Abstract
Of 368 acute otitis media (AOM) cases among 7-valent pneumococcal conjugate (PCV-7) vaccinated children, 43.5% were colonized by multiple otopathogens in the nasopharynx but only 7.1% experienced polymicrobial AOM. When co-colonization occurred, Haemophilus influenzae predominated over all Streptococcus pneumonia strains except 19A strains to cause AOM. Haemophilus influenzae and Streptococcus pneumonia both predominated over Moraxella catarrhalis to cause AOM.
Keywords: Streptococcus pneumoniae (Spn), Haemophilus influenzae (Hflu), Moraxella catarrhalis (Mcat), polymicrobial colonisation, acute otitis media
Introduction
The prevalence of Streptococcus pneumoniae (Spn) as a bacterial pathogen decreased following the introduction of the 7-valent pneumococcal conjugate vaccine (PCV7); however, the relative increase in proportion of non-PCV7 serotypes following PCV7 introduction soon resulted in non-PCV7 serotypes, mainly serotype 19A, becoming a major otopathogen (1). A serotype 19A strain of Spn was isolated from children in Rochester NY that was resistant to all 18 antibiotics approved for the acute otitis media (AOM) treatment indication in children (2).
Spn, Haemophilus influenzae (Hflu), and Moraxella catarrhalis (Mcat) are the most common bacterial pathogens associated with AOM in the US. The initial step of AOM pathogenesis is nasopharyngeal (NP) colonization by otopathogenic bacteria. Although otopathogenic bacteria often coexist in the NP, knowledge is limited regarding which otopathogens are more likely to predominate in causing AOM when multiple otopathogens colonize the NP. We hypothesized that polymicrobial otopathogen NP colonization generally results in a predominant species progressing from the NP via the eustachian tube to the middle ear. In this study we sought to determine which otopathogens successfully emerged from polymicrobial NP colonization and caused AOM in PCV7-vaccined children. Based on our previous work describing the success of Spn expressing serotype 19A as a frequent otopathogen in the PCV7 vaccine era, we especially sought to determine the outcome of polymicrobial NP colonization by 19A strains compared with non-19A strains.
Material and Methods
Study design
This study is a secondary analysis of data collected from June 2006 to end of 2010 from children enrolled in a 5-year prospective study supported by the National Institute on Deafness and Communication Disorders. Healthy children without previous episodes of AOM were enrolled at 6 months of age from a middle class, suburban sociodemographic pediatric practice in Rochester, NY. When AOM was suspected, a tympanocentesis was performed to confirm the diagnosis as previously described (1). At the time of an AOM diagnosis, middle ear fluid (MEF), nasal swab (NS) and oropharyngeal (OP) samples were obtained for bacterial pathogen cultures. Hereafter NP results include those obtained from both NS and OP samples. All of the children received age-appropriate standard vaccinations including the PCV-7 (Prevnar, Wyeth Pharmaceuticals, Collegeville, PA). The demographic characteristics of the children were similar to those described previously (1). This study was approved by the Institutional Review Board of University of Rochester and Rochester General Hospital, and written informed consent was obtained from parents of all children prior to enrollment in the study.
Sample collection and microbiology
Sample collection, the tympanocentesis procedure and microbiology methods were as previously described (1). All Spn isolates were serotyped by Quellung reaction.
Statistics
Statistical significance was determined by repeated measures logistic regression with generalized estimating equations and Fisher's exact test. All p values were two tailed and a p value of < 0.05 was considered significant.
Results
We analyzed NP and MEF samples obtained during 368 AOM visits (including 198 first AOM and 170 recurrent AOM) from 231 children aged 6-30 months. There were no significant differences in culture positive rates of multiple otopathogens according to subject age, gender, or between first and recurrent AOM (data not shown). Overall, the culture positive rate of multiple otopathogens in the NP was 43.5% (160/368), which was significantly higher than the frequency of polymicrobial AOM (7.1%, 26/368, p<0.0001). The NP positive rate of the combination Spn-Mcat (16.0%, 59/368) was significantly higher than Hflu-Mcat (7.1%, 26/368, p=0.002), and Spn-Hflu-Mcat (8.7%, 32/368, p=0.003), but not higher than Spn-Hflu (11.7%, 43/368, p=0.1). The MEF culture positive rate of combinations Spn-Mcat (4.3%, 16/368) was significantly higher than Hflu-Mcat (0.3%, 1/368, p=0.0002), and than Spn-Hflu-Mcat (0%, 0/368, p<0.0001), but not higher than Spn-Hflu (2.4%, 9/368, p=0.2). Each combination had higher positive rates in NP than in MEF (all p values <0.0001).
To investigate which species was more likely to cause AOM when multiple otopathogens were in the NP, we analyzed the culture outcomes of each otopathogen in MEF when multiple otopathogens were present in the NP. Spn isolates with the same serotype in NP and MEF were considered identical. Since there is no serotyping for Hlfu, we used multilocus sequencing typing (MLST) data from the children as previously reported (3). The Hflu isolates with same MLST in NP and MEF were considered identical. Among the 43 AOM cases when both Spn and Hflu were simultaneously detected in the NP, Hflu AOM cases (65.1%) were significantly higher than Spn AOM cases (25.6%, p=0.0005 Figure 1A). Among 59 AOM cases when both Spn and Mcat were simultaneously detected in the NP, Spn AOM (64.4%) were significantly higher than Mcat AOM cases (39.0%) (p=0.01, Figure 1B). Among 26 AOM cases when both Hflu and Mcat were simultaneously detected in the NP, Hflu AOM cases (69.2%) were significantly higher than Mcat cases (23.1%) (p=0.002, Fig.1C). Among the 32 AOM cases that were culture positive for all three otopathogens in the NP, 50.0% were culture positive in MEF for Hflu, 28.1% for Spn, and 3.1% for Mcat (Fig.1D).
Figure 1. The predominant otopathogen to cause AOM when multiple otopathogens were simultaneously present in the NP.
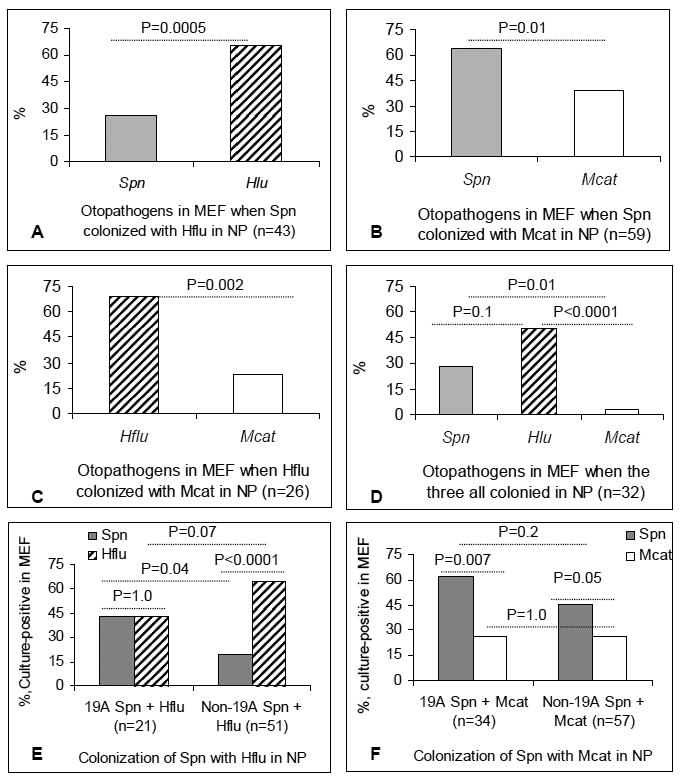
When multiple otopathogens were simultaneously present in the NP at the time of AOM the culture-positive rate in MEF was compared using logistic regression with repeated measures and Fisher's exact test. The percentage of each otopathogen causing AOM when co-colonization occurred involving, A, Spn with Hflu; B, Spn with Mcat; C, Hflu with Mcat; D, all three otopathogens; E, 19A or non-19A strains of Spn with Hflu; B, 19A or non-19A strains of Spn with Mcat. NP: Nasopharynx; MEF: middle ear fluid; Spn, S. pneumonia; Hflu: H. influenzae; Mcat: M.catarrhalis.
To assess whether strains of Spn expressing particular serotypes were more likely to cause AOM, we analyzed the proportion of each individual serotype that was present in MEF when detected in the NP. Overall, we found that 19A was the most frequent serotype, followed by serotype 15 in both the NP and MEF. Among 188 visits that were culture-positive in the NP for Spn, 36.2% expressed the 19A serotype, and 16.5% expressed serotype 15. Among 99 Spn AOM episodes, 43.4% were caused by 19A strains and 16.2% were caused by serotype 15 strains. The subtypes of serotype 15 were not determined. When present in NP, 19A strains (63.2%, 43/68) were more likely than non-19A strains than (46.3%, 57/123) to be present in MEF to cause AOM (p=0.03).
At the time of AOM, when Spn co-colonized with Hflu in the NP, the culture-positive rate of Hflu predominated over non-19A strains (64.7%, 33/51 vs. 19.6%, 10/51, p<0.0001), but not 19A strains (42.9%, 9/21 vs. 42.9%, 9/21, p=1.0);The culture-positive rate of 19A strains (42.9%, 9/21) in MEF was significant higher than that of non-19A strains (19.6%, 10/51, p=0.04, Figure 1E). When Spn co-colonized with Mcat in the NP, both 19A (61.8%, 21/34) and non-19A strains of Spn (45.6%, 26/57) predominated over Mcat (26.5%, 9/34, p=0.007, 26.32%, 15/57, p=0.05, respectively), but there was no difference in the culture-positive rate in MEF between 19A (61.8%, 21/34) and non-19A strains (45.6%, 26/57, p=0.2, Figure 1F).
Discussion
This study investigated the predominant otopathogens causing AOM when multiple otopathogens co-colonize the NP. We found that: (1) culture-positive rates of multiple otopathogens in NP were significantly higher than those in MEF. The combination of Spn-Mcat was the most frequent multiple otopathogen combination in both the NP and MEF; (2) when multiple otopathogens co-colonized the NP, Hflu predominated over Mcat and non-19A but not 19A Spn strains. Hflu and Spn predominated over Mcat to cause AOM; (3) Spn expressing the 19A capsular serotype of Spn colonized the NP more frequently than any other capsular types, and 19A strains were more likely than non-19A strains to cause AOM when they colonized the NP.
There are previous studies involving AOM describing rates of multiple bacterial pathogens in MEF (4, 5), however the results are inconsistent probably due to differences in population dynamics, PCV-7 vaccine use, frequency of viral upper respiratory tract infections, antibiotics use, otopathogen culture or detection methods, study designs, diagnostic method s (tympanostomy or otorrhea) and others. The rate in this study (7.1% of AOM cases) is consistent with other prior studies in the US (6).
Current knowledge is limited regarding which otopathogens have a greater capacity to cause AOM when multiple bacterial otopathogens co-colonize the NP. Similar to our results, Syrijanen et al (7) found that if both Spn and Hflu were in the NP, Hflu was more likely cultured from MEF. However, we also found that the predominance of Hflu over Spn was dependent on the capsular serotype. While Hflu predominated over Spn expressing non-19A serotypes, this wan not the case for 19A Spn stains. The implications of these findings may be that elimination of Spn strains expressing the 19A serotype by vaccination with PCV-13 results in an increased predominance of Hflu as an otopathogen. Since we found that both Hflu and Spn appear to predominate over Mcat when co-colonization occurs it seems less likely that Mcat will emerge as a more important otopathogen in the near future.
Interactions among bacterial optothogens may influence species persistence in the NP. Negative associations have been observed between otopathogens in previous report(8). Host innate immune responses during concurrent colonization by otopathogens may play an important role in the outcome of interspecies interactions at mucosal surfaces (9). PCV-7 vaccination changes the environment in the NP by eliminating vaccine-strains. That in turn changes the balance between strains, reflecting their absolute fitness, ultimately shifting their abundance (10). Our previous studies have shown that a proportional reduction in NP colonization and AOM caused by Spn following introduction of PCV-7 vaccine was subsequently followed by a gradual and steady resurgence of Spn strains (1), led by serotype 19A capsular organisms (2). In the long-term, disease caused by non PCV-7 strains could partially undermine the impact of the vaccine.
Limitations of this study are that we did not employ quantitative detection methods to evaluate the impact of bacterial loads on culture outcomes, nor did we assess viral-bacterial interactions and impact of antibiotic treatment on culture outcomes.
Acknowledgments
The authors thank Sally Thomas, LPN, CCRC, nurses and staff at Legacy Pediatrics, collaborating pediatricians from Long Pond Pediatrics, Rainbow Pediatrics, Lewis Pediatrics, and the parents who consented to the long and challenging study. We also thank Diana Adlowitz and Jennifer Wills for assistance with pathogen identification and Anthony Almudevar for data analysis.
Funding: This work was supported by NIH, NIDCD RO1 08671 and the Thrasher Research Fund (Salt Lake, UT) award no. 02823-2 and by an investigator- initiated research grant from Wyeth Vaccines, now Pfizer Vaccines (Collegeville, PA) (all to M.E. Pichichero).
Footnotes
The authors declare no conflicts of interest.
References
- 1.Casey JR, Adlowitz DG, Pichichero ME. New patterns in the otopathogens causing acute otitis media six to eight years after introduction of pneumococcal conjugate vaccine. Pediatr Infect Dis J. 2010;29(4):304–9. doi: 10.1097/INF.0b013e3181c1bc48. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Pichichero ME, Casey JR. Emergence of a multiresistant serotype 19A pneumococcal strain not included in the 7-valent conjugate vaccine as an otopathogen in children. Jama. 2007;298(15):1772–8. doi: 10.1001/jama.298.15.1772. [DOI] [PubMed] [Google Scholar]
- 3.Kaur R, Chang A, Xu Q, Casey JR, Pichichero ME. Phylogenetic relatedness and diversity of non-typable Haemophilus influenzae in the nasopharynx and middle ear fluid of children with acute otitis media. J Med Microbiol. 2011;60(Pt 12):1841–8. doi: 10.1099/jmm.0.034041-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Ruohola A, Meurman O, Nikkari S, Skottman T, Heikkinen T, Ruuskanen O. The dynamics of bacteria in the middle ear during the course of acute otitis media with tympanostomy tube otorrhea. Pediatr Infect Dis J. 2007;26(10):892–6. doi: 10.1097/INF.0b013e31812e4b6c. [DOI] [PubMed] [Google Scholar]
- 5.Kilpi T, Herva E, Kaijalainen T, Syrjanen R, Takala AK. Bacteriology of acute otitis media in a cohort of Finnish children followed for the first two years of life. Pediatr Infect Dis J. 2001;20(7):654–62. doi: 10.1097/00006454-200107000-00004. [DOI] [PubMed] [Google Scholar]
- 6.Pichichero ME, Casey JR, Hoberman A, Schwartz R. Pathogens causing recurrent and difficult-to-treat acute otitis media” 2003-2006. Clin Pediatr. 2008;47:901–906. doi: 10.1177/0009922808319966. [DOI] [PubMed] [Google Scholar]
- 7.Syrjanen RK, Herva EE, Makela PH, Puhakka HJ, Auranen KJ, Takala AK, et al. The value of nasopharyngeal culture in predicting the etiology of acute otitis media in children less than two years of age. Pediatr Infect Dis J. 2006;25(11):1032–6. doi: 10.1097/01.inf.0000241097.37428.1d. [DOI] [PubMed] [Google Scholar]
- 8.Pettigrew MM, Gent JF, Revai K, Patel JA, Chonmaitree T. Microbial interactions during upper respiratory tract infections. Emerg Infect Dis. 2008;14(10):1584–91. doi: 10.3201/eid1410.080119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Lysenko ES, Ratner AJ, Nelson AL, Weiser JN. The role of innate immune responses in the outcome of interspecies competition for colonization of mucosal surfaces. PLoS Pathog. 2005;1(1):e1. doi: 10.1371/journal.ppat.0010001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Martcheva M, Bolker BM, Holt RD. Vaccine-induced pathogen strain replacement: what are the mechanisms? J R Soc Interface. 2008;5(18):3–13. doi: 10.1098/rsif.2007.0236. [DOI] [PMC free article] [PubMed] [Google Scholar]


