Abstract
Nanoparticle-based delivery has become an important strategy to advance therapeutic oligonucleotides into clinical reality. Delivery by nanocarriers can enhance access of oligonucleotides to their pharmacological targets within cells; preferably, targeting ligands are incorporated into nanoparticles for targeting oligonucleotides to disease sites, often by conjugation to delivery carriers. In this study, a splice-switching oligonucleotide (SSO) was conjugated to a bivalent RGD peptide, and then, the RGD-SSO conjugate was formulated into polyplexes with a cationic polymer polyethylenimine. The resultant polyplexes of RGD-oligonucleotide conjugate demonstrated dramatic increase in the pharmacological response of splicing correction compared to free RGD-SSO conjugate or the polyplexes of unconjugated SSO, through integrin-mediated endocytosis and rapid endosomal release. This study has shown that coupling a targeting ligand to cargo oligonucleotide can maintain the integrin targeting ability after the peptide-oligonucleotide conjugate is complexed with cationic polymer. Preliminary study also revealed that integrin targeting redirects intracellular trafficking of the polyplexes to caveolar pathway and thereby generates greater effectiveness of the oligonucleotide. This study provides a new platform technology to construct multifunctional delivery systems of therapeutic oligonucleotides.
Keywords: Antisense, RGD peptide, Integrin, Nanoparticles, Polyethylenimine, Polyplexes, Splice-switching oligonucleotide, Targeted delivery
INTRODUCTION
Antisense and siRNA oligonucleotides have shown mounting promise to become mainstream therapeutic entities. As of 2010, 43 therapeutic oligonucleotides were in clinical trials.1,2 For example, a splice-switching oligonucleotide (SSO), capable of altering alternative splicing of pre-mRNA in the nucleus, has shown to restore dystrophin function in patients with Duchenne muscular dystrophy in a phase II clinical trial.3 In spite of the promise, full potential of oligonucleotide-based therapeutics has not been unleashed, as oligonucleotides can theoretically regulate expression of any gene and there are over 1000 disease-related genes in the human genome and even more in human pathogens.4,5 The underlying hurdle is the enormous difficulty for oligonucleotides, which are hydrophilic and often charged macromolecules, to cross cellular membranes and then traffic to their sites of action in the cytosol or nucleus.6–8 One strategy to overcome this barrier is to complex oligonucleotides with lipids,9 polymers,10 or cell-penetrating peptides,11 and then the resultant nanoparticles allow intracellular delivery of the oligonucleotides to their intracellular sites of action. Preferably, targeting ligands are incorporated into these nanoparticles to improve delivery to target cells in disease and minimize side effects to irrelevant cells. For example, polymer-based nanoparticles coated with transferrin were able to deliver siRNAs to solid tumors overexpressing transferrin receptors and cause gene specific RNAi activity in humans;12 this clinical study highlights the importance of targeted delivery for siRNA therapeutics.
Arginine-glycine-aspartic Acid (RGD) peptide has been widely used to modify nanoparticles for targeting gene and drugs to tumors, as it specifically binds to integrin αvβ3, a cell surface glycoprotein that is preferentially expressed in angiogenic endothelia and in certain tumor types including melanomas.13 In these applications, RGD peptide is often conjugated to nanocarriers including cationic lipids and polymers, and then the RGD-carrier conjugates are complexed with negatively charged nucleic acids to form nanoparticles.13 For example, polyethylenimine (PEI) was conjugated directly to a RGD peptide, and then the RGD-PEI was condensed with plasmid DNA into nanoparticles that demonstrated greater in vitro transfection efficiency.14 In vivo behaviors of an integrin-targeted nanoparticle can be improved by incorporating polyethylene glycol (PEG) to decrease non-specific binding.15–17 Thus, in the latter study, RGD-PEG-PEI particles selectively delivered VEGFR2 siRNA to integrin-expressed tumor tissues and suppressed tumor growth and angiogenesis in tumor-bearing mice.17
Delivery of gene-specific SSOs into tumor cells can redirect alternative splicing of Bcl-x18 and STAT319 and cause apoptosis of the tumor cells. Thus, coupling tumor-targeting ligands such as RGD peptide with antitumor SSOs in nanoparticles will provide a novel strategy for targeted tumor therapy. In the current study, a SSO, which is designed to correct splicing of an aberrant intron inserted into an eGFP reporter gene, was used as model oligonucleotide. It was conjugated to a bivalent RGD peptide, and then, the RGD-SSO conjugate was formulated with PEI into nanoparticles. Functional delivery of the targeted polyplexes was tested in integrin αvβ3 expressing A375SM cells that had been stably transfected with the eGFP gene interrupted by the abnormally spliced intron. Successful delivery of the SSO to the cell nucleus would lead to up-regulation of eGFP expression, providing a positive read-out. The results showed that the polyplexes of RGD-SSO conjugate produced greater and faster reporter gene induction compared to free RGD-SSO conjugate, or polyplexes of free SSO, through integrin-mediated endocytosis and efficient endosomal release. Preliminary study also demonstrated that integrin-targeting changes intracellular trafficking of the nanoparticles and thereby produces greater effectiveness of the oligonucleotide.
MATERIALS AND METHODS
Syntheses of RGD-oligonucleotide Conjugate
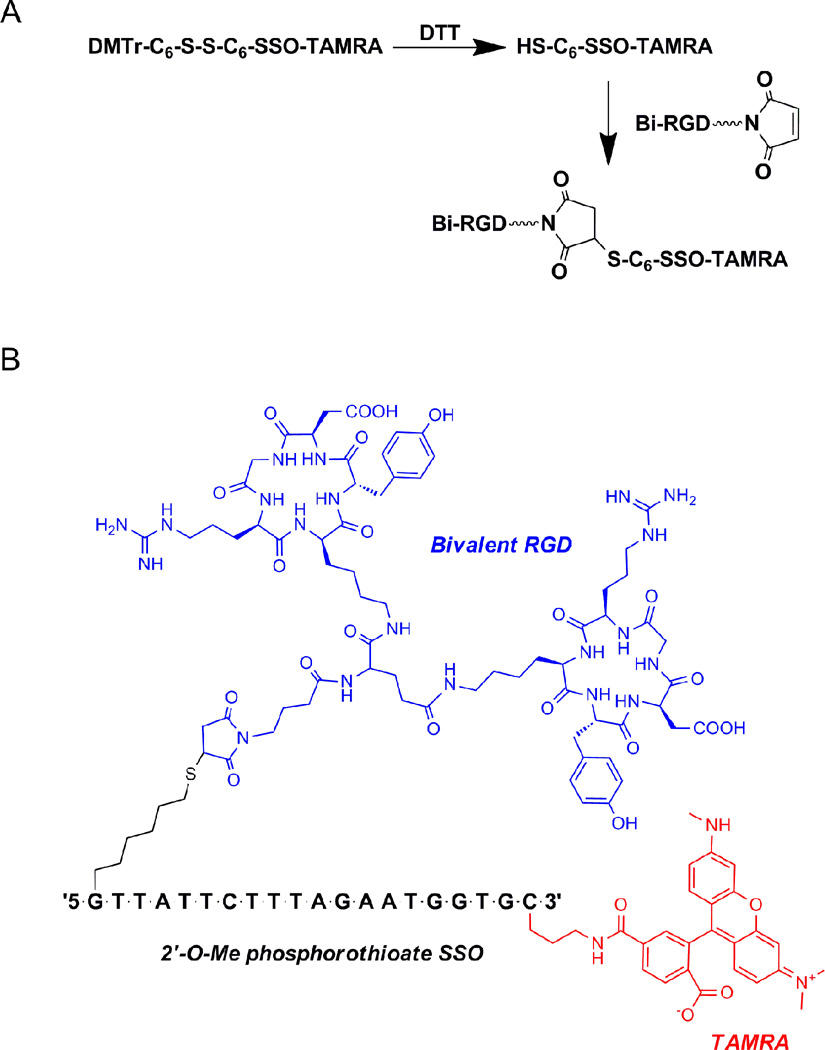
Peptide-oligonucleotide conjugates were constructed as described previously.20 Briefly, the SSO (5'-GTTATTCTTTAGAATGGTGC-3') is a 2'-O-Me oligonucleotide with phosphorothioate linkages. The 3’-TAMRA conjugates were synthesized using phosphoramidites of the ultraMILD-protected bases on CPG supports (Glen Research, Sterling, VA, USA) in a AB 3400 DNA synthesizer (Applied Biosystems, Foster City, CA, USA) and a thiol linker was introduced at the 5′-end of the oligonucleotide. The 5′-thiol functionality was generated by treating the disulfide bond of the oligonucleotide with 100 mM of aqueous DTT (Fig 1A). The 5'-thiol oligonucleotide was reacted with the maleimide-containing bivalent cyclic RGD peptides for 3 h to prepare peptide conjugate (Fig 1A). The reaction mixture was then purified by HPLC using a 1 mL Resource Q column (GE Healthcare, Uppsala, Sweden). The purified conjugate was dialyzed versus milli-Q water, and analyzed by MALDI-TOF (Applied Biosystem).
Fig. 1.
(A) Synthetic scheme for the conjugation of oligonucleotide to the bivalent RGD peptide.20 (B) Chemical structure of the RGD-SSO-TAMRA.
Cell Lines
Integrin αvβ3 expressing A375SM cells were cultured in DMEM medium (Invitrogen, Carlsbad, CA, USA) supplemented with 10% fetal bovine serum (FBS) (Sigma, St. Louis, MO, USA). The A375SM cells were stably transfected with eGFP gene inserted by an aberrant intron as described previously,21 and were referred to as A375/eGFP654 cells. They were cultured in DMEM medium containing 10% FBS and 200 µg/mL hygromycin B (Roche, Mannheim, Germany). M21+ melanoma cells with high expression of integrin αvβ3, as well as M21− cells that lack this integrin, were obtained from Dr. D. Cheresh (University of California, San Diego)22 and were cultured in DMEM medium (Invitrogen) supplemented with 10% FBS (Sigma).
Preparation of Polyplexes
To prepare polyplexes at different N/P ratios, variable amounts of jetPEI™ (Polyplus, Illkirch, France) were diluted into 100 µl of 150 mM NaCl solution and were then mixed by vortex with an equal volume of NaCl solution containing 120 pmoles of the SSO or RGD-SSO. The resultant dose solution contained 600 nM SSO or RGD-SSO. After 20-min incubation at room temperature, the polyplexes were then added to the cells. N/P ratio was calculated as a ratio between the positively charged amine groups of PEI and negatively charged phosphate groups of SSO. N/P molar ratios were calculated based on the fact that there are 7.5 nitrogen residues per µl of jetPEI™ solutions as per manufacturers’ instructions.
Particle Size and Zeta Potential Measurement
The average particle sizes of the polyplexes in 150 mM NaCl solution were determined using a Coulter N5 Plus Sub-Micron Particle Sizer (Beckman Coulter, Miami, FL, USA) at a fixed angle of 90° and a temperature of 25°C. Light scattering intensity was maintained within the required range of the instrument (5 × 104 to 1 × 106 counts/sec) in all the measurements. Each sample was analyzed in triplicate. The same polyplexes were placed in the dip cell of Zetasizer Nano Z (Malvern Instruments, Westborough, MA, USA) to determine zeta potential. Each sample was analyzed in quintuplicate.
Cellular Transfection by Polyplexes
A375/eGFP654 cells were seeded on 24-well plates at 6 × 104 cells per well in various experiments. The following day, 100 µl of the polyplexes at the concentration of 600 nM were then added to each well containing 500 µl Opti-MEM I medium (Invitrogen), which made up the 0.6 mL dose solution containing 100 nM SSO. Following the 4-h treatment at 37 °C, the cells were subsequently washed with PBS and were then replenished with fresh media for another 20-h culture. Then functional delivery was examined by measuring eGFP expression with flow cytometry. In one experiment, A375/eGFP654 cells were treated with the polyplexes of SSO or RGD-SSO (25 nM, N/P = 6) in the absence or presence of 10 µM cyclic RGD peptide RGDfV (Peptides International Inc., Louisville, KY, USA). In another experiment, the polyplexes were added to the wells containing DMEM media or the same media plus 10% FBS in order to examine serum effect on transfection.
In a dose-dependence experiment, A375/eGFP654 cells were treated with increasing concentrations of the PEI/SSO or PEI/RGD-SSO polyplexes (N/P = 6) for 4 h. After another 20-h culture, eGFP expression was measured by flow cytometry. In a time-dependence experiment, A375/eGFP654 cells were dosed with the PEI/SSO or PEI/RGD-SSO polyplexes (25 nM, N/P = 6) at the same time. Then, the dose solution was removed and changed to fresh media at various times and eGFP expression was measured by flow cytometry 24 h after dosing, so that the total time of treatment and following culture remained constant for all the time point groups.
Flow Cytometric Analysis
Total cellular uptake of the TAMRA-labeled SSO and eGFP expression induced by the SSO were measured by flow cytometry using a LSR II cell analyzer (Becton-Dickenson, San Jose, CA, USA). After treatment with the polyplexes and further culture, the cells were trypsinized and analyzed by flow cytometry, with a 488 nm laser coupled with a 525/50 filter for eGFP and a 561 nm laser coupled with a 582/15 emission filter for TAMRA. In two-color flow cytometry, the cells containing a single fluorophore were used as controls to set up compensation.
Confocal Fluorescence Microscopy
Intracellular distribution of the oligonucleotides and induced eGFP expression in living cells were examined using an Olympus Confocal FV300 fluorescent microscope with 60× oil immersion objectives. A375/eGFP654 cells (for induction experiment) or A375SM (for colocalization experiment) were plated in 35 mm glass bottom microwell dishes (MatTek, Ashland, MA, USA) and were then transfected with the polyplexes of SSO-TAMRA or RGD-SSO-TAMRA (100 nM, N/P = 6). Cellular accumulation of the TAMRA-labeled oligonucleotides and eGFP induction were visualized after 4-h transfection plus 20-h culture. Colocalization of the oligonucleotides (100 nM) with Alexa 488-labeled transferrin (Tfn) or cholera toxin B (CTB) (Invitrogen), as markers for clathrin-coated vesicles or lipid rafts, respectively, was also accomplished via confocal microscopy. Cellular distribution of the oligonucleotides was observed by confocal microscopy after 2-h transfection, and transferrin (20 µg/mL) and cholera toxin B (4 µg/mL) were used for 15 and 30 min, respectively, prior to live cell imaging.
Cytotoxicity Assay
The cytotoxicity of polyplexes was measured with the Alamar Blue assay.23 In brief, A375/eGFP654 cells were seeded in 96-well plates at 3000 cells/well. After 24 hours, the cells were exposed to different concentrations of polyplexes (N/P = 6) for 4 h. Polyplex-containing medium was replaced with fresh medium, and cells were incubated for another 20 h. Alamar Blue reagent was added and incubated for 2 hour. The samples were read in a FLUOstar Omega microplate reader (BMG LABTECH, Cary, NC, USA) set at 540 nm excitation wavelength and 590 nm emission wavelength.
Data Analysis
Data are expressed as mean ± SD from three measurements unless otherwise noted. Statistical significance was evaluated using t-test for two-sample comparison or ANOVA followed by Dunnet’s test for multiple comparisons. The data were analyzed with GraphPad Prism 5 (GraphPad Software, Inc., La Jolla, CA, USA).
RESULTS
Syntheses and Characterization
The chemical structure of the final oligonucleotide conjugate is shown in Figure 1B. A TAMRA fluorophore was introduced at the 3'-end of the oligonucleotide. The bivalent RGD peptide with maleimide functionality was coupled with the 5'-thiol oligonucleotide. After purification by chromatography and dialysis, the identity of the final products were confirmed by MALDI-TOF mass spectroscopy (SSO-TAMRA: calculated mass = 7692.0 (M+H)+ and mass found = 7691.8 (M+H)+; RGD-SSO-TAMRA: calculated mass = 9427.3 (M+H)+ and mass found = 9427.4 (M+H)+).
Particle Sizes and Zeta Potentials of Polyplexes
Average particle sizes and zeta potentials for the polyplexes at the N/P value of 6 are summarized in Table 1. The diameters for the PEI/SSO and PEI/RGD-SSO polyplexes were 352.5 and 325.4 nm, respectively, and were not statistically different, indicating that incorporating RGD peptide does not change the polyplex formation. Polydispersity index (P.I.) values were small (< 0.15), indicating uniform and monodispersed nanoparticles. Both PEI/SSO and PEI/RGD-SSO polyplexes showed positive zeta potential around 20 mV.
Table 1.
Particle sizes of the polyplexesa
| Particle Size (nm) | P.I. | Zeta Potential (mV) | |
|---|---|---|---|
| PEI/SSO | 352.5 ± 8.3 | 0.136 ± 0.136 | 18.5 ± 2.4 |
| PEI/R-SSO | 325.4 ± 20.5 | 0.117 ± 0.249 | 23.4 ± 1.5 |
Results are expressed as mean ± SD (n = 3 or 5)
Functional Delivery of Polyplexes
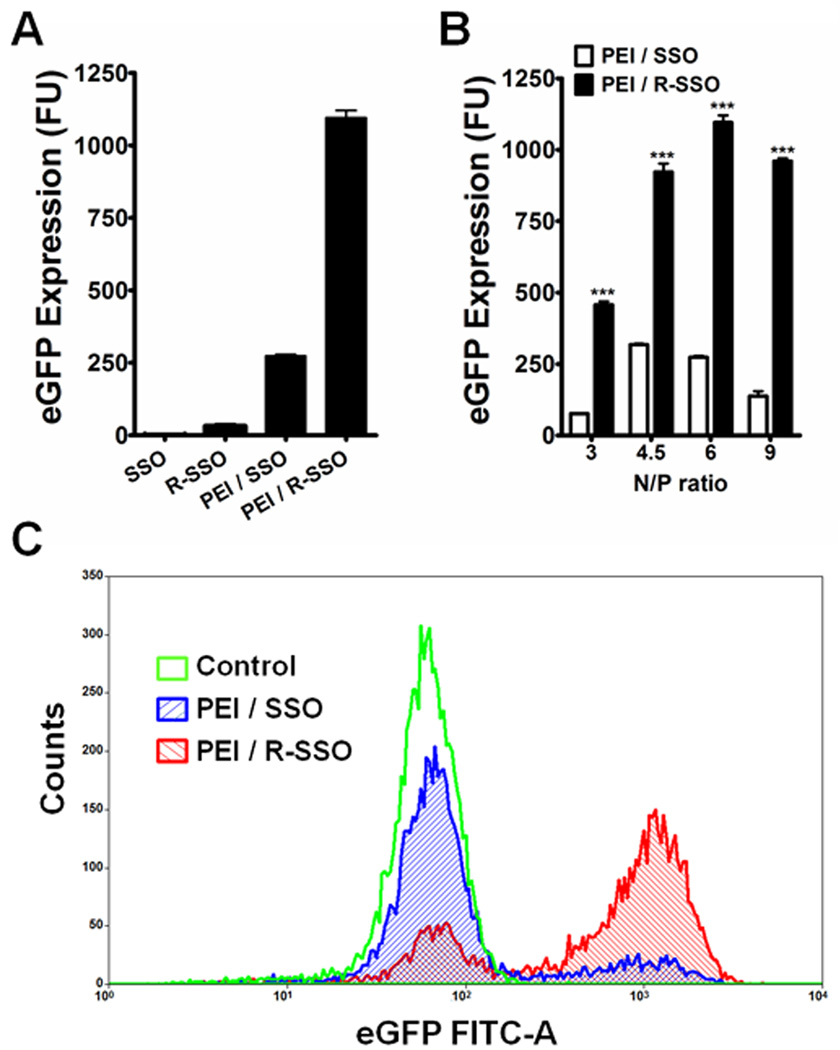
Treatment of the polyplexes of SSO or RGD-SSO at the concentration of 100 nM for 4 h and further 20-h culture produced dramatic increases in eGFP expression, whereas direct treatment with free SSO or RGD-SSO under the same condition failed to produce induction at similar levels (Fig 2A). In all the ratios of the amounts of PEI and the SSO tested, the RGD-SSO polyplexes produced significant higher transfection than the PEI/SSO polyplexes, and the optimal transfection with the polyplexes was achieved when the N/P ratio was 6 (Fig 2B). At this ratio, PEI/RGD-SSO produced 4.1-folded higher expression levels than PEI/SSO, and the transfection efficiency of PEI/RGD-SSO was 65% while that of PEI/SSO was 17% (Fig 2C). The decrease of eGFP induction at the N/P ratio of 9 (Fig 2B) might be due to the cytotoxicity of the polyplexes to the transfected cells at the high N/P ratio.
Fig. 2.
Functional delivery of polyplexes of SSO or RGD-SSO. A. Comparison of eGFP induction by the treatments of free SSO, RGD-SSO, and their PEI polyplexes with the N/P ratio of 6. B. Induction of eGFP by PEI/SSO or PEI/RGD-SSO polyplexes prepared with the N/P ratios at 3, 4.5, 6 and 9. Data represent mean ± S.D. of a representative experiment in triplicate. Statistical significance between the PEI/SSO and PEI/RGD-SSO treatments was evaluated using unpaired t tests. *** indicates p < 0.001. C. Flow cytometry of the control A375/eGFP654 cells and those treated by the PEI/SSO or PEI/RGD-SSO polyplexes with the N/P ratio of 6.
Integrin-Mediated Cellular Delivery and Serum Effect
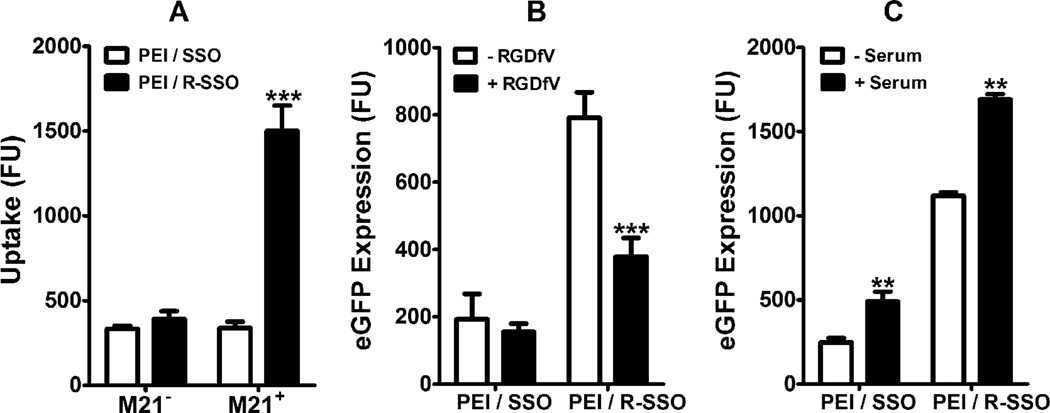
In order to test whether the superior transfection of the PEI/RGD-SSO polyplexes is due to receptor-mediated endocytosis involving integrin αvβ3, cellular uptake of the polyplexes was evaluated in integrin αvβ3-expressing M21+ cells as well as M21− cells that lack this integrin. Uptake of the PEI/RGD-SSO polyplexes was 4-fold higher in M21+ cells than that in M21− cells, whereas uptake of the PEI/SSO polyplexes showed similar uptake in these two cell lines (Fig 3A). This indicated that the PEI/RGD-SSO polyplexes undergo integrin-mediated endocytosis to enter the cells. To further confirm this mechanism, the biological effect of the polyplexes was tested under coincubation with excess amounts (10 µM) of a cyclic RGD peptide (RGDfV) that is known to be a selective inhibitor of integrin αvβ3. Co-incubation with this peptide led to an inhibition of the effect of PEI/RGD-SSO on eGFP expression but did not affect that of PEI/SSO (Fig 3B). This observation supports the concept that the effect of PEI/RGD-SSO on splicing largely depends on its initial uptake via the integrin receptor.
Fig. 3.
Integrin-mediated cellular delivery and serum effect. A. Uptake of PEI/SSO or PEI/RGD-SSO polyplexes (25 nM SSO) was compared in αvβ3 positive M21+ cells and M21− cells that do not express this integrin. B. Free RGDfV peptide at the concentration of 10 µM was added to the cells 30 min, prior to transfection with either PEI/SSO or PEI/RGD-SSO polyplexes at the concentrations of 25 nM SSO for 4 h. C. A375/eGFP654 cells were treated with either PEI/SSO or PEI/RGD-SSO polyplexes in the absence or presence of 10% FBS for 4 h. Data represent mean ± S.D. of a representative experiment in triplicate. Statistical significance between the control and the treatments was evaluated using unpaired t tests. ** indicates p < 0.01 and *** indicates p < 0.001.
The presence of 10% FBS in polyplex-containing media increased eGFP induction by 198% and 151% for PEI/SSO and PEI/RGD-SSO, respectively, when compared to those in serum-free media (Fig 3C). This indicated that both PEI/SSO and PEI/RGD-SSO polyplexes are stable in the presence of the serum and that the serum does not interfere with the integrin targeting of the PEI/RGD-SSO polyplexes. The effects of serum on nanoparticle-mediated gene transfection are variable, largely depending on carrier property, cell type, and complex stability,24 but this result supported the notion that serum proteins can enhance PEI-mediated gene delivery.25
Dose- and Time-Response Studies
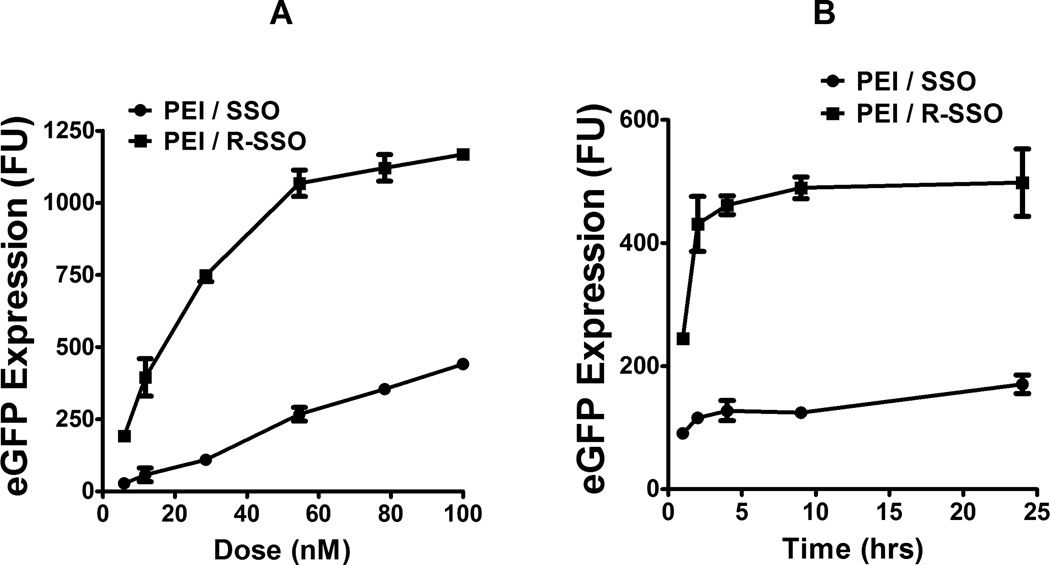
Functional delivery of the polyplexes was evaluated as a function of concentration with 4-h treatment followed by 20-h culture. The eGFP induction by PEI/RGD-SSO as a function of concentration was saturable, while that by PEI/SSO was linear up to 100 nM (Fig 4A), supporting the notion that PEI/RGD-SSO utilizes a saturable receptor-mediated mechanism for functional delivery.26
Fig. 4.
Dose- and time-dependent responses to the polyplexes. A. In the dose-dependence experiment, A375/eGFP654 cells were treated with increasing concentrations of the PEI/SSO or PEI/RGD-SSO polyplexes for 4 h followed by another 20-h culture. B. In the time-dependence experiment, A375/eGFP654 cells were dosed with the PEI/SSO or PEI/RGD-SSO polyplexes at the concentration of 25 nM. Then the dose solution was changed to fresh media at various times and eGFP expression was measured 24 h after dosing by flow cytometry.
The time-dependence response of eGFP induction to the polyplexes was examined by removing the dose solution at various times. The result in Fig 4B showed a quick onset for both the PEI/SSO and PEI/RGD-SSO polyplexes. Both polyplexes showed dramatic increase of transfection at the first 2 h, and further treatment only produced moderate increase in the effect, which might result from the rapid endosomal release of the PEI polyplexes.
Cytotoxicity
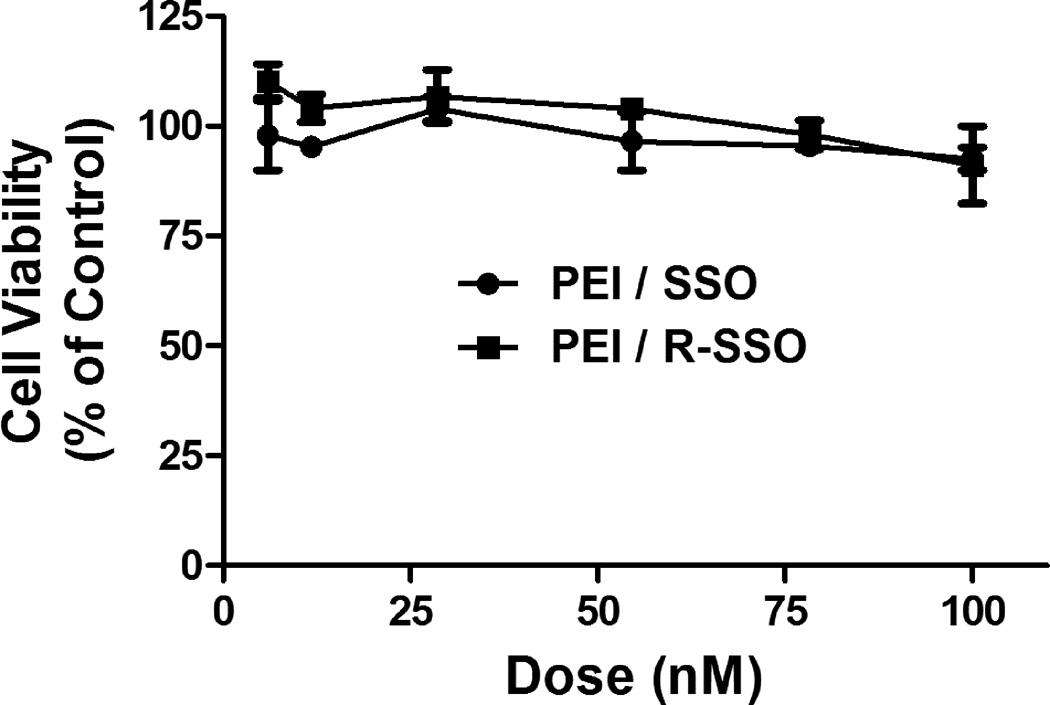
There was little toxicity associated with the use of the polyplexes of SSO or RGD-SSO (N/P = 6) at the concentrations examined. Thus in the Alamar Blue assay, the viability of the cells treated with up to 100 nM oligonucleotides in polyplexes were over 90% of the control cells (Fig 5). In the dose-response and time-response studies, the cells treated by the polyplexes showed similar profiles in the forward scattering and side scattering as compared to the untreated control cells in flow cytometry analysis (data not shown), and additionally, the cells treated with the polyplexes maintained normal morphology as illustrated in the confocal images of Figs 6B and 7.
Fig. 5.
Cytotoxicity of the polyplexes. The cytotoxicity of the PEI/SSO and PEI/RGD-SSO polyplexes (N/P = 6) to A375/eGFP654 cells was measured in 96-well plates with the Alamar Blue assay.
Fig. 6.
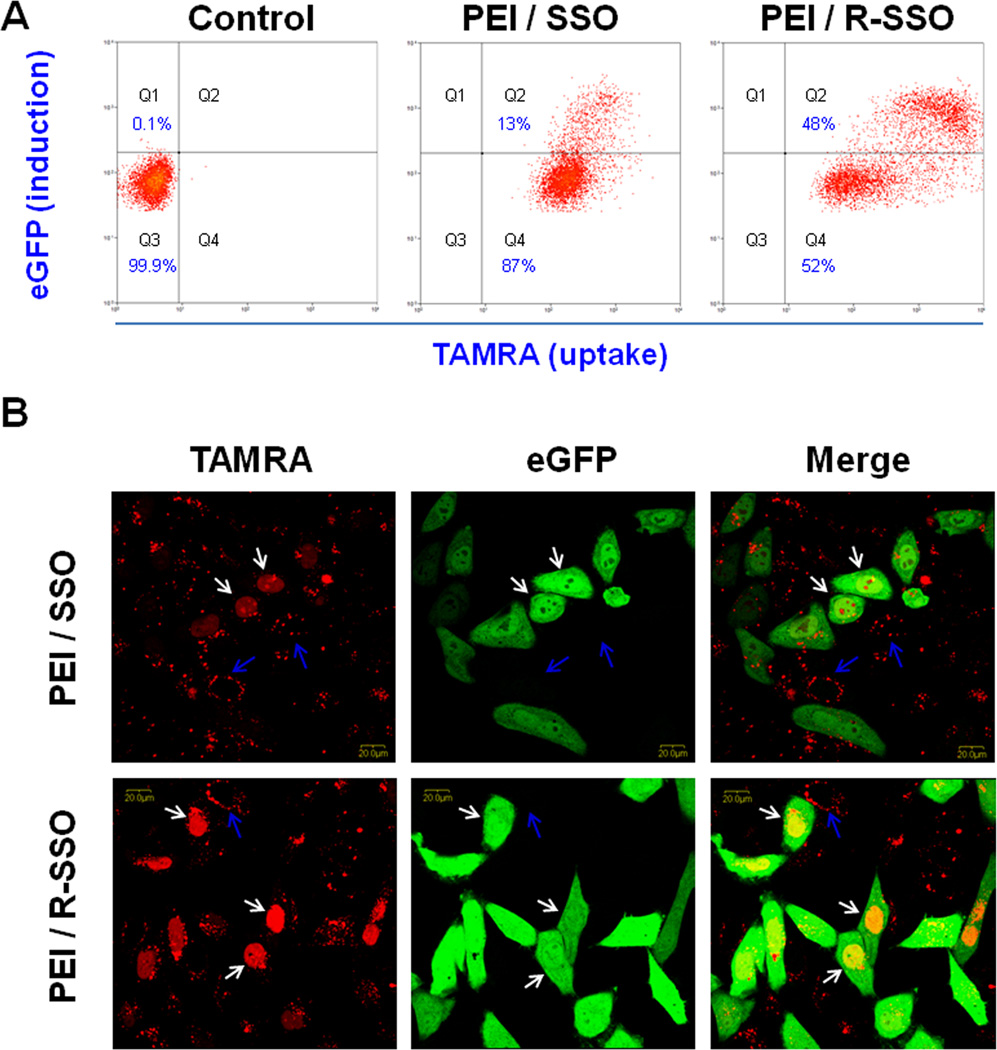
Relationship between cellular uptake and functional induction of the polyplexes. A. A375/eGFP654 cells were transfected for 4 h with the PEI polyplexes of SSO or RGD-SSO that were labeled with TAMRA. After another 20-h culture, the accumulation of SSO-TAMRA (abscissa) and induction of eGFP (ordinate) were analyzed with flow cytometry. The population of A375/eGFP654 cells without any treatment (the control cells) only showed in Q3 in the flow cytometry graph. The cell populations showing in Q2 and Q4 had higher TAMRA fluorescence and had taken up SSO-TAMRA; however, only the cells in Q2 had higher eGFP expression levels, which resulted from functional delivery of SSO to their pharmacological sites in the nucleus. B. A375/eGFP654 cells were seeded on the cover glass and then transfected with the polyplexes of SSO-TAMRA or RGD-SSO-TAMRA. After 20-h culture, the distribution of SSO-TAMRA (shown in red) and induction of eGFP (shown in green) were observed with a confocal microscope. Representative cells showing nuclear staining of SSO-TAMRA were marked with white arrows, whereas the cells without nuclear entry of SSO-TAMRA were labeled with blue arrows.
Fig. 7.
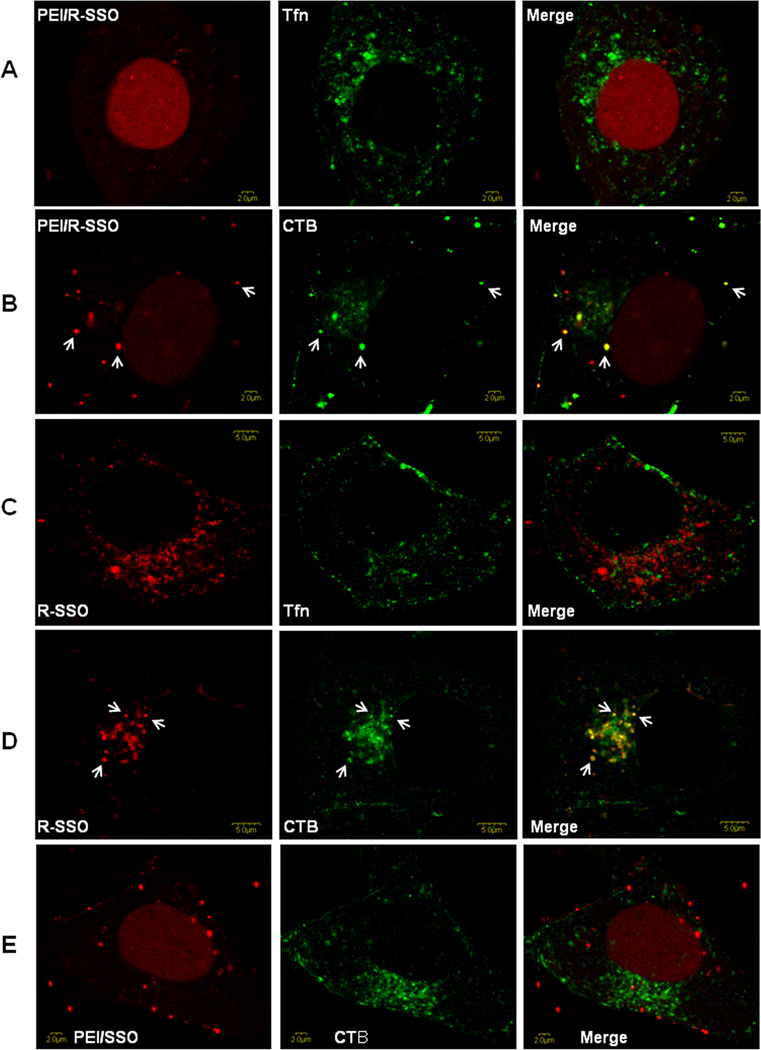
Subcellular localization of the polyplexes. A375SM were treated with PEI/SSO-TAMRA, PEI/RGD-SSO-TAMRA, or RGD-SSO-TAMRA at the concentration of 100 nM for 2 h. Alexa 488-labeled Tfn (20 µg/mL) or CTB (4 µg/mL), was added to the dose solutions 15 and 30 min, respectively, prior to live cell imaging. Colocalization of the TAMRA-labeled SSO with endocytosis markers was observed by confocal fluorescence microscopy as described in the materials and methods section. A. PEI/RGD-SSO with Tfn; B. PEI/RGD-SSO with CTB; C. RGD-SSO with Tfn; D. RGD-SSO with CTB; E. PEI/SSO with CTB. Representative cells showing co-localization were marked with white arrows.
Functional Delivery of SSO Depends on Its Nuclear Entry
In order to examine the correlation between cellular uptake of the SSO polyplexes and their pharmacological responses, two-color flow cytometry was used to simultaneously measure uptake of the polyplexes of TAMRA labeled oligonucleotide (the abscissa) and the functional induction of eGFP reporter (the ordinate) (Fig 6A). The population of A375/eGFP654 cells without any treatment (the control cells) only appeared in Q3 in the flow graph. The cell populations in Q2 and Q4 had higher TAMRA fluorescence, indicating that SSO-TAMRA was taken up by these cells; however, only the cells in Q2 expressed higher eGFP levels, indicating successful delivery of SSO to their site of action in the nucleus. Transfection with the SSO formulated into the polyplexes led to functional eGFP induction due to correct splicing of an aberrant intron inserted into this reporter gene (Fig 6A). The result indicated that functional induction requires high accumulation of SSO in the cells since the cells with higher eGFP expression in Q2 also had higher accumulation of SSO-TAMRA. The result also showed that 48% of the cell population transfected with the PEI/RGD-SSO polyplexes was in Q2, comparing to 13% of the cells transfected with the PEI/SSO polyplexes (Fig. 6A). In addition, the level of TAMRA-labeled oligonucleotide in Q2 was higher for the PEI/RGD-SSO polyplexes than that for the PEI/SSO polyplexes, which may partially explain the greater efficacy of the PEI/RGD-SSO polyplexes.
In Fig 6B, after transfection and following culture, A375/eGFP654 cells were observed with confocal microscopy for the intracellular distribution of SSO-TAMRA (shown in red) and functional eGFP expression (shown in green). The results demonstrated a correlation of nuclear entry of the oligonucleotide and functional eGFP induction. The cells with the SSO-TAMRA in the nucleus (labeled with white arrows) tended to show strong eGFP expression, indicating that the ability of oligonucleotide to traffic to the nucleus, the site of splicing correction, determines its effectiveness as a therapeutic oligonucleotide. The cells treated with PEI/RGD-SSO showed stronger nuclear accumulation of SSO, and thus the eGFP expression was higher compared to that of PEI/SSO (Fig 6B), indicating that greater nuclear entry of oligonucleotide by targeted polyplexes is responsible for their superior effectiveness.
Subcellular Localization
As seen in Figs 7A and 7B, RGD-SSO-TAMRA delivered by the PEI polyplexes entered the nucleus in large amount within 2 h, while some of the material continued to be vesicular and some modest cytosolic fluorescence was discernible. RGD-SSO-TAMRA without complexation with PEI displayed substantial uptake into vesicular structures in the cytosol (Figs 7C and 7D), these presumably being various types of endosomes, indicating that low eGFP induction by the oligonucleotide conjugate is due to poor endosomal release. In order to more precisely identify the endomembrane compartments involved in the uptake and trafficking of the polyplexes, A375SM cells were coincubated with the TAMRA-labeled oligonucleotide (red) in the polyplexes and Alexa 488 (green) labeled markers for well-known endocytotic pathways. Thus, transferrin is known to be internalized via the clathrin coated pit pathway, while cholera toxin B is substantially internalized via the caveolar pathway or other lipid raft dependent pathways.27 As seen in Fig 7A, at 2 h there was virtually no overlap of the labels for transferrin and RGD-SSO-TAMRA delivery by PEI; in contrast there was substantial overlap of the cholera toxin B and the RGD-SSO-TAMRA in the vesicles (Fig 7B). Similar colocalization was observed for free RGD-SSO-TAMRA without PEI complexation (Figs 7C and 7D). This suggests that the initial uptake pathway for RGD-SSO-TAMRA, either complexed with PEI or not, involves caveolae; this observation is consistent with literature on the mechanism of internalization of integrin αvβ3.28 In contrast, there was no overlap of the labels for cholera toxin B and SSO-TAMRA delivered by the PEI polyplexes (Fig 7E), indicating that the SSO polyplexes enter the cells by the endocytotic pathways that do not involve caveolae, and incorporating RGD peptide to oligonucleotide in the polyplexes changes the uptake pathway of the polyplexes. This change of the uptake pathway may contribute to the greater effectiveness of the targeted polyplexes.
DISCUSSION
There are mounting efforts in maturing antisense and siRNA oligonucleotides into therapeutic entities as evidenced by the 43 siRNA and antisense drugs in clinical trials as of 2010.1,2 In most of these clinical trials, oligonucleotides are given as ‘free’ molecules, and this has led to suboptimal efficacy and limited tissue distribution.8 Receptor-targeted delivery systems not only improve cellular uptake, but also allow preferential delivery of therapeutics to particular tissues and cell types of interest.8 One simple approach for targeted delivery is to covalently link therapeutic oligonucleotide to receptor ligand, so that the resultant conjugate can bind to the receptor at disease sites and undergo endocytosis for intracellular delivery.29 Antisense and siRNA oligonucleotides have been conjugated to ligands with high affinity for integrin αvβ3,20,30,31 for a G protein-coupled receptor,26 or for the sigma receptor.32 Although peptide-oligonucleotide conjugates demonstrate much higher effectiveness than free oligonucleotides in cellular models, they often need long treatment time for optimal response to occur.26,31 As it is more difficult to achieve long retention time of the conjugates around the target cells in animal models, “free” RGD-SSO only showed moderate activity in vivo (unpublished data). Persistent endosomal localization of the conjugates in the cells suggests that endosomal release may occur at a slow rate and may present the rate-limiting step for the pharmacological action.26,31
This study aimed to increase the endosomal release of oligonucleotides by coupling an endosomal releasing agent to the targeted conjugates. Gene delivery by PEI polyplexes has proven a successful strategy thanks to the effective endosomal release property of PEI via the proton sponge effect.33 Therefore, PEI has been chosen to be complexed with RGD-oligonucleotide conjugate. The results from this study demonstrated that PEI/RGD-SSO showed dramatic increase of eGFP induction with only 2-h treatment (Fig 4B), and the pharmacological response was significantly higher than the polyplexes of free SSO. Free SSO and RGD-SSO were equally effective in splicing correction when delivered directly to the cytosol by scrape-loading or electroporation,30 indicating that there is no difference in the intrinsic efficacy or nuclear entry rate between them. In fact, nuclear entry is not the rate-limiting step for monomeric oligonucleotides in light of the observation that oligonucleotide rapidly accumulates in the nucleus within minutes after being microinjected into the cytoplasm of mammalian cells.34 Thus, the enhancement in transfection should be due to the different cellular uptake and the followed intracellular transport processes. The enhancement of induction by PEI/RGD-SSO can be reversed by coincubation with an excess amount of RGDfV peptides, whereas these RGD peptides do not affect the transfection by PEI/SSO. Collectively, the enhancement in functional delivery of the RGD-SSO polyplexes is due to integrin αvβ3-mediated endocytosis.
RGD-surface-modified nanoparticles, including polyplexes and liposomes, have been utilized to deliver therapeutic genes for a decade.13 RGD peptide is often chemically conjugated to cationic carriers such as PEI; however, the different conjugation methods have produced variable outcomes in functional delivery of nucleic acids. The simplest conjugation method is to directly conjugate PEI with a RGD peptide, and RGD-PEI complexes of plasmid DNA demonstrated enhanced in vitro transfection efficiency.14 PEG has been used as a spacer between PEI and RGD peptide in order to overcome nonspecific binding of PEI polyplexes.16,17 However, another study indicated that using PEG as spacer impairs gene transfection, possibly by shielding RGD-integrin binding,15 while a recent study indicated that RGD-PEI-PEG is a better construct than RGD-PEG-PEI for gene delivery.35 The disparity may rise from the complicated chemical reactions involving heterogeneous polymers, in which the targeting peptides may be inactivated.36 This study showed that conjugating RGD peptide to the cargo oligonucleotide can maintain the integrin targeting ability after the peptide-oligonucleotide conjugate is complexed with PEI. Conjugation of targeting peptides to oligonucleotides involves relatively simple chemical reactions and generates the monomeric ligand-oligonucleotide conjugates with defined molecular weight,20,26 which may provide an advantage in drug development. In addition, multiple functionalities, including targeting ligands and endosomal release agents, are required to be incorporated into the delivery systems for superior transfection efficiency. Thus, the method of coupling the targeting ligands to oligonucleotides provides flexibility for constructing multifunctional nanoparticles.
The results in this study also revealed that the RGD modified polyplexes may utilize an internalization pathway that is different from the unmodified polyplexes. Previous study has shown that the SSO polyplexes may arrive at the nucleus, the site of action, via a combination of multiple noncaveolar and clathrin-independent endocytosis pathways.21 In this study, a larger amount of RGD-SSO delivered by PEI enters the nucleus and thus produces greater pharmacological response compared to the PEI/SSO polyplexes (Fig 6B). Further colocalization of the polyplexes to endocytosis markers demonstrated that the RGD modified polyplexes utilize the caveolar pathway, whereas the nontargeted polyplexes do not (Fig 7). This may indicate that the caveolar pathway is a productive mechanism for functional delivery of nucleic acids. In another study, RGD-PEG-block-poly(lysine) was shown to improve plasmid transfection dramatically in Hela cells expressing integrin αvβ3.37 Initial studies supported that the presence of RGD peptide changed the intracellular trafficking route of the polyplexes to the caveolar pathway followed by trafficking to the perinuclear region, thereby avoiding the lysosomal degradation of delivered genes.37 Despite these exciting preliminary observations, the enormous complexity of intracellular trafficking of therapeutic oligonucleotides has just begun to be elucidated38 and more mechanism studies are warranted to advance the understanding of the key cellular steps underlying targeted delivery of nucleic acids.
In conclusion, the study showed that coupling a targeting ligand to a cargo oligonucleotide can maintain the integrin targeting ability after the peptide-oligonucleotide conjugate is complexed with cationic polymer. A preliminary study also demonstrated that integrin targeting alters intracellular trafficking of the nanoparticles and thus leads to superior effectiveness of the oligonucleotide. This study provides a novel platform technology to construct multifunctional delivery systems of therapeutic oligonucleotides. As SSOs have demonstrated antitumor activity,18,19 integrin-targeted delivery system will bring oligonucleotide-based therapeutics even closer to clinic reality.
ACKNOWLEDGMENTS
The authors gratefully acknowledge Dr. Rudy L. Juliano (University of North Carolina at Chapel Hill, USA) for invaluable discussion on the study and proof-reading of the manuscript. This work was supported by NIH grants 1R01CA151964 and UL1RR025747.
Abbreviations
- CTB
cholera toxin B
- eGFP
enhanced green fluorescence protein
- PEI
polyethylenimine
- P.I.
polydispersity index
- RGD
Arginine-Glycine-Aspartic Acid
- SSO
splice-switching oligonucleotide
- Tfn
transferrin
REFERENCES
- 1.Vaishnaw AK, Gollob J, Gamba-Vitalo C, Hutabarat R, Sah D, Meyers R, de Fougerolles T, Maraganore J. A status report on RNAi therapeutics. Silence. 2010;1(1):14. doi: 10.1186/1758-907X-1-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Bennett CF, Swayze EE. RNA targeting therapeutics: molecular mechanisms of antisense oligonucleotides as a therapeutic platform. Annu Rev Pharmacol Toxicol. 2010;50:259–293. doi: 10.1146/annurev.pharmtox.010909.105654. [DOI] [PubMed] [Google Scholar]
- 3.Cirak S, Arechavala-Gomeza V, Guglieri M, Feng L, Torelli S, Anthony K, Abbs S, Garralda ME, Bourke J, Wells DJ, Dickson G, Wood MJ, Wilton SD, Straub V, Kole R, Shrewsbury SB, Sewry C, Morgan JE, Bushby K, Muntoni F. Exon skipping and dystrophin restoration in patients with Duchenne muscular dystrophy after systemic phosphorodiamidate morpholino oligomer treatment: an open-label, phase 2, dose-escalation study. Lancet. 2011;378(9791):595–605. doi: 10.1016/S0140-6736(11)60756-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Paolini GV, Shapland RH, van Hoorn WP, Mason JS, Hopkins AL. Global mapping of pharmacological space. Nat Biotechnol. 2006;24(7):805–815. doi: 10.1038/nbt1228. [DOI] [PubMed] [Google Scholar]
- 5.Overington JP, Al-Lazikani B, Hopkins AL. How many drug targets are there? Nat Rev Drug Discov. 2006;5(12):993–996. doi: 10.1038/nrd2199. [DOI] [PubMed] [Google Scholar]
- 6.Juliano R, Bauman J, Kang H, Ming X. Biological barriers to therapy with antisense and siRNA oligonucleotides. Mol Pharm. 2009;6(3):686–695. doi: 10.1021/mp900093r. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Whitehead KA, Langer R, Anderson DG. Knocking down barriers: advances in siRNA delivery. Nat Rev Drug Discov. 2009;8(2):129–138. doi: 10.1038/nrd2742. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Ming X. Cellular delivery of siRNA and antisense oligonucleotides via receptor-mediated endocytosis. Expert Opin Drug Deliv. 2011;8(4):435–449. doi: 10.1517/17425247.2011.561313. [DOI] [PubMed] [Google Scholar]
- 9.Tseng YC, Mozumdar S, Huang L. Lipid-based systemic delivery of siRNA. Adv Drug Deliv Rev. 2009;61(9):721–731. doi: 10.1016/j.addr.2009.03.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Kim WJ, Kim SW. Efficient siRNA delivery with non-viral polymeric vehicles. Pharm Res. 2009;26(3):657–666. doi: 10.1007/s11095-008-9774-1. [DOI] [PubMed] [Google Scholar]
- 11.Said Hassane F, Saleh AF, Abes R, Gait MJ, Lebleu B. Cell penetrating peptides: overview and applications to the delivery of oligonucleotides. Cell Mol Life Sci. 2010;67(5):715–726. doi: 10.1007/s00018-009-0186-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Davis ME, Zuckerman JE, Choi CH, Seligson D, Tolcher A, Alabi CA, Yen Y, Heidel JD, Ribas A. Evidence of RNAi in humans from systemically administered siRNA via targeted nanoparticles. Nature. 2010;464(7291):1067–1070. doi: 10.1038/nature08956. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Juliano RL, Ming X, Nakagawa O, Xu R, Yoo H. Integrin targeted delivery of gene therapeutics. Theranostics. 2011;1:211–219. doi: 10.7150/thno/v01p0211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Erbacher P, Remy JS, Behr JP. Gene transfer with synthetic virus-like particles via the integrin-mediated endocytosis pathway. Gene Ther. 1999;6(1):138–145. doi: 10.1038/sj.gt.3300783. [DOI] [PubMed] [Google Scholar]
- 15.Kunath K, Merdan T, Hegener O, Haberlein H, Kissel T. Integrin targeting using RGD-PEI conjugates for in vitro gene transfer. J Gene Med. 2003;5(7):588–599. doi: 10.1002/jgm.382. [DOI] [PubMed] [Google Scholar]
- 16.Woodle MC, Scaria P, Ganesh S, Subramanian K, Titmas R, Cheng C, Yang J, Pan Y, Weng K, Gu C, Torkelson S. Sterically stabilized polyplex: ligand-mediated activity. J Control Release. 2001;74(1–3):309–311. doi: 10.1016/s0168-3659(01)00339-x. [DOI] [PubMed] [Google Scholar]
- 17.Schiffelers RM, Ansari A, Xu J, Zhou Q, Tang Q, Storm G, Molema G, Lu PY, Scaria PV, Woodle MC. Cancer siRNA therapy by tumor selective delivery with ligand-targeted sterically stabilized nanoparticle. Nucleic Acids Res. 2004;32(19):e149. doi: 10.1093/nar/gnh140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Bauman JA, Li SD, Yang A, Huang L, Kole R. Anti-tumor activity of splice-switching oligonucleotides. Nucleic Acids Res. 2010;38(22):8348–8356. doi: 10.1093/nar/gkq731. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Zammarchi F, de Stanchina E, Bournazou E, Supakorndej T, Martires K, Riedel E, Corben AD, Bromberg JF, Cartegni L. Antitumorigenic potential of STAT3 alternative splicing modulation. Proc Natl Acad Sci U S A. 2011;108(43):17779–17784. doi: 10.1073/pnas.1108482108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Alam MR, Dixit V, Kang H, Li ZB, Chen X, Trejo J, Fisher M, Juliano RL. Intracellular delivery of an anionic antisense oligonucleotide via receptor-mediated endocytosis. Nucleic Acids Res. 2008;36(8):2764–2776. doi: 10.1093/nar/gkn115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Ming X, Sato K, Juliano RL. Unconventional internalization mechanisms underlying functional delivery of antisense oligonucleotides via cationic lipoplexes and polyplexes. J Control Release. 2011;153(1):83–92. doi: 10.1016/j.jconrel.2011.04.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Felding-Habermann B, Mueller BM, Romerdahl CA, Cheresh DA. Involvement of integrin alpha V gene expression in human melanoma tumorigenicity. J Clin Invest. 1992;89(6):2018–2022. doi: 10.1172/JCI115811. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Ming X, Ju W, Wu H, Tidwell RR, Hall JE, Thakker DR. Transport of dicationic drugs pentamidine and furamidine by human organic cation transporters. Drug Metab Dispos. 2009;37(2):424–430. doi: 10.1124/dmd.108.024083. [DOI] [PubMed] [Google Scholar]
- 24.Audouy S, Molema G, de Leij L, Hoekstra D. Serum as a modulator of lipoplex-mediated gene transfection: dependence of amphiphile, cell type and complex stability. J Gene Med. 2000;2(6):465–476. doi: 10.1002/1521-2254(200011/12)2:6<465::AID-JGM141>3.0.CO;2-Z. [DOI] [PubMed] [Google Scholar]
- 25.Carrabino S, Di Gioia S, Copreni E, Conese M. Serum albumin enhances polyethylenimine-mediated gene delivery to human respiratory epithelial cells. J Gene Med. 2005;7(12):1555–1564. doi: 10.1002/jgm.799. [DOI] [PubMed] [Google Scholar]
- 26.Ming X, Alam MR, Fisher M, Yan Y, Chen X, Juliano RL. Intracellular delivery of an antisense oligonucleotide via endocytosis of a G protein-coupled receptor. Nucleic Acids Res. 2010;38(19):6567–6576. doi: 10.1093/nar/gkq534. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Khalil IA, Kogure K, Akita H, Harashima H. Uptake pathways and subsequent intracellular trafficking in nonviral gene delivery. Pharmacol Rev. 2006;58(1):32–45. doi: 10.1124/pr.58.1.8. [DOI] [PubMed] [Google Scholar]
- 28.Caswell PT, Norman JC. Integrin trafficking and the control of cell migration. Traffic. 2006;7(1):14–21. doi: 10.1111/j.1600-0854.2005.00362.x. [DOI] [PubMed] [Google Scholar]
- 29.Juliano RL, Ming X, Nakagawa O. The chemistry and biology of oligonucleotide conjugates. Acc Chem Res. 2012 doi: 10.1021/ar2002123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Alam MR, Ming X, Dixit V, Fisher M, Chen X, Juliano RL. The biological effect of an antisense oligonucleotide depends on its route of endocytosis and trafficking. Oligonucleotides. 2010;20(2):103–109. doi: 10.1089/oli.2009.0211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Alam MR, Ming X, Fisher M, Lackey JG, Rajeev KG, Manoharan M, Juliano RL. Multivalent cyclic RGD conjugates fort targeted delivery of small interfering RNA. Bioconjugate chemistry. 2011;22(8):1673–1681. doi: 10.1021/bc200235q. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Nakagawa O, Ming X, Huang L, Juliano RL. Targeted intracellular delivery of antisense oligonucleotides via conjugation with small-molecule ligands. J Am Chem Soc. 2010;132(26):8848–8849. doi: 10.1021/ja102635c. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Boussif O, Lezoualc'h F, Zanta MA, Mergny MD, Scherman D, Demeneix B, Behr JP. A versatile vector for gene and oligonucleotide transfer into cells in culture and in vivo: polyethylenimine. Proc Natl Acad Sci U S A. 1995;92(16):7297–7301. doi: 10.1073/pnas.92.16.7297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Fisher TL, Terhorst T, Cao X, Wagner RW. Intracellular disposition and metabolism of fluorescently-labeled unmodified and modified oligonucleotides microinjected into mammalian cells. Nucleic acids research. 1993;21(16):3857–3865. doi: 10.1093/nar/21.16.3857. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Merkel OM, Germershaus O, Wada CK, Tarcha PJ, Merdan T, Kissel T. Integrin alphaVbeta3 targeted gene delivery using RGD peptidomimetic conjugates with copolymers of PEGylated poly(ethylene imine) Bioconjug Chem. 2009;20(6):1270–1280. doi: 10.1021/bc9001695. [DOI] [PubMed] [Google Scholar]
- 36.Zeng J, Wang X, Wang S. Self-assembled ternary complexes of plasmid DNA, low molecular weight polyethylenimine and targeting peptide for nonviral gene delivery into neurons. Biomaterials. 2007;28(7):1443–1451. doi: 10.1016/j.biomaterials.2006.11.015. [DOI] [PubMed] [Google Scholar]
- 37.Oba M, Aoyagi K, Miyata K, Matsumoto Y, Itaka K, Nishiyama N, Yamasaki Y, Koyama H, Kataoka K. Polyplex micelles with cyclic RGD peptide ligands and disulfide cross-links directing to the enhanced transfection via controlled intracellular trafficking. Mol Pharm. 2008;5(6):1080–1092. doi: 10.1021/mp800070s. [DOI] [PubMed] [Google Scholar]
- 38.Juliano RL, Ming X, Nakagawa O. Cellular uptake and intracellular trafficking of antisense and siRNA oligonucleotides. Bioconjugate chemistry. 2012;23(2):147–157. doi: 10.1021/bc200377d. [DOI] [PMC free article] [PubMed] [Google Scholar]