About 1000 cells were counted in each of three embryonic spleens for each genotype.
Human ICA case report
Both parents, I.1 and I.2 (Figure 6A), originate from Congo-Brazzaville, Africa, and their first 3 children died between 8 and 12 months of age of fulminant sepsis. The index case II.4, born after the parents emigrated to France, died of sepsis caused by a Streptococcus α hemolyticus at 23 months. 99mTechnetium-labelled red blood cell scintigraphy (Bearn et al., 1992) revealed asplenia in II.4 and necropsy confirmed ICA with normal heart and viscera disposition. The parents report that the course of disease in the 4 children was strikingly similar. Although no medical work-up was performed for the first 3 children, a diagnosis of ICA is likely. The parents and their fifth child (II.5) were screened and ICA was diagnosed when II.5 was 2 months old and the father 35 years old. Child II.5, now 13 years old, is under antibiotic prophylaxis and has received appropriate immunizations. He is overweight (BMI=28.2 kg/m2) and has autism. Despite the lack of antibiotic prophylaxis or vaccinations, the father never experienced significant infections. Indeed, incomplete clinical penetrance of autosomal dominant ICA is rare but has been reported (Lindor et al., 1995).
IRB approval
This study was approved by the local institutional review board (IRB #JCA-0689). Written informed consent for participation in the study was obtained from all patients and family members studied.
Massively parallel sequencing
DNA (3µg) extracted from EBV-B cells from patients I.1 and II.5 or from tissues conserved from the necropsy of II.4 was sheared with a S2 Ultrasonicator (Covaris). An adapter-ligated library was prepared with the Paired-End Genomic DNA Sample kit (Illumina). The Sure Select Human All Exon kit 38Mb (Agilent Technologies) was used for exome capture. Single-end sequencing was performed on a Genome Analyzer IIx (Illumina), generating 72-base reads.
Sequence alignment, variant calling, and annotation
BWA aligner (Li and Durbin, 2009) was used to align the obtained sequences to the human reference genome (hg 19 build). Downstream processing was performed with the Genome Analysis Toolkit (GATK; McKenna et al., 2010), SAM tools (Li and Durbin, 2009), and Picard Tools (http://picard.sourceforge.net). Substitution calls were made with a GATK UnifiedGenotyper and indel calls with a GATK IndelGenotyperV2. All calls with a read coverage 2× and a Phred-scaled SNP quality of 20 were filtered out. No other GATK filters were applied. Variants were annotated and analyzed with software developed in-house (SQL database query-driven system; Liu et al., 2011). Polymorphisms reported in the National Center for Biotechnology Information (NCBI) dbSNP build 134 and 1,000 Genomes Project databases (http://www.1000genomes.org), or identified in our own database of 400 exomes, were filtered out.
Highlights.
Pbx1 deficiency causes organ growth defects including asplenia
A Pbx/Nkx2-5/p15Ink4b module controls spleen mesenchymal proliferation
Loss of p15Ink4b rescues expansion and morphogenesis in Pbx1 mutant spleens
The P236H substitution in NKX2-5 is associated with isolated congenital asplenia
Supplementary Material
ACKNOWLEDGEMENTS
We thank Dr. M. Barbacid and Dr. J. Burch for p15 and Wt1Cre mice, respectively; Dr. J. Massague for p15 constructs; Dr. M. Cleary for α-Pbx abs; Dr. T. Rabbitts for Tlx1 probes; Dr. S. Rafii for support and insight; Drs. E. Lacy, A. Koff, D. Herzlinger, A. Foley, and F. Lupu for discussions; C. Fiorese for help with Nkx2-5 mutant construct preparation; Dr. A. Puel and Dr. A. Abhyankar for help in WES analysis; and Dr. J-F. Emile for patient II.4 gDNA. M.K. was a Cohenca Fellowship recipient. Work supported by the NIH (HD43997, HD061403, and DE18031 to L.S. and HL085345 to C.P.C.); The March of Dimes and Birth Defects Foundation (6-FY03-071 to L.S.); Associazione Italiana Ricerca Cancro (AIRC; Start-Up 4780 to A.B.); Marie Curie Foundation (IRG-2007 208932 to A.B); St. Giles Foundation, Rockefeller University Center for Clinical and Translational Science Grant (UL1RR024143 to J-L.C). L.S. is a Hirschl Scholar.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
SUPPLEMENTARY INFORMATION
7 Figures, 5 Tables, and Supplementary Experimental Procedures.
REFERENCES
- Adzhubei IA, Schmidt S, Peshkin L, Ramensky VE, Gerasimova A, Bork P, Kondrashov AS, Sunyaev SR. A method and server for predicting damaging missense mutations. Nat. Methods. 2010;7:248–249. doi: 10.1038/nmeth0410-248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alcaïs A, Quintana-Murci L, Thaler DS, Schurr E, Abel L, Casanova JL. Life-threatening infectious diseases of childhood: single-gene inborn errors of immunity? Ann. NY Acad. Sci. 2010;1214:18–33. doi: 10.1111/j.1749-6632.2010.05834.x. [DOI] [PubMed] [Google Scholar]
- Baldwin HS, Shen HM, Yan HC, DeLisser HM, Chung A, Mickanin C, Trask T, Kirschbaum NE, Newman PJ, Albelda SM, et al. Platelet endothelial cell adhesion molecule-1 (PECAM-1/CD31): alternatively spliced, functionally distinct isoforms expressed during mammalian cardiovascular development. Development. 1994;120:2539–2553. doi: 10.1242/dev.120.9.2539. [DOI] [PubMed] [Google Scholar]
- Bearn P, Persad R, Wilson N, Flanagan J, Williams T. 99mTechnetium-labelled red blood cell scintigraphy as an alternative to angiography in the investigation of gastrointestinal bleeding: clinical experience in a district general hospital. Ann. R. Coll. Surg. Engl. 1992;74:192–199. [PMC free article] [PubMed] [Google Scholar]
- Berthelsen J, Zappavigna V, Ferretti E, Mavilio F, Blasi F. The novel homeoprotein Prep1 modulates Pbx-Hox protein cooperativity. EMBO. J. 1998;17:1434–1445. doi: 10.1093/emboj/17.5.1434. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Biben C, Weber R, Kesteven S, Stanley E, McDonald L, Elliott DA, Barnett L, Köentgen F, Robb L, Feneley M, et al. Cardiac septal and valvular dysmorphogenesis in mice heterozygous for mutations in the homeobox gene Nkx2-5. Circ. Res. 2000;87:888–895. doi: 10.1161/01.res.87.10.888. [DOI] [PubMed] [Google Scholar]
- Bjerke GA, Hyman-Walsh C, Wotton D. Cooperative transcriptional activation by Klf4, Meis2 and Pbx1. Mol. Cell Biol. 2011;31:3723–3733. doi: 10.1128/MCB.01456-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bolze A, Byun M, McDonald D, Morgan NV, Abhyankar A, Premkumar L, Puel A, Bacon CM, Rieux-Laucat F, Pang K, et al. Whole-exome-sequencing-based discovery of human FADD deficiency. Am. J. Hum. Genet. 2010;87:873–881. doi: 10.1016/j.ajhg.2010.10.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brendolan A, Ferretti E, Salsi V, Moses K, Quaggin S, Blasi F, Cleary ML, Selleri L. A Pbx1-dependent genetic and transcriptional network regulates spleen ontogeny. Development. 2005;132:3113–3126. doi: 10.1242/dev.01884. [DOI] [PubMed] [Google Scholar]
- Brendolan A, Rosado MM, Carsetti R, Selleri L, Dear TN. Development and function of the mammalian spleen. Bioessays. 2007;29:166–177. doi: 10.1002/bies.20528. [DOI] [PubMed] [Google Scholar]
- Burn SF, Boot MJ, de Angelis C, Doohan R, Arques CG, Torres M, Hill RE. The dynamics of spleen morphogenesis. Dev. Biol. 2008;318:303–311. doi: 10.1016/j.ydbio.2008.03.031. [DOI] [PubMed] [Google Scholar]
- Byun M, Abhyankar A, Lelarge V, Plancoulaine S, Palanduz A, Telhan L, Boisson B, Picard C, Dewell S, Zhao C, et al. Whole-exome sequencing-based discovery of STIM1 deficiency in a child with fatal classic Kaposi sarcoma. J. Exp. Med. 2010;207:2307–2312. doi: 10.1084/jem.20101597. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Capellini TD, Di Giacomo G, Salsi V, Brendolan A, Ferretti E, Srivastava D, Zappavigna V, Selleri L. Pbx1/Pbx2 requirement for distal limb patterning is mediated by the hierarchical control of Hox gene spatial distribution and Shh expression. Development. 2006;133:2263–2273. doi: 10.1242/dev.02395. [DOI] [PubMed] [Google Scholar]
- Elliott DA, Solloway MJ, Wise N, Biben C, Costa MW, Furtado MB, Lange M, Dunwoodie S, Harvey RP. A tyrosine-rich domain within homeodomain transcription factor Nkx2-5 is an essential element in the early cardiac transcriptional regulatory machinery. Development. 2006;133:1311–1322. doi: 10.1242/dev.02305. [DOI] [PubMed] [Google Scholar]
- Ferretti E, Li B, Zewdu R, Wells V, Hebert JM, Karner C, Anderson MJ, Williams T, Dixon J, Dixon MJ, et al. A conserved Pbx-Wnt-p63-Irf6 regulatory module controls face morphogenesis by promoting epithelial apoptosis. Dev. Cell. 2011;21 doi: 10.1016/j.devcel.2011.08.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harvey RP, Lai D, Elliott D, Biben C, Solloway M, Prall O, Stennard F, Schindeler A, Groves N, Lavulo L, et al. Homeodomain factor Nkx2-5 in heart development and disease. Cold Spring Harb. Symp. Quant. Biol. 2002;67:107–114. doi: 10.1101/sqb.2002.67.107. [DOI] [PubMed] [Google Scholar]
- Hecksher-Sørensen J, Watson RP, Lettice LA, Serup P, Eley L, De Angelis C, Ahlgren U, Hill RE. The splanchnic mesodermal plate directs spleen and pancreatic laterality, and is regulated by Bapx1/Nkx3.2. Development. 2004;131:4665–4675. doi: 10.1242/dev.01364. [DOI] [PubMed] [Google Scholar]
- Herzer U, Crocoll A, Barton D, Howells N, Englert C. The Wilms tumor suppressor gene wt1 is required for development of the spleen. Curr. Biol. 1999;9:837–840. doi: 10.1016/s0960-9822(99)80369-8. [DOI] [PubMed] [Google Scholar]
- Hooper M, Hardy K, Handyside A, Hunter S, Monk M. HPRT-deficient (Lesch-Nyhan) mouse embryos derived from germline colonization by cultured cells. Nature. 1987;326:292–295. doi: 10.1038/326292a0. [DOI] [PubMed] [Google Scholar]
- Kanzler B, Dear TN. Hox11 acts cell autonomously in spleen development and its absence results in altered cell fate of mesenchymal spleen precursors. Dev. Biol. 2001;234:231–243. doi: 10.1006/dbio.2001.0239. [DOI] [PubMed] [Google Scholar]
- Kawabe T, Muslin AJ, Korsmeyer SJ. HOX11 interacts with protein phosphatases PP2A and PP1 and disrupts a G2/M cell-cycle checkpoint. Nature. 1997;385:454–458. doi: 10.1038/385454a0. [DOI] [PubMed] [Google Scholar]
- Kim SK, Selleri L, Lee JS, Zhang AY, Gu X, Jacobs Y, Cleary ML. Pbx1 inactivation disrupts pancreas development and in Ipf1-deficient mice promotes diabetes mellitus. Nat. Genet. 2002;30:430–435. doi: 10.1038/ng860. [DOI] [PubMed] [Google Scholar]
- Latres E, Malumbres M, Sotillo R, Martín J, Ortega S, Martín-Caballero J, Flores JM, Cordón-Cardo C, Barbacid M. Limited overlapping roles of P15(INK4b) and P18(INK4c) cell cycle inhibitors in proliferation and tumorigenesis. EMBO. J. 2000;19:3496–3506. doi: 10.1093/emboj/19.13.3496. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lettice LA, Purdie LA, Carlson GJ, Kilanowski F, Dorin J, Hill RE. The mouse bagpipe gene controls development of axial skeleton, skull, and spleen. Proc. Natl. Acad. Sci. USA. 1999;96:9695–9700. doi: 10.1073/pnas.96.17.9695. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lewandoski M, Martin GR. Cre-mediated chromosome loss in mice. Nat. Genet. 1997;17:223–225. doi: 10.1038/ng1097-223. [DOI] [PubMed] [Google Scholar]
- Li H, Durbin R. Fast and accurate short read alignment with Burrows-Wheeler transform. Bioinformatics. 2009;25:1754–1760. doi: 10.1093/bioinformatics/btp324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li JM, Nichols MA, Chandrasekharan S, Xiong Y, Wang XF. Transforming growth factor beta activates the promoter of cyclin-dependent kinase inhibitor p15INK4B through an Sp1 consensus site. J. Biol. Chem. 1995;270:26750–26753. doi: 10.1074/jbc.270.45.26750. [DOI] [PubMed] [Google Scholar]
- Lindor NM, Smithson WA, Ahumada CA, Michels VV, Opitz JM. Asplenia in two father-son pairs. Am. J. Med. Genet. 1995;56:10–11. doi: 10.1002/ajmg.1320560104. [DOI] [PubMed] [Google Scholar]
- Liu L, Okada S, Kong XF, Kreins AY, Cypowyj S, Abhyankar A, Toubiana J, Itan Y, Audry M, Nitschke P, et al. Gain-of-function human STAT1 mutations impair IL-17 immunity and underlie chronic mucocutaneous candidiasis. J. Exp. Med. 2011;208:1635–1648. doi: 10.1084/jem.20110958. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lu J, Chang P, Richardson JA, Gan L, Weiler H, Olson EN. The basic helix-loop-helix transcription factor capsulin controls spleen organogenesis. Proc. Natl. Acad. Sci. USA. 2000;97:9525–9530. doi: 10.1073/pnas.97.17.9525. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lyons I, Parsons LM, Hartley L, Li R, Andrews JE, Robb L, Harvey RP. Myogenic and morphogenetic defects in the heart tubes of murine embryos lacking the homeo box gene Nkx2-5. Genes Dev. 1995;9:1654–1666. doi: 10.1101/gad.9.13.1654. [DOI] [PubMed] [Google Scholar]
- Mahlaoui N, Minard-Colin V, Picard C, Bolze A, Ku CL, Tournilhac O, Gilbert-Dussardier B, Pautard B, Durand P, Devictor D, et al. Isolated congenital asplenia: a French nationwide retrospective survey of 20 cases. J. Pediatr. 2011;158:142–148. doi: 10.1016/j.jpeds.2010.07.027. [DOI] [PubMed] [Google Scholar]
- McKenna A, Hanna M, Banks E, Sivachenko A, Cibulskis K, Kernytsky A, Garimella K, Altshuler D, Gabriel S, Daly M, et al. The Genome Analysis Toolkit: a MapReduce framework for analyzing next-generation DNA sequencing data. Genome Res. 2010;20:1297–1303. doi: 10.1101/gr.107524.110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miller JP, Yeh N, Vidal A, Koff A. Interweaving the cell cycle machinery with cell differentiation. Cell Cycle. 2007;6:2932–2938. doi: 10.4161/cc.6.23.5042. [DOI] [PubMed] [Google Scholar]
- Moens CB, Selleri L. Hox cofactors in vertebrate development. Dev. Biol. 2006;291:193–206. doi: 10.1016/j.ydbio.2005.10.032. [DOI] [PubMed] [Google Scholar]
- Moore KM, Persaud TVN. The Developing Human: Clinically Oriented Embryology. Philadelphia: Saunders Elsevier; 2007. p. 223. [Google Scholar]
- Pabst O, Zweigerdt R, Arnold HH. Targeted disruption of the homeobox transcription factor Nkx2-3 in mice results in postnatal lethality and abnormal development of small intestine and spleen. Development. 1999;126:2215–2225. doi: 10.1242/dev.126.10.2215. [DOI] [PubMed] [Google Scholar]
- Patterson KD, Drysdale TA, Krieg PA. Embryonic origins of spleen asymmetry. Development. 2000;127:167–175. doi: 10.1242/dev.127.1.167. [DOI] [PubMed] [Google Scholar]
- Porembka M, Doyle M, Chapman W. Disorders of the spleen. In: Greer JP, Foerster J, Rodgers GM, Paraskevas F, Glader B, Arber DA, Means RT Jr., editors. Wintrobe's Clinical Hematology. Philadelphia: Wolters Kluwer Health/Lippincott Williams & Wilkins; 2008. pp. 1637–1654. [Google Scholar]
- Prall OW, Menon MK, Solloway MJ, Watanabe Y, Zaffran S, Bajolle F, Biben C, McBride JJ, Robertson BR, Chaulet H, et al. An Nkx2-5/Bmp2/Smad1 negative feedback loop controls heart progenitor specification and proliferation. Cell. 2007;128:947–959. doi: 10.1016/j.cell.2007.01.042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reecy JM, Li X, Yamada M, DeMayo FJ, Newman CS, Harvey RP, Schwartz RJ. Identification of upstream regulatory regions in the heart-expressed homeobox gene Nkx2-5. Development. 1999;126:839–849. doi: 10.1242/dev.126.4.839. [DOI] [PubMed] [Google Scholar]
- Reynisdottir I, Massague J. The subcellular locations of p15(Ink4b) and p27(Kip1) coordinate their inhibitory interactions with cdk4 and cdk2. Genes Dev. 1997;11:492–503. doi: 10.1101/gad.11.4.492. [DOI] [PubMed] [Google Scholar]
- Roberts CW, Shutter JR, Korsmeyer SJ. Hox11 controls the genesis of the spleen. Nature. 1994;368:747–749. doi: 10.1038/368747a0. [DOI] [PubMed] [Google Scholar]
- Sasaki K, Matsumura G. Spleen lymphocytes and haemopoiesis in the mouse embryo. J Anat. 1988;160:27–37. [PMC free article] [PubMed] [Google Scholar]
- Selleri L, Depew MJ, Jacobs Y, Chanda SK, Tsang KY, Cheah KS, Rubenstein JL, O'Gorman S, Cleary ML. Requirement for Pbx1 in skeletal patterning and programming chondrocyte proliferation and differentiation. Development. 2001;128:3543–3557. doi: 10.1242/dev.128.18.3543. [DOI] [PubMed] [Google Scholar]
- Sock E, Rettig SD, Enderich J, Bösl MR, Tamm ER, Wegner M. Gene targeting reveals a widespread role for the high-mobility-group transcription factor Sox11 in tissue remodeling. Mol. Cell Biol. 2004;24:6635–6644. doi: 10.1128/MCB.24.15.6635-6644.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Soriano P. Generalized lacZ expression with the ROSA26 Cre reporter strain. Nat. Genet. 1999;21:70–71. doi: 10.1038/5007. [DOI] [PubMed] [Google Scholar]
- Staller P, Peukert K, Kiermaier A, Seoane J, Lukas J, Karsunky H, Möröy T, Bartek J, Massagué J, Hänel F, et al. Repression of p15INK4b expression by Myc through association with Miz-1. Nat. Cell Biol. 2001;3:392–399. doi: 10.1038/35070076. [DOI] [PubMed] [Google Scholar]
- Stanley EG, Biben C, Elefanty A, Barnett L, Koentgen F, Robb L, Harvey RP. Efficient Cre-mediated deletion in cardiac progenitor cells conferred by a 3'UTR-ires-Cre allele of the homeobox gene Nkx2-5. Int. J. Dev. Biol. 2002;46:431–439. [PubMed] [Google Scholar]
- Todaro GJ, Green H. Quantitative studies of the growth of mouse embryo cells in culture and their development into established lines. J. Cell Biol. 1963;17:299–313. doi: 10.1083/jcb.17.2.299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vannucchi AM, Paoletti F, Linari S, Cellai C, Caporale R, Ferrini PR, Sanchez M, Migliaccio G, Migliaccio AR. Identification and characterization of a bipotent (erythroid and megakaryocytic) cell precursor from the spleen of phenylhydrazine-treated mice. Blood. 2000;95:2559–2568. [PubMed] [Google Scholar]
- Wilm B, Ipenberg A, Hastie ND, Burch JB, Bader DM. The serosal mesothelium is a major source of smooth muscle cells of the gut vasculature. Development. 2005;132:5317–5328. doi: 10.1242/dev.02141. [DOI] [PubMed] [Google Scholar]
- Zhu L, Belmont JW, Ware SM. Genetics of human heterotaxias. Eur. J. Hum. Genet. 2006;14:17–25. doi: 10.1038/sj.ejhg.5201506. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.