Abstract
Although amyloid fibers are found in neurodegenerative diseases, evidence points to soluble oligomers of amyloid-forming proteins as the cytotoxic species. Here, we establish that our preparation of toxic amyloid-β1–42 (Abeta42) fibrillar oligomers (TABFOs) shares with mature amyloid fibrils the cross-β structure, in which adjacent β-sheets adhere by interpenetration of protein side chains. We study the structure and properties of TABFOs by powder X-ray diffraction, EM, circular dichroism, FTIR spectroscopy, chromatography, conformational antibodies, and celluar toxicity. In TABFOs, Abeta42 molecules stack into short protofilaments consisting of pairs of helical β-sheets that wrap around each other to form a superhelix. Wrapping results in a hole along the superhelix axis, providing insight into how Abeta may form pathogenic amyloid pores. Our model is consistent with numerous properties of Abeta42 fibrillar oligomers, including heterogenous size, ability to seed new populations of fibrillar oligomers, and fiber-like morphology.
Keywords: Abeta oligomers, toxic oligomers, Alzheimer's disease, domain swapping, protein aggregation
Several neurodegenerative diseases are correlated with amyloid fibrillar deposits (1). For a number of these diseases, it has been postulated that amyloid fibers may not play the primary causative role (2). Rather, soluble aggregates of the amyloidogenic proteins are likely the relevant etiological agents (2, 3). The most prevalent of these neurodegenerative diseases, Alzheimer's disease (4), is strongly linked to the presence of soluble aggregates of amyloid-β (Abeta) (5). Abeta aggregates have been shown to impair neurite function (6), synaptic morphology (7), cognitive function (8), and cell viability (9). In the prion conditions, also classed as amyloid diseases (10), small oligomers have also been identified as the toxic species (11). Recently, the availability of structure-specific antibodies has provided a means to group oligomers into two broad antigenic categories known as prefibrillar and fibrillar oligomers (12).
Fibrillar oligomers are recognized by the OC antibody isolated from rabbits immunized with Abeta fibers (13), suggesting that Abeta fibrillar oligomers share surface features with Abeta fibers. In addition to fiber-like morphology, fibrillar oligomers are similar to fibers in that fibrillar oligomers can seed new populations of fibrillar oligomers (14). The ability to seed suggests that, like fibers, fibrillar oligomers are organized into a repeating array or lattice of monomers, wherein the monomers have identical structures. Fibrillar oligomers likely have a distinct lattice from fibers, because Abeta fibrillar oligomers do not seed Abeta fiber formation (14). Here, we characterize a particular preparation of fibrillar oligomers that we term toxic Abeta1–42 (Abeta42) fibrillar oligomers (TABFOs).
The structure of amyloid fibers may provide insight into the structure of fibrillar oligomers. Fiber diffraction studies of chemically pure amyloid display a cross-β diffraction pattern that includes a 4.7 Å meridional reflection (parallel to the fiber direction) and an ∼10 Å equatorial reflection (perpendicular to the fiber direction) (15, 16). The meridional reflection arises from the hydrogen-bonded stacking of β-strands perpendicular to the fiber axis. The equatorial reflection arises from adjacent β-sheets that run the length of the fibers that are held together by tight, zipper-like side chain interactions between the β-sheets (17–20). We suggest the term cross-β structure for oligomers or fibers made of adjacent β-sheets that adhere to each other by interpenetration of protein side chains.
Abeta has a strong propensity to form fibers with cross-β structure (19), and current models for the fiber structure of Abeta are consistent with the concept of a steric zipper (21–23). In the prevailing fiber models, Abeta forms a U-turn, where both inter- and intramolecular side chain contacts satisfy the requirement for a steric zipper. In fibers, the U-turns align parallel to stack into two adjacent β-sheets that extend along the fiber axis.
A fundamental question is how Abeta can form both fibers and fibrillar oligomers, despite potential architectural differences between these two species. Here, we address this question by using X-ray powder diffraction and X-ray–constrained molecular modeling, buttressed by information from other experimental methods, to develop a model for TABFOs. Our model is consistent with much of what is known about Abeta fibrillar oligomers, including reactivity with the OC antibody, ability to seed new populations of fibrillar oligomers, strong EPR spin coupling of Abeta42 residues 27–35, and fiber-like morphology (13, 14). Furthermore, our model suggests a structural basis for pore formation and by extension, a possible mechanism for fibrillar oligomer-mediated toxicity.
Results
Preparation of TABFOs.
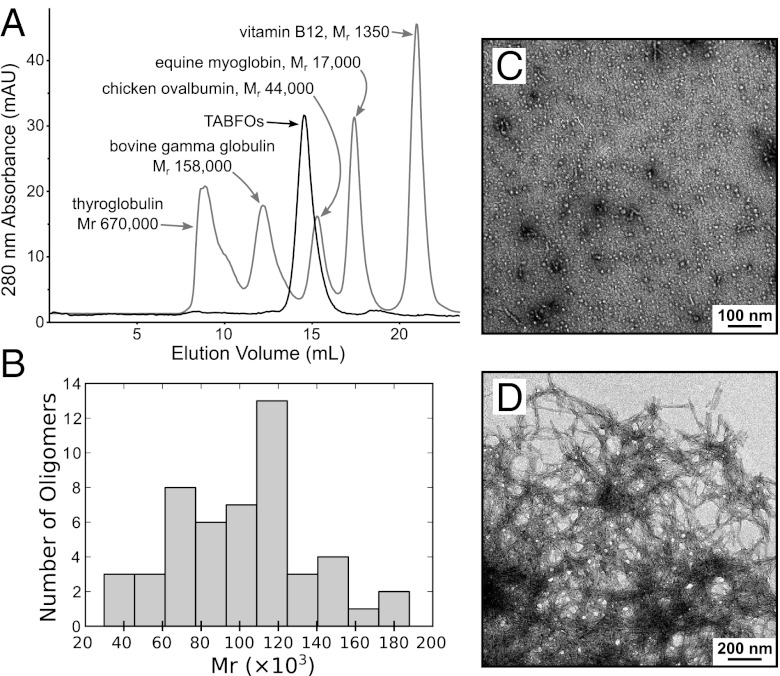
We prepare recombinant WT Abeta42 as described in Materials and Methods (24). We dissolve Abeta42 in hexafluoroisopropanol (HFIP) to disrupt any aggregates. HFIP is then removed by evaporation. Abeta42 is resuspended in dilute ammonium hydroxide, which prevents fiber formation and allows TABFOs to form. TABFOs are stable in size exclusion chromatography (Fig. 1A), which is our final step of purification. Observed in negative-stain transmission EM (TEM), TABFOs appear similar to fibrillar oligomers (14) (Fig. 1C).
Fig. 1.
TABFOs have an Mr of ∼80,000 (∼19 Abeta42 monomers), with morphology distinct from the morphology of Abeta42 fibers. (A) TABFOs have an Mr of 60,000 ± 4,200 by SEC. Two SEC runs are superposed. Dark trace, TABFOs; light trace, protein calibration standards. The peaks of the protein standards are labeled with the corresponding molecular masses. A graphical analysis from these runs is shown in Fig. S1B. (B) TABFOs have an Mr of 100,000 ± 3,500 by TEM. Shown is the distribution of measured Mr for a randomly selected sample of 50 TABFO particles from the TEM image in C. The mean of the SEC and TEM Mr measurements is 80,000 ± 20,000. (C) TABFOs imaged by TEM. (Scale bar: 100 nm.) (D) Fibers from TABFOs grown overnight at 37 °C in PBS. (Scale bar: 200 nm.)
TABFOs convert into fibers in PBS, suggesting that TABFOs are not dead-end aggregates. Fibers grown from TABFOs have typical amyloid appearance by TEM (Fig. 1D). These fibers also cause thioflavin T (ThT) fluorescence in fiber growth kinetics assay (Fig. S1A). Fiber formation from TABFOs has a 2,155 ± 301 s tenth time, which is the time to attain one-tenth of the maximum signal. This time is longer than the ∼720 s tenth time reported for monomeric Abeta42 prepared in a similar manner (24), suggesting that TABFOs may undergo conformational changes during conversion to fibers.
Size of TABFOs.
From analytical size exclusion chromatography (SEC), we measure the mean relative molecular mass (Mr) of TABFOs to be 60,000 ± 4,200 (Fig. 1A and Fig. S1B). TABFOs display a range of sizes by TEM, varying from 13 to 28 nm in the longest dimension (Fig. 1C), similar to fibrillar oligomers (14). To estimate the size from TEM, we randomly choose 50 TABFO particles from the micrograph shown in Fig. 1C modeled as cylinders with a partial specific volume of 0.73 cm3/g (25). The distribution of molecular masses is shown in Fig. 1B. From these data, we measure the mean Mr of TABFOs to be 100,000 ± 3,500 (SEM), which is larger than the apparent size by SEC. This difference may have multiple origins, such as nonideal elution by SEC, overlap in TEM, misestimation of partial specific volume, or deviation from cylindrical shape, which we use in the volume calculation. The average of the two size estimates gives a mean Mr of 80,000 ± 20,000. Our measurement for the mean Mr is equivalent to 19 ± 4 Abeta42 monomers, suggesting that TABFOs are slightly larger by mass than Abeta42 fibrillar oligomers of 3–10 monomers previously identified (14).
TABFOs Are Identified by Antigenic Recognition by the OC Antibody.
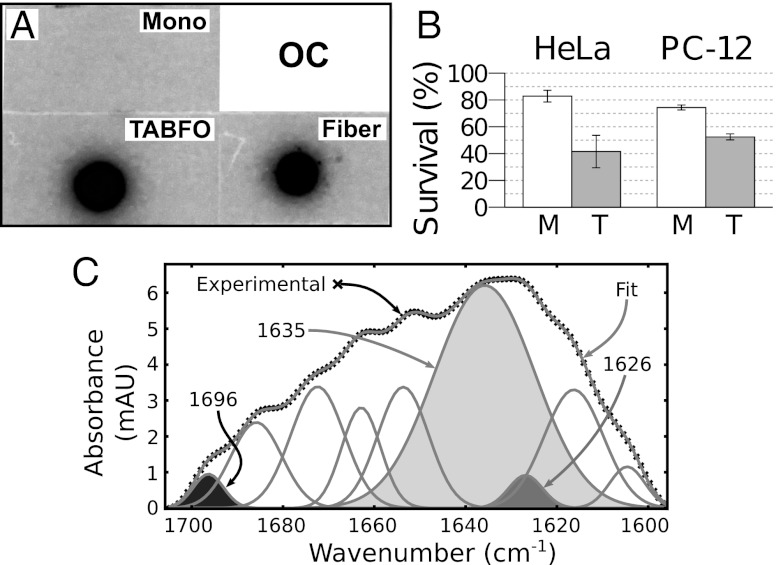
To determine whether TABFOs are fibrillar oligomers, we probe dot blots of TABFOs with OC antibodies (13) that recognize fibrillar oligomers. We find that OC antibodies recognize TABFOs (Fig. 2A), showing that TABFOs may have structural features shared by fibers (14). Blotting a replicate membrane with 6E10 antibodies shows that the differences are not explained by loading differences (Fig. S1C).
Fig. 2.
TABFOs react with the fiber-specific OC antibody, are toxic, and have antiparallel β-sheet structure. (A) Immunoblot with fiber-specific OC antibody. Fiber, fibers grown from TABFOs; Mono, Abeta42 monomers spotted immediately on resuspension into PBS after HFIP treatment. (B) MTT assay shows that Abeta42 TABFOs are significantly more toxic to mammalian cells than Abeta42 monomers. TABFOs and monomers are applied to HeLa and PC-12 cell lines at 500 nM (concentration by monomer equivalent), and survival assayed as described in Materials and Methods. M, monomeric Abeta42; T, TABFOs. (C) TABFOs have antiparallel β-sheets by FTIR. The peaks from a Gaussian deconvolution of the FTIR spectrum are shown. Shading is explained in the text.
TABFOs Are Toxic to Neuronal Cells.
MTT [3-(4,5-Dimethylthiazol-2-yl)-2,5-diphenyltetrazolium bromide] cell survival assays (26) show that TABFOs are toxic to mammalian cells (Fig. 2B). In these assays, we find that TABFOs are toxic to HeLa cells and PC-12 neuronal cells at concentrations as low as 2.25 μg/mL (0.5 μM of Abeta42 monomer equivalent). Monomeric Abeta42 is significantly less toxic than oligomers, but it does show some toxicity, suggesting that monomeric Abeta42 may convert to toxic species during the assay.
TABFOs Are β-Rich with Antiparallel β-Sheets.
To characterize the structure of TABFOs, we perform circular dichroism spectroscopy (CD). We analyze the data with two different software packages: K2D2 (27) and CDSSTR from the CDPro Suite (28). As seen in Table 1, TABFOs are similar to fibers in that both have significant β-structure and negligible α-helical content. Despite these similarities, the CD spectra of fibers and TABFOs are somewhat different (Fig. S1D). TABFOs have a minimum at about 200 nm combined with negative Δε for wavelengths between 200 and 240 nm, suggesting that TABFOs are mostly disordered but have some antiparallel β-structure (29). Fibers have a maximum and minimum at about 200 and 220 nm, respectively, consistent with a significant component of β-sheets (29).
Table 1.
Abeta42 fibers and TABFOs have similar β-sheet content
| Fibers |
TABFOs |
|||
| K2D2 (%) | CDPro (%) | K2D2 (%) | CDPro (%) | |
| α-Helix | 9.8 | 3.5 | 6.3 | 1.9 |
| β-Sheet | 39.5 | 41.3 | 32.4 | 34.2 |
| Unstructured | 50.7 | 55.2 | 61.3 | 63.9 |
We also use FTIR to assess the secondary structure content of TABFOs. As seen in Fig. 2C, the major component of the FTIR spectrum is centered around 1,630 cm−1, suggesting that TABFOs have β-sheet content (30). To determine whether the β-sheets are antiparallel, we use Fourier self-deconvolution to identify peak centers followed by Gaussian fitting to quantify the deconvolved peaks of the FTIR (31). For TABFOs, we measure the ratio of the 1,696-cm−1 peak (area of the black peak in Fig. 2C) to the peaks around 1,630 cm−1 (combined areas of the 1,635-cm−1 light gray and 1,626-cm−1 dark gray peaks in Fig. 2C) to be 0.044, agreeing closely with the 1,695 to 1,630 cm−1 ratio of ∼0.056 predicted for an infinite antiparallel β-sheet (32). From this agreement, we conclude that the β-sheets of TABFOs are antiparallel.
TABFOs Have Fiber-Like Cross-β Architecture.
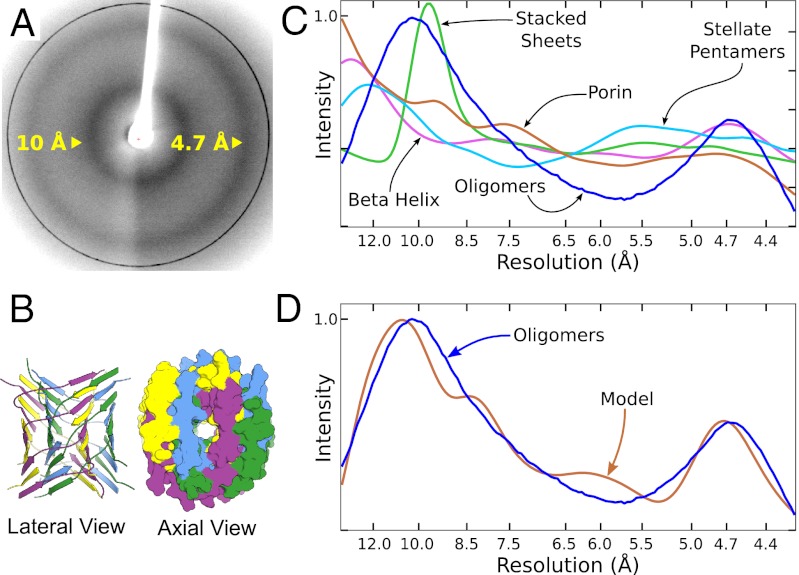
To probe the architecture of TABFOs, we perform X-ray powder diffraction of lyophilized samples. As shown in Fig. 3A, TABFOs produce broad diffraction peaks at 4.7 and ∼10 Å. Although the TABFOs are not oriented in this experiment, these peaks are consistent with a spherically averaged cross-β pattern (15–17). This pattern does not arise from the presence of mature amyloid fibers, because EM micrographs of these oligomers do not reveal mature fibers (Fig. 1C).
Fig. 3.
A domain-swapped, cross-β model for TABFOs. (A) The TABFO experimental powder diffraction shows concentric rings centered at 4.7 and 10 Å. (B) Cartoon drawing of a representative model for TABFOs. This model has a 40° crossing angle, is 10-layers long, and is composed of 20 ordered Abeta monomers. (Left) Ribbon representation (lateral view). (Right) Space filling representation (axial view). The oligomer has two runaway domain swaps. The first swap is green/magenta, where the green monomers run down from the N to C terminus and the magenta monomers run up. In the second swap (yellow/blue), the yellow monomers run down, and the blue monomers run up. (C) Simulated diffraction patterns of various models for TABFOs (Fig. S2A). Blue, radially averaged experimental TABFO pattern; brown, OmpF porin [Protein Data Bank (PDB) ID code 2ZFG] (50); green, an interpretation of oligomers characterized in the work by Sandberg et al. (35) (40 GAIIGLL strands arranged in idealized stacked dimers making antiparallel sheets); cyan, an interpretation of oligomers characterized in the work by Mastrangelo et al. (51) [monomers of PDB ID code 2OTK (34) arranged as stacked pentamers]; magenta, the β-helix of spruce budworm antifreeze protein, PDB ID code 1M8N (52). (D) The scaled, radially averaged pattern (blue) from C is superimposed on the simulated powder diffraction pattern (brown) of the model in B.
To arrive at an atomic model for TABFOs, we develop a quantitative test of the fit for each of various models to our observations. We start by limiting tested models to those models containing antiparallel β-structure, which is demanded by our CD and FTIR data. Each model is then evaluated by comparing its simulated powder X-ray diffraction pattern with the experimental pattern (Fig. 3A) using a measurement of fit (Ro2) similar to the Rwp previously described (33), except that we normalize Ro2 such that it is zero for a perfect fit and 1.0 for the worst possible fit (SI Materials and Methods).
Of the models that we test, none lacking cross-β structure fit the experimental data with a value for Ro2 smaller than the threshold of 0.30. This finding is consistent with conclusions from simulations of fiber diffraction previously reported (18). Models giving values for Ro2 greater than 0.30 include β-barrels, stacked β-sheets, β-helices, and more divergent structures (Fig. 3C and Fig. S2A).
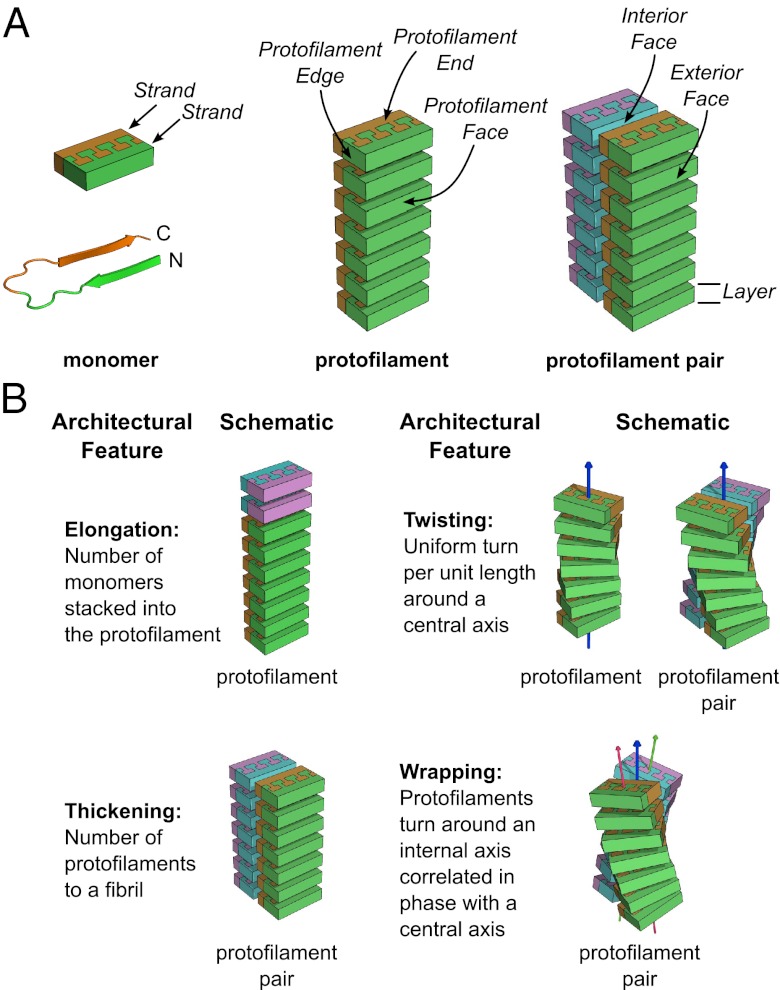
To model cross-β TABFOs, we begin by using models with 20 Abeta42 monomers, close to the mean of 19 monomers estimated from the combination of SEC and EM. We then stack monomers to create antiparallel β-sheets using the U-turn structure of fibrillar Abeta42 determined from solid-state NMR (22) as the monomer subunit. The monomer model that we use omits the N-terminal segment (residues 1–12) that is disordered in both fibrillar (22, 23) and nonfibrillar Abeta (34, 35). The hydrogen bonding of the two closely associated β-sheets (formed by the two arms of the U) runs the length of the stack, similar to a model for antiparallel Abeta fibers previously described (21, 36). We term this stack of monomers a protofilament (Fig. 4A and Fig. S2B). Protofilaments have ends at the extremes of the stacking direction, faces made of the side chains pointing from the sheets, and edges formed by the residues in the hinge of the U and polypeptide termini (Fig. 4A).
Fig. 4.
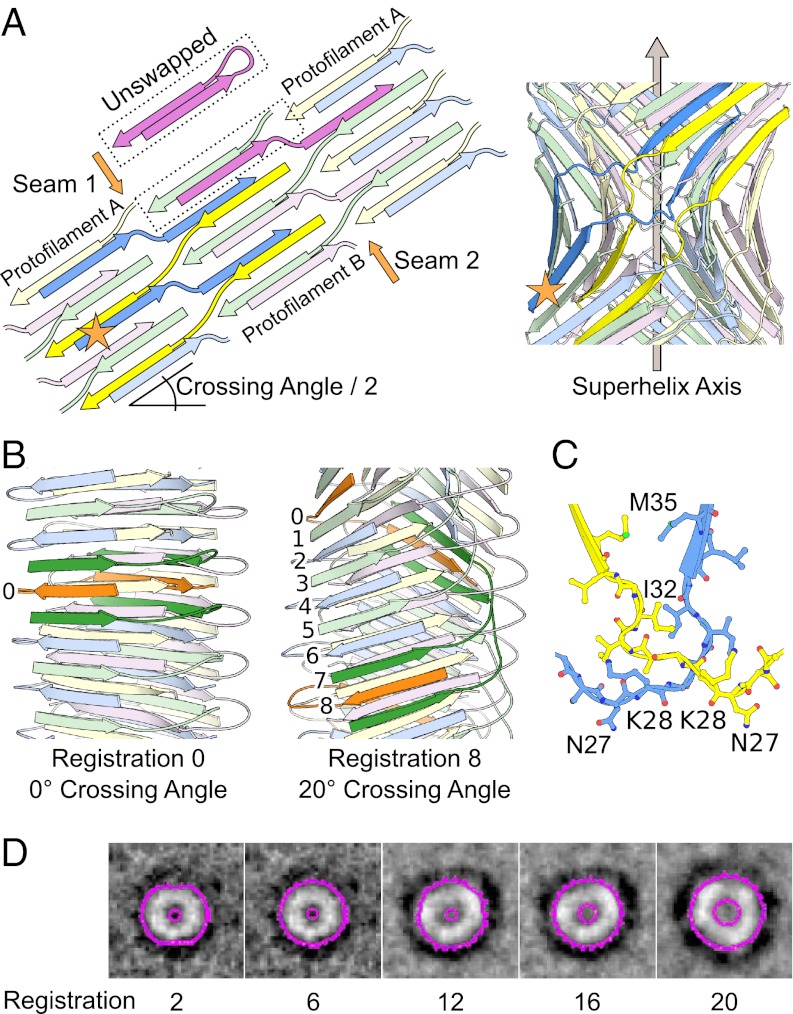
Protofilaments are antiparallel stacks of Abeta42 monomers with varying geometries. (A) A protofilament pair is made of two protofilaments that are antiparallel stacks of Abeta42 monomers. (Left) An Abeta42 monomer is shown in Lower as a cartoon with a U-turn (22) and schematized in Upper as a brick with interdigitated side chains. The N-terminal β-strand (N) is green, and the C-terminal β-strand (C) is orange. (Center) The monomers stack antiparallel such that all of the N-terminal strands make a sheet on one face of the protofilament. Likewise, all of the C-terminal strands make a sheet on the other face of the protofilament. Two monomers form the two ends of the protofilament. Edges are made of the alternating turns and termini of the stacked monomers. (Right) A protofilament pair consists of two protofilaments that interact through the faces made by C-terminal β-strands. The C-terminal face of the cyan–magenta protofilament is cyan. This arrangement of protofilaments allows the disordered N-terminal residues (1–12) to point to the exterior of the protofilament pair in domain-swapped structures. A layer of a protofilament is one Abeta42 monomer thick in the stacking direction. (B) Models vary by elongation (stacking more layers of monomers to the protofilaments), thickening from a protofilament to a protofilament pair, twisting around a central axis, or wrapping around a central axis to create protofilaments with internal helical axes that are in phase with the central helical axis (41). Color differences indicate growth by elongation and thickening. Helical symmetry axes are shown as arrows. Wrapping geometry results in two additional helical symmetry axes that run through the individual protofilaments.
In our simulations, we vary this initial model in four ways (Fig. 4B) using Ro2 as a guide: (i) elongation (addition or removal of monomers at the protofilament ends, producing more or less elongated protofilaments, respectively), (ii) thickening (adjoining two protofilaments along their faces to make a protofilament pair) (Fig. 4A and Fig. S2C), (iii) twisting (rotation of successive layers of the protofilaments around a common helical axis), and (iv) wrapping (correlated twist of a protofilament pair to make a superhelical fibril). Twisting and wrapping are mutually exclusive geometries. Twisting is measured by relative twist, which is the rotation between neighboring layers in the sheet (20). Wrapping is measured by the crossing angle of the protofilament pair (Fig. S2D), which is the angle the protofilament axes make when projected along a line perpendicular to both. A model that is either twisted or wrapped is said to have helicity, because it contains at least one helical axis (Fig. 4B).
In generating our initial wrapped models, we find that, despite their better fit to the experimental diffraction pattern, the two protofilaments do not contact each other significantly. The reason is that, at higher crossing angles, two wrapped protofilaments are unable to interact through their interior faces, because these faces have opposed curvature. To create contact, and thus binding energy between the two wrapped protofilaments, we introduce runaway domain swapping interactions that join the two protofilaments at the seams created by the protofilament edges. Runaway swapping interactions have been observed in fiber assembly (37–39). A TABFO model with runaway domain swapping is shown schematically in Fig. 5A and illustrated in Movie S1.
Fig. 5.
The registration of runaway domain swapping is related to TABFO geometry. (A) Two runaway domain swaps (blue–yellow and magenta–green) extend the length of TABFOs, tightly stitching the two seams between the protofilaments. In our model, Tycko/Riek U interactions (22, 23) are present in intermolecular, domain-swapped form, rather than as intramolecular interactions. (Left) The orange star marks the equivalent monomer in the schematic in Left and the model in Right. Shown for comparison is a schematic of a monomer that is not swapped (bright magenta, labeled unswapped). (Right) Cartoon representation of the TABFO model schematized in Left. The orange star has the same meaning as in Left. (B) The registration between the two swaps (blue–yellow and magenta–green) is defined operationally; beginning with an N-terminal β-strand of protofilament I (colored orange and labeled 0 in B), follow this first monomer to its C-terminal strand in protofilament II. Then, follow the second and third monomers (green) that border this C-terminal strand back to protofilament I. These two green monomers border a fourth monomer (colored orange and labeled 0 in Left and 8 in Right) in protofilament I. The registration is the number of layer interfaces between the N-terminal strand of the first monomer and the N-terminal strand of the fourth monomer. In the case of no wrapping (zero registration) the first and fourth monomers are the same. (C) Close-up view of a cross-over (residues 27–35) from the model in B is shown in ball and stick representation. The distances between identical residues in the two monomers are consistent with previously reported EPR data (14). (D) TABFOs get wider with larger holes as the registration of runaway domain swapping increases. Axial projections of the molecular surface of several TABFOs with different registrations (indicated by integers below) are shown in magenta inside outlines (holes) and outside outlines for clarity (compare with Fig. 3B, right side). The outlines are superposed on reproduced images of projection averages of amyloid pores formed by E22GAbeta1–40 observed in the work by Lashuel et al. (45). (Adapted by permission from Macmillan Publishers Ltd: Nature, ref. 45, copyright 2002.)
Detailed Models.
An initial protofilament model of 20 monomers (Fig. S3A, single protofilament no helicity) fits the diffraction pattern reasonably well (Ro2 = 0.290). For comparison, Ro2 is 0.383 for active phosphofructokinase (PDB ID 4PFK) (40), a typical mixed α/β-protein. From the initial protofilament model, halving the layers and thickening to a protofilament pair improve the fit slightly (Ro2 = 0.135) (Fig. S3B, protofilament pair no helicity). The fit is further improved by using wrapped models with runaway swapping, where the crossing angle is between 20° and 70° inclusively. Wrapped models are robust to variation (Fig. S3 and Tables S1 and S2). The best fit is with a 40° crossing angle (Ro2 = 0.072) (Table S1). The simulated powder diffraction patterns of wrapped models with runaway domain swapping generally fit the experimental pattern better than equivalent models without swapping. For example, the Ro2 for the swapped model in Fig. 3B is 0.072, whereas the Ro2 for the equivalent model without swapping is 0.116. We use the model in Fig. 3B as a reference for comparison in several simulations herein. Fig. 3D shows the simulated powder diffraction of the reference model superimposed with the experimental powder diffraction pattern of TABFOs.
Geometry of TABFOs Is Related to the Registration of the Runaway Domain Swap.
The crossing angle is related to the registration of the runaway domain swap, which is defined operationally in Fig. 5B (Movie S2). Registration is a quantized measurement limited to even integers. Our models show that the crossing angle increases with registration (Fig. S4A and Table S1). Although a set of TABFOs may have differing registrations, the hydrogen bonding in the sheets will be nearly identical as well as interactions between sheets of the same protofilament. Thus, within a set of TABFOs that differs by registration, the local chemical environment of every monomer will be identical, except for small differences in the hinge (residues 24–31) and side chains of the interior face.
Discussion
Structure of TABFOs.
Our data from CD, FTIR, and X-ray powder diffraction reveal that TABFOs are ordered aggregates with cross-β architecture. However, TABFOs are not simply short protofilaments. Using the power that X-ray powder diffraction offers of comparing observed diffraction patterns with patterns rigorously calculated from atomic models, we find that TABFOs are likely composed of thickened (laterally associated) protofilaments that twist around internal helical axes. These internal axes wrap around a common superhelical axis in a geometry that we term wrapping (Fig. 4B). When a protofilament is wrapped around a helical axis in this manner, it obtains a twist that is in phase with the fiber helix. Such matching of phases has been called correlated twist in the context of amyloid fibers (41). Protofilaments that wrap around a common helical axis retain a constant interface along the length of the fiber (41). Another consequence of wrapping is that the hydrogen bonding geometry along the β-sheets is optimized (42).
In our domain-swapped model for TABFOs, the intramolecular interaction between the two arms of the U-turn (Figs. 4A and 5A, unswapped) is replaced by an intermolecular interaction between one arm of the first Abeta molecule and the other arm swapped from a second molecule. Swapping in our models is enabled by the flexible hinge (residues 24–31) (22, 23, 34, 35) near the cross-over of the swap (residues 27–35). The resulting interactions in the cross-over (Fig. 5C) are consistent with published EPR data of FOs that suggest that the 27–35 segment is in proximity to another or other copies of itself (14). Although exact side chain positions cannot be determined from EPR data, our model is consistent with trends in this EPR data. For example, I32 and M35 show the strongest EPR spin coupling and are also closest in our model. Moreover, the EPR spin coupling gradually diminishes from the cross-over in a manner consistent with the increasing distances in our models.
Our model for TABFOs agrees with predictions that mature amyloid fibers and fibrillar oligomers share common surface features but likely have different lattices (13, 14). As we show here, the TABFO architecture produces simulated powder diffraction consistent with cross-β structure (Fig. 3 and Fig. S3). This fiber-like architecture may underlie the specificity of OC antibody for fibrillar oligomers and mature amyloid fibers (Fig. 2) (12). Additionally, our modeling suggests that seeding by fibrillar oligomers likely occurs by addition to the ends of the fibrillar oligomers (14).
Size Limitation.
An upper limit to the size of TABFOs may arise from increasing stress on new monomers as they are layered onto the ends of the protofilaments. In wrapping, the β-sheet hydrogen bonding distances of the outermost sheet must be longer than the distances of the innermost sheet to maintain native side chain contacts between swapping partners. This stretching is exaggerated at high crossing angles but may be compensated by increased distortion of monomers as they are layered, allowing them to retain native side chain interactions while also maintaining optimal hydrogen bonding distances with neighbors in the sheet. Growth may stop when the energy cost to distort a monomer balances the energy gained from hydrogen bonding.
This situation is analogous to the opposition of forces that limits the size of amylodogenic peptide microcrystals (43). In these microcrystals, the propensity for an intrinsic β-sheet twist opposes the requirement for aligned hydrogen bonding within the crystal lattice, resulting in a buildup of strain as layers add to a growing microcrystal.
In Fig. S4B, we depict a TABFO with a high crossing angle of 70° to illustrate how retaining the monomer conformation opposes optimal hydrogen bonding. One prediction of this energetic tradeoff is that oligomers with low crossing angles will have a propensity to grow longer than oligomers with high crossing angles, a prediction similar to one arising from studies of macrocyclic β-sheet mimics (44). Additionally, long oligomers may appear filamentous by EM.
Structure–Toxicity Relationship.
Our modeling shows that higher crossing angles are related to greater curvature and increasingly large holes in the oligomers, suggesting that TABFO toxicity may be related to the potential for TABFOs to form pores. Pore formation has been implicated in the toxicity of Abeta oligomers (45, 46). At high crossing angles, opposing curvature of the two protofilaments dictates that they can interact only at their edges (defined in Fig. 4), creating a hole along the helical axis of the TABFO (Fig. 3B, Right). Viewed along the central axis, TABFOs are similar in appearance to projection averages of amyloid pores formed by E22GAbeta1–40 (45). In Fig. 5D, we show axial projections of TABFOs with a range of crossing angles. These projections are superimposed on reproduced images of E22GAbeta1–40 amyloid pores observed previously (45). The superposition shows that the TABFO models that agree well with experimental powder diffraction data (Ro2 < 0.10) (Table S1) have hole and outside diameters consistent with amyloid pores of E22GAbeta1–40. Notably, the E22G mutation is near the hinge (residues 24–31), suggesting that the increased flexibility in this part of Abeta may facilitate pore formation.
Conclusion
We develop atomic models for a type of toxic Abeta42 oligomer that we call TABFOs. From powder X-ray diffraction, TEM, CD, and FTIR, we show that TABFOs are short, wrapped, fiber-like structures (Fig. 3B), where the energy that joins protofilaments is explained by runaway domain swapping (Fig. 5A). The fiber-like architecture of TABFOs explains their reactivity with the fiber-specific OC antibody (13) and the ability to seed new populations of TABFOs (14). A range of TABFO structures differing in the crossing angle of the two protofilaments agree well with our experimental data, with the best fits being for TABFOs that have a 40° crossing angle. The crossing angle is related to the registration of the runaway domain swapping that, after it is established, would determine the overall geometry growing oligomer. Models of TABFOs with crossing angles of at least 40° have a hole along the central axis, suggesting the potential for pore-like activity that may explain TABFO toxicity.
Materials and Methods
Preparation of Abeta42.
Abeta42 is incorporated into a fusion construct with 19 Asn-Ala-Asn-Pro repeats N-terminal to a Tev protease site ending with an aspartate. After Tev cleavage, this aspartate remains on the cleavage product as the first residue of the Abeta42 sequence. In our final steps of the preparation of TABFOs, we elute the Tev cleavage product from a reverse-phase column followed by lyophilization and resuspension in HFIP to disrupt any aggregates (47). We then eliminate the HFIP by evaporation.
SEC.
For sizing, TABFOs are concentrated to 1 mg/mL and run over a Superdex 200 HR 10/30 column (17–1088-01; GE Healthcare) in 50 mM ammonium hydroxide.
Preparation of Samples for TEM.
Fibers for TEM are grown from TABFOs in PBS overnight at 37 °C with shaking at 225 rpm. TABFOs are lyophilized for X-ray powder diffraction and then resuspended in 50 mM NH4OH at 1 mg/mL for optimal distribution on the grids.
ThT Fiber Assay Reaction Conditions.
Concentrated TABFOs are diluted to 10 μM in PBS and immediately filtered through 0.1-μm filters before starting the assay. The reaction conditions are 10 μM ThT in PBS at 37 °C.
Sample Preparation for Powder Diffraction.
Lyophilized TABFOs are applied by stabbing with the jagged ends of broken glass capillaries. Samples are sufficiently large to accommodate a 50-μm X-ray beam without incurring scatter from the capillary.
Simulated Powder Diffraction Patterns.
Simulated powder diffraction patterns are generated from the models using the simulated powder diffraction function of XPREPX (48) using structure factors and phases calculated by CCP4 (49).
Supplementary Material
Acknowledgments
We thank Drs. Todd Yeates, Lukasz Salwinski, Rebecca Nelson, and Zhefeng Guo for discussions; the laboratory of Charles Glabe for help with immunoblots; and Prof. Arnold Berk and Dr. Dawei Guo for help with tissue culture experiments. This work was supported by National Institutes of Health Grants AG 029430 and FGM077789A, the Department of Energy, National Science Foundation Grant MCB 0958111, and the Howard Hughes Medical Institute.
Footnotes
The authors declare no conflict of interest.
*This Direct Submission article had a prearranged editor.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1203193109/-/DCSupplemental.
References
- 1.Selkoe DJ. Folding proteins in fatal ways. Nature. 2003;426:900–904. doi: 10.1038/nature02264. [DOI] [PubMed] [Google Scholar]
- 2.Kirkitadze MD, Bitan G, Teplow DB. Paradigm shifts in Alzheimer’s disease and other neurodegenerative disorders: The emerging role of oligomeric assemblies. J Neurosci Res. 2002;69:567–577. doi: 10.1002/jnr.10328. [DOI] [PubMed] [Google Scholar]
- 3.Bucciantini M, et al. Inherent toxicity of aggregates implies a common mechanism for protein misfolding diseases. Nature. 2002;416:507–511. doi: 10.1038/416507a. [DOI] [PubMed] [Google Scholar]
- 4.Ewbank DC. Deaths attributable to Alzheimer’s disease in the United States. Am J Public Health. 1999;89:90–92. doi: 10.2105/ajph.89.1.90. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Irvine GB, El-Agnaf OM, Shankar GM, Walsh DM. Protein aggregation in the brain: The molecular basis for Alzheimer’s and Parkinson’s diseases. Mol Med. 2008;14:451–464. doi: 10.2119/2007-00100.Irvine. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Li S, et al. Soluble oligomers of amyloid Beta protein facilitate hippocampal long-term depression by disrupting neuronal glutamate uptake. Neuron. 2009;62:788–801. doi: 10.1016/j.neuron.2009.05.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Calabrese B, et al. Rapid, concurrent alterations in pre- and postsynaptic structure induced by naturally-secreted amyloid-beta protein. Mol Cell Neurosci. 2007;35:183–193. doi: 10.1016/j.mcn.2007.02.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Martins IC, et al. Lipids revert inert Abeta amyloid fibrils to neurotoxic protofibrils that affect learning in mice. EMBO J. 2008;27:224–233. doi: 10.1038/sj.emboj.7601953. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Barghorn S, et al. Globular amyloid beta-peptide oligomer - a homogenous and stable neuropathological protein in Alzheimer’s disease. J Neurochem. 2005;95:834–847. doi: 10.1111/j.1471-4159.2005.03407.x. [DOI] [PubMed] [Google Scholar]
- 10.Westermark P, et al. A primer of amyloid nomenclature. Amyloid. 2007;14:179–183. doi: 10.1080/13506120701460923. [DOI] [PubMed] [Google Scholar]
- 11.Caughey B, Lansbury PT. Protofibrils, pores, fibrils, and neurodegeneration: Separating the responsible protein aggregates from the innocent bystanders. Annu Rev Neurosci. 2003;26:267–298. doi: 10.1146/annurev.neuro.26.010302.081142. [DOI] [PubMed] [Google Scholar]
- 12.Glabe CG. Structural classification of toxic amyloid oligomers. J Biol Chem. 2008;283:29639–29643. doi: 10.1074/jbc.R800016200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Kayed R, et al. Fibril specific, conformation dependent antibodies recognize a generic epitope common to amyloid fibrils and fibrillar oligomers that is absent in prefibrillar oligomers. Mol Neurodegener. 2007;2:18. doi: 10.1186/1750-1326-2-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Wu JW, et al. Fibrillar oligomers nucleate the oligomerization of monomeric amyloid beta but do not seed fibril formation. J Biol Chem. 2010;285:6071–6079. doi: 10.1074/jbc.M109.069542. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Bonar L, Cohen AS, Skinner MM. Characterization of the amyloid fibril as a cross-beta protein. Proc Soc Exp Biol Med. 1969;131:1373–1375. doi: 10.3181/00379727-131-34110. [DOI] [PubMed] [Google Scholar]
- 16.Eanes ED, Glenner GG. X-ray diffraction studies on amyloid filaments. J Histochem Cytochem. 1968;16:673–677. doi: 10.1177/16.11.673. [DOI] [PubMed] [Google Scholar]
- 17.Astbury WT, Dickinson S, Bailey K. The X-ray interpretation of denaturation and the structure of the seed globulins. Biochem J. 1935;29:2351–2360. doi: 10.1042/bj0292351. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Jahn TR, et al. The common architecture of cross-beta amyloid. J Mol Biol. 2010;395:717–727. doi: 10.1016/j.jmb.2009.09.039. [DOI] [PubMed] [Google Scholar]
- 19.Kirschner DA, Abraham C, Selkoe DJ. X-ray diffraction from intraneuronal paired helical filaments and extraneuronal amyloid fibers in Alzheimer disease indicates cross-beta conformation. Proc Natl Acad Sci USA. 1986;83:503–507. doi: 10.1073/pnas.83.2.503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Sunde M, et al. Common core structure of amyloid fibrils by synchrotron X-ray diffraction. J Mol Biol. 1997;273:729–739. doi: 10.1006/jmbi.1997.1348. [DOI] [PubMed] [Google Scholar]
- 21.Colletier J-P, et al. Molecular basis for amyloid-beta polymorphism. Proc Natl Acad Sci USA. 2011;108:16938–16943. doi: 10.1073/pnas.1112600108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Lührs T, et al. 3D structure of Alzheimer’s amyloid-beta(1-42) fibrils. Proc Natl Acad Sci USA. 2005;102:17342–17347. doi: 10.1073/pnas.0506723102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Petkova AT, et al. A structural model for Alzheimer’s beta-amyloid fibrils based on experimental constraints from solid state NMR. Proc Natl Acad Sci USA. 2002;99:16742–16747. doi: 10.1073/pnas.262663499. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Finder VH, Vodopivec I, Nitsch RM, Glockshuber R. The recombinant amyloid-beta peptide Abeta1-42 aggregates faster and is more neurotoxic than synthetic Abeta1-42. J Mol Biol. 2010;396:9–18. doi: 10.1016/j.jmb.2009.12.016. [DOI] [PubMed] [Google Scholar]
- 25.Mok Y-F, Howlett GJ. Sedimentation velocity analysis of amyloid oligomers and fibrils. Methods Enzymol. 2006;413:199–217. doi: 10.1016/S0076-6879(06)13011-6. [DOI] [PubMed] [Google Scholar]
- 26.Mosmann T. Rapid colorimetric assay for cellular growth and survival: Application to proliferation and cytotoxicity assays. J Immunol Methods. 1983;65:55–63. doi: 10.1016/0022-1759(83)90303-4. [DOI] [PubMed] [Google Scholar]
- 27.Perez-Iratxeta C, Andrade-Navarro MA. K2D2: Estimation of protein secondary structure from circular dichroism spectra. BMC Struct Biol. 2008;8:25. doi: 10.1186/1472-6807-8-25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Johnson WC. Analyzing protein circular dichroism spectra for accurate secondary structures. Proteins. 1999;35:307–312. [PubMed] [Google Scholar]
- 29.Kelly SM, Price NC. The use of circular dichroism in the investigation of protein structure and function. Curr Protein Pept Sci. 2000;1:349–384. doi: 10.2174/1389203003381315. [DOI] [PubMed] [Google Scholar]
- 30.Kong J, Yu S. Fourier transform infrared spectroscopic analysis of protein secondary structures. Acta Biochim Biophys Sin (Shanghai) 2007;39:549–559. doi: 10.1111/j.1745-7270.2007.00320.x. [DOI] [PubMed] [Google Scholar]
- 31.Susi H, Byler DM. Fourier deconvolution of the amide I Raman band of proteins as related to conformation. Appl Spectrosc. 1988;42:819–826. [Google Scholar]
- 32.Chirgadze YN, Nevskaya NA. Infrared spectra and resonance interaction of amide-I vibration of the antiparallel-chain pleated sheet. Biopolymers. 1976;15:607–625. doi: 10.1002/bip.1976.360150402. [DOI] [PubMed] [Google Scholar]
- 33.Harris KDM, Johnston RL, Kariuki BM. The genetic algorithm: Foundations and applications in structure solution from powder diffraction data. Acta Crystallogr A. 1998;54:632–645. [Google Scholar]
- 34.Hoyer W, Grönwall C, Jonsson A, Ståhl S, Härd T. Stabilization of a β-hairpin in monomeric Alzheimer’s amyloid-β peptide inhibits amyloid formation. Proc Natl Acad Sci USA. 2008;105:5099–5104. doi: 10.1073/pnas.0711731105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Sandberg A, et al. Stabilization of neurotoxic Alzheimer amyloid-beta oligomers by protein engineering. Proc Natl Acad Sci USA. 2010;107:15595–15600. doi: 10.1073/pnas.1001740107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Tycko R, Sciarretta KL, Orgel JPRO, Meredith SC. Evidence for novel beta-sheet structures in Iowa mutant beta-amyloid fibrils. Biochemistry. 2009;48:6072–6084. doi: 10.1021/bi9002666. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Guo Z, Eisenberg D. Runaway domain swapping in amyloid-like fibrils of T7 endonuclease I. Proc Natl Acad Sci USA. 2006;103:8042–8047. doi: 10.1073/pnas.0602607103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Liu C, Sawaya MR, Eisenberg D. β₂-microglobulin forms three-dimensional domain-swapped amyloid fibrils with disulfide linkages. Nat Struct Mol Biol. 2011;18:49–55. doi: 10.1038/nsmb.1948. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Sambashivan S, Liu Y, Sawaya MR, Gingery M, Eisenberg D. Amyloid-like fibrils of ribonuclease A with three-dimensional domain-swapped and native-like structure. Nature. 2005;437:266–269. doi: 10.1038/nature03916. [DOI] [PubMed] [Google Scholar]
- 40.Evans PR, Farrants GW, Hudson PJ. Phosphofructokinase: Structure and control. Philos Trans R Soc Lond B Biol Sci. 1981;293:53–62. doi: 10.1098/rstb.1981.0059. [DOI] [PubMed] [Google Scholar]
- 41.Jiménez JL, et al. The protofilament structure of insulin amyloid fibrils. Proc Natl Acad Sci USA. 2002;99:9196–9201. doi: 10.1073/pnas.142459399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Wang J, Gülich S, Bradford C, Ramirez-Alvarado M, Regan L. A twisted four-sheeted model for an amyloid fibril. Structure. 2005;13:1279–1288. doi: 10.1016/j.str.2005.06.010. [DOI] [PubMed] [Google Scholar]
- 43.Nelson R, et al. Structure of the cross-beta spine of amyloid-like fibrils. Nature. 2005;435:773–778. doi: 10.1038/nature03680. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Liu C, et al. Characteristics of amyloid-related oligomers revealed by crystal structures of macrocyclic β-sheet mimics. J Am Chem Soc. 2011;133:6736–6744. doi: 10.1021/ja200222n. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Lashuel HA, Hartley D, Petre BM, Walz T, Lansbury PT., Jr Neurodegenerative disease: Amyloid pores from pathogenic mutations. Nature. 2002;418:291. doi: 10.1038/418291a. [DOI] [PubMed] [Google Scholar]
- 46.Zhu YJ, Lin H, Lal R. Fresh and nonfibrillar amyloid beta protein(1-40) induces rapid cellular degeneration in aged human fibroblasts: Evidence for AbetaP-channel-mediated cellular toxicity. FASEB J. 2000;14:1244–1254. doi: 10.1096/fasebj.14.9.1244. [DOI] [PubMed] [Google Scholar]
- 47.Stine WB, Jr, Dahlgren KN, Krafft GA, LaDu MJ. In vitro characterization of conditions for amyloid-beta peptide oligomerization and fibrillogenesis. J Biol Chem. 2003;278:11612–11622. doi: 10.1074/jbc.M210207200. [DOI] [PubMed] [Google Scholar]
- 48.Sheldrick GM. A short history of SHELX. Acta Crystallogr A. 2008;64:112–122. doi: 10.1107/S0108767307043930. [DOI] [PubMed] [Google Scholar]
- 49.Collaborative Computational Project, Number 4 The CCP4 suite: Programs for protein crystallography. Acta Crystallogr D Biol Crystallogr. 1994;50:760–763. doi: 10.1107/S0907444994003112. [DOI] [PubMed] [Google Scholar]
- 50.Yamashita E, Zhalnina MV, Zakharov SD, Sharma O, Cramer WA. Crystal structures of the OmpF porin: Function in a colicin translocon. EMBO J. 2008;27:2171–2180. doi: 10.1038/emboj.2008.137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Mastrangelo IA, et al. High-resolution atomic force microscopy of soluble Abeta42 oligomers. J Mol Biol. 2006;358:106–119. doi: 10.1016/j.jmb.2006.01.042. [DOI] [PubMed] [Google Scholar]
- 52.Leinala EK, et al. A beta-helical antifreeze protein isoform with increased activity. Structural and functional insights. J Biol Chem. 2002;277:33349–33352. doi: 10.1074/jbc.M205575200. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.