Abstract
Background
Lymph node metastasis is an important indicator of oncologic outcome for patients with rectal cancer. Identifying predictive biomarkers of lymph node metastasis could therefore be clinically useful.
Objective
To assess whether chromosomal copy number alterations can assist in predicting lymph node metastasis in patients with locally advanced rectal cancer treated with pre-operative chemoradiation therapy.
Design
Non-randomized, prospective Phase II study.
Setting
Multi-institutional.
Patients
95 patients with stage II (cT3-4, cN0) or stage III (any cT, cN1-2) rectal cancer.
Intervention
Patients were treated with pre-operative chemoradiation therapy (CRT) followed by total mesorectal excision. Pretreatment biopsy tumor DNA and surgical margin control DNA was extracted and analyzed by oligonucleotide array-based comparative genomic hybridization. Chromosomal copy number alterations were correlated with lymph node metastasis. Finally, a model for predicting lymph node metastasis was built.
Main outcome measures
To determine if chromosomal copy number alterations are associated with lymph node metastasis in patients with rectal cancer, and to assess the accuracy of oligonucleotide array-based comparative genomic hybridization for predicting lymph node metastasis.
Results
Twenty-five of 95 (26%) patients had lymph node metastasis after chemoradiation. Losses of 28 chromosomal regions, most notably in chromosome 4, were significantly associated with lymph node metastasis. Our predictive model contained 65 probes and predicted lymph node metastasis with 68% sensitivity, 93% specificity, and positive and negative predictive values of 77% and 89%. Using this model lymph node status (positive or negative) after CRT was predicted accurately in 82 out of 95 patients (86%).
Limitations
The patient cohort was not completely homogeneous which may have influenced their clinical outcome. Additionally, while we performed rigorous statistically sound internal validation, external validation will be important to further corroborate our findings.
Conclusions
Copy number alterations can help identify rectal cancer patients at risk of lymph node metastasis after chemoradiation.
Keywords: Rectal cancer, lymph node metastasis, array-based comparative genomic hybridization (aCGH), copy number alteration (CNA)
Introduction
Preoperative chemoradiation therapy (CRT) followed by total mesorectal excision (TME) is the current standard of care for locally advanced rectal cancer.1 This treatment provides excellent local tumor control and long-term survival.2, 3 However, TME is a formidable operation associated with some mortality, significant morbidity, and long-lasting sequelae that permanently impair quality of life.4, 5 Patient response to CRT is also variable. While a proportion of rectal cancer patients treated with CRT have no detectable cancer cells in the bowel wall or mesorectal lymph nodes and achieve a pathologic complete response (pCR), other patients have only a partial response and have persistent cancer cells in their surgical specimens.
It is well-established that patients with a pCR have better oncologic outcomes compared to non-pCR patients, with lower local recurrence, less distant metastasis and improved overall survival.2, 3 However, there is currently no accurate way to predict the presence of residual cancer cells in the mesorectal lymph nodes before surgery.6 Histopathological analysis following TME remains the most reliable way to identify patients with tumor deposits in the mesorectal lymph nodes at risk of metastatic disease.
The human genome is a patchwork of DNA segments with a high degree of variability in copy number, order, and orientation between individuals and populations. Studies have shown that of the various structural variations that occur, the physical gain or loss of DNA segments from chromosomes, known as copy number alterations (CNAs), is a common occurrence in many genetic diseases,7 and many studies have examined the role that these DNA additions (gains) or deletions (losses) play in disease.8 The presence of DNA CNAs is also a cardinal feature of cancer, characterized by the gain and loss of entire chromosomes or chromosomal segments (known as aneuploidy). This includes colorectal cancer.9, 10 Gains and losses of chromosomal segments lead to changes of gene expression in oncogenes and tumor suppressor genes that are important for the progression of colorectal cancer.
The hybridization of tumor DNA to normal metaphase chromosomes, a technique known as comparative genomic hybridization (CGH), allows for the simultaneous screening and mapping of gains and losses of specific chromosomal segments.11, 12 New high-throughput approaches, such as array-based comparative genomic hybridization (aCGH) using oligonucleotides allow genome-wide detection of chromosomal CNAs at a higher resolution compared to metaphase-based CGH. Oligonucleotide aCGH is therefore one of the preferred methods to characterize the degree of chromosomal complexity in cancer.
Emerging evidence suggests that specific CNAs are associated with distant metastasis in advanced colorectal cancer,12–14 but no studies have examined CNAs and persistent lymph node metastasis specifically in rectal cancer. Given that lymph node metastasis is a critical indicator of clinical outcome, it would be clinically useful if high-risk patients could be accurately identified before surgery as this may help direct therapy.
We used high density whole genome oligonucleotide aCGH to identify CNAs in patients with locally advanced rectal cancer to determine whether a specific CNA profile associates with persistent lymph node metastasis. Here, we present the CNA profiles for 95 rectal cancer patients treated with CRT and TME, compare CNAs between lymph node positive and negative patients, and describe the value of this molecular approach for predicting persistent lymph node metastasis in patients with rectal cancer.
Materials and Methods
Patients
This study included patients with clinical stage II (cT3-4, cN0) or stage III (any cT, cN1-2) invasive adenocarcinoma of the rectum with a distal tumor border within 12 cm of the anal verge, as measured on rigid proctoscopic exam, who were enrolled in the Timing of Rectal Cancer Response to Chemoradiation study, a multi-institutional clinical trial investigating the effect of increasing the CRT-to-surgery interval, and adding chemotherapy, modified FOLFOX-6 (mFOLFOX-6) during the waiting period (ClinicalTrials.org Identifier: NCT00335816). This trial was designed in a series of sequential Phase II trials or study groups (SGs), each with a progressively longer CRT-to-surgery interval and increasing cycles of pre-operative mFOLFOX-6. This study was approved by an Institutional Review Board (IRB) at each participating institution as well as a central IRB, and informed written consent was obtained from each patient prior to enrollment in the trial. Patients included in the present study were pooled from SG1 (n=43) and SG2 (n=52). Further details of patient eligibility for this trial are presented elsewhere.15
Treatment protocol
Patients in both groups were treated with CRT; 5-Fluorouracil (FU) and a total of 50.4Gy of radiation as described previously.15 Patients in SG1 underwent TME an average of 6 weeks after completing CRT (standard of care). Following CRT, patients in SG2 with no evidence of stable disease received 2 cycles of additional chemotherapy (mFOLFOX-6) as described previously.15 Patients in SG2 underwent TME an average of 11 weeks after completing CRT. The clinical outcomes for these patients are presented elsewhere.15
Tumor response and lymph node status
Patients were assessed for tumor response (pCR versus non-pCR) and lymph node status (ypN1-2 versus ypN0) after TME. Pathologic complete response was defined as the complete absence of tumor cells from the bowel wall and mesorectal lymph nodes. An average of 14 lymph nodes was collected from each patient. Tumor response and lymph node status were evaluated by two independent pathologists according to the recommendations of the American Joint Committee of Cancer (AJCC).16
Sample preparation, whole genome amplification, and oligonucleotide aCGH
Tumor DNA from each patient was obtained from pretreatment biopsy tissue and control DNA was obtained following treatment from paired normal surgical tissue from the proximal resection margin. To extract DNA, 10-20 slides per patient sample from formalin-fixed paraffin-embedded (FFPE) tumor biopsy and normal tissue were de-paraffinized, hydrated, and stained with 0.2% methylene blue. A 27.5 gauge needle was then used to manually micro-dissect cells under inverted microscopy. This method ensured that the purity of the tumor cells in each extracted DNA specimen was at least 85%. DNA was quantified by measuring absorbance, and 100–200ng of DNA was amplified using the GenomePlex Complete Whole Genome Amplification (WGA)-2 kit (Sigma Cor., Cream Ridge, NJ). WGA-DNA was purified with the GenElute PCR CleanUp kit (Sigma Cor., Cream Ridge, NJ) and quantified. DNA quality was determined by running 2μl of WGA-DNA on a 2% agarose gel. If the size of any DNA amplicon was less than 200 base pairs in size (a marker of poor quality), the sample was removed from the study because it was deemed unsuitable for aCGH. Only samples >200 base pairs in size were included in the current study.
The Agilent microarray platform was used for oligonucleotide aCGH (Human Genomic CGH 244A Microarrays), with 8.9kb overall median probe-spacing covering more than 236,000 coding and non-coding human DNA sequences. aCGH assays were conducted according to manufacturer’s instructions (Agilent Cor., Santa Clara, CA). Briefly, for each sample, 2μg of WGA-DNA was labeled with the non-enzymatic Universal Linkage System (ULS). Equal amounts of tumor biopsy and paired normal surgical specimen DNA were labeled with ULS-Cy5 and ULS-Cy3, respectively. The labeled samples were purified using Agilent-KREApure columns, and then combined with the hybridization mixture in a SureHyb chamber. Hybridization of arrays was carried out at 65°C for 40 hours. Arrays were then washed in Wash Buffer-1 and Wash Buffer-2.
Scanning and image analysis were performed on an Agilent scanner. Agilent Feature Extraction Software (v.9.5) was used for data extraction from raw microarray image files.
Statistical analysis
Patient characteristics
To determine differences in clinicopathological features between cN1-2 and cN0 patients, and between ypN1-2 and ypN0 patients, Student’s t test was used to compare means of continuous variables, and the two-sided Fisher’s exact test or the Fisher-Freeman-Halton exact test using Monte Carlo was chosen for categorical variables.
Characterizing chromosomal CNAs
Nexus copy-number software (v.5.0) (BioDiscovery Inc., El Segundo, CA), was used to identify chromosomal CNAs using the rank segmentation algorithm, a modified version of the circular binary segmentation (CBS) algorithm, as described previously.17 Copy-number alterations were determined for each sample, and the fraction of genome alteration (FGA) was calculated to reflect the degree of genomic instability. The FGA was determined by dividing the overall altered segment size by the genome size using the NCBI hg18 database (Build 36.1) comprising 3,080,436,051 base-pairs.
Identifying CNA signatures
Unsupervised hierarchical cluster analysis was used to analyze the distribution of whole-genome CNA profiles. The CNAs that differed between cN1-2 and cN0 patients, and between ypN1-2 and ypN0 patients were identified using Nexus copy-number software (v.5.0) (BioDiscovery Inc., El Segundo, CA) (p <0.05, differential threshold >25%). To assess CNA differences between cN1-2 and cN0 patients, and between ypN1-2 and ypN0 patients, the two-sided Fisher’s exact test was used and Q-bound was utilized to correct for multiple testing by performing false discovery rate (FDR) analysis, defined as the proportion of false positives among all positives.18, 19 A Q-bound score of <0.05 was considered statistically significant to minimize the false negative rate.19
Identification of functionally relevant genes in ypN1-2 and ypN0 patients
The genes within CNA regions that differed between ypN1-2 and ypN0 patients were identified using Fisher’s Exact test (p-value <0.05, differential threshold >25%), and potential gene candidates for human cancer were prioritized among the genes relevant to persistent lymph node metastasis using Ingenuity Pathway Analysis (IPA) biomarker filter analysis.
Predicting persistent lymph node metastases after neoadjuvant CRT
Predictive biomarkers for persistent lymph node metastasis were derived using a combination of methods including feature (candidate biomarker) selection, classification model-fitting, and cross-validation, as described previously.20, 21 Differential features were selected from all probes on the aCGH array based on two criteria: FDR - the adjusted p-values using the Benjamini and Hochberg method18 based on the p-values of the Student’s t test between ypN1-2 and ypN0 patients, and probe-signal fold-change between ypN1-2 and ypN0 patients. The features were fed into the linear-kernel Support Vector Machine (SVM) classifier to train the classification model based on the selected features.22 Leave-one-out cross-validation was performed to evaluate the classification performance on test samples. ROCR package23 was used to calculate performance measures such as sensitivity, specificity, positive and negative predictive values, and to plot the performance measures using the receiver operating characteristic (ROC) curve. The area under the ROC curve (AUC) was calculated to quantitatively summarize the performance of the model. The final predictive model contained 65 features using the following feature selection cutoffs: FDR threshold ≤0.15; Absolute log2 fold-change ≥0.55.
Results
Patient characteristics, lymph node status, and tumor response
The clinical and pathological characteristics of the 95 patients are shown overall and stratified by pretreatment clinical stage (Table 1) and pathologic lymph node status (Table 2). Before treatment, 75% of patients were cN1-2 by endorectal ultrasound (ERUS) or MRI. Clinical stage (cN1-2 versus cN0) was not significantly associated with pretreatment patient demographics, cT-stage, ypT status, ypN status, or the number of retrieved lymph nodes, but was associated with histological tumor grade (Table 1). After CRT, 25 out of 95 (26%) patients were ypN1-2 (Table 2); 11 out of 43 patients (26%) in SG1 and 14 out of 52 patients (27%) in SG2. The total number of lymph nodes examined for the entire patient cohort differed significantly between ypN1-2 and ypN0 patients, with more lymph nodes retrieved in the ypN1-2 group (p=0.006) (Table 2).
Table 1.
Patient characteristics overall and stratified by pretreatment clinical stage
| Characteristic/demographic | Overall n=95 | cN1-2 n=71 | cN0 n=24 | p-value |
|---|---|---|---|---|
| Age, mean ± SD | 56±12 | 56±13 | 57±11 | 0.96 |
| Gender, n (%) | 0.06 | |||
| Male | 54 (57%) | 36 (51%) | 18 (75%) | |
| Female | 41 (43%) | 35 (49%) | 6 (25%) | |
| Histological tumor grade, n (%) | 0.02 * | |||
| Well differentiated | 17 (18%) | 8 (11%) | 9 (38%) | |
| Moderately differentiated | 74 (78%) | 59 (83%) | 15 (62%) | |
| Poorly differentiated | 4 (4%) | 4 (6%) | 0 (0%) | |
| Pretreatment T stage, n (%) | 0.18 | |||
| cT2 | 8 (8%) | 8 (11%) | 0 (0%) | |
| cT3 | 86 (91%) | 62 (88%) | 24 (100%) | |
| cT4 | 1 (1%) | 1 (1%) | 0 (0%) | |
| Study Groupψ | 0.35 | |||
| SG1 | 43 (45%) | 30 (42%) | 13 (54%) | |
| SG2 | 52 (55%) | 41 (58%) | 11 (46%) | |
| Pathologic tumor stage, n (%) | 0.12 | |||
| ypT0 | 29 (31%) | 25 (35%) | 4 (17%) | |
| ypT1-4 | 66 (69%) | 46 (65%) | 20 (83%) | |
| Pathologic lymph node stage, n (%) | 0.29 | |||
| ypN0 | 70 (74%) | 50 (70%) | 20 (83%) | |
| ypN1-2 | 25 (26%) | 21 (30%) | 4 (17%) | |
| Total lymph nodes, range (median) | 2–31 (12) | 2–31 (12) | 2–26 (12) | 0.24 |
Statistically significant (p <0.05).
Patients in SG1 had surgery a mean of 5.7 weeks ±1 week after the completion of chemoradiation; SG2 had surgery a mean of 11 weeks ±2 weeks after the completion of chemoradiation. cN: Pretreatment clinical stage; SD: standard deviation; cT: Pretreatment T-stage; ypT: T-stage after neoadjuvant therapy (y) at histopathological evaluation (p); ypN: Nodal stage after neoadjuvant therapy (y) at histopathological evaluation (p); ypN1-2: pathologic lymph node positive after chemoradiation; ypN0: pathologic lymph node negative after chemoradiation.
Table 2.
Patient characteristics overall and stratified by pathologic lymph node status
| Characteristic/demographic | Overall n=95 | ypN1-2 n=25 | ypN0 n=70 | p-value |
|---|---|---|---|---|
| Age, mean ± SD | 56±12 | 53±10 | 58±12 | 0.08 |
| Gender, n (%) | 0.06 | |||
| Male | 54 (57%) | 10 (40%) | 44 (63%) | |
| Female | 41 (43%) | 15 (60%) | 26 (37%) | |
| Histological tumor grade, n (%) | 0.07 | |||
| Well differentiated | 17 (18%) | 5 (20%) | 12 (17%) | |
| Moderately differentiated | 74 (78%) | 17 (68%) | 57 (81%) | |
| Poorly differentiated | 4 (4%) | 3 (12%) | 1 (1%) | |
| Pretreatment T stage, n (%) | 1.00 | |||
| cT2 | 8 (8%) | 2 (8%) | 6 (9%) | |
| cT3 | 86 (91%) | 23 (92%) | 63 (90%) | |
| cT4 | 1 (1%) | 0 | 1 (1%) | |
| Pretreatment clinical stage, n (%) | 0.59 | |||
| cN0 | 24 (25%) | 5 (20%) | 19 (27%) | |
| cN1-2 | 71 (75%) | 20 (80%) | 51 (73%) | |
| Study Groupψ | 1.00 | |||
| SG1 | 43 (100%) | 11 (26%) | 32 (74%) | |
| SG2 | 52 (100%) | 14 (27%) | 38 (73%) | |
| Pathologic tumor stage, n (%) | 0.009 * | |||
| ypT0 | 27 (28%) | 2 (8%) | 25 (36%) | |
| ypT1-4 | 68 (72%) | 23 (92%) | 45 (64%) | |
| Total lymph nodes, range (median) | 2–31 (12) | 7–31 (16) | 2–30 (11) | 0.006 * |
Statistically significant (p <0.05).
Patients in SG1 had surgery a mean of 5.7 weeks ±1 week after the completion of chemoradiation; SG2 had surgery a mean of 11 weeks ±2 weeks after the completion of chemoradiation. cN: Pretreatment clinical stage; SD: standard deviation; cT: Pretreatment T-stage; ypT: T-stage after neoadjuvant therapy (y) at histopathological evaluation (p); ypN: Nodal stage after neoadjuvant therapy (y) at histopathological evaluation (p); ypN1-2: pathologic lymph node positive after chemoradiation; ypN0: pathologic lymph node negative after chemoradiation.
Twenty seven out of 95 (28%) patients were ypT0 and only 2 of these patients were ypN1-2. The remaining 25 ypT0 patients were ypN0, indicating that persistent lymph node metastasis significantly associates with ypT1-4 after CRT (p=0.009).
Chromosomal CNA gains and losses overall and stratified by pretreatment clinical stage and lymph node status after CRT
We found no difference in copy number gains and losses between cN1-2 and cN0 patients (data not shown). The mean number of copy number gains and losses overall and stratified by lymph node status after CRT are presented in Table 3. Patients who were ypN1-2 had a higher number of total gains and losses, a higher number of single copy gains and losses, a higher number of high copy gains, a higher FGA and a lower number of high copy losses compared to ypN0 patients. These differences showed a strong trend towards statistical significance.
Table 3.
Mean numbers of CNA events
| CNA event | Overall n=95 | ypN1-2 n=25 | ypN0 n=70 | p-value |
|---|---|---|---|---|
| FGA | 0.30±0.16 | 0.35±0.17 | 0.29±0.15 | 0.08 |
| Total gains and losses | 117±103 | 156±152 | 103±75 | 0.10 |
| Single copy gain | 52±66 | 69±66 | 46±33 | 0.10 |
| High copy gain | 91±16 | 14±26 | 8±9 | 0.21 |
| Single copy loss | 55±47 | 72±63 | 49±39 | 0.09 |
| High copy loss | 0.9±2 | 0.6±1 | 1±2 | 0.19 |
CNA: Copy number alteration; FGA: Fraction of genomic alteration; ypN1-2: pathologic lymph node positive after chemoradiation; ypN0: pathologic lymph node negative after chemoradiation. Mean ± standard deviation shown.
Specific chromosomal CNAs overall and stratified by pretreatment clinical stage and lymph node status after CRT
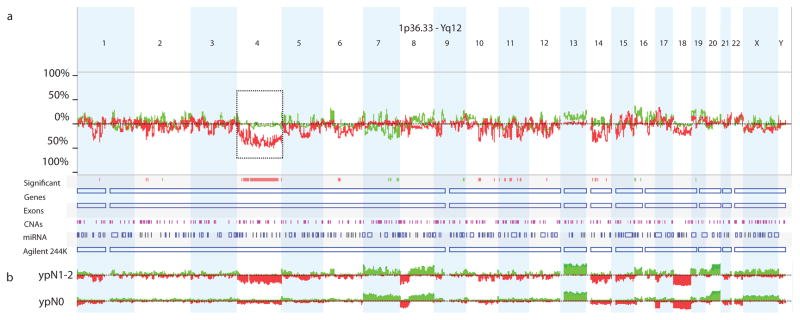
In the entire patient cohort, overall copy number gains most frequently affected chromosomal regions 20q11.21-q13.33 (68%), 13q11-34 (55%), 7p22.3-p22.2 (36%), and 8q23.3-q24.3 (36%), while losses were most frequently observed in chromosomal regions 18q11.32-q23 (60%), 17p13.3-q11.1 (39%), 10q23.1 (38%), and 4q32.1-q32.3 (37%) (Fig. 1A).
Figure 1.
Copy number alterations detected by aCGH (a) overall and (b) stratified by lymph node status (ypN1-2, n=25 versus ypN0, n=70). Loss of chromosome 4 depicted in the dashed box was found to be the most significantly different CNA region between lymph node positive (ypN1-2) and lymph node negative (ypN0) patients. Sample identification - Red: Loss; Green: Gain.
A total of 207 chromosomal regions were found to be different between cN1-2 and cN0 patients, but when the p-value was corrected by multiple testing (FDR), none of the differences reached statistical significance (Supplementary Table 1).
Chromosomal regions associated with persistent lymph node metastasis were identified by comparing the CNA profiles of ypN1-2 and ypN0 patients (Fig. 1B). A total of 1,023 chromosomal regions were found to be different (gained or lost) between ypN1-2 and ypN0 patients with a minimal differential threshold of >25% and a p-value of <0.05 (Supplementary Table 2). When p-values were corrected for multiple testing and a Q-bound of <0.05 was applied, loss of 270 regions remained associated with ypN1-2 disease (Supplementary Table 3). Notably, 248 out of the 270 (92%) differential regions associated with persistent lymph node metastasis were in chromosome 4; and over half of these (135 out of 248 regions [54%] on chromosome 4) were found to be the most significantly different between ypN1-2 and ypN0 patients.
Given the extensive sequence overlap provided by high density arrays, most of the differential regions were close to one another (with an internal distance of <1 mega-base) and were combined to form a number of large consecutive sequences. After combining the consecutive CNAs, loss of 28 chromosomal regions in chromosomes 1, 2, 4, 6, 10, 11, and 14 occurred significantly more frequently in ypN1-2 patients compared to ypN0 patients (Table 4).
Table 4.
CNA regions that differ significantly between ypN1-2 and ypN0 patients*
| Chromosome Loss | Frequency of occurrence | p-value | Q-bound | |
|---|---|---|---|---|
| ypN1-2 n=25 | ypN0 n=70 | |||
| 4q31.3-q32.3 | 68% | 17% | 0.000005 | 0.01 |
| 4q21.1-q21.22 | 64% | 16% | 0.00001 | 0.01 |
| 4q26-q31.3 | 68% | 20% | 0.00003 | 0.01 |
| 4q25-q26 | 76% | 29% | 0.00004 | 0.01 |
| 4q13.3-q21.1 | 52% | 10% | 0.00004 | 0.01 |
| 4q12-q13.2 | 72% | 24% | 0.00005 | 0.01 |
| 4p13-p11 | 64% | 20% | 0.0001 | 0.02 |
| 4q13.2-q13.3 | 56% | 14% | 0.0001 | 0.02 |
| 4q21.23-q23 | 60% | 17% | 0.0002 | 0.02 |
| 4p15.1 | 72% | 29% | 0.0003 | 0.02 |
| 10q25.3 | 56% | 17% | 0.0004 | 0.03 |
| 4q35.2 | 64% | 24% | 0.0005 | 0.03 |
| 6q12 | 36% | 6% | 0.0006 | 0.03 |
| 6q13 | 36% | 6% | 0.0006 | 0.03 |
| 11q21-q22.1 | 52% | 14% | 0.0006 | 0.03 |
| 4q23-q24 | 56% | 19% | 0.0007 | 0.03 |
| 14q21.1 | 72% | 31% | 0.0008 | 0.03 |
| 1p21.3 | 60% | 21% | 0.0008 | 0.03 |
| 11q11 | 44% | 11% | 0.001 | 0.04 |
| 2p16.3 | 44% | 11% | 0.001 | 0.04 |
| 4p15.1-p14 | 64% | 26% | 0.001 | 0.04 |
| 11p13 | 36% | 7% | 0.001 | 0.04 |
| 11q14.1 | 36% | 7% | 0.001 | 0.046 |
| 4q32.3 | 68% | 30% | 0.001 | 0.049 |
| 4q33-q34.1 | 64% | 27% | 0.001 | 0.049 |
| 4q34.1 | 64% | 27% | 0.001 | 0.049 |
| 4p15.32 | 64% | 27% | 0.001 | 0.049 |
| 10q21.1 | 56% | 20% | 0.001 | 0.049 |
A Q-bound score of <0.05 was considered statistically significant. CNA: Copy number alteration; ypN1-2: pathologic lymph node positive after chemoradiation; ypN0: pathologic lymph node negative after chemoradiation.
Genes that differed between ypN1-2 and ypN0 patients
The chromosomal regions that differed between ypN1-2 and ypN0 patients contain 342 genes (Supplementary Table 3). To identify genes that may play a role in persistent lymph node metastasis, we used IPA biomarker filter analysis to search for the genes relevant to persistent lymph node metastasis. We identified 22 out of 342 potential biomarker candidates associated with cancer prognosis, disease progression, therapeutic efficacy, response to therapy, and diagnosis (Table 5). All of these genes were lost more frequently in ypN1-2 patients compared to ypN0 patients.
Table 5.
Candidate biomarkers differentially expressed between ypN1-2 and ypN0 patients
| Symbol | Gene Name | Location | Type | Application |
|---|---|---|---|---|
| ABCG2 | ATP-binding cassette, sub-family G, member 2 | Membrane | Transporter | Diagnosis, Efficacy, Prognosis |
| ANTXR2 | Anthrax toxin receptor 2 | Membrane | Other | Diagnosis |
| ANXA5 | Annexin A5 | Membrane | Other | Diagnosis |
| BMP3 | Bone morphogenetic protein 3 | Extracellular | Growth factor | Diagnosis |
| BMPR1B | Bone morphogenetic protein receptor, type IB | Membrane | Kinase | Unspecified |
| CCNA2 | Cyclin A2 | Nucleus | Other | Efficacy, Prognosis |
| DCK | deoxycytidine kinase | Nucleus | Kinase | Diagnosis, Efficacy, Response |
| FGA | Fibrinogen alpha chain | Extracellular | Other | Diagnosis |
| FGF2 | Fibroblast growth factor 2 | Extracellular | Growth factor | Diagnosis, Prognosis Progression, Efficacy |
| HPGD | Hydroxyprostaglandin dehydrogenase 15-(NAD) | Cytoplasm | Enzyme | Prognosis |
| IBSP | Integrin-binding sialoprotein | Extracellular | Other | Progression, Efficacy |
| IL15 | Interleukin 15 | Extracellular | Cytokine | Diagnosis, Progression, Efficacy |
| IL2 | Interleukin 2 | Extracellular | Cytokine | Diagnosis, Progression, Efficacy |
| PLK4 | Polo-like kinase 4 | Cytoplasm | Kinase | Diagnosis |
| PRSS12 | Protease, serine, 12 | Extracellular | Peptidase | Diagnosis |
| SFRP2 | secreted frizzled-related protein 2 | Membrane | Receptor | Diagnosis |
| SPARCL1 | SPARC-like 1 (hevin) | Extracellular | Other | Progression |
| SPP1 | secreted phosphoprotein 1 | Extracellular | Cytokine | Diagnosis, Unspecified |
| SSTR1 | somatostatin receptor 1 | Membrane | G-protein receptor | Diagnosis, Prognosis |
| UCP1 | uncoupling protein 1 (mitochondrial, proton carrier) | Cytoplasm | Transporter | Efficacy, Unspecified |
| UGT2B17 | UDP glucuronosyltransferase 2 family, polypeptide B17 | Cytoplasm | Enzyme | Diagnosis |
| UGT2B7 | UDP glucuronosyltransferase 2 family, polypeptide B7 | Cytoplasm | Enzyme | Efficacy, Prognosis |
ypN1-2: pathologic lymph node positive after chemoradiation; ypN0: pathologic lymph node negative after chemoradiation.
Predicting persistent lymph node metastases after neoadjuvant CRT
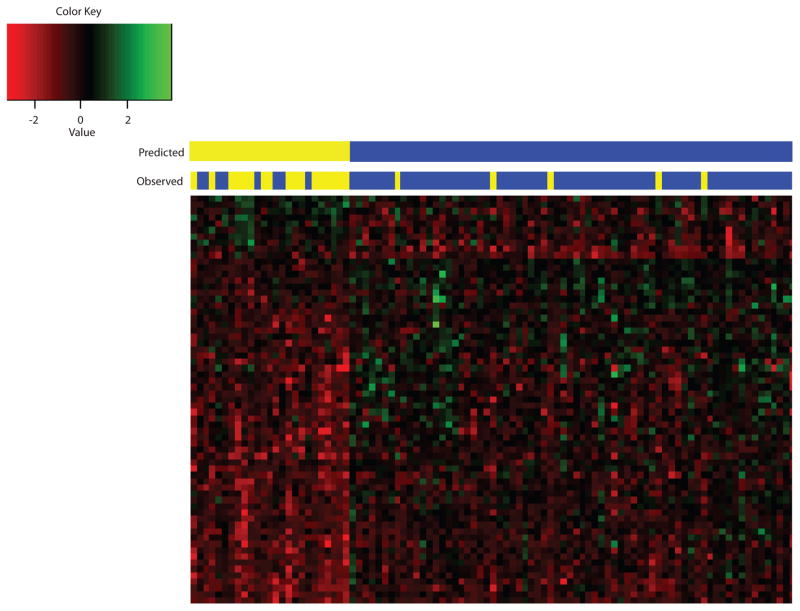
To determine how accurately oligonucleotide aCGH can predict persistent lymph node metastasis in rectal cancer patients, a biomarker model was built using SVM. Sixty-five probes were selected using FDR and fold-change cutoffs (FDR threshold ≤0.15; log2 fold-change ≥0.55) (Supplementary Table 4). Using leave-one-out cross-validation, aCGH predicted persistent lymph node metastasis with a sensitivity of 68%, specificity of 93%, positive predictive value of 77%, and negative predictive value of 89%. Lymph node status (ypN1-2 or ypN0) was predicted accurately in 82 out of 95 patients (86%) with this model (Fig. 2), and the performance of the model was plotted as an ROC curve with an AUC value of 0.91.
Figure 2.
Predictive CNA model for persistent lymph node metastasis. 65 probes were used to build a predictive model using Support Vector Machine (SVM) to predict persistent lymph node metastasis. Sample identification - Red: Loss; Green: Gain. Yellow: Lymph node positive (ypN1-2); Blue: Lymph node negative (ypN0).
Discussion
In our study we showed that CNAs detected by oligonucleotide aCGH may help identify rectal cancer patients likely to have persistent lymph node metastasis following CRT. We found that ypN1-2 patients had more overall copy gains and losses compared to ypN0 patients, and that losses of 28 chromosomal regions, predominantly in chromosomes 4, 10, 11, 14, and 6 occurred significantly more frequently in ypN1-2 patients compared to ypN0 patients. We also found that persistent lymph node metastasis significantly associated with tumor response after CRT, and that patients with ypT0 tumors were significantly less likely to have persistent lymph node metastasis compared with ypT1-4 tumors. Finally, we built a persistent lymph node metastasis prediction model that contained 65 probes and predicted ypN1-2 status with a high degree of accuracy. Conversely, we found no correlation between pretreatment clinical stage and pathologic lymph node stage; and there were no CNA differences between clinical node positive and negative patients. This likely reflects the inaccuracy of current clinical staging methods such as imaging technology in identifying nodal metastasis.
Only one previous study has focused on identifying a specific correlation between CNAs in primary tumors with lymph node metastasis in colorectal cancer.24 In this study, the CNA profiles of 50 colorectal patients were determined, and the authors found that gain of chromosome 8q23-24 may be predictive of lymph node metastasis. Gain of chromosome 8q24.3 also occurred significantly more frequently in ypN1-2 versus ypN0 patients in our study (p<0.02) (Supplementary Table 2). However, when the data was corrected for FDR (Q-bound), this CNA was no longer significant (Q-bound >0.44). Additional independent studies have also suggested that 8q24.3 may play a role in distant metastasis.14 Therefore, gain of this chromosomal region may indeed be an indicator of persistent lymph node metastasis in rectal cancer after CRT, and the possible false negative we observed may be due to our statistical threshold (Q-bound) being set at a more stringent level (<0.05) to correct for multiple testing.
Of the 28 CNAs significantly associated with persistent lymph node metastasis in our study, over half (61%) were located in chromosome 4. Other studies have also shown an association between loss of chromosome 4p or 4q and metastasis in colorectal cancer.25–27 Diep et al. analyzed genome-wide CNAs in 373 colorectal tumors and 102 liver metastases, and showed that loss of chromosome 4p and 4q were significantly associated with disease progression.25 Arribas et al. showed that loss of heterozygosity of 4p14-16 was indicative of a shorter disease-free survival period in 143 colorectal cancer patients.26 Bardi et al. performed cytogenetic analysis of 150 primary colorectal cancers and demonstrated that loss of chromosome 4 was significantly correlated with shorter disease-free survival.27 Our data is largely consistent with previous studies and suggests that loss of chromosome 4 may play an important role in tumor progression, persistent lymph node metastasis and decreased overall survival. Thus, screening for this CNA using aCGH in rectal cancer patients may help identify at-risk patients and direct appropriate treatment.
There is substantial evidence suggesting that genomic alterations can result in corresponding gene expression changes in colorectal cancer, as shown by integrating gene expression and DNA copy number profiles. We built a predictive model of persistent lymph node metastasis using SVM comprising 65 probes which contain 34 genes, and performed leave-one-out cross-validation. Majority of these genes are contained on chromosome 4, which was most commonly lost in ypN1-2 patients. This suggests that some of these genes may represent tumor suppressors and that their direct loss may in fact promote lymph node metastasis in these patients. However, it will be important to examine these individual genes in the future to determine if their expression is indeed altered and to establish their clinical significance, if any.
While we showed aCGH to be an effective predictor of persistent lymph node metastasis in rectal cancer, it is notable that we found tumor response to CRT to be the best predictor of persistent lymph node metastasis. In our patient cohort, all but two ypT0 patients (93%) were ypN0 suggesting that patients with excellent tumor response are highly unlikely to be ypN1-2 and therefore unlikely to develop distant disease. This is clinically significant as it suggests that if we can identify a subset of patients with a pCR to CRT who are also highly unlikely to have metastasis by aCGH screening, these patients may ultimately benefit from an organ preservation approach.
There are a number of limitations to our study that deserve mention. While our aCGH analysis was based on the largest rectal cancer cohort reported thus far, the patient group was not completely homogeneous; there were two treatment arms in our study, and while all patients received pre-operative CRT, some patients also received additional chemotherapy that may have influenced their clinical outcome. Additionally, while we performed rigorous statistically-sound internal validation, external validation of our predictive model will be important to further corroborate our findings. Finally, we compared DNA from pretreatment tumor to post-treatment DNA from normal surgical margins. This raises the issue of whether the post-treatment control tissue may have been affected by the CRT and additional neoadjuvant chemotherapy. However, the control tissue was obtained from the proximal resection margin, which is usually outside the radiation field, and there is no data indicating that CNAs arise following chemotherapy treatment of normal tissue. Furthermore, we have recently shown that mutations in KRAS and TP53, two genes which play a role in the pathogenesis of rectal cancer, remain largely unchanged after CRT in rectal cancer patients.28
In conclusion, we showed that specific CNAs are significantly associated with persistent lymph node metastasis following CRT in locally advanced rectal cancer, in particular loss of chromosome 4. These findings are clinically relevant and support the use of aCGH to identify those patients who are more likely to have persistent lymph node metastasis and direct them to receive appropriate systemic therapy. This approach may also help identify those patients who are highly unlikely to have lymph node metastasis after CRT, particularly if they have a pCR. These patients may ultimately benefit from an organ preservation approach.
Supplementary Material
Acknowledgments
The authors thank Nicola Solomon, PhD, for assistance in writing and editing the manuscript.
Footnotes
Disclosures: This study was supported by the National Institutes of Health (NIH), National Cancer Institute (NCI) R01 Grant CA090559 (JGA). ClinicalTrials.org Identifier: NCT00335816. The authors declare no conflicts of interest associated with this manuscript.
Meeting presentation: Presented at the American Society of Colon and Rectal Surgeons (ASCRS) Annual Meeting, Vancouver, Canada, May 14-18, 2011. Awarded Best Rectal Cancer Poster provided by Richard Wolf Medical Instruments Corporation in memory of Dr. Gerhard F. Buess.
Author contributions: Conception and design - ZC, ZL, XD, JGA; Acquisition of data -ZC, WL; Analysis and interpretation of data - ZC, ZL, XD, CW, JGA; Drafting the article or revising it critically for important intellectual content - ZC, ZL, XD, CW, WL, JGA; Final approval of the version to be published - ZC, ZL, XD, CW, WL, JGA.
References
- 1.Sauer R, Becker H, Hohenberger W, et al. Preoperative versus postoperative chemoradiotherapy for rectal cancer. N Engl J Med. 2004;351:1731–1740. doi: 10.1056/NEJMoa040694. [DOI] [PubMed] [Google Scholar]
- 2.Ciccocioppo A, Stephens JH, Hewett PJ, Rieger NA. Complete pathologic response after preoperative rectal cancer chemoradiotherapy. ANZ J Surg. 2009;79:481–484. doi: 10.1111/j.1445-2197.2009.04950.x. [DOI] [PubMed] [Google Scholar]
- 3.Maas M, Nelemans PJ, Valentini V, et al. Long-term outcome in patients with a pathological complete response after chemoradiation for rectal cancer: a pooled analysis of individual patient data. Lancet Oncol. 2010;11:835–844. doi: 10.1016/S1470-2045(10)70172-8. [DOI] [PubMed] [Google Scholar]
- 4.Peeters KC, van de Velde CJ, Leer JW, et al. Late side effects of short-course preoperative radiotherapy combined with total mesorectal excision for rectal cancer: increased bowel dysfunction in irradiated patients-a Dutch colorectal cancer group study. J Clin Oncol. 2005;23:6199–206. doi: 10.1200/JCO.2005.14.779. [DOI] [PubMed] [Google Scholar]
- 5.Hendren SK, O’Connor BI, Liu M, et al. Prevalence of male and female sexual dysfunction is high following surgery for rectal cancer. Ann Surg. 2005;242:212–23. doi: 10.1097/01.sla.0000171299.43954.ce. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Evans J, Patel U, Brown G. Rectal cancer: primary staging and assessment after chemoradiotherapy. Semin Radiat Oncol. 2011;21:169–77. doi: 10.1016/j.semradonc.2011.02.002. [DOI] [PubMed] [Google Scholar]
- 7.Frazer KA, Murray SS, Schork NJ, Topol EJ. Human genetic variation and its contribution to complex traits. Nat Rev Genet. 2009;10:241–51. doi: 10.1038/nrg2554. [DOI] [PubMed] [Google Scholar]
- 8.Redon R, Ishikawa S, Fitch KR, et al. Global variation in copy number in the human genome. Nature. 2006;444:444–54. doi: 10.1038/nature05329. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Lengauer C, Kinzler KW, Vogelstein B. Genetic instabilities in human cancers. Nature. 1998;396:643–649. doi: 10.1038/25292. [DOI] [PubMed] [Google Scholar]
- 10.Teixeira MR, Heim S. Multiple numerical chromosome aberrations in cancer: what are their causes and what are their consequences? Semin Cancer Biol. 2005;15:3–12. doi: 10.1016/j.semcancer.2004.09.006. [DOI] [PubMed] [Google Scholar]
- 11.Grade M, Gaedcke J, Wangsa D, et al. Chromosomal copy number changes of locally advanced rectal cancers treated with preoperative chemoradiotherapy. Cancer Genet Cytogenet. 2009;193:19–28. doi: 10.1016/j.cancergencyto.2009.03.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Postma C, Koopman M, Buffart TE, et al. DNA copy number profiles of primary tumors as predictors of response to chemotherapy in advanced colorectal cancer. Ann Oncol. 2009;20:1048–1056. doi: 10.1093/annonc/mdn738. [DOI] [PubMed] [Google Scholar]
- 13.Cardoso J, Boer J, Morreau H, Fodde R. Expression and genomic profiling of colorectal cancer. Biochim Biophys Acta. 2007;1775:103–137. doi: 10.1016/j.bbcan.2006.08.004. [DOI] [PubMed] [Google Scholar]
- 14.Diep CB, Kleivi K, Ribeiro FR, Teixeira MR, Lindgjaerde OC, Lothe RA. The order of genetic events associated with colorectal cancer progression inferred from meta-analysis of copy number changes. Genes Chromosomes Cancer. 2006;45:31–41. doi: 10.1002/gcc.20261. [DOI] [PubMed] [Google Scholar]
- 15.Garcia-Aguilar J, Smith DD, Avila K, Bergsland EK, Chu P, Krieg RM Timing of Rectal Cancer Response to Chemoradiation Consortium. Optimal timing of surgery after chemoradiation for advanced rectal cancer: preliminary results of a multicenter, nonrandomized phase II prospective trial. Ann Surg. 2011;254:97–102. doi: 10.1097/SLA.0b013e3182196e1f. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Edge SB, Byrd DR, Compton CC, Fritz AG, Greene FL, Trotti A. American Joint Committee on Cancer (AJCC) cancer staging manual. 7. Chicago: Springer, Inc; 2010. [Google Scholar]
- 17.Chen Z, Liu Z, Li W, et al. Chromosomal copy number alterations are associated with tumor response to chemoradiation in locally advanced rectal cancer. Genes Chromosomes Cancer. 2011;50:689–99. doi: 10.1002/gcc.20891. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Benjamini YH, Hochberg Y. Controlling the false discovery rate: a practical and powerful approach to multiple testing. J R Stat Soc Ser B Statistical Methodology. 1995;57:289–300. [Google Scholar]
- 19.Storey JD, Tibshirani R. Statistical significance for genomewide studies. Proc Natl Acad Sci USA. 2003;100:9440–9445. doi: 10.1073/pnas.1530509100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Shi L, Reid LH, Jones WD, et al. The MicroArray Quality Control (MAQC) project shows inter- and intraplatform reproducibility of gene expression measurements. Nat Biotechnol. 2006;24:1151–1161. doi: 10.1038/nbt1239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Deng X, Campagne F. Introduction to the development and validation of predictive biomarker models from high-throughput data sets. Methods Mol Biol. 2010;620:435–470. doi: 10.1007/978-1-60761-580-4_15. [DOI] [PubMed] [Google Scholar]
- 22.Bang H, Davidian M. Experimental statistics for biological sciences. Methods Mol Biol. 2010;620:3–104. doi: 10.1007/978-1-60761-580-4_1. [DOI] [PubMed] [Google Scholar]
- 23.Sing T, Sander O, Beerenwinkel N, Lengauer T. ROCR: visualizing classifier performance in R. Bioinformatics. 2005;21:3940–3941. doi: 10.1093/bioinformatics/bti623. [DOI] [PubMed] [Google Scholar]
- 24.Ghadimi BM, Grade M, Liersch T, Langer C, Siemer A, Fuzesi L, Becker H. Gain of chromosome 8q23-24 is a predictive marker for lymph node positivity in colorectal cancer. Clin Cancer Res. 2003;9:1808–1814. [PubMed] [Google Scholar]
- 25.Diep CB, Teixeira MR, Thorstensen L, et al. Genome characteristics of primary carcinomas, local recurrences, carcinomatoses, and liver metastases from colorectal cancer patients. Mol Cancer. 2004;23:3–6. doi: 10.1186/1476-4598-3-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Arribas R, Ribas M, Risques RA, et al. Prospective assessment of allelic losses at 4p14-16 in colorectal cancer: two mutational patterns and a locus associated with poorer survival. Clin Cancer Res. 1999;5:3454–3459. [PubMed] [Google Scholar]
- 27.Bardi G, Fenger C, Johansson B, Mitelman F, Heim S. Tumor karyotype predicts clinical outcome in colorectal cancer patients. J Clin Oncol. 2004;22:2623–2634. doi: 10.1200/JCO.2004.11.014. [DOI] [PubMed] [Google Scholar]
- 28.Chen Z, Duldulao MP, Li W, Lee W, Kim J, Garcia-Aguilar J. Molecular Diagnosis of Response to Neoadjuvant Chemoradiation Therapy in Patients with Locally Advanced Rectal Cancer. J Am Coll Surg. 2011;212:1008–1017.e1. doi: 10.1016/j.jamcollsurg.2011.02.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.