Abstract
Partition systems are responsible for the process whereby large and essential plasmids are accurately positioned to daughter cells during bacterial division. They are typically made of three components: a centromere-like DNA zone, an adaptor protein, and an assembling protein that is either a Walker-box ATPase (type I) or an actin-like ATPase (type II). A recently described type III segregation system has a tubulin/FtsZ-like protein, called TubZ, for plasmid movement. Here, we present the 2.3 Å structure and dynamic assembly of a TubZ tubulin homolog from a bacteriophage and unravel the Clostridium botulinum phage c-st type III partition system. Using biochemical and biophysical approaches, we prove that a gene upstream from tubZ encodes the partner TubR and localize the centromeric region (tubS), both of which are essential for anchoring phage DNA to the motile TubZ filaments. Finally, we describe a conserved fourth component, TubY, which modulates the TubZ-R-S complex interaction.
Keywords: DNA segregation, cytomotive filaments, virulence, plasmid partitioning
Virulence factors of pathogenic bacteria are often carried on mobile extrachromosomal elements that have a low copy number to reduce the genetic load requirements of the host. The correct segregation after replication and during bacterial cell division is of special importance to inherit features that facilitate bacterial invasion and infection. This function is carried out by plasmid-encoded partition systems, which typically require only three elements: a centromere-like DNA sequence, a centromere-binding protein, and a partition motor NTPase (1). Based on the nature of the protein responsible for the movement during segregation, these systems have been classified into type I (contains an ATPase with a variation of the Walker A-type fold), type II (uses actin-like ATPases), and type III (uses the GTPase tubulin/FtsZ-like protein TubZ).
Tubulin/FtsZ-like proteins form a family of GTPases with a common fold and display filament formation induced by GTP (2). They assemble into protofilaments, where the nucleotide sits at the monomer-monomer longitudinal interface. The C-terminal GTPase domain of the upper subunit complements the GTP-binding pocket (N-terminal domain) of the lower subunit, providing residues for GTP hydrolysis (3). Protofilaments coalesce into larger structures through lateral interaction and disassemble upon hydrolysis due to a conformational change that is not completely understood. TubZ was recently identified as a tubulin/FtsZ-like protein involved in low-copy plasmid segregation in Bacillus anthracis (pXO1) and Bacillus thuringiensis (pBtoxis) (4–6). It shows only a 10–20% sequence identity with either tubulin or FtsZ, but its crystal structure showed an overall folding related to the family (7, 8) in which the C-terminal domain is less conserved. Structures diverge especially at the C termini of this domain, where TubZ forms a single, long helix and a flexible tail that is involved in the interaction with its segregation partner TubR (7). TubZ assembles into double-helical filaments (8) that hydrolyze GTP (9).
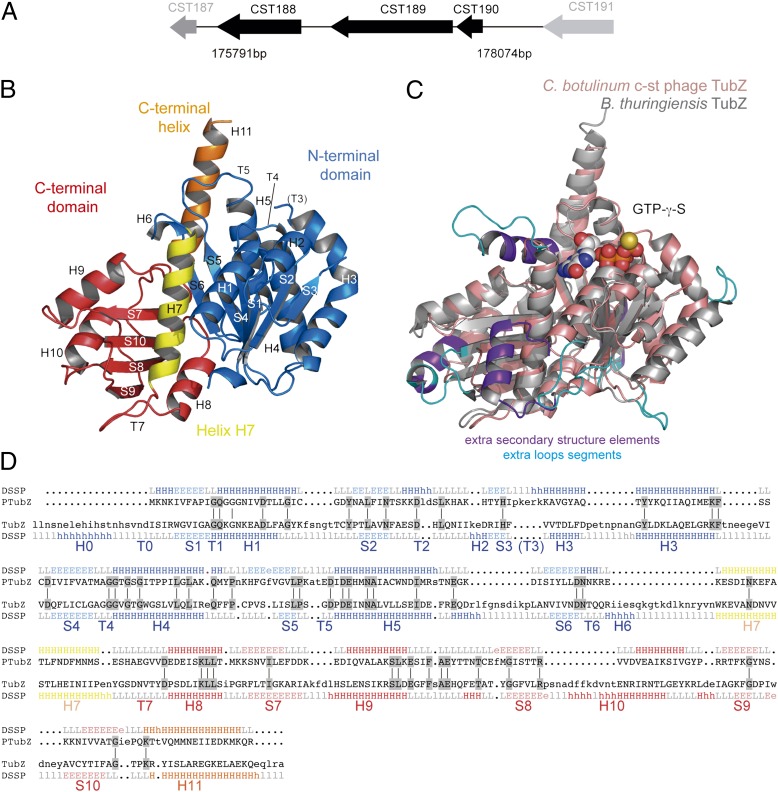
The ORF CST189 near the putative replication origin in the genome of the Clostridium botulinum neurotoxin-carrying c-st bacteriophage encodes a tubulin/FtsZ-like protein (10). Phage c-st is a linear 185.7-kb dsDNA Siphoviridae with no RNA stage (11). The phage DNA circularizes as a plasmid prophage in the host cell instead of integrating into the host chromosome. Otherwise, the phage conserves its propagation and infection mechanism through a typical phage capsid structure (10). Interestingly, the adjacent locus CST190 encodes a putative DNA-binding protein, and upstream, there is a long intergenic region enriched in directly repeated DNA sequences (iterons), which is a typical feature of centromere-like sequences (12). We thought it possible that these elements (Fig. 1A) constitute the prophage c-st segregation system. Here, we report the crystal structure and filament assembly of a phage-encoded tubulin homolog, the TubZ from the C. botulinum phage c-st. We show that CST190 encodes the DNA-binding protein TubR; identify the centromere-like region at 114 bp upstream from tubR; and describe a fourth partition system component encoded by CST188, called here TubY, that modulates the assembly of the TubZ-R-S complex.
Fig. 1.
C. botulinum phage c-st partition operon and TubZ structure. (A) CST188–190 cluster structure and localization in phage c-st genome. (B) Cartoon representation of the apo TubZ-T100A structure. Secondary structure elements are labeled according to Nogales et al. (2). The nucleotide-binding domain (N-terminal domain) with an empty nucleotide-binding site is shown in blue, the GTPase domain (C-terminal domain) is shown in red, the central helix H7 is shown in yellow, and the long C-termini TubZ helix H11 is shown in orange. (C) Superimposition of phage TubZ (salmon) onto B. thuringiensis plasmid TubZ (gray, displaying bound nucleotide). Regions with extra secondary structure elements in plasmid TubZ are highlighted in purple, and those with extra secondary structure elements in loops are highlighted in blue. (D) 3D alignment of protein structures from phage (PTubZ) and bacteria (TubZ) with reference to secondary structure elements (E, strand; H, helix; L, loop) and highlighting conserved residues (gray). Colors are the same as in B.
Results and Discussion
Phage Tubulin Homolog Structure Reveals a TubZ.
We have solved the structure of the full-length (358 residues) single-mutant (T100A) CST189 protein at a resolution of 2.3 Å. This mutant formed more stable crystals than the WT. The structure was solved by molecular replacement using the N-terminal domain of Bacillus subtilis FtsZ [Protein Data Bank (PDB) ID code 2VAM] with BALBES (13) and building the C-terminal domain with Auto-Rickshaw (14). There is one molecule in the crystallographic asymmetrical unit, and the model consists of residues 1–53 and 60–306, with an Rwork/Rfree of 0.17/0.22 (Table S1). The overall folding is related to the tubulin/FtsZ family and is highly homologous to plasmid TubZ despite the low sequence identity (16%), consisting of an N-terminal domain of the Rossman fold type connected to the C-terminal domain by the core central helix H7 and a single C-terminal long H11 helix (Fig. 1B). The model gap sequence localizes at the T3 loop, which is involved in nucleotide binding and is often very flexible, as shown previously in FtsZ and TubZ (7, 8, 15). The most conserved regions are those involved in nucleotide binding and hydrolysis, whereas main differences with the plasmid TubZ include the absence of initial helix H0, the very much shorter version of helix H6, and substantially shorter surface loops (Fig. 1 C and D). As in plasmid TubZ, the C-terminal domain is also rotated and tilted outward relative to the N-terminal domain, which suggests that the interdomain wedge provided by H0 (16) is not required for domain rotation. Otherwise, both proteins superimpose very well, with an rmsd of 2.2 Å, and as previously shown for plasmid TubZ, the phage’s protein superimposes optimally with rmsds of 3.0 Å and 3.1 Å with B. subtilis FtsZ and bovine α-tubulin (7), despite their very low sequence identity (15% and 10%, respectively).
Phage TubZ Double-Helical Filament Assembly and GTPase.
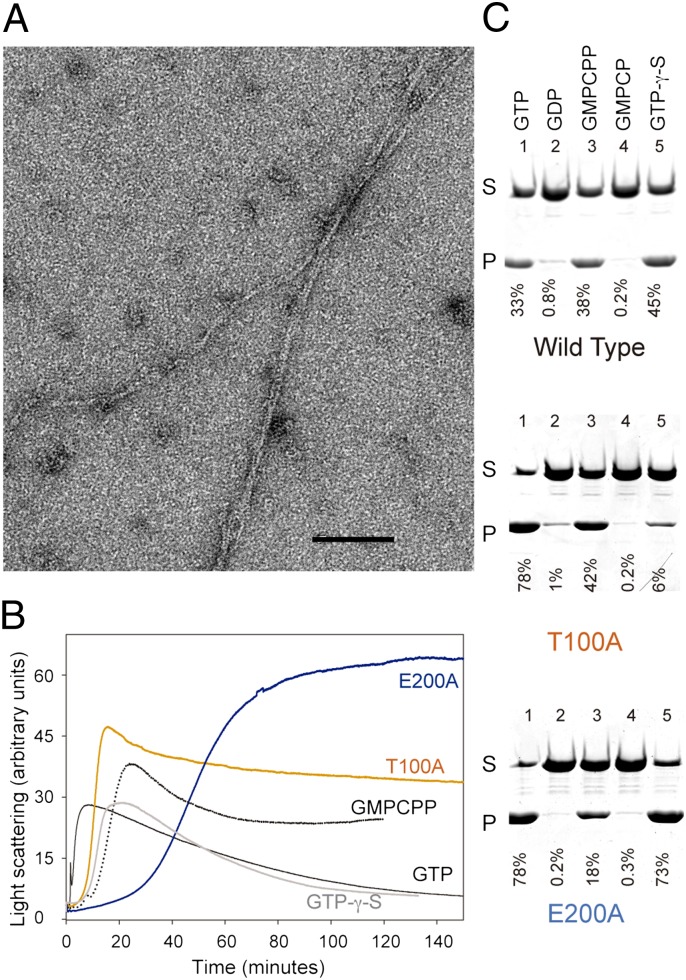
Phage TubZ purified as a monomer, as shown by analytical ultracentrifugation (AUC) at increasing protein concentrations, even in the presence of nucleotide (GTP or GDP) or Mg2+, with a sedimentation coefficient of s20,w = 3.4S (Fig. S1 A and B). We found that the purified protein, which had 0.45 nucleotide bound per TubZ molecule (89% GDP), assembles on addition of both GTP and Mg2+ into double-helical filaments (Fig. 2A) that coalesced into bundles, as described for plasmid TubZ (8, 9). Phage TubZ also shows a cooperative assembly, wherein all the protein polymerizes above a critical concentration of ∼1 μM (Fig. S2B). The nucleotide content of the filaments is mainly GDP (96%; Table S2), and they disassemble upon GTP hydrolysis (Fig. 2B), supporting a microtubule-like end-capping mechanism similar to that of the closely related plasmid TubZ (9).
Fig. 2.
Assembly properties of phage TubZ. (A) Electron micrograph of 10 μM TubZ two-stranded helical filaments assembled in PKE buffer [50 mM piperazine-1,4-bis-2-ethanesulfonic acid–potassium hydroxide, 100 mM potassium acetate, 1 mM EDTA (pH 6.5)] with 1 mM GTP and 6 mM Mg2+. (Scale bar: 100 nm.) (B) TubZ assembly monitored by 90° light scattering. WT TubZ (10 μM) polymerized in PKE buffer with 6 mM Mg2+ and 1 mM GTP (black), 0.1 mM GMPCPP (dotted black line), or 0.1 mM GTP-γ-S (gray). TubZ-T100A (10 μM; orange) or TubZ-E200A (10 μM; blue) was assembled in PKE with 6 mM Mg2+ and 1 mM GTP. There is an increase/decrease on the signal attributable to the formation/disassembly of polymers. (C) SDS/PAGE of sedimentation assays of 10 μM TubZ (WT and mutants) assembled in PKE buffer with 6 mM Mg2+ and 1 mM GTP (line 1), 2 mM GDP (line 2), 0.1 mM GMPCPP (line 3), 0.1 mM GMPCP (line 4), and 0.1mM GTP-γ-S (line 5). P, pellet; S, supernatant. Nucleotide diphosphates GDP and GMPCP do not induce filament formation.
Interestingly, the cocatalytic T7 loop of phage TubZ diverges from the highly conserved sequence (DXXD/E) typical of tubulin-like proteins and, instead, presents the very acidic motif DEDE. Here, Asp/Glu residues are key to complementing the N-terminal GTP-binding domain for the GTP-γ phosphate hydrolysis. The two extra negatively charged residues might be the reason for the reduced GTPase hydrolysis rate that we observe (0.97 ± 0.2 min−1; Table S2) compared with plasmid TubZ [16–25 min−1 (9)]. GTPase activity is related to filament dynamics; thereby, the low hydrolysis rate has a marked effect on the depolymerization of phage TubZ filaments, which stay in solution in vitro for longer than their plasmid homologs.
To determine how TubZ’s cocatalytic T7 loop and the tubulin signature motif in loop T4 are involved in nucleotide binding and hydrolysis, we introduced typical tubulin/FtsZ GTPase inactivating mutations in phage TubZ and built E200A (at the T7 loop) and T100A (located at T4). Mutant T100A purified with no nucleotide bound and E200A purified with 0.4 guanine nucleotide per TubZ, but both retain the ability to polymerize on addition of GTP and Mg2+. The equivalent mutation in the T7 loop of plasmid B. thuringiensis BtTubZ (D269A) had shown a substantial reduction in critical concentration and treadmilling dynamics in vivo (6), and a related mutation in the T4 loop of B. anthracis BaTubZ (T125A) showed an important decrease of the GTPase activity (17). Consistent with these data, we found that our phage mutant proteins TubZ-E200A and TubZ-T100A, which still assemble cooperatively into the double-helical filaments (Fig. S2 B–D), have markedly reduced GTPase rates, lower critical concentration values, and a smaller proportion of polymer GDP than WT TubZ (Table S2). Consequently, they polymerize, forming more stable polymers with even slower dynamics and longer lag phases (Fig. 2B).
We decided to investigate the role of GTP hydrolysis in phage TubZ filament dynamics further using GTP analogs. Guanosine-5′-[(alpha,beta)-methyleno] triphosphate (GMPCPP) is a slowly hydrolyzable nucleotide that enhances tubulin (18) and FtsZ assembly (19), whereas GTP-γ-S stabilizes FtsZ polymers (20). Both analogs support plasmid TubZ assembly and slow down disassembly (9). We found that phage TubZ also polymerized into filaments with GMPCPP and GTP-γ-S (Fig. 2B), forming double filaments as with GTP but with a reduced critical concentration and longer lag time (Table S2). Phage TubZ hydrolyses GMPCPP very slowly, and filament disassembly is thus reduced; however, surprisingly, the protein hydrolyzes GTP-γ-S more rapidly, which results in the complete disassembly of the filaments (Fig. 2B). The GTPase mutants show different polymerization properties with the two analogs: GMPCPP better supports the assembly of TubZ-T100A, and GTP-γ-S better supports the polymerization of TubZ-E200A (Fig. 2C), indicating that the upper and lower TubZ interacting surfaces display different interactions with these nucleotides.
Phage TubZ’s GTPase activity, and thus polymer dynamics, is highly dependent on monovalent and divalent cations. Replacing K+ with Na+ and Mg2+ with Ca2+ induced no variations in the critical concentration or polymer GDP content but led to a longer assembly lag phase and a decrease in the GTPase rate (Table S2), accompanied by a slower disassembly. Including both Na+ and Ca2+ under the same experimental conditions practically abolished the GTPase-related polymer dynamics, because we did not observe filament disassembly (Fig. S1C). Our results suggest that even having a peculiar cocatalytic T7 loop, which might be responsible for decreasing the nucleotide hydrolysis rate, and thus the dynamics, phage TubZ’s biochemical properties are related to those of its plasmid homolog (9). Still, phage TubZ has the peculiarity, with respect to tubulin/FtsZ, of showing no significant changes in dynamics attributable to ion strength, neither in K+- nor Na+-containing buffers (Fig. S1D), which also might be related to the highly acidic loop T7.
Phage TubR Protein Binds Centromeric tubS DNA.
Segregation systems require a DNA-binding adaptor protein that tethers the plasmid to the cytoskeletal protein to mediate separation. Loci immediately upstream (CST190) and downstream (CST188) of the tubZ gene (Fig. 1A) include HTH motifs present in many DNA-binding proteins. However, because of the smaller size (81 residues; Fig. S3A) and its gene localization similar to plasmid tubR, the CST190 protein is the stronger candidate to be the TubR adaptor of the phage. To test this hypothesis, we expressed and purified this protein, and analyzed its ability to bind DNA and to interact with TubZ filaments.
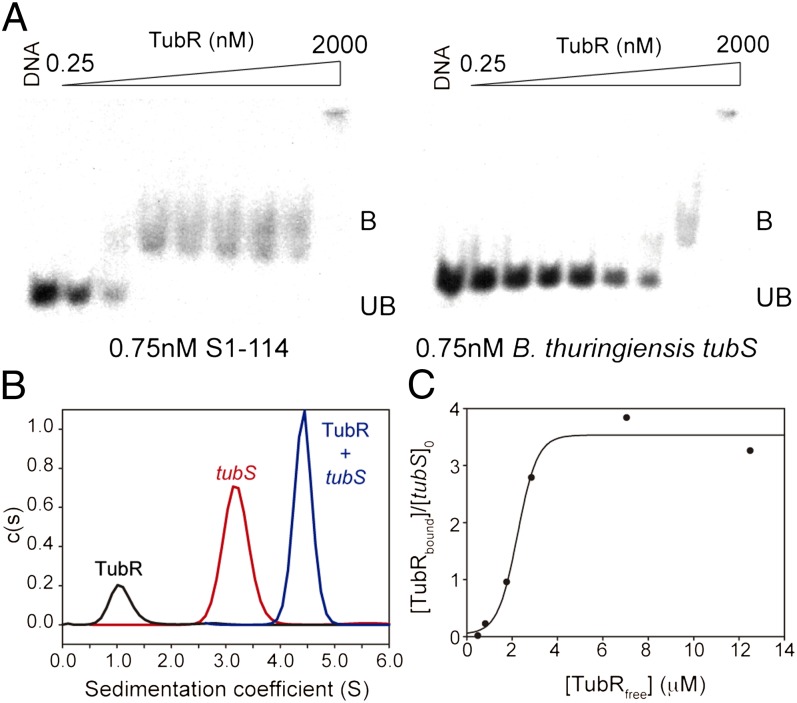
Unlike plasmid TubR, which is a dimer in solution (7), phage TubR purified as a monomer, without signs of the presence of additional species in a concentration range of 10–100 μM and with a sedimentation coefficient of s020,w = 1.3S (Fig. S3 B and C). We determined the DNA-binding specificity of this protein with DNA EMSAs. The centromere-like site is typically located either upstream or downstream of the partition operon; thus, our initial mapping approach included ∼200-bp radiolabeled fragments covering intergenic adjacent regions upstream of CST190 (fragment S1) and downstream of CST188 (fragments S2 and S3), which contain directed and inverted repeated sequences (Fig. S4 A and B). TubR shows specific binding at nanomolar concentrations only to fragment S1 (Fig. S4C). To analyze the centromere-like sequence further, we divided this fragment into three subfragments of 44 bp, avoiding the first 26 bp upstream from the start codon that contains the Shine–Dalgarno sequence (Fig. S4D). TubR bound with the highest specificity to fragment S1-114, which covers the sequence 114–157 bp upstream from the tubR start codon (Fig. 3A and Fig. S4E). We selected this region as a representative phage tubS centromeric sequence for further experiments because, in addition, its distance from the tubR gene is compatible with that found in B. thuringiensis tubS (12). We observed that TubR binds very weakly to shuffled fragment S1-114 (Fig. S4E), showing that the nucleotide sequence is essential for recognition, as with B. thuringiensis tubS (Fig. 3A), indicating a lack of promiscuity between the centromeric sequences. Curiously, plasmid iterons have the phage TTTGAC sequence repetition inverted (Fig. S4D), but phage TubR only binds with high affinity to its own centromere-like sequence, which is indicative of high recognition specificity. We studied the TubR-tubS interaction with AUC sedimentation velocity experiments, where TubR (1.3S), tubS (3.2S), and their complex (4.6S) were unambiguously resolved (Fig. 3B). Because tubS (S1-114) includes four iterons (Fig. S4D), there could be several binding sites. AUC sedimentation equilibrium titration experiments indicated that TubR binds to tubS, reaching binding saturation at four molecules per tubS (Fig. 3C). Although the structure of the complex is unknown, this result differs from the previously described plasmid TubR dimer binding (7).
Fig. 3.
TubR-tubS interaction. (A) EMSAs using purified TubR and γ-32P–labeled DNA subfragment S1-114 (Left) and B. thuringiensis centromere-like sequence tubS (Right). The DNA band shift (B, bound; UB, unbound) attributable to binding of TubR is indicated. The TubR concentrations increase fourfold in successive lanes from left to right. (B) AUC velocity sedimentation experiments, where each curve correspond to an independent experiment of 10 μM TubR (black; monitored at 280 nm), 1.2 μM subfragment S1-114 (tubS; red), or 1.2 μM tubS plus 10 μM TubR (blue), with the latter two monitored at 260 nm. The sedimentation peaks from protein (1.2S) and DNA (3.2S) are clearly distinguishable from the complex (4.6S). (C) Titration of tubS with increasing TubR concentrations. Binding was determined by the increment molecular mass of the complexes measured by sedimentation equilibrium AUC. The binding of protein to DNA increases with protein concentration to near-saturation at a stoichiometry of 3.55 ± 0.21 TubR per tubS DNA molecule, according to a cooperative model fit indicated by the line.
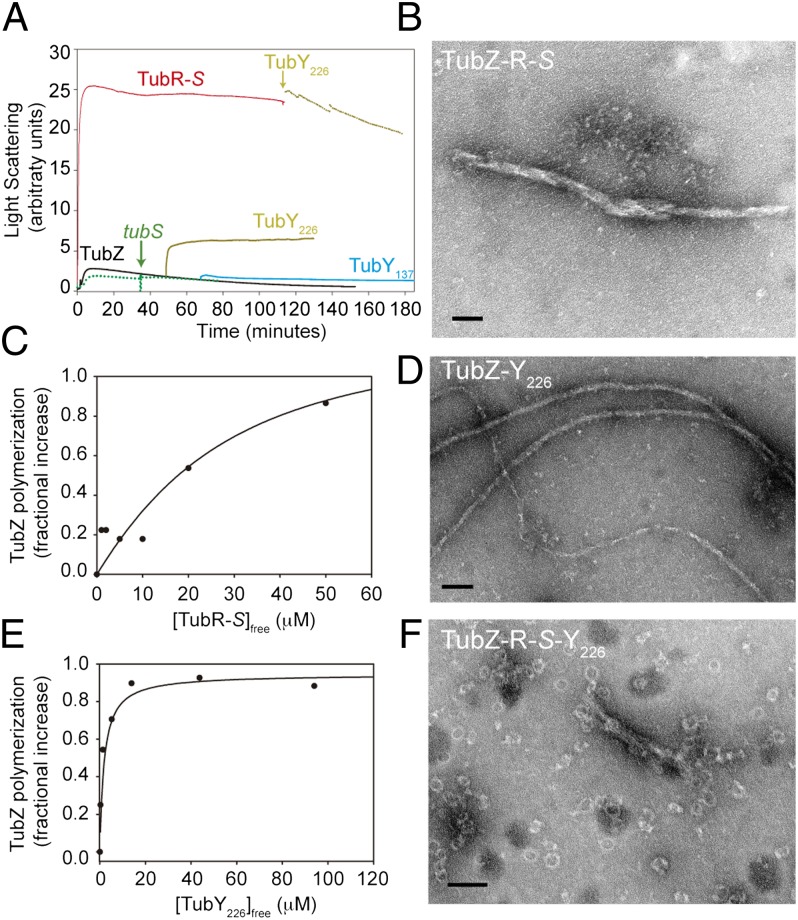
TubR also binds to TubZ filaments but only when it is in complex with tubS (Fig. S5), possibly because it then exposes the correct surface for TubZ interaction. TubR-S has a dramatic effect on TubZ assembly, with a large increase in the light scattering signal (Fig. 4A) and a 40% decline in the GTPase activity, which are associated with the coalescence of the TubZ double-helical filament into huge bundles (Fig. 4B). Furthermore, TubR-S complex may bind along the TubZ filament, because titration results were compatible with one binding site for the complex per TubZ molecule, with an apparent dissociation constant (Kd) of ∼40 μM (Fig. 4C). These results are compatible with the known plasmid TubR interaction with filaments through TubZ C-termini and the tramming segregation mechanism proposed for type III partition systems (7, 21).
Fig. 4.
TubR and TubY interaction with TubZ filaments. (A) TubZ (10 μM) polymerization monitored by 90° light scattering (black) shows the effect of the partners’ addition during assembly. Additions are 10 μM TubR-5 μM tubS complex (red), 10 μM TubY226 (yellow), 10 μM TubY137 (blue), and 5 μM tubS (dotted green line). (B) Electron micrograph of TubZ bundles induced by interaction with TubR-S. (C) Titration by sedimentation of the increase in TubZ polymerization vs. TubR–tubS complex concentration. The solid line is a model fit for approximately 1:1 binding with Kd = 39 μM. (D) TubZ filaments in the presence of TubY226. (E) Titrations by sedimentation of the increase in TubZ polymerization vs. TubY226 concentration; the line is a model fit with 1:1 binding (n = 1) and Kd = 0.9 μM. Models with n < 1 were equally compatible with the data, whereas models with n > 1 strongly deviated from the data. (F) Reshaping of the TubZ-R-S complex bundles into rings when TubY226 is added. (Scale bars: 100 nm.)
TubY, a Regulatory Partner Encoded by CST188.
Downstream of tubZ, the ORF CST188 (Fig. 1A) encodes a 239-residue protein, here called TubY, that includes an HTH motif and a C-terminal domain with a 90-residue-long predicted coiled-coil region (similar to the MerR family of transcriptional regulators), as well as a conserved short tail of 13 hydrophobic and positively charged residues at the C-termini, NNKKGFFGKLFKR (Fig. S6A), which could fold into an amphipathic helix. To determine whether TubY is able to interact with the TubZ-R-S complex, we purified two soluble and folded truncated versions (Fig. S6B), because the full-length protein was insoluble. TubY226 excludes only the last 13 residues, and TubY137 also excludes the coiled-coil.
TubY137 purified as a monomer with a s020,w = 1.5S, with a weak tendency to self-associate (Fig. S6 C and D). TubY226 predominantly formed s20,w = 8.9S octamers (Fig. S6 C and E), probably attributable to the long coiled-coil, which is a secondary structure element typically involved in protein-protein interactions. Our EMSA assays showed nonspecific DNA binding by TubY137 and TubY226 at micromolar concentrations to fragments S1, S2, S3, and B. thuringiensis tubS. TubY226 binds directly to TubZ filaments through its coiled-coil region (TubY137 does not interact; Fig. S5A), with a micromolar affinity and one or fewer binding sites per TubZ molecule (Fig. 4E). TubY226 oligomers should disassemble when binding to TubZ polymers, where TubY226 induces stabilization and a 33% decrease of the GTPase activity (Fig. 4A), although we have not observed any gross bundling of the filaments (Fig. 4D) with respect to TubZ alone (Fig. 2A). However, we found that TubY226 also interacts with TubZ filaments when they are in complex with TubR-S, inducing depolymerization (Fig. 4A) and a remarkable reshaping of the huge TubZ-R-S complex bundles into rings of variable diameter (30–40 nm; Fig. 4F), which is a good indication of its modulatory activity on this in vitro system.
Interestingly, we have found tubY homologous sequences (Fig. 5A) in other TubZ-coding extrachromosomal elements from C. botulinum; in chromosomal TubZ-encoding gene clusters of Clostridium acetobutylicum, Clostridium kluyveri, Clostridium cellulovorans, and Clostridium butyricum (with the exception of Clostridium sticklandii); and in some Bacillus plasmids, such as pOX1 from B. anthracis, pBtoxis from B. thuringiensis, and pBc239 from Bacillus cereus. In Bacillus, the tubY homolog sequences are at longer intergenic distances and frequently in opposite orientations (Fig. 5A), which may argue against its cotranscription. However, the clustering of genes in bacteria contributes to the attainment of high local protein concentrations close to their encoding genes (because transcription and translation are simultaneous); thus, they may selectively interact even when they are not cotranscribed (22, 23). Furthermore, the Par protein, responsible for the stabilization of plasmid pSK1 in Staphylococcus spp. (24), could be a TubY remote homolog because it shares both the HTH and coiled-coil motifs and 45% identity at the C-terminal tail with TubY, but this plasmid lacks TubZ-R homologs.
Fig. 5.
Gene organization and evolution of TubZ-based segrosomes. (A) Comparative schematic diagram of the type III segregation system cluster in different replicons from Clostridium and two Bacillus species. The positions of the genes tubZ (orange), tubR (red), and the unique component described here tubY (yellow) are shown. (B) Phylogenetic tree of TubZ shows relationships between phage (red), plasmid (blue), and chromosomal (gray) tubZ sequences. Bootstrap values representing confidence levels are indicated for selected branches. (Right) Presence or absence of tubR (R) and tubY (Y) near tubZ is shown. Question marks indicate the presence of putative proteins that share with TubR only an HTH-motif and a small size or include only long coiled-coil motifs like TubY.
TubZ-Based Phage and Plasmid Partition Systems.
Botulinum neurotoxins are among the most poisonous known substances because of their effect of blocking the acetylcholine receptors in the neuromuscular junction. There are seven types of toxins (A, B, C1, and D–G) produced by Clostridium, and types C1 and D are carried by bacteriophages. The c-st phage encodes botulism toxin but does not carry any essential genes for C. botulinum survival. Therefore, the acquisition of a partition system for phage perpetuation appears surprising. However, it is crucial to maintaining bacterial virulent capacity.
Phage c-st recircularizes as a plasmid-like entity instead of inserting into the host chromosome (10). The structural and biochemical features of its TubZ-R-S complex cluster, which is only 1 kb away from the replication origin, suggest that it constitutes a type III segregation system of this large genome, similar to those described for Bacillus plasmids (4, 5). Other prophages are known to contain plasmid segregation systems (e.g., the well-characterized type Ia system in P1) (25, 26), albeit with none sharing a tubulin-like assembly machine. There are several options to explain the acquisition of this gene cluster. Considering that this family of phages is genetically very unstable (27), probably with a high tendency to recombine with other DNA molecules, Clostridium species could get these genes from a current Bacillus species, or vice versa, but it is equally possible that both organisms could have acquired them from an ancestral extrachromosomal molecule shared between the related Bacillus and Clostridium genera. Likewise, although type III partition systems have been described only in Bacillus species, we cannot rule out their existence in other distant species. A phylogenetic analysis of TubZ indicates at least five clades (Fig. 5B): (i) Clostridium homologs that further subdivide into phage, plasmid, and chromosomal versions; (ii) distant plasmid homologs from Bacillus species; (iii) extrachromosomal homologs from Bacillus megaterium; (iv) phage homologs from Pseudomonas; and (v) putative chromosomal tubZ in Archaebacteria, which are only vaguely related to Bacillus tubZ. Clostridium and Bacillus extrachromosomal elements show all cluster components (TubZ, TubR, and TubY), and its topology (Fig. 5A) and localization (close to the origin of replication) are similar. However, it is difficult to determine the origin and function of these genes in Clostridium chromosomes, because segregation likely relies on the ParAB homologs Soj and Spo0J (28). Here, the cluster is far from the origin of replication, and there is probably no selective pressure, which may cause loss of elements (Fig. 5A). Interestingly, the extrachromosomal elements of B. megaterium and phages from Pseudomonas aeruginosa include TubZ, a longer TubR, and a protein that contains a long coiled-coil similar to that of TubY (Fig. 5B). Archaea include up to five different FtsZ homologs: FtsZ1 and FtsZ2 are FtsZ-like, whereas FtsZ3, FtsZ4, and FtsZ5 are divergent FtsZs related to TubZ. Very interesting is the archaea Haloarcula marismortui, whose genetic material includes genes encoding all five FtsZ homologs and is dispersed into nine replicons, five in the range of 50–410 kb (29). There are no TubR-like proteins in the archaea analyzed apart from H. marismortui, where we find a gene encoding a small HTH motif close to ftsZ3. Despite the fact that there are no TubY-like proteins either, the other adjacent gene to ftsZ3 encodes a predicted protein of similar length to TubY with no HTH motif but with a long coiled-coil (Fig. 5B).
Notice that sequence identity between different TubZs is unusually low, even between very close species, although the overall structure and the filaments assembled are very similar; therefore, their sequence divergence may relate to the specificity of these segregation machines (30). Otherwise, there could be interferences between segregation systems through nonspecific protein-protein and protein-DNA interactions when there are several extrachromosomal elements in the same organism, as happens in C. botulinum D-1873, which has a phage D-1873 (31) and a plasmid pCLG2, both with a complete TubZ-R-S complex cluster. In fact, we found a lack of high-affinity binding of phage TubR to the B. thuringiensis tubS, which supports the high specificity of these systems.
Therefore, a possible situation is that these large DNA entities (plasmids or phages) were all originally phages that, on evolution, have been perpetuated in bacteria to retain special virulence features. Partition systems were apparently selected as the segregation machineries of large, low-copy extrachromosomal replicons, whereas smaller ones may simply perpetuate by passive diffusion. We have shown that the tubulin/FtsZ family of cytomotive GTPases extends to bacteriophages, describing the properties and interactions of the components of a putative partition system in the botulinum c-st phage. TubR bound to c-st phage DNA through tubS iterons may interact with TubZ filaments, probably through the flexible TubZ C-terminal region (7) that is exposed in the surface of TubZ filaments (8). Treadmilling filaments can travel around the cell poles (6), although there is no evidence of a direct interaction between filaments and the membrane. TubZ filaments may transport the phage DNA to the cell poles, where TubR–DNA complexes are somehow released from TubZ, perhaps by the bending force generated at the poles (7). In addition, we have shown that phage c-st TubY binds to TubZ filaments and reshapes TubZ-R-S complex bundles into rings. Therefore, we propose that TubY has a regulatory function. It may be envisaged that TubY carries out the disassembly of the segrosome complex, leading to the release of the phage DNA, but TubY could also prevent TubZ-R-S complex bundling in vivo, and thus support correct segregation dynamics. It remains to be studied how the TubZ-R-S + Y system assembles and performs phage DNA segregation during bacterial cell division.
Materials and Methods
Detailed information is provided in SI Materials and Methods. Briefly, TubZ, TubZ-T100A, and TubZ-E200A were expressed in Escherichia coli and purified by affinity chromatography, followed by His tag removal and gel filtration. TubR and TubY were codon-optimized for E. coli expression, purified by ammonium sulfate precipitation, ion exchange, and gel filtration. TubZ-T100A crystals were grown with ammonium acetate and PEG 10,000, and the structure was solved using molecular replacement. Detailed methods for AUC, TubZ assembly, GTPase activity, polymer-bound nucleotide, EM, EMSA, and phylogenetic analysis are provided in SI Materials and Methods.
Supplementary Material
Acknowledgments
We thank staff at beamlines ID14eh4 European Synchrotron Radiation Facility (ESRF) and PROXIMA 1 (SOLEIL) for their support. We also thank K. Oguma (Okayama University) for support in the initial stages of this project; J. Löwe (Medical Research Council Laboratory of Molecular Biology) for helpful crystallography discussion; A. Romero, F. J. Medrano, and F. M. Ruiz (Consejo Superior de Investigaciones Cientificas-Centro de Investigaciones Biológicas) for crystallography facilities and support; R. Giraldo and G. del Solar (Consejo Superior de Investigaciones Cientificas-CIB) for advice on DNA-binding proteins; C. Alfonso (Consejo Superior de Investigaciones Cientificas-CIB) for AUC analysis; and D. Juan (Consejo Superior de Investigaciones Cientificas-CIB) for technical assistance. M.A.O. was supported by a Juan de la Cierva contract; A.J.M.-G. was supported by a Juan de la Cierva contract, a Junta de Ampliación de Estudios-Doc contract, and a Miguel Servet contract. This work was supported by grants from Plan Nacional de Investigación BFU2008-00013, BFU2011-23416, and Comunidad de Madrid S2010/BMD-2353 (to J.M.A.).
Footnotes
The authors declare no conflict of interest.
Data deposition: The atomic coordinates and structure factors reported in this paper have been deposited in the Protein Data Bank, www.pdb.org (PDB ID code 3V3T).
This article is a PNAS Direct Submission. L.A.A. is a guest editor invited by the Editorial Board.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1121546109/-/DCSupplemental.
References
- 1.Gerdes K, Møller-Jensen J, Bugge Jensen R. Plasmid and chromosome partitioning: Surprises from phylogeny. Mol Microbiol. 2000;37:455–466. doi: 10.1046/j.1365-2958.2000.01975.x. [DOI] [PubMed] [Google Scholar]
- 2.Nogales E, Downing KH, Amos LA, Löwe J. Tubulin and FtsZ form a distinct family of GTPases. Nat Struct Biol. 1998;5:451–458. doi: 10.1038/nsb0698-451. [DOI] [PubMed] [Google Scholar]
- 3.Oliva MA, Cordell SC, Löwe J. Structural insights into FtsZ protofilament formation. Nat Struct Mol Biol. 2004;11:1243–1250. doi: 10.1038/nsmb855. [DOI] [PubMed] [Google Scholar]
- 4.Tinsley E, Khan SA. A novel FtsZ-like protein is involved in replication of the anthrax toxin-encoding pXO1 plasmid in Bacillus anthracis. J Bacteriol. 2006;188:2829–2835. doi: 10.1128/JB.188.8.2829-2835.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Tang M, Bideshi DK, Park HW, Federici BA. Minireplicon from pBtoxis of Bacillus thuringiensis subsp. israelensis. Appl Environ Microbiol. 2006;72:6948–6954. doi: 10.1128/AEM.00976-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Larsen RA, et al. Treadmilling of a prokaryotic tubulin-like protein, TubZ, required for plasmid stability in Bacillus thuringiensis. Genes Dev. 2007;21:1340–1352. doi: 10.1101/gad.1546107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Ni L, Xu W, Kumaraswami M, Schumacher MA. Plasmid protein TubR uses a distinct mode of HTH-DNA binding and recruits the prokaryotic tubulin homolog TubZ to effect DNA partition. Proc Natl Acad Sci USA. 2010;107:11763–11768. doi: 10.1073/pnas.1003817107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Aylett CH, Wang Q, Michie KA, Amos LA, Löwe J. Filament structure of bacterial tubulin homologue TubZ. Proc Natl Acad Sci USA. 2010;107:19766–19771. doi: 10.1073/pnas.1010176107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Chen Y, Erickson HP. In vitro assembly studies of FtsZ/tubulin-like proteins (TubZ) from Bacillus plasmids: Evidence for a capping mechanism. J Biol Chem. 2008;283:8102–8109. doi: 10.1074/jbc.M709163200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Sakaguchi Y, et al. The genome sequence of Clostridium botulinum type C neurotoxin-converting phage and the molecular mechanisms of unstable lysogeny. Proc Natl Acad Sci USA. 2005;102:17472–17477. doi: 10.1073/pnas.0505503102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Eklund MW, Poysky FT, Reed SM, Smith CA. Bacteriophage and the toxigenicity of Clostridium botulinum type C. Science. 1971;172:480–482. doi: 10.1126/science.172.3982.480. [DOI] [PubMed] [Google Scholar]
- 12.Tang M, Bideshi DK, Park HW, Federici BA. Iteron-binding ORF157 and FtsZ-like ORF156 proteins encoded by pBtoxis play a role in its replication in Bacillus thuringiensis subsp. israelensis. J Bacteriol. 2007;189:8053–8058. doi: 10.1128/JB.00908-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Long F, Vagin AA, Young P, Murshudov GN. BALBES: A molecular-replacement pipeline. Acta Crystallogr D Biol Crystallogr. 2008;64:125–132. doi: 10.1107/S0907444907050172. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Panjikar S, Parthasarathy V, Lamzin VS, Weiss MS, Tucker PA. Auto-rickshaw: An automated crystal structure determination platform as an efficient tool for the validation of an X-ray diffraction experiment. Acta Crystallogr D Biol Crystallogr. 2005;61:449–457. doi: 10.1107/S0907444905001307. [DOI] [PubMed] [Google Scholar]
- 15.Díaz JF, et al. Activation of cell division protein FtsZ. Control of switch loop T3 conformation by the nucleotide gamma-phosphate. J Biol Chem. 2001;276:17307–17315. doi: 10.1074/jbc.M010920200. [DOI] [PubMed] [Google Scholar]
- 16.Nogales E. When cytoskeletal worlds collide. Proc Natl Acad Sci USA. 2010;107:19609–19610. doi: 10.1073/pnas.1014665107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Anand SP, Akhtar P, Tinsley E, Watkins SC, Khan SA. GTP-dependent polymerization of the tubulin-like RepX replication protein encoded by the pXO1 plasmid of Bacillus anthracis. Mol Microbiol. 2008;67:881–890. doi: 10.1111/j.1365-2958.2007.06100.x. [DOI] [PubMed] [Google Scholar]
- 18.Hyman AA, Salser S, Drechsel DN, Unwin N, Mitchison TJ. Role of GTP hydrolysis in microtubule dynamics: Information from a slowly hydrolyzable analogue, GMPCPP. Mol Biol Cell. 1992;3:1155–1167. doi: 10.1091/mbc.3.10.1155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Oliva MA, et al. Assembly of archaeal cell division protein FtsZ and a GTPase-inactive mutant into double-stranded filaments. J Biol Chem. 2003;278:33562–33570. doi: 10.1074/jbc.M303798200. [DOI] [PubMed] [Google Scholar]
- 20.Scheffers DJ, den Blaauwen T, Driessen AJ. Non-hydrolysable GTP-gamma-S stabilizes the FtsZ polymer in a GDP-bound state. Mol Microbiol. 2000;35:1211–1219. doi: 10.1046/j.1365-2958.2000.01791.x. [DOI] [PubMed] [Google Scholar]
- 21.Schumacher MA. Bacterial plasmid partition machinery: A minimalist approach to survival. Curr Opin Struct Biol. 2012;22(1):72–79. doi: 10.1016/j.sbi.2011.11.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Lawrence J. Selfish operons: The evolutionary impact of gene clustering in prokaryotes and eukaryotes. Curr Opin Genet Dev. 1999;9:642–648. doi: 10.1016/s0959-437x(99)00025-8. [DOI] [PubMed] [Google Scholar]
- 23.Montero Llopis P, et al. Spatial organization of the flow of genetic information in bacteria. Nature. 2010;466:77–81. doi: 10.1038/nature09152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Simpson AE, Skurray RA, Firth N. A single gene on the staphylococcal multiresistance plasmid pSK1 encodes a novel partitioning system. J Bacteriol. 2003;185:2143–2152. doi: 10.1128/JB.185.7.2143-2152.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Hayes F, Barillà D. The bacterial segrosome: A dynamic nucleoprotein machine for DNA trafficking and segregation. Nat Rev Microbiol. 2006;4(2):133–143. doi: 10.1038/nrmicro1342. [DOI] [PubMed] [Google Scholar]
- 26.Dunham TD, Xu W, Funnell BE, Schumacher MA. Structural basis for ADP-mediated transcriptional regulation by P1 and P7 ParA. EMBO J. 2009;28:1792–1802. doi: 10.1038/emboj.2009.120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Sakaguchi Y, et al. Molecular analysis of an extrachromosomal element containing the C2 toxin gene discovered in Clostridium botulinum type C. J Bacteriol. 2009;191:3282–3291. doi: 10.1128/JB.01797-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Lee PS, Grossman AD. The chromosome partitioning proteins Soj (ParA) and Spo0J (ParB) contribute to accurate chromosome partitioning, separation of replicated sister origins, and regulation of replication initiation in Bacillus subtilis. Mol Microbiol. 2006;60:853–869. doi: 10.1111/j.1365-2958.2006.05140.x. [DOI] [PubMed] [Google Scholar]
- 29.Baliga NS, et al. Genome sequence of Haloarcula marismortui: A halophilic archaeon from the Dead Sea. Genome Res. 2004;14:2221–2234. doi: 10.1101/gr.2700304. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Fothergill TJ, Barillà D, Hayes F. Protein diversity confers specificity in plasmid segregation. J Bacteriol. 2005;187:2651–2661. doi: 10.1128/JB.187.8.2651-2661.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Fujii N, Oguma K, Yokosawa N, Kimura K, Tsuzuki K. Characterization of bacteriophage nucleic acids obtained from Clostridium botulinum types C and D. Appl Environ Microbiol. 1988;54:69–73. doi: 10.1128/aem.54.1.69-73.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.