Summary
Alignment of chromosomes at the metaphase plate is a signature of cell division in metazoan cells, yet the mechanisms controlling this process remain ambiguous. Here we use a combination of quantitative live cell imaging and reconstituted dynamic microtubule assays to investigate the molecular control of mitotic centromere movements. We establish that Kif18A (kinesin-8) attenuates centromere movement by directly promoting microtubule pausing in a concentration-dependent manner. This activity provides the dominant mechanism for restricting centromere movement to the spindle midzone. Furthermore, polar ejection forces spatially confine chromosomes via position-dependent regulation of kinetochore tension and centromere switch rates. We demonstrate that polar ejection forces are antagonistically modulated by chromokinesins. These pushing forces depend on Kid (kinesin-10) activity and are antagonized by Kif4A (kinesin-4), which functions to directly suppress microtubule growth. These data support a model in which Kif18A and polar ejection forces synergistically promote centromere alignment via spatial control of kinetochore-microtubule dynamics.
Introduction
Alignment of chromosomes at the equator of the mitotic spindle is highly conserved and is widely believed to ensure the equal segregation of replicated chromosomes during cell division. In vertebrate cells, aligned chromosomes are not static. Rather, the chromosomes are constantly moving in an oscillatory pattern, with paired chromosomes displaying coordinated movements (Skibbens et al., 1993). Therefore, to promote and maintain chromosome alignment, cells must confine these oscillatory movements to a region around the spindle equator. To accomplish this, we hypothesize that bi-oriented, congressing chromosomes must preferentially reverse direction if they approach too closely to either spindle pole. We set out to test this hypothesis and identify the molecular machinery responsible for controlling directional switches.
To understand the spatial control of chromosome movement, one must differentiate between processes controlling attachment versus those governing alignment and retention at the midzone. Attachment is an important early step in congression but it is mechanistically distinct from alignment (Cheng et al., 2011; Kim et al., 2010). Interfering with this process affects a stochastic proportion of the total number of chromosomes. We, therefore, confined our analysis to bioriented chromosomes to specifically study the spatial information that confines centromeres at the metaphase plate. Purturbation of this process would, by necessity, simultaneously affect all chromosomes.
Presently, there are three popular models for congression of bioriented chromosomes, each of which depends on the activity of kinesins: 1) direct regulation of kinetochore fiber (K-fiber) end dynamics by kinesin-8 motors (Du et al., 2010; Garcia et al., 2002; Gupta et al., 2006; Mayr et al., 2007; Stumpff et al., 2011; Stumpff et al., 2008; Varga et al., 2006; Wargacki et al., 2010; West et al., 2002); 2) polar ejection forces (microtubule-dependent pushing forces acting on chromosome arms, which are commonly presumed to be supplied by the chromokinesins Kid and Kif4A) (Bieling et al., 2010a; Brouhard and Hunt, 2005; Cassimeris et al., 1994; Funabiki and Murray, 2000; Ke et al., 2009; Levesque and Compton, 2001; Rieder et al., 1986; Rieder and Salmon, 1994); 3) plus-end directed motility along microtubules via CENP-E (Kapoor et al., 2006; Kim et al., 2010). Published data unequivocally demonstrates that alignment of bioriented chromosomes is highly dependent on the activity of the kinesin-8 motor, Kif18A (Zhu et al., 2005), making it a good candidate for controlling position-dependent switching. In contrast, the majority of chromosomes successfully congress in the absence of either CENP-E or chromokinesins (Levesque and Compton, 2001; Mazumdar et al., 2004; Vernos et al., 1995; Yen et al., 1991; Zhu et al., 2005). In the case of CENP-E depletion, those chromosomes that do not congress exhibit a defect in biorientation. Recent studies have mechanistically defined the role of CENP-E in promoting the alignment of chromosomes prior to biorientation (Cai et al., 2009; Kapoor et al., 2006; Kim et al., 2010) and demonstrated that it does not affect the switching of bioriented kinetochores (Jaqaman et al., 2010). In contrast, the functions of chromokinesins in metaphase alignment are unclear, as chromosomes will readily congress in the absence of Kid (Levesque and Compton, 2001). Similarly, there are conflicting reports regarding Kif4A’s effects on alignment (Mazumdar et al., 2004; Vernos et al., 1995; Zhu et al., 2005). This is puzzling because the polar ejection force represents a potentially powerful, positionally encoded force to direct chromosomes away from spindle poles (Cassimeris et al., 1994; Inoue and Salmon, 1995; Rieder et al., 1986; Rieder and Salmon, 1994).
We used a combination of quantitative cell imaging assays and analyses of purified, dynamic microtubules to investigate the mechanistic contributions of Kif18A and chromokinesins to the positional control of centromere movements. We find that the equatorial position of centromere pairs is maintained via spatial regulation of centromere switch rates. This control is primarily dependent on the direct suppression of K-fiber dynamics by Kif18A. However, we demonstrate that chromokinesins and polar ejection forces also contribute to this spatial control by inducing position-dependent increases in interkinetochore tension. Only one chromokinesin, Kid, is the source of the polar ejection force and, surprisingly, this Kid-dependent force gradient alone is sufficient for aligning centromeres at the metaphase plate. The second chromokinesin, Kif4A, unexpectedly antagonizes centromere alignment by suppressing microtubule polymerization. These data support a model in which centromere alignment is controlled by two independent mechanisms that function to promote position-dependent switching and confine centromere movements.
Results
Kid and Kif4A oppositely affect centromere alignment
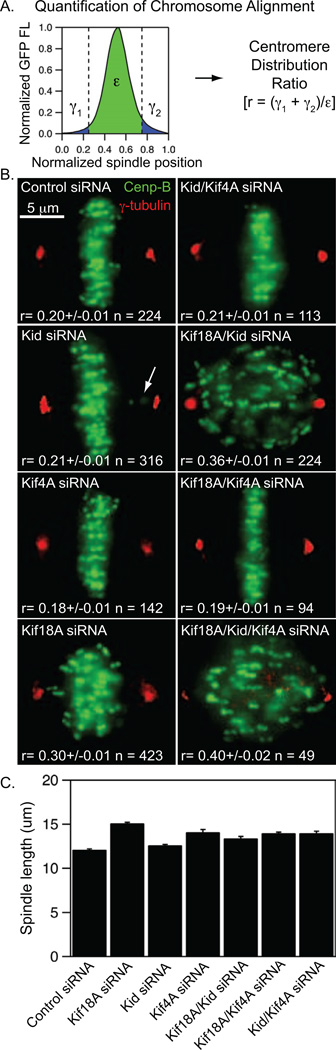
To quantitatively evaluate the effects of Kif18A, Kid and Kif4A on mitotic centromere alignment in a large population of cells, we developed an assay to measure the spatial distribution of centromeres along the pole-to-pole axis of the spindle. HeLa cells expressing EGFP-CENP-B to label centromeres were treated with siRNAs against single or multiple kinesins, which resulted in specific and effective depletion of the targeted motors (Figure S1A). The distribution of fluorescently labeled centromeres along the pole-to-pole axis was then measured in all cells with paired, bi-oriented centromeres (Figure 1 A–B and Figure S1B). Spindle lengths were normalized to account for the spindle length increase caused by Kif18A or Kif4A depletion (Figure 1C). GFP-distribution measurements were then used to calculate a centromere-distribution ratio (r) for each cell based on the following formula:
“γ1” and “γ2” are the sums of the EGFP-CENP-B fluorescence in the two quarter spindles nearest the poles and “ε” is the sum of the EGFP-CENP-B fluorescence measured in the middle 50% of the spindle (Figure 1A). In this assay, a cell with well-aligned centromeres will have a centromere-distribution ratio of 0.15 – 0.2, while a cell with a completely random distribution of centromeres is predicted to have a ratio of 0.60 – 0.65 based on the geometry of the spindle.
Figure 1.
Kif18A and chromokinesins collectively regulate centromere alignment. (A) Method used to quantitatively analyze centromere alignment. The distribution of GFP fluorescence (FL) along the normalized pole-to-pole axis was measured in HeLa cells expressing EGFP-CENP-B (green) and stained for γ-tubulin (red). A centromere distribution ratio (r) was calculated for each cell using the indicated formula. (B) Representative images of cells treated with the indicated siRNAs. The mean ± SEM for centromere distribution ratios measured from the indicated number of mitotic cells (n) are reported for each cell type. p-values calculated from comparison to control siRNA cells are 4.9 × 10−29 (Kif18A), 0.23 (Kid), 0.003 (Kif4A), 1.2 × 10−35 (Kif18A/Kid), 0.14 (Kif18A/Kif4A), 0.46 (Kid/Kif4A) and 1.0 × 10−12 (Kif18A/Kid/Kif4A). Arrow indicates an example of a centromere pair near the spindle pole in a Kid-depleted cell. (C) Plot of average spindle length in HeLa cells depleted of the indicated kinesins. Spindle lengths were measured as the distance between γ-tubulin labeled centrosomes in fixed cells. Error bars are SEM.
Kif18A-depletion significantly increased centromere distribution (r = 0.30 ± 0.01 and p = 4.9 × 10−29 compared to controls, Figure 1B and Figure S1B), consistent with previous findings (Mayr et al., 2007; Stumpff et al., 2008; Zhu et al., 2005). The distributions of centromeres in cells depleted of Kid were not significantly different than those in control siRNA treated cells (r = 0.21 ± 0.01 and p = 0.23, Figure 1B and Figure S1B). However, consistent with previous studies (Levesque and Compton, 2001), we found that Kid-depleted cells often had one or more centromeres near the spindle poles, which led to a shoulder in the distribution of r values (Figure 1B and Figure S1B). Surprisingly, we found that depletion of Kif4A induces a small but significant decrease in the distribution of centromeres (r = 0.18 ± 0.01 and p = 0.003, Figure 1B and Figure S1B) compared to controls. This decrease was not correlated with changes in spindle length (Pearson correlation coefficient = 0.12), but was dependent on the presence of Kid, as cells co-depleted of Kid and Kif4A displayed centromere distributions similar to those measured in Kid-depleted cells (r = 0.21 ± 0.01 and p = 0.46 compared to controls, Figure 1B and Figure S1B).
Kid and Kif4A also had opposite effects on centromere alignment when depleted in combination with Kif18A. In cells depleted of both Kif18A and Kid, centromere alignment defects were more severe than those measured in cells depleted of Kif18A alone (r = 0.36 ± 0.01 p = 1.2 × 10−35 compared to controls, Figure 1B and Figure S1B). In contrast, cells depleted of Kif18A and Kif4A had less severe alignment defects than those depleted of Kif18A alone (r = 0.19 ± 0.01 and p = 0.14 compared to controls, Figure 1B and Figure S1B), confirming that Kif4A-depletion increases centromere alignment. Interestingly, cells co-depleted of Kif18A and Kif4A were also more capable of progressing through cell division than those depleted of Kif18A alone (Figure S1C). Cells depleted of all three kinesins displayed centromere distributions similar to those seen in Kif18A-Kid co-depleted cells, consistent with the idea that the increase in alignment caused by Kif4A-depletion requires Kid activity (r = 0.40 ± 0.02 and p = 1.0 × 10−12 compared to controls, Figure 1B and Figure S1B). These data indicate that Kid acts synergistically with Kif18A to promote centromere alignment. In contrast, Kif4A antagonizes centromere alignment through a Kid-dependent mechanism.
Kif18A attenuates centromere movements
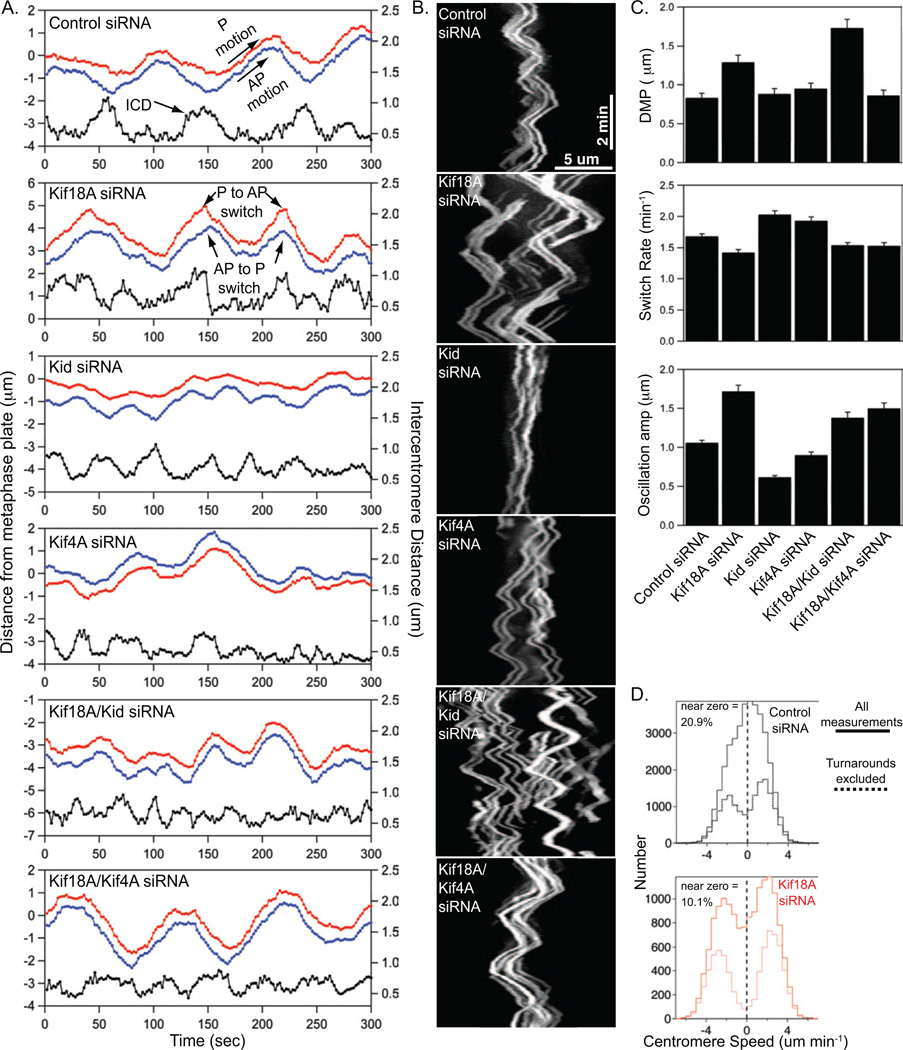
To determine how Kif18A, Kid and Kif4A affect centromere movements, we imaged and tracked the movements of EGFP-CENP-B labeled centromeres with high temporal resolution (2 second intervals) in cells treated with siRNAs specifically targeting these motors (Figure 2 A–B, Figure S1A and supplemental movies S1–S2). EGFP-CENP-B has been well-characterized as a marker for centromere movements in HeLa cells (Shelby et al., 1996), and we find that the movements of fluorescent CENP-B foci closely mirror those of the kinetochore protein Nuf2 (Figure S2A). Consistent with our previous work, centromeres in cells depleted of Kif18A displayed large oscillations, characterized by faster velocities and reduced rates of directional switching, at positions further away from the metaphase plate than those in control cells (Figure 2 A–C, Figure S2B and Table 1) (Stumpff et al., 2008). Additionally, we observed that Kif18A-depleted cells exhibited a two-fold decrease in attenuated centromere movements when all velocities were evaluated and a four-fold decrease when velocity measurements in the vicinity (+/− 10 seconds) of turnarounds were excluded (Figure 2D). These data demonstrate that Kif18A suppresses centromere movements and increases the proportion of time that centromeres spend in a slow velocity state during both directional switches and persistent movement.
Figure 2.
Kif18A and chromokinesins differentially impact mitotic centromere movements. (A) Plots of centromere movement as a function of time. The red and blue traces show the movements of a pair of sister centromeres. The black traces show changes in the distance between the red and blue centromeres, called intercentromere distance (ICD), over time. Examples of poleward (P) motion, away from pole (AP) motion, poleward-to-away from pole switches (P to AP) and away from pole-to-poleward switches are indicated with arrows. (B) Kymographs of EGFP-CENP-B movements in HeLa cells treated with siRNAs targeting the indicated kinesins. Horizontal scale bar is 5 µm, vertical scale bar is 2 minutes. (C) Plots of average distance from the metaphase plate (DMP), switch rates, and oscillation amplitudes for centromeres tracked in cells depleted of the indicated kinesins. Error bars indicate SEM. (D) Histograms of centromere speeds measured using a five point (10 second) sliding regression fit to plots of centromere movement. Negative values represent speeds during P movement, while positive values are speeds measured during AP movement. The solid lines display all speed measurements made from the data set described in Table 1, while the dashed lines represent the same data set with the exclusion of points that occured within 10-seconds of a directional switch. The percentage of near-zero speeds (between −0.5 and 0.5 µm/min) measured in control and Kif18A-depleted cells is reported for the unfiltered data set (solid lines).
Table 1.
Fluorescent centromeres in live HeLa cells were tracked and the average amplitude (Amp), away-from-pole velocity (AP vel), poleward velocity (P vel), switch rate, distance from metaphase plate (DMP) and intercentromere distance (ICD) were measured for each centromere. The total number of cells and centromeres (Cen) analyzed for each siRNA treatment are reported. Parameter values are reported as the mean +/− SEM.
| siRNA | # cells/ # Cen |
Amplitude (um) |
AP vel (um/min) |
P vel (um/min) |
Switch (min-1) |
DMP (um) |
ICD (um) |
|---|---|---|---|---|---|---|---|
| Control | 20/ 236 | 1.06 +/− 0.03 | 1.71 +/− 0.04 | 1.96 +/− 0.05 | 1.68 +/− 0.04 | 0.83 +/− 0.06 | 0.67 +/− 0.01 |
| Kif18A | 11/ 108 | 1.72 +/− 0.08 | 2.59 +/− 0.06 | 2.81 +/− 0.06 | 1.42 +/− 0.05 | 1.29 +/− 0.09 | 0.67 +/− 0.01 |
| Kid | 13/ 110 | 0.62 +/− 0.02 | 1.07 +/− 0.05 | 1.19 +/− 0.05 | 2.03 +/− 0.06 | 0.88 +/− 0.07 | 0.74 +/− 0.01 |
| Kif4A | 11/ 108 | 0.90 +/− 0.04 | 1.69 +/− 0.06 | 1.83 +/− 0.07 | 1.93 +/− 0.06 | 0.95 +/− 0.07 | 0.66 +/− 0.01 |
| Kif18A/Kid | 15/ 154 | 1.38 +/− 0.07 | 1.93 +/− 0.06 | 2.28 +/− 0.07 | 1.54 +/− 0.04 | 1.73 +/− 0.11 | 0.72 +/− 0.01 |
| Kif18A/Kif4A | 8/ 86 | 1.50 +/− 0.07 | 2.40 +/− 0.10 | 2.88 +/− 0.12 | 1.53 +/− 0.05 | 0.86 +/− 0.07 | 0.69 +/− 0.01 |
Kif18A and Kid synergistically confine centromere movements
Consistent with previous studies, we observed that centromeres in cells depleted of Kid alone displayed smaller oscillations than those in control cells (Levesque and Compton, 2001). These movements were characterized by reduced velocities and increased rates of directional switching (Figure 2 A–C, Figure S2B and Table 1). Interestingly, we found that the opposite effects of Kif18A and Kid on centromere movements are additive, leading to velocities and switch rates intermediate to those of either single knockdown (Figure 2C, Figure S2B and Table 1). Strikingly, however, centromeres made oscillatory movements that were centered at positions further away from the spindle equator in the double-depleted cells compared to those of either single knockdown (Figure 2C and Table 1). Thus, despite having opposite effects on centromere movement parameters, Kif18A and Kid synergistically promote the positioning of bioriented centromeres near the spindle equator.
Depletion of Kif4A spatially confines centromere movements in the absence of Kif18A
Centromeres in cells depleted of Kif4A displayed slightly reduced oscillatory movements to those seen in control-depleted cells, resulting from an increase in the directional switch rate (Figure 2C and Table 1). Co-depletion of Kif18A and Kif4A led to oscillatory movements that were similar to those observed in Kif18A-depleted cells, but these oscillations were on average positioned significantly closer to the metaphase plate compared to those in cells depleted of Kif18A alone (Figure 2 A–C and Table 1). Thus, abrogation of Kif4A function effectively confines centromere movements in the absence of Kif18A, indicating that Kid and Kif4A oppositely affect the positioning of bioriented centromeres near the metaphase plate.
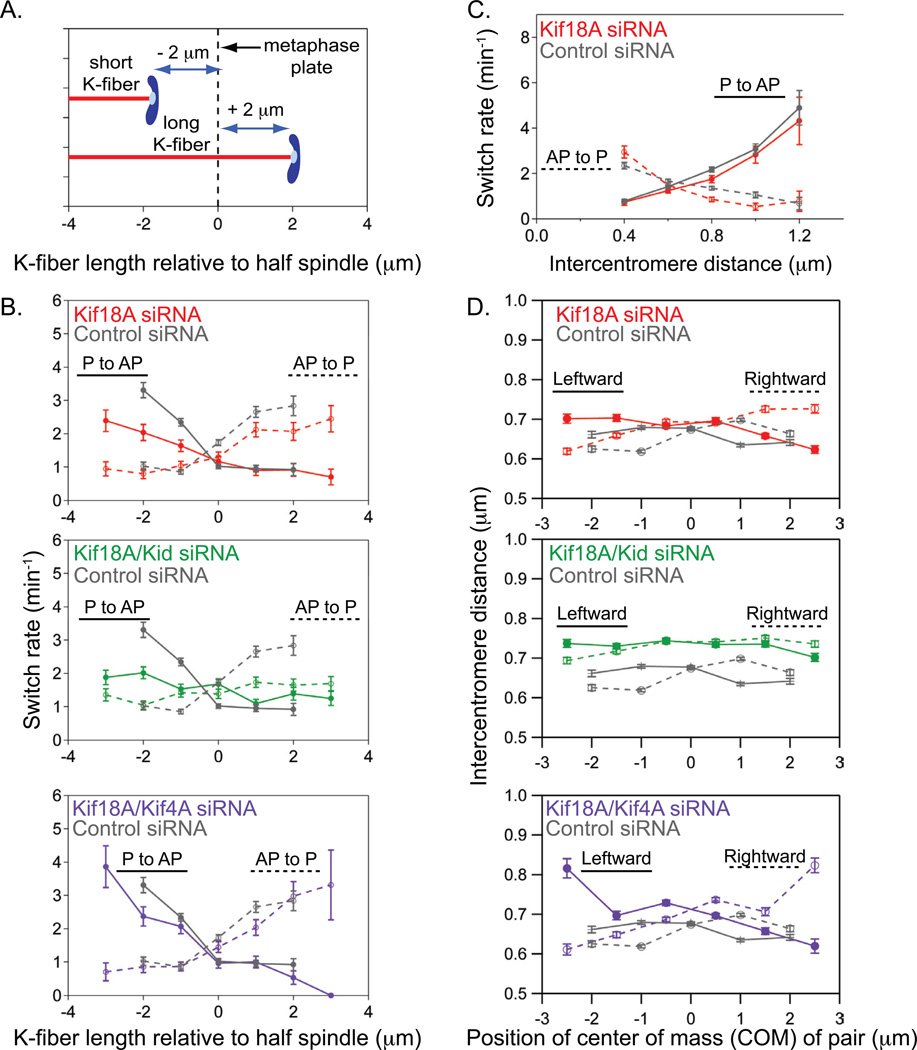
Centromere directional switches vary with position
The positioning of centromeres at the spindle equator likely requires spatial regulation of centromere directional switch rates (Skibbens et al., 1993). To test this hypothesis, we measured centromere switch rates as a function of their position within the spindle (Figure 3A). These analyses reveal that the rate at which the centromere changes direction from poleward (P) to away from pole (AP) movement or vice versa increases as centromeres move further away from the metaphase plate (Figure 3B), suggesting that directional switches are spatially regulated within the spindle. Consistent with studies in PtK1 cells (Tirnauer et al., 2002), we found that AP to P switches in HeLa cells occur concaminantly with K-fiber catastrophes, while P to AP switches occur with K-fiber rescues, suggesting that directional switches are also coupled to K-fiber dynamics (Figure S2 C–D and supplemental movie S3).
Figure 3.
Kid and Kif4A oppositely tune centromere switch rates and intercentromere distance as a function of position in the absence of Kif18A activity. (A) Schematic showing how centromere positions were assigned via measuring K-fiber length relative to the half spindle. Centromeres attached to short K-fibers, which are on the same side of the metaphase plate as the pole they are attached to, were assigned negative centromere-to-metaphase plate values. Centromeres attached to long K-fibers were assigned positive centromere-to-metaphase plate values. (B) Plots of switch rate for both poleward to away-from-pole (P to AP) and away-from-pole to poleward (AP to P) switches as a function of position on the spindle, expressed as K-fiber length relative to the half spindle. Switch rates were measured from the populations of kinesin-depleted cells described in Table 1. Error bars represent uncertainty due to counting statistics. (C) Plots of switch rate as a function of intercentromere distance for both P to AP and AP to P switches in control and Kif18A-depleted cells. None of the kinesin depletions tested changed this correlation (see also Figure S3 C–D). Error bars represent uncertainty due to counting statistics. (D) Plots of intercentromere distance as a function of the position of the center of mass of the centromere pair relative to the metaphase plate during motion of the pair towards the left pole (solid line) and towards the right pole (dashed line). Positions of centromere pairs were determined essentially as described in (A), but the distance between the metaphase plate and the point midway between the two sister centromeres, center of mass (COM) position, was measured. Error bars indicate SEM.
Kif18A, Kid and Kif4A spatially regulate centromere switch rates
The spatial correlation of switch rates was significantly reduced by depletion of either Kif18A or chromokinesins (Figure 3B and Figure S3 A–B). In both Kid and Kif4A-depleted cells, the reduced slopes of the switch rate versus position graphs are due to an increase in the rate of switching near the spindle equator (Figure S3 A–B). This effect can explain the hyper-alignment observed in Kif4A-depleted cells (Figure 1B) and the suppressed centromere oscillations seen in Kid-depleted cells (Figure 2 and Table 1). In contrast, Kif18A-depletion led to a reduction in the switch rate at positions away from the spindle equator, an effect that would reduce the alignment of centromeres (Figure 3B and Figure S3B). In cells depleted of both Kif18A and Kid, the correlation between switch rate and position was nearly completely removed, indicating that the probability of a centromere making a directional switch was equal regardless of its position on the spindle (Figure 3B and Figure S3B). Thus, under these conditions, nearly all spatial regulation of centromere switch rates was lost, consistent with the severe centromere alignment defect observed in these cells (Figure 1B). In contrast, co-depleting cells of Kif18A and Kif4A restored the correlation between switch rate and position to a level similar to that observed in control-depleted cells (Figure 3B and Figure S3B). Taken together these data suggest that Kif18A and Kid increase the spatial regulation of centromere directional switches, while Kif4A suppresses it.
Switch rates are strongly correlated with intercentromere distance
Previous studies suggest that tension can influence microtubule dynamics and affect chromosome movements (Akiyoshi et al., 2010; Civelekoglu-Scholey et al., 2006; Franck et al., 2007; Gardner et al., 2005; Khodjakov and Rieder, 1996; Nicklas, 1988; Skibbens et al., 1995; Skibbens and Salmon, 1997). Thus, influencing tension at the kinetochore could be one mechanism to control centromere switch rates. To address this possibility, we analyzed switch rates as a function of intercentromere distance (Waters et al., 1996). Plots of intercentromere distance and centromere movement indicate that directional switches are more likely to occur at extremes of both low and high intercentromere distance (Figure 2A). Statistical analyses confirmed a clear correlation between switch rate and intercentromere distance that was unaffected by depletion of Kif18A, Kid or Kif4A (Figure 3C and Figure S3 C–D). These data suggest that variations in kinetochore tension may influence the probability of centromere directional switches, but the sensitivity of directional switches to changes in force is independent of Kif18A, Kid and Kif4A activity.
Kid and Kif4A oppositely affect position-dependent increases in intercentromere distance
If changes in kinetochore tension contribute to the spatial regulation of centromere movements, we expect intercentromere distance to vary with position within the spindle (Shelby et al., 1996; Waters et al., 1996). To test this hypothesis, we scored intercentromere distance during the coordinated movement of sister centromeres as a function of the position of the pair. Centromere pair position was defined as the distance of the center of mass (COM) of the pair from the metaphase plate. In control, Kid and Kif4A-depleted cells, we observed little to no variation in intercentromere distance as COM position changed (Figure 3D and Figure S3 E–F). In the absence of Kif18A, however, we found that intercentromere distance significantly increased when centromere pairs were close to a spindle pole and moving towards it but not when centromere pairs were close to a pole and moving away from it (Figure 3D). Co-depleting cells of Kif18A and Kid eliminated this direction-dependent increase in intercentromere distance, while co-depletion of Kif18A and Kif4A enhanced it (Figure 3D). These data suggest that Kid induces a position-dependent increase in tension at the kinetochore, which in turn specifically increases the switch rates of centromeres as they move towards a pole. In contrast, Kif4A antagonizes this activity.
Kid and Kif4A oppositely regulate the polar ejection force
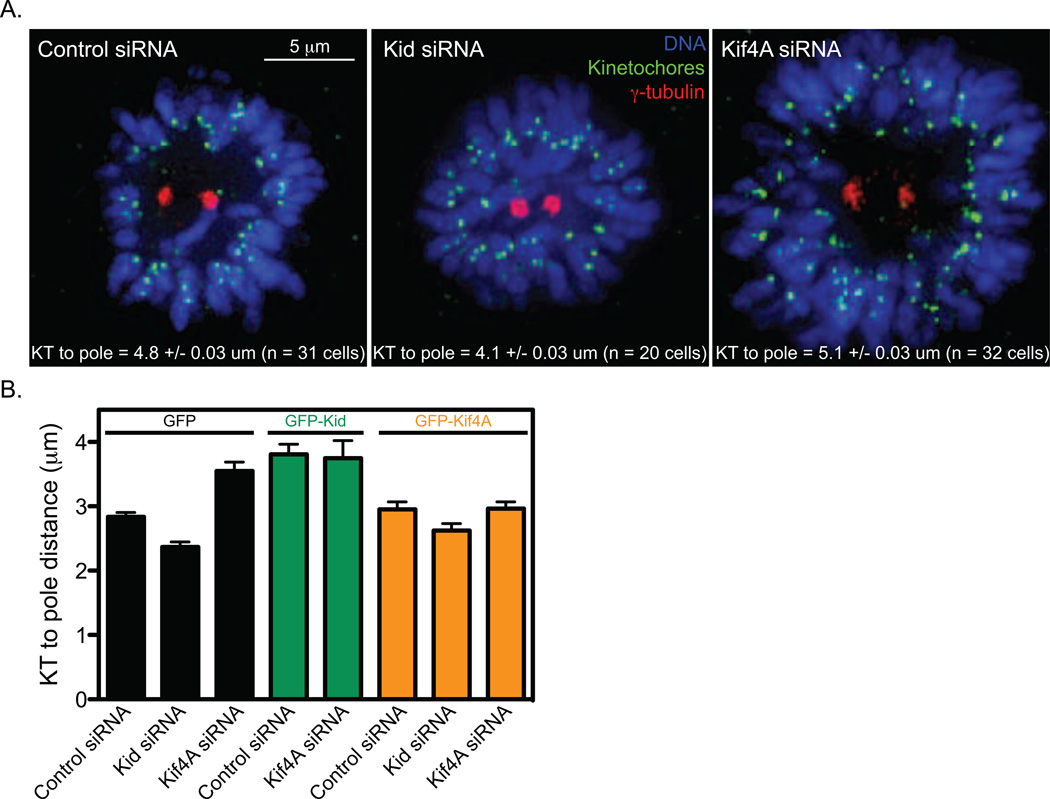
We hypothesize that Kid and Kif4A may be influencing the spatial regulation of centromere switch rates via modulation of the polar ejection force. However, the opposite effects of Kid and Kif4A on centromere movements are surprising since both motors display similar localization to chromosome arms, and both have been proposed to positively contribute to the polar ejection force (Antonio et al., 2000; Bieling et al., 2010a; Brouhard and Hunt, 2005; Funabiki and Murray, 2000; Levesque and Compton, 2001; Mazumdar and Misteli, 2005). To resolve this question, we directly compared the effects of Kid and Kif4A-depletion on the positioning of chromosomes around monopolar asters. We found that Kid and Kif4A oppositely affect the distance between kinetochores and centrosomes in both HeLa and HCT116 cells. The average kinetochore-to-pole distance was decreased in Kid-depleted cells and was increased in Kif4A-depleted cells compared to controls (Figure 4 A–B and Figure S4). Furthermore, cells expressing GFP-Kid displayed increased kinetochore-to-pole distances (Figure 4B). While expression of an siRNA-resistant GFP-Kif4A did not reduce the average kinetochore-to-pole distance alone, it was able to rescue the increase caused by Kif4A-depletion (Figure 4B). In contrast to previous assumptions, these results indicate that Kid enhances the polar ejection force while Kif4A antagonizes it.
Figure 4.
Kid and Kif4A oppositely tune the polar ejection force. (A) Following a 2-hour treatment with 100 µM monastrol, HeLa cells pre-treated with the indicated siRNAs were fixed and processed for immunofluorescence. DNA was stained with DAPI (blue), kinetochores were visualized with Crest serum (green) and centrosomes were localized with anti-γ-tubulin antibodies. Scale bar is 5 µm. Kinetochore (KT)-to-pole distances measured in cells treated with the indicated siRNAs and the mean ± SEM from the analysis of the indicated number of cells (n) is reported for each distribution. Distributions are displayed in Figure S4. p-values calculated from comparison to control siRNA cells are 0.0001 (Kid siRNA) and 0.03 (Kif4A siRNA). (B) Graph of KT-to-pole distances measured in monopolar HCT116 cells expressing GFP (black bars), GFP-Kid (green bars) or GFP-Kif4A (orange bars) and treated with the indicated siRNAs. Error bars are SEM. The following data sets are significantly different (p < 0.001) from the GFP/control-siRNA data set in a two-tailed t-test comparison: GFP/Kid-siRNA, GFP/Kif4A-siRNA, GFP-Kid/control-siRNA and GFP-Kid/Kif4A-siRNA. GFP/Kif4A-siRNA is also significantly different from GFP-Kif4A/Kif4A-siRNA (p = 0.001).
Kif4A functions to suppress spindle microtubule dynamics during chromosome alignment
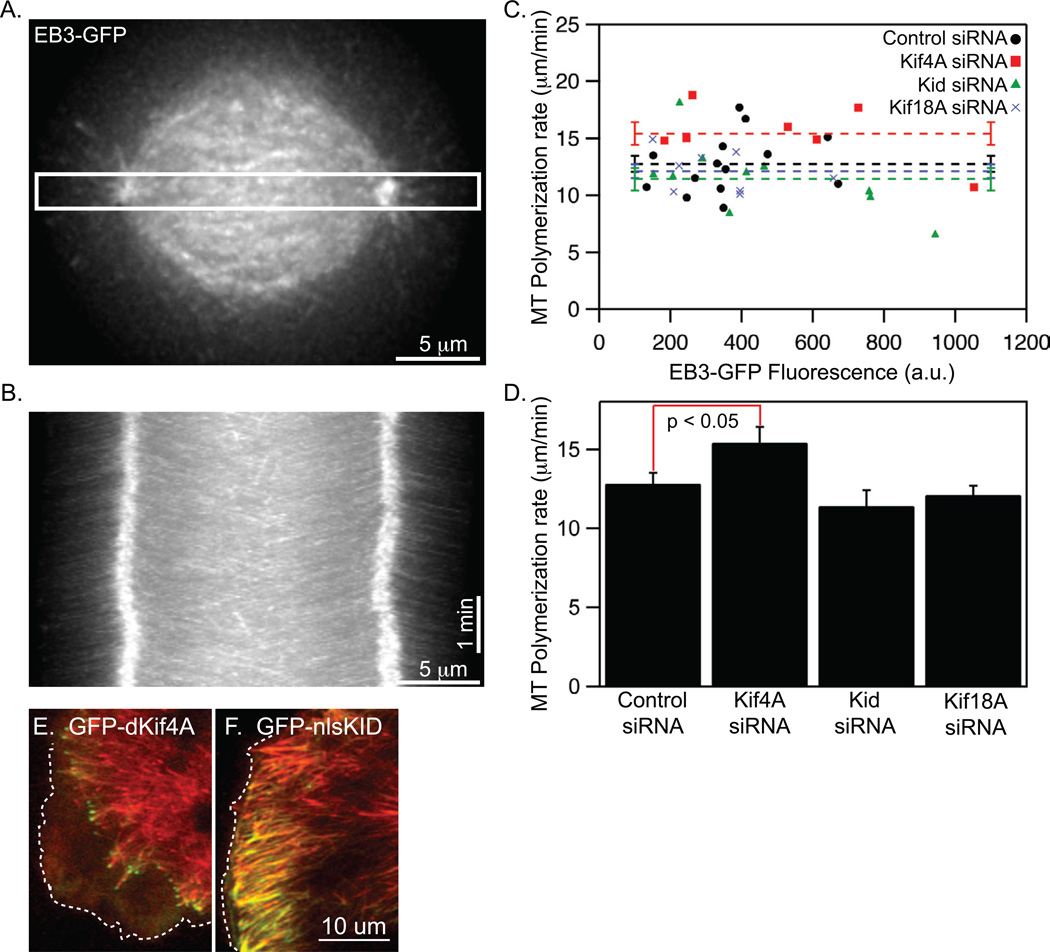
The fact that Kif4A suppresses the polar ejection force is inconsistent with the proposed idea that it uses its plus-end directed motility to “carry” chromosomes towards the spindle equator. An alternative idea is suggested by studies indicating that kinesin-4 motors may function to suppress microtubule growth (Bieling et al., 2010b; Bringmann et al., 2004; Castoldi and Vernos, 2006; Hu et al., 2011; Vernos et al., 1995). However, it is not known whether Kif4A affects microtubule polymerization within the spindle during chromosome alignment. To address this question we measured the movements of the microtubule tip-tracking protein EB3-GFP in mitotic cells (Figure 5 A–D). Polymerization rates did not positively correlate with EB3-GFP fluorescence, indicating that EB3-GFP did not measurably increase microtubule growth on its own within the range of expression levels we evaluated (Figure 5C). However, the polymerization rates of non-kinetochore microtubules were significantly faster in cells depleted of Kif4A relative to those in control, Kid or Kif18A depleted cells (Figure 5D). Consistent with these results, we also found that over-expression of Kif4A but not Kid suppressed the dynamics of interphase microtubules (Figure 5 E–F and supplemental movie S4). For these experiments, we constructed mutant versions of Kif4A (GFP-dKif4A) and Kid (GFP-Kid-NLS) that disrupt nuclear localization of the motors (Tahara et al., 2008). Taken together, these data indicate that Kif4A reduces microtubule polymerization in vivo and that it carries out this function in pre-anaphase mitotic cells.
Figure 5.
Kif4A suppresses microtubule dynamics in cells. (A) An EB3-GFP expressing cell that was imaged to measure microtubule polymerization rates. Scale bar is 5 µm. (B) Kymograph of EB3-GFP generated from the region indicated by the white box in (A). Horizontal scale bar is 5 µm, vertical scale bar is 1 minute. (C) Plot of microtubule (MT) polymerization rates as a function of EB3-GFP fluorescence. Horizontal lines indicate means and error bars indicate SEM. r = 0.27 from a Pearson correlation analysis of control cells, indicating that EB3-GFP expression is not strongly correlated with microtubule polymerization rate in the analyzed cells. (D) Average microtubule (MT) polymerization rates measured in cells treated with siRNAs targeting the indicated kinesins. p-values calculated from comparison to control siRNA measurements (n = 14 cells) are 0.048 (Kif4A siRNA, n = 7 cells), 0.29 (Kid siRNA, n = 10 cells) and 0.49 (Kif18A siRNA, n = 8 cells). Error bars indicate SEM. (E) A live interphase cell expressing a truncated version of Kif4A (GFP-dKif4A) that accumulates in the cytoplasm on microubule plus-ends. The edge of the cell membrane is indicated (dotted line). (F) A live interphase cell expressing a mutated version of Kid (GFP-nlsKid) that accumulates on cytoplasmic microtubules. In contrast to GFP-dKif4A, GFP-Kid-NLS primarily localizes to the microtuble lattice rather than plus-ends.
Kif18A and Kif4A directly suppress microtubule plus-end dynamics
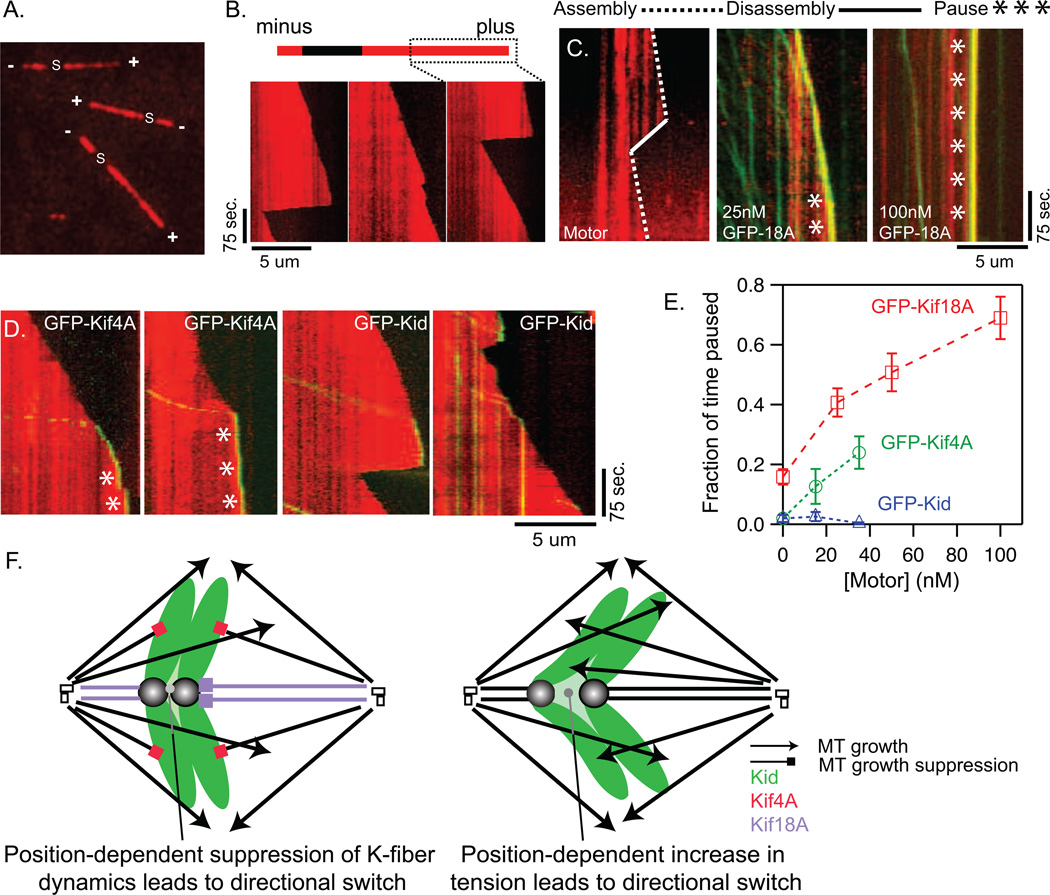
We next investigated whether the suppression of K-fiber dynamics and non-kinetochore microtubules by Kif18A and Kif4A, respectively, is due to direct activities of these motors. The effects of purified GFP-Kif18A, GFP-Kif4A and GFP-Kid (Figure S5) on dynamic microtubules were evaluated using an in vitro total internal reflection fluorescence (TIRF) microscopy assay (Figure 6 A–B). All three motors displayed ATP-dependent plus-end directed motility in these assays (Figure 6 C–D). In the absence of motor or in the presence of GFP-Kid, microtubules spent the majority of the observed time in a growth state and rarely paused (Figure 6E and supplemental movies S5–S7). In contrast, Kif18A and Kif4A robustly suppressed microtubule dynamics and promoted microtubule pausing in a concentration-dependent manner (Figure 6E and supplemental movies S5–S7). Microtubule growth suppression was accompanied by a visible accumulation of GFP-Kif18A or GFP-Kif4A at the plus-ends of microtubules (Figure 6 C–D and supplemental movies S5–S7). In cases where microtubules switched to depolymerization in the presence of motor, we did not observe GFP-foci tracking with the tips of shortening microtubules (supplemental movies S6–S7), suggesting that Kif18A, Kif4A and Kid do not remain bound to the ends of microtubules during depolymerization. These data indicate that Kif18A and Kif4A can directly suppress microtubule growth.
Figure 6.
Kif18A and Kif4A suppress microtubule dynamics by promoting plus-end pausing while Kid does not. (A) Live Alexa561-labeled microtubules polarity marked with unlabeled, unstabilized seeds (s). Plus-ends (+) and minus-ends are indicated (−). (B) Representative kymographs of live microtubule plus-ends at 37°C in the absence of motor. (C) Kymographs showing microtubule plus-end dynamics (red) in the presence of no motor (left panel); 25 nM GFP-Kif18A (green, middle panel) or 100nM GFP-Kif18A (green, right panel). Characteristically microtubules will still assemble in 25nM GFP-Kif18A but transitions to a pause state are common (*). In 100 nM GFP-Kif18A microtubules are often paused through the entire 300 sec. observation time. (D) Kymographs showing microtubule plus-end dynamics (red) in the presence of 35 nM GFP-Kif4A (left two panels) or GFP-Kid (right two panels). Microtubules appear to pause frequently (*) in the presence of GFP-Kif4A, while they remain dynamic in the presence of GFP-Kid. (E) Plot of the fraction of time microtubules spent in a paused state as a function of motor concentration ([motor]). GFP-Kif18A and GFP-Kif4A increased the proportion of time microtubules spent paused in a dose dependent manner. P<0.0001 for all concentrations. In contrast, GFP-Kid did not significantly increase the proportion of time microtubules spent paused. Note that it was necessary to analyze Kif18A and chromokinesins under slightly different buffer conditions to optimize for motor solubility (see Experimental Procedures for details). Error bars indicate SEM. (F) Model for control of chromosome movement by Kid, Kif4A and Kif18A. In control cells (top) switch rates are strictly dependent on position within the spindle and are controlled via direct suppression of K-fiber dynamics by Kif18A. In Kif18A/Kif4A depleted cells (bottom) switch rates are also dependent on position, but switching is now influenced by kinetochore tension.
Discussion
Recent evidence suggests that the majority of chromosomes in mammalian cells are laterally attached to microtubules near the center of the mitotic spindle at the start of prometaphase (Kitajima et al., 2011; Magidson et al., 2011). In order to maintain this position as paired chromosomes establish attachments to the ends of K-fibers, the dynamics of K-fibers must be spatially controlled. We demonstrate here that this control depends on the spatial regulation of centromere switch rates provided by Kif18A, Kid and Kif4A (Figure 6).
Centromere movements are predominantly confined by the activity of Kif18A. In the absence of Kif18A centromeres spend less time in an attenuated or slow velocity state, and purified Kif18A directly induces microtubule pausing in a concentration-dependent manner. We propose that Kif18A utilizes its highly processive, plus-end directed motility to accumulate in a length-dependent manner at K-fiber plus-ends, where it directly suppresses their dymanics to trigger the directional reversal of centromere pairs (Mayr et al., 2007; Stumpff et al., 2011; Stumpff et al., 2008). This mechanism, which operates independently of chromokinesins and interkinetochore tension, is the primary way human cells prevent chromosomes from straying too far from the spindle midzone (Figure 6F).
Kid and Kif4A also contribute to the spatial control of centromere movements. In contrast to Kif18A, our data indicate that Kid promotes and Kif4A antagonizes an increase in interkinetochore tension as centromeres move away from the spindle midzone. Our analyses of intercentromere distance and directional switch rates suggest that this increase in tension promotes a change in the direction of centromere movement back towards the spindle equator (Figure 6F). As this relationship between tension, spindle position and switching is masked in the presence of Kif18A, it has not been described previously. This has profound implications for the tension-sensing checkpoint and may explain why cells have evolved a second dominant mechanism to control position independent of interkinetochore tension. However, the fact that spindle length, centromere movements and centromere alignment are altered in cells lacking Kif4A or Kid activity indicates that chromokinesins are indeed important for proper spindle function. In future studies, it will be interesting to determine if the relative contribution of Kif18A and chromokinesins to chromosome alignment varies among different mammalian cell types and whether these mechanisms are necessary to maintain genomic stability.
Our finding that Kid and Kif4A oppositely regulate the polar ejection force was unexpected. Both motors display plus-end directed motility and were previously proposed to utilize this activity to transport chromosomes towards the plus-ends of microtubules, away from the spindle poles (Bieling et al., 2010a; Bringmann et al., 2004; Brouhard and Hunt, 2005; Yajima et al., 2003). However, our studies of Kif4A contradict the idea that kinesin-4 motors positively contribute to chromosome alignment and the polar ejection force (Bieling et al., 2010a; Mazumdar et al., 2004; Sekine et al., 1994; Vernos et al., 1995). Instead, our data suggest that Kif4A negatively regulates the polar ejection force by reducing microtubule growth rates. We demonstrate that human Kif4A directly suppresses microtubule dynamics, similar to the Xenopus kinesin-4, XKlp1 (Bieling et al., 2010b; Bringmann et al., 2004). This activity leads to a small but significant decrease in microtubule polymerization and reduces spindle length. Since Kif4A is primarily localized to mitotic chromatin (Lee et al., 2001), we speculate that the global change in microtubule polymerization rates we measured after Kif4A depletion could be due to the elimination of a suppressive effect Kif4A exerts on a subpopulation of microtubules that contact chromosome arms. Such a change could alter the overall tubulin dimer to polymer homeostasis, as has been previously reported for other local modulators of microtubule dynamics (Logue et al., 2011; Mimori-Kiyosue et al., 2005).
The fact that Kif4A’s effects on centromere alignment require the presence of Kid suggests that Kif4A and Kid antagonistically regulate the same polar ejection mechanism. These data support a model in which Kif4A functions to decrease the average length of microtubules within the spindle, and in doing so, limits the magnitude of the polar ejection force by reducing the number of Kid-microtubule interactions. Furthermore, through this antagonistic control of polar ejection forces, chromokinesins spatially modulate interkinetochore tension and centromere switch rates (Figure 6F).
Collectively, these data provide a molecular model for the spatial control of centromere movements and highlight an important and underappreciated role for the suppression of microtubule dynamics in this process. Kif18A suppresses the dynamics of K-fibers while Kif4A suppresses the dynamics of non-kinetochore microtubules. Interestingly, the similar biochemical activities of these two kinesin-like motors, when applied to distinct populations of spindle microtubules, oppositely contribute to the spatial constraint of centromere movements within the spindle.
Experimental Procedures
Cell culture and Transfections
HeLa cells were cultured in MEM-alpha (Gibco) with 10% fetal bovine serum (Hyclone) as previously described (Maney et al., 1998). HCT116 cells were cultured in RPMI-1640 (Gibco) media containing 10% FBS. For DNA transfections, cells were electroporated in a Nucleofector II (Lonza) according to the manufacturer’s instructions. Following electroporation, cells were either plated on glass coverslips (for fixed cell assays) or poly-L-lysine-coated glass bottom dishes (for live cell assays, Maktek corporation).
Twelve hours later, siRNAs complexed with Lipofectamine RNAiMAX (Invitrogen) were added according to manufacturer’s instructions. Each siRNA was added to a final concentration of 20 nM. For experiments involving double-depletions, control cells were treated with 40 nM negative control siRNA. Cells were fixed, imaged or lysed 36–48 hours after siRNA treatment. For monopolar spindle experiments, cells were treated with 100 µM monastrol for 2 hours prior to fixation.
Plasmids and Reagents
DNA plasmids for EGFP-CENP-B, mRFP-CENP-B, Nuf2-GFP, EB3-GFP, EB3-mRFP and Venus-centrin (a gift from Benjamin Major) were previously described (Grigoriev et al., 2008; Stepanova et al., 2003; Stumpff et al., 2008; Sundin et al., 2011). Human Kif4A and Kid genes were cloned into pEGFP-C1 (Clontech). An siRNA-resistant Kif4A gene was constructed by replacing the sequence TCCAATGTGCTCAGACGT with AGTAACGTACTCCGGAGG. dKif4A was constructed by cutting the unique XhoI site in the siRNA-resistant plasmid, filling in the 5' overhang and religating to alter the reading frame such that w.t. Kif4A translation ends at R995, followed by SSEELAWKRN. The Kid-NLS clone was made as previously described (Tahara et al., 2008). Kinesins were targeted for siRNA-mediated depletion using Silencer Validated siRNAs (Ambion) against Kif18A (GCCAAUUCUUCGUAGUUUUTT or GCUGGAUUUCAUAAAGUGGTT), Kid (GCUGCUCUCUAGAGAUUGCTT or GCAAGAUUGGAGCUACUCGTT) and Kif4A (CCAAUGUGCUCAGACGUAATT or CCAAAUCAUUUGCCGAG). Control siRNA cells were treated with Silencer Negative Control #1 siRNA (Ambion).
Cell Fixation and Immunofluorescence
HeLa or HCT116 cells were fixed for 10 minutes in −20° C methanol containing 1% paraformaldehyde. Cells were incubated for 1 hour at room temperature with primary antibodies diluted in Antibody Dilution Buffer (Tris buffered saline pH 7.4, 1% BSA, 0.1% Triton-X 100 and 0.1% sodium azide): mouse-anti-γ-tubulin (Sigma, 1:1000) or human-CREST serum (a kind gift from Bill Brinkley, 1:500). Cells were labeled for 1 hour at room temperature with anti-mouse or anti-human secondary antibodies conjugated to rhodamine or fluorescein (Jackson Laboratories). Cells were mounted in Vectashield containing DAPI (Vector) and imaged with either a Nikon equipped with a Sensys CCD camera and 60X 1.4 NA lens (Nikon) or a Deltavision system equipped with a Coolsnap HQ CCD camera (Photometrics), 60X 1.42 NA lens (Olympus) and Softworx software (Applied Precision).
Western Blots
HeLa cells were lysed in RIPA buffer (50 mM Tris-HCl pH 7.4, 150 mM NaCl, 2 mM EDTA, 1% NP-40 and 0.1% SDS) and boiled for 10 minutes after addition of Laemmli buffer to 1X. Lysates were separated on 4–12% Bis-Tris gels and transferred to nitrocellulose. Blots were probed with rabbit-anti-Kif18A (Bethyl, 1:2000), rabbit-anti-Kid (Bethyl, 1:2000), rabbit-anti-Kif4A (Bethyl, 1:2000) or mouse-anti-GAPDH (Calbiochem, 1:1000) primary antibodies and anti-rabbit or anti-mouse secondary antibodies conjugated to HRP (Jackson laboratories). Proteins were visualized by chemiluminescence using a CCD camera-based gel imaging station (Alpha Innotech).
Live Cell Imaging
Cells were cultured on poly-L-lysine coated glass bottom dishes and switched to 37° C CO2-independent media immediately prior to imaging. Cells were imaged with a Deltavision system equipped with a Coolsnap HQ CCD camera (Photometrics), Softworx software (Applied Precision) and either a 60X 1.42 NA or 60X 1.49 NA objective (Olympus). For time-lapse images of centromere movements, Z-stacks containing five optical sections were collected at 2-second intervals. Prior to analysis, images were deconvolved and projected in Softworx (Applied precision). For analyses of interphase microtubules, single-focal plane TIRF images were collected at 2 or 5 second intervals.
Quantitative Analyses of Centromere Distribution
Images of EGFP-CENP-B expressing cells were rotated such that the pole-to-pole axis was horizontal. The sum of the EGFP fluorescence in every pixel column in a rectangular region of interest (ROI) was then measured and background corrected. The height of the ROI was fixed at 20 pixels while the length of the ROI stretched between the centroids of each centrosomal γ-tubulin focus. To calculate the centromere distribution ratio (r), the spindle length was divided into quarters and the sum of the EGFP fluorescence in the two quarters nearest the poles was divided by the sum of the fluorescence in the two quarters adjacent to the spindle equator. Statistical analyses were performed using a two-tailed student t-test.
Quantitative Analyses of Monopolar Spindles
Images of monastrol treated cells were analyzed using the Point Picker plug-in for ImageJ. The midpoint between the two γ-tubulin spots in each monopolar spindle was used as the reference point for distance measurements. Kinetochore-to-pole distances were then calculated and plotted in Igor (Wavemetrics). Statistical analyses were performed using a two-tailed student t-test.
Measurement of Microtubule Polymerization Rates in Cells
Time-lapse images of EB3-GFP expressing cells were rotated such that the pole-to-pole axis was horizontal and kymographs of EB3-GFP labeled microtubule tips were generated from a rectangular ROI centered on the centrosomes. The height of the ROI was fixed at 30 pixels and the length was defined by the diameter of the cell. Microtubule polymerization rates were calculated by regression fits to each individual EB3-GFP track that could be distinguished as previously described (Tirnauer et al., 2002). All measurements from a single cell were averaged to obtain a mean microtubule polymerization rate for that cell. Similar results were obtained from analyses of spindle microtubules polymerizing towards the equator and astral microtubules polymerizing towards the cell periphery. Data from analyses of astral microtubules are reported in Figure 5. Integrated GFP fluorescence levels were quantified in each cell using ImageJ. Statistical analyses were performed using a two-tailed student t-test.
Quantitative Analyses of Centromere Movements
Bi-oriented centromeres in late prometaphase or metaphase cells were tracked using the MtrackJ plug-in (Erik Meijering, Biomedical Imaging Group Rotterdam) for ImageJ (NIH). A focal plane at the center of the spindle was chosen and only cells with spindles oriented parallel to the plane of focus were analyzed. We and others have previously determined that the movements of centromeres at the extreme periphery of the spindle differ from those along the pole-to-pole axis, so only pairs oscillating close to the long axis of the spindle were analyzed (Canman et al., 2002; Cimini et al., 2004; Stumpff et al., 2008). Kymographs of centromere movements were generated in ImageJ (NIH). The metaphase plate for each cell was objectively determined by generating a projection of all the images from a time-lapse and applying a threshold to eliminate all fluorescence except the EGFP-CENP-B signal in a single cell. A line fit to this adjusted projection in ImageJ (NIH) was defined as the metaphase plate. Centromere movement parameters and intercentromere distances were quantified from centromere tracks in Igor Pro 6.0 (Wavemetrics). Velocity measurements were made using linear regression analysis over a 5-point sliding window. The measurements made on a single centromere track were then averaged to calculate the mean velocities for a single centromere. Data points within 10-seconds of a change in direction were excluded from velocity analyses. Directional switch points were defined as points when a centromere moved consistently in one direction for at least 8 seconds (4 frames) then changed directions and moved consistently towards the opposite pole for at least 8 seconds. Oscillation amplitudes were calculated by measuring the distance moved between directional switch points. For spatially resolved measurements, centromere position was defined by the distance between the centromere and the metaphase plate. The sign of the distance was determined by a centromere’s position relative to the pole it was attached to. The distance was negative if a centromere was on the same side of the metaphase plate as the pole it was attached to and positive if a centromere was on the opposite side of the metaphase plate as the pole it was attached to. For determining the position of a pair of centromeres, the distance between the metaphase plate and the center of mass (COM) of the pair was calculated. COM is defined as the position exactly halfway between the two sister centromeres. All statistical analyses were performed using a two-tailed student t-test.
Live Microtubule Assay
GFP-Kif18A-6XHis, GFP-Kif4A-6XHis and GFP-Kid-6XHis proteins were expressed and purified from sf9 cells as previously described (Stumpff et al., 2011). The effects of these motors on the behavior of dynamic microtubules was determined using a live microtubule TIRF assay as previously described (Stumpff et al., 2011). Briefly, dynamic microtubules attached to PEG-silane coated glass coverslips via rigor-kinesin were imaged at 2-second intervals in the presence or absence of motor in either chromokinesin buffer (45 mM KCl, 36 mM PIPES-K pH 6.9, 90 mM Acetate-K, 22 mM imidazole-HCl, 5 mM MgCl2, 1 mM EGTA, 1% glycerol, 0.05% methylcellulose, 0.06% Brij-35 and 100 µg/ml kappa-casein) or Kif18A buffer (60 mM KCl, 15 mM PIPES-K pH 6.9, 12 mM imidazole-HCl, 2 mM MgCl2, 0.2 mM EGTA, 0.5% glycerol, 0.1% methycellulose and 700 µg/ml kappa-casein) supplemented with 10 mM glucose, 2 mM DTT, 200 µg/ml Glucose Oxidase, 15 µg/ml Catalase, 1 mM ATP, 1 mM GTP and 1 mg/ml bovine brain tubulin (1/120 conjugated to Alexa568). Imaging was carried out using a Personal Deltavision microscope (Applied Precision) outfitted with 4-laser TIRF capabilities, Olympus 60X, 1.49 NA TIRF objective and Ultimate focus (Applied Precision) at 37°C. The plus-ends of individual microtubules were tracked using SoftWoRx Explorer 1.3.0 (Applied Precision) and kymographs were prepared from representative microtubules in ImageJ 1.42q (National Institutes of Health). A pause was defined as no measurable lengthening for 10 frames (20 seconds).
HIGHLIGHTS.
Spatial control of centromere switch rates is predominantly provided by Kif18A
Kid and Kif4A spatially regulate kinetochore tension to control switch rates
The chromokinesins Kid and Kif4A oppositely tune the polar ejection force
Kif18A and Kif4A suppress the growth of distinct subsets of spindle microtubules
Supplementary Material
Acknowledgements
The authors thank Carol Huseby for technical assistance; Dr. Jennifer DeLuca, Dr. Anna Akhmanova and Dr. Benjamin Major for reagents; and members of the Wordeman, Biggins and Asbury labs for critical reading of the manuscript. This work was supported by a National Institutes of Health grant (GM69429) and National Science Foundation grant (1041173) to L.W.; a National Institutes of Health grant (GM79373) to C.A.; and a Ruth L. Kirschstein National Service Award (GM778572) and Leukemia and Lymphoma Society Special Fellow Award (3652-11) to J.S. Implementation of TIRF capabilities on the Personal DV microscope was provided by Applied Precision, Issaquah, WA.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Akiyoshi B, Sarangapani KK, Powers AF, Nelson CR, Reichow SL, Arellano-Santoyo H, Gonen T, Ranish JA, Asbury CL, Biggins S. Tension directly stabilizes reconstituted kinetochore-microtubule attachments. Nature. 2010;468:576–579. doi: 10.1038/nature09594. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Antonio C, Ferby I, Wilhelm H, Jones M, Karsenti E, Nebreda AR, Vernos I. Xkid, a chromokinesin required for chromosome alignment on the metaphase plate. Cell. 2000;102:425–435. doi: 10.1016/s0092-8674(00)00048-9. [DOI] [PubMed] [Google Scholar]
- Bieling P, Kronja I, Surrey T. Microtubule Motility on Reconstituted Meiotic Chromatin. Curr Biol. 2010a doi: 10.1016/j.cub.2010.02.067. [DOI] [PubMed] [Google Scholar]
- Bieling P, Telley IA, Surrey T. A Minimal Midzone Protein Module Controls Formation and Length of Antiparallel Microtubule Overlaps. Cell. 2010b;142:420–432. doi: 10.1016/j.cell.2010.06.033. [DOI] [PubMed] [Google Scholar]
- Bringmann H, Skiniotis G, Spilker A, Kandels-Lewis S, Vernos I, Surrey T. A kinesin-like motor inhibits microtubule dynamic instability. Science. 2004;303:1519–1522. doi: 10.1126/science.1094838. [DOI] [PubMed] [Google Scholar]
- Brouhard GJ, Hunt AJ. Microtubule movements on the arms of mitotic chromosomes: polar ejection forces quantified in vitro. Proc Natl Acad Sci U S A. 2005;102:13903–13908. doi: 10.1073/pnas.0506017102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cai S, O'Connell CB, Khodjakov A, Walczak CE. Chromosome congression in the absence of kinetochore fibres. Nature cell biology. 2009;11:832–838. doi: 10.1038/ncb1890. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Canman JC, Salmon ED, Fang G. Inducing precocious anaphase in cultured mammalian cells. Cell Motil Cytoskeleton. 2002;52:61–65. doi: 10.1002/cm.10032. [DOI] [PubMed] [Google Scholar]
- Cassimeris L, Rieder CL, Salmon ED. Microtubule assembly and kinetochore directional instability in vertebrate monopolar spindles: implications for the mechanism of chromosome congression. J Cell Sci. 1994;107(Pt 1):285–297. doi: 10.1242/jcs.107.1.285. [DOI] [PubMed] [Google Scholar]
- Castoldi M, Vernos I. Chromokinesin Xklp1 contributes to the regulation of microtubule density and organization during spindle assembly. Mol Biol Cell. 2006;17:1451–1460. doi: 10.1091/mbc.E05-04-0271. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cheng L, Zhang J, Ahmad S, Rozier L, Yu H, Deng H, Mao Y. Aurora B regulates formin mDia3 in achieving metaphase chromosome alignment. Developmental cell. 2011;20:342–352. doi: 10.1016/j.devcel.2011.01.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cimini D, Cameron LA, Salmon ED. Anaphase spindle mechanics prevent mis-segregation of merotelically oriented chromosomes. Curr Biol. 2004;14:2149–2155. doi: 10.1016/j.cub.2004.11.029. [DOI] [PubMed] [Google Scholar]
- Civelekoglu-Scholey G, Sharp DJ, Mogilner A, Scholey JM. Model of chromosome motility in Drosophila embryos: adaptation of a general mechanism for rapid mitosis. Biophys J. 2006;90:3966–3982. doi: 10.1529/biophysj.105.078691. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Du Y, English CA, Ohi R. The kinesin-8 Kif18A dampens microtubule plus-end dynamics. Curr Biol. 2010;20:374–380. doi: 10.1016/j.cub.2009.12.049. [DOI] [PubMed] [Google Scholar]
- Franck AD, Powers AF, Gestaut DR, Gonen T, Davis TN, Asbury CL. Tension applied through the Dam1 complex promotes microtubule elongation providing a direct mechanism for length control in mitosis. Nat Cell Biol. 2007;9:832–837. doi: 10.1038/ncb1609. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Funabiki H, Murray AW. The Xenopus chromokinesin Xkid is essential for metaphase chromosome alignment and must be degraded to allow anaphase chromosome movement. Cell. 2000;102:411–424. doi: 10.1016/s0092-8674(00)00047-7. [DOI] [PubMed] [Google Scholar]
- Garcia MA, Koonrugsa N, Toda T. Two kinesin-like Kin I family proteins in fission yeast regulate the establishment of metaphase and the onset of anaphase A. Curr Biol. 2002;12:610–621. doi: 10.1016/s0960-9822(02)00761-3. [DOI] [PubMed] [Google Scholar]
- Gardner MK, Pearson CG, Sprague BL, Zarzar TR, Bloom K, Salmon ED, Odde DJ. Tension-dependent regulation of microtubule dynamics at kinetochores can explain metaphase congression in yeast. Mol Biol Cell. 2005;16:3764–3775. doi: 10.1091/mbc.E05-04-0275. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grigoriev I, Gouveia SM, van der Vaart B, Demmers J, Smyth JT, Honnappa S, Splinter D, Steinmetz MO, Putney JW, Jr, Hoogenraad CC, et al. STIM1 is a MT-plus-end-tracking protein involved in remodeling of the ER. Current biology : CB. 2008;18:177–182. doi: 10.1016/j.cub.2007.12.050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gupta ML, Jr, Carvalho P, Roof DM, Pellman D. Plus end-specific depolymerase activity of Kip3, a kinesin-8 protein, explains its role in positioning the yeast mitotic spindle. Nat Cell Biol. 2006;8:913–923. doi: 10.1038/ncb1457. [DOI] [PubMed] [Google Scholar]
- Hu CK, Coughlin M, Field CM, Mitchison TJ. KIF4 regulates midzone length during cytokinesis. Current biology : CB. 2011;21:815–824. doi: 10.1016/j.cub.2011.04.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Inoue S, Salmon ED. Force generation by microtubule assembly/disassembly in mitosis and related movements. Mol Biol Cell. 1995;6:1619–1640. doi: 10.1091/mbc.6.12.1619. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jaqaman K, King EM, Amaro AC, Winter JR, Dorn JF, Elliott HL, McHedlishvili N, McClelland SE, Porter IM, Posch M, et al. Kinetochore alignment within the metaphase plate is regulated by centromere stiffness and microtubule depolymerases. J Cell Biol. 2010;188:665–679. doi: 10.1083/jcb.200909005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kapoor TM, Lampson MA, Hergert P, Cameron L, Cimini D, Salmon ED, McEwen BF, Khodjakov A. Chromosomes can congress to the metaphase plate before biorientation. Science. 2006;311:388–391. doi: 10.1126/science.1122142. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ke K, Cheng J, Hunt AJ. The distribution of polar ejection forces determines the amplitude of chromosome directional instability. Curr Biol. 2009;19:807–815. doi: 10.1016/j.cub.2009.04.036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Khodjakov A, Rieder CL. Kinetochores moving away from their associated pole do not exert a significant pushing force on the chromosome. J Cell Biol. 1996;135:315–327. doi: 10.1083/jcb.135.2.315. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim Y, Holland AJ, Lan W, Cleveland DW. Aurora kinases and protein phosphatase 1 mediate chromosome congression through regulation of CENP-E. Cell. 2010;142:444–455. doi: 10.1016/j.cell.2010.06.039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kitajima TS, Ohsugi M, Ellenberg J. Complete kinetochore tracking reveals error-prone homologous chromosome biorientation in mammalian oocytes. Cell. 2011;146:568–581. doi: 10.1016/j.cell.2011.07.031. [DOI] [PubMed] [Google Scholar]
- Lee YM, Lee S, Lee E, Shin H, Hahn H, Choi W, Kim W. Human kinesin superfamily member 4 is dominantly localized in the nuclear matrix and is associated with chromosomes during mitosis. The Biochemical journal. 2001;360:549–556. doi: 10.1042/0264-6021:3600549. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Levesque AA, Compton DA. The chromokinesin Kid is necessary for chromosome arm orientation and oscillation, but not congression, on mitotic spindles. J Cell Biol. 2001;154:1135–1146. doi: 10.1083/jcb.200106093. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Logue JS, Whiting JL, Tunquist B, Sacks DB, Langeberg LK, Wordeman L, Scott JD. AKAP220 protein organizes signaling elements that impact cell migration. The Journal of biological chemistry. 2011;286:39269–39281. doi: 10.1074/jbc.M111.277756. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Magidson V, O'Connell CB, Loncarek J, Paul R, Mogilner A, Khodjakov A. The spatial arrangement of chromosomes during prometaphase facilitates spindle assembly. Cell. 2011;146:555–567. doi: 10.1016/j.cell.2011.07.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maney T, Hunter AW, Wagenbach M, Wordeman L. Mitotic centromere-associated kinesin is important for anaphase chromosome segregation. J Cell Biol. 1998;142:787–801. doi: 10.1083/jcb.142.3.787. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mayr MI, Hummer S, Bormann J, Gruner T, Adio S, Woehlke G, Mayer TU. The human kinesin Kif18A is a motile microtubule depolymerase essential for chromosome congression. Curr Biol. 2007;17:488–498. doi: 10.1016/j.cub.2007.02.036. [DOI] [PubMed] [Google Scholar]
- Mazumdar M, Misteli T. Chromokinesins: multitalented players in mitosis. Trends Cell Biol. 2005;15:349–355. doi: 10.1016/j.tcb.2005.05.006. [DOI] [PubMed] [Google Scholar]
- Mazumdar M, Sundareshan S, Misteli T. Human chromokinesin KIF4A functions in chromosome condensation and segregation. J Cell Biol. 2004;166:613–620. doi: 10.1083/jcb.200401142. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mimori-Kiyosue Y, Grigoriev I, Lansbergen G, Sasaki H, Matsui C, Severin F, Galjart N, Grosveld F, Vorobjev I, Tsukita S, et al. CLASP1 and CLASP2 bind to EB1 and regulate microtubule plus-end dynamics at the cell cortex. The Journal of cell biology. 2005;168:141–153. doi: 10.1083/jcb.200405094. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nicklas RB. The forces that move chromosomes in mitosis. Annual review of biophysics and biophysical chemistry. 1988;17:431–449. doi: 10.1146/annurev.bb.17.060188.002243. [DOI] [PubMed] [Google Scholar]
- Rieder CL, Davison EA, Jensen LC, Cassimeris L, Salmon ED. Oscillatory movements of monooriented chromosomes and their position relative to the spindle pole result from the ejection properties of the aster and half-spindle. J Cell Biol. 1986;103:581–591. doi: 10.1083/jcb.103.2.581. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rieder CL, Salmon ED. Motile kinetochores and polar ejection forces dictate chromosome position on the vertebrate mitotic spindle. J Cell Biol. 1994;124:223–233. doi: 10.1083/jcb.124.3.223. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sekine Y, Okada Y, Noda Y, Kondo S, Aizawa H, Takemura R, Hirokawa N. A novel microtubule-based motor protein (KIF4) for organelle transports, whose expression is regulated developmentally. J Cell Biol. 1994;127:187–201. doi: 10.1083/jcb.127.1.187. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shelby RD, Hahn KM, Sullivan KF. Dynamic elastic behavior of alpha-satellite DNA domains visualized in situ in living human cells. The Journal of cell biology. 1996;135:545–557. doi: 10.1083/jcb.135.3.545. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Skibbens RV, Rieder CL, Salmon ED. Kinetochore motility after severing between sister centromeres using laser microsurgery: evidence that kinetochore directional instability and position is regulated by tension. J Cell Sci. 1995;108(Pt 7):2537–2548. doi: 10.1242/jcs.108.7.2537. [DOI] [PubMed] [Google Scholar]
- Skibbens RV, Salmon ED. Micromanipulation of chromosomes in mitotic vertebrate tissue cells: tension controls the state of kinetochore movement. Exp Cell Res. 1997;235:314–324. doi: 10.1006/excr.1997.3691. [DOI] [PubMed] [Google Scholar]
- Skibbens RV, Skeen VP, Salmon ED. Directional instability of kinetochore motility during chromosome congression and segregation in mitotic newt lung cells: a push-pull mechanism. J Cell Biol. 1993;122:859–875. doi: 10.1083/jcb.122.4.859. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stepanova T, Slemmer J, Hoogenraad CC, Lansbergen G, Dortland B, De Zeeuw CI, Grosveld F, van Cappellen G, Akhmanova A, Galjart N. Visualization of microtubule growth in cultured neurons via the use of EB3-GFP (end-binding protein 3-green fluorescent protein) J Neurosci. 2003;23:2655–2664. doi: 10.1523/JNEUROSCI.23-07-02655.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stumpff J, Du Y, English CA, Maliga Z, Wagenbach M, Asbury C, Wordeman L, Ohi R. A Tethering Mechanism Controls the Processivity and Kinetochore-Microtubule Plus-End Enrichment of the Kinesin-8 Kif18A. Mol Cell. 2011 doi: 10.1016/j.molcel.2011.07.022. in press. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stumpff J, von Dassow G, Wagenbach M, Asbury C, Wordeman L. The kinesin-8 motor Kif18A suppresses kinetochore movements to control mitotic chromosome alignment. Dev Cell. 2008;14:252–262. doi: 10.1016/j.devcel.2007.11.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sundin LJ, Guimaraes GJ, Deluca JG. The NDC80 complex proteins Nuf2 and Hec1 make distinct contributions to kinetochore-microtubule attachment in mitosis. Molecular biology of the cell. 2011 doi: 10.1091/mbc.E10-08-0671. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tahara K, Takagi M, Ohsugi M, Sone T, Nishiumi F, Maeshima K, Horiuchi Y, Tokai-Nishizumi N, Imamoto F, Yamamoto T, et al. Importin-beta and the small guanosine triphosphatase Ran mediate chromosome loading of the human chromokinesin Kid. J Cell Biol. 2008;180:493–506. doi: 10.1083/jcb.200708003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tirnauer JS, Canman JC, Salmon ED, Mitchison TJ. EB1 targets to kinetochores with attached, polymerizing microtubules. Mol Biol Cell. 2002;13:4308–4316. doi: 10.1091/mbc.E02-04-0236. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Varga V, Helenius J, Tanaka K, Hyman AA, Tanaka TU, Howard J. Yeast kinesin-8 depolymerizes microtubules in a length-dependent manner. Nat Cell Biol. 2006;8:957–962. doi: 10.1038/ncb1462. [DOI] [PubMed] [Google Scholar]
- Vernos I, Raats J, Hirano T, Heasman J, Karsenti E, Wylie C. Xklp1, a chromosomal Xenopus kinesin-like protein essential for spindle organization and chromosome positioning. Cell. 1995;81:117–127. doi: 10.1016/0092-8674(95)90376-3. [DOI] [PubMed] [Google Scholar]
- Wargacki MM, Tay JC, Muller EG, Asbury CL, Davis TN. Kip3, the yeast kinesin-8, is required for clustering of kinetochores at metaphase. Cell Cycle. 2010;9 doi: 10.4161/cc.9.13.12076. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Waters JC, Skibbens RV, Salmon ED. Oscillating mitotic newt lung cell kinetochores are, on average, under tension and rarely push. J Cell Sci. 1996;109(Pt 12):2823–2831. doi: 10.1242/jcs.109.12.2823. [DOI] [PubMed] [Google Scholar]
- West RR, Malmstrom T, McIntosh JR. Kinesins klp5(+) and klp6(+) are required for normal chromosome movement in mitosis. J Cell Sci. 2002;115:931–940. doi: 10.1242/jcs.115.5.931. [DOI] [PubMed] [Google Scholar]
- Yajima J, Edamatsu M, Watai-Nishii J, Tokai-Nishizumi N, Yamamoto T, Toyoshima YY. The human chromokinesin Kid is a plus end-directed microtubule-based motor. Embo J. 2003;22:1067–1074. doi: 10.1093/emboj/cdg102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yen TJ, Compton DA, Wise D, Zinkowski RP, Brinkley BR, Earnshaw WC, Cleveland DW. CENP-E, a novel human centromere-associated protein required for progression from metaphase to anaphase. The EMBO journal. 1991;10:1245–1254. doi: 10.1002/j.1460-2075.1991.tb08066.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu C, Zhao J, Bibikova M, Leverson JD, Bossy-Wetzel E, Fan JB, Abraham RT, Jiang W. Functional Analysis of Human Microtubule-based Motor Proteins, the Kinesins and Dyneins, in Mitosis/Cytokinesis Using RNA Interference. Mol Biol Cell. 2005;16:3187–3199. doi: 10.1091/mbc.E05-02-0167. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.