Abstract
Most extant genus-level radiations in gymnosperms are of Oligocene age or younger, reflecting widespread extinction during climate cooling at the Oligocene/Miocene boundary [∼23 million years ago (Ma)]. Recent biogeographic studies have revealed many instances of long-distance dispersal in gymnosperms as well as in angiosperms. Acting together, extinction and long-distance dispersal are likely to erase historical biogeographic signals. Notwithstanding this problem, we show that phylogenetic relationships in the gymnosperm family Cupressaceae (162 species, 32 genera) exhibit patterns expected from the Jurassic/Cretaceous breakup of Pangea. A phylogeny was generated for 122 representatives covering all genera, using up to 10,000 nucleotides of plastid, mitochondrial, and nuclear sequence per species. Relying on 16 fossil calibration points and three molecular dating methods, we show that Cupressaceae originated during the Triassic, when Pangea was intact. Vicariance between the two subfamilies, the Laurasian Cupressoideae and the Gondwanan Callitroideae, occurred around 153 Ma (124–183 Ma), when Gondwana and Laurasia were separating. Three further intercontinental disjunctions involving the Northern and Southern Hemisphere are coincidental with or immediately followed the breakup of Pangea.
Keywords: ancestral areas reconstruction, molecular clock
Between the Early Triassic and the Middle Jurassic, virtually all continents were joined to form the supercontinent Pangea (1–3). Around 160–138 million years ago (Ma) (1, 3), Pangea broke up into two supercontinents: Laurasia, comprising land that eventually gave rise to North America, Europe, and much of Asia, and Gondwana, made up of land that subsequently gave rise to South America, Africa, India, Antarctica, and Australia. Biostratigraphic data suggest that Late Triassic and Early Jurassic Pangea had a warm and equable climate without glaciation or sea ice and that it lacked significant geographic barriers from pole to pole (4). However, because of Pangea’s great latitudinal expanse, faunal provinces already had developed before its break-up, and dated molecular phylogenies of reptiles, amphibians, and mammals have made clear that subsequent lineage divergence within these groups matches the separation and fragmentation of Laurasia and Gondwana (5–10). Until now, there has been no equivalent evidence for any plant family.
The fossil record shows that gymnosperms dominated the vegetation of Pangea but declined in dominance and abundance from the Mid-Cretaceous onwards (11, 12). Perhaps because of the extinction of entire clades, molecular-clock studies of gymnosperms consistently have inferred young, usually Oligocene, ages for the crown groups of living genera, e.g., Phyllocladus (13), Gnetum (14), Cedrus (15), Agathis (16, 17), Ephedra (18), Juniperus (19), Pseudotsuga (20), Podocarpus, Nageia, Dacrydium, Dacrycarpus (21), and Pinus subgenera Pinus and Strobus (22, 23). Radiations are especially young in the cycads (24–26).
Among the few spermatophyte clades that still may reflect events related to the break-up of Pangea is the conifer family Cupressaceae (including the former Taxodiaceae) (27–31). Cupressaceae occur on all continents except Antarctica and comprise 162 species in 32 genera (see Table S2 for subfamilies, genera, and species numbers). The family has a well-studied fossil record going back to the Jurassic (32–36). Using ancient fossils to calibrate genetic distances in molecular phylogenies can be problematic, because the older a fossil is, the more likely it is to represent an extinct lineage that diverged somewhere along the line leading to the extant taxon with which it is being compared (37). However, probability distributions on fossil calibration ages allow some manipulation of this uncertainty (38), and judicious use of multiple fossils also may help circumvent calibration pitfalls (39).
Here we present a phylogeny for 122 species from the 32 genera of Cupressaceae (plus 22 species representing relevant outgroups) and use 16 fossil calibration points and three dating approaches to estimate divergence times in the Cupressaceae. We then perform ancestral area reconstructions (AARs) using maximum likelihood based on datasets with or without incorporated fossils. Possible changes in diversification rates were inferred with an approach that accounts for nonrandom taxon sampling in molecular phylogenies (40). Nonrandom sampling arises when phylogenies include at least one species per genus but not all congenerics, thereby overrepresenting deep nodes (diversification events) in the tree. Experiments have confirmed the theoretical expectation that such sampling leads to the erroneous inference of diversification rate downturns (41, 42). We aimed to test the hypothesis that, given their fossil record, the deepest Cupressaceae divergences should reflect the break-up of Pangea and that evolution of the family then continued on the separating continental landmasses.
Results
Cupressaceae Phylogenetics.
After sequence alignment and removal of ambiguous regions, we obtained two datasets, one of 56 taxa and 10,472 aligned nucleotides from plastid, mitochondrial, and nuclear DNA, the other of 144 taxa and 7,171 nucleotides from plastid DNA only. Maximum likelihood, parsimony, and Bayesian optimization inferred similar topologies from both datasets. Support values for major groups are high, with three-quarters of the genus-level nodes having >95% posterior probability (Fig. S1).
Divergence Times.
Bayesian coestimation of topology and divergence time (BEAST, using uniform prior distributions on calibration ages) (43) generally gave the oldest ages, and Penalized likelihood (44, 45) gave the youngest (Table S3). An alternative Bayesian approach, which used a fixed topology (MULTIDIVTIME) (46), yielded ages for short-branched nodes (most nodes within Cupressoideae; Fig. S1) that were similar to or younger than those obtained with BEAST; ages for long-branched nodes (most nodes within Callitroideae; Fig. S1) were similar to or older than those obtained with BEAST. Confidence intervals around estimates from the two Bayesian approaches overlapped (Table S3). With all three dating approaches, the more densely sampled 144-taxon dataset produced slightly older age estimates (compare Fig. S2 A and B), a result that is consistent with the effects of undersampling observed elsewhere (47). Because BEAST allows more complex nucleotide-substitution models than do the other two dating approaches, and because dates from the 56-taxon matrix might be less accurate because of undersampling, the following discussion focuses on the results obtained with the 144-taxon matrix analyzed using BEAST (Fig. 1 and Fig. S2B).
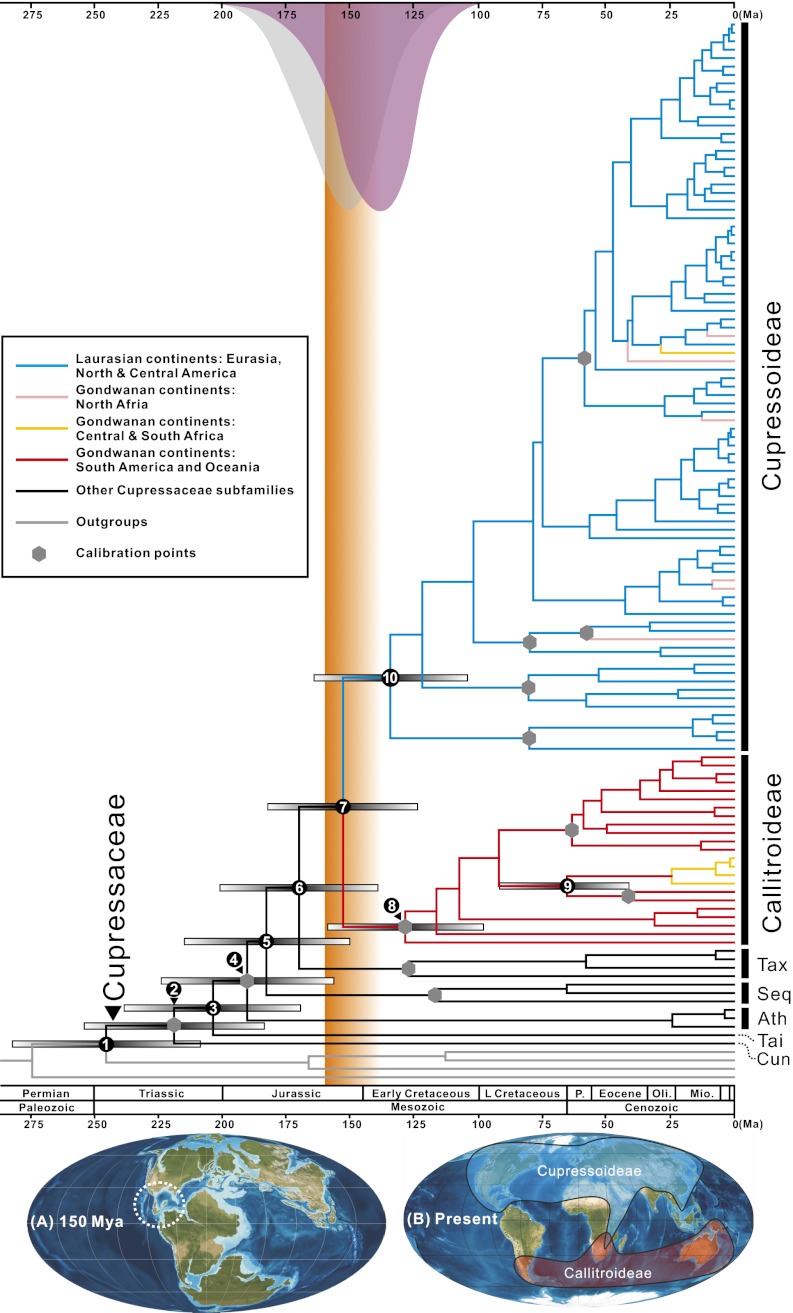
Fig. 1.
(Upper) Chronogram for 122 Cupressaceae species and 22 outgroups based on an alignment of >7,000 nucleotides of plastid DNA (144-taxon dataset). A geological time scale is shown at the bottom (48). Blue lines represent Cupressoideae restricted to the area of Laurasian continents. Red lines represent Callitroideae restricted to Gondwanan continents. Pink lines represent species occurring in Africa in and north of the Sahara. Yellow lines represent species occurring in Africa south of the Sahara. Gray hexagons represent calibration points. Gray bars represent 95% HPD intervals for nodes 1–10. Gray (run 1) and purple (run 7) normal distributions represent the posterior for the BEAST age estimate of node 7 when uniform or lognormal priors were applied to calibration points. Orange shading indicates the period of decreasing feasibility of floristic exchange between Laurasia and Gondwana. Divergence times of nodes 5, 6, 7, 8, and 10 overlap with the fragmentation of Pangea. (Lower) Maps show (A) a paleocontinent reconstruction at 150 Ma and (B) the current distribution of Callitroideae and Cupressoideae. The stippled circle in A emphasizes island chains between North and South America; Ath, Athrotaxidoideae; Cun, Cunninghamioideae; Seq, Sequoioideae; Tai, Taiwanioideae; Tax, Taxodioideae. Reprinted with permission from Ron Blakey, Colorado Plateau Geosystems.
Cupressaceae split from their sister lineage during the late Permian and early Triassic (209–282 Ma; node 1 in Fig. 1, Table 1, and Table S3) and began to diversify into seven major lineages (commonly ranked as subfamilies) during the Triassic (184–254 Ma; node 2 in Fig. 1, Table 1, and Table S3). The genera belonging to each subfamily are shown in Fig. S1. The stem lineages of Cunninghamioideae, Taiwanioideae, Athrotaxidoideae, Sequoioideae, and Taxodioideae appeared around 184–254 Ma, 170–238 Ma, 157–224 Ma, 150–215 Ma, and 140–201 Ma, respectively (nodes 2–6 in Fig. 1, Table 1, and Table S3). The youngest subfamilies are the Cupressoideae and Callitroideae, which diverged from each other 124–183 Ma (node 7 in Fig. 1, Table 1, and Table S3). Most cupressaceous genera with two or more species diversified after the Cretaceous/Tertiary boundary (65.5 ± 0.3 Ma) (48) (Fig. S2B); the only exception is Chamaecyparis, which is dated to 61–108 Ma (node 23 in Fig. S2B; note that this is the crown age for the Chamaecyparis-Fokienia clade).
Table 1.
Divergence times for the Cupressaceae obtained under a Bayesian relaxed clock as implemented in the program BEAST
| Ages (Ma)* |
|||
| Uniform priors (P, subset VND)† | Lognormal priors (P, subset VND)‡ | ||
| Node | Node description | Uniform priors (subset NVND) |
|
| 1 | Stem lineage of Cupressaceae | 245 (209–282) | 242 (194–293) |
| 2 | Crown lineage of Cupressaceae | 219 (184–254) | 211 (168–259) |
| (Stem of Cunninghamioideae) | |||
| 3 | Stem of Taiwanioideae | 204 (170–238) | 195 (157–240) |
| 4 | Stem of Athrotaxoideae | 190 (157–224) | 182 (145–222) |
| 5 | Stem of Sequoioideae | 183 (150–215) | 174 (139–213) |
| 6 | Stem of Taxodioideae | 170 (140–201) | 159 (128–194) |
| 7 | Divergence between Cupressoideae and Callitroideae | 153 (124–183) | 143 (114–175) |
| 8 | Crown lineage of Callitroideae | 128 (98–159) | 121 (92–152) |
| 9 | Stem lineage of Widdringtonia (endemic in southern Africa) | 65 (42–92) | 62 (40–86) |
| 10 | Crown lineage of Cupressoideae | 134 (104–164) | 123 (93–154) |
The nodes are numbered as in Fig. 1 (144-taxon dataset: 144 taxa and 7,171 nucleotides).
*Million year ranges in parentheses denote the 95% HPD.
BEAST analyses with different uniform distribution priors on the calibration closest to the root (calibration point P; Figs. S2 and S3) yielded largely overlapping 95% highest posterior density (HPD) age ranges for all nodes of interest, indicating that the chosen maximum constraint (the only such constraint used in the analysis) had no overly strong effect on the remaining dates (Fig. S3; compare run 1 with runs 2–4). BEAST analyses with different subsets of calibration points, all with uniform priors, showed that calibration P plus calibrations A, B, E, F, G, J, K, and L (subset VND; SI Text and Table S4) (Fig. S3, run 5) yielded node ages similar to those obtained with calibrations A through P (Table S4) (Fig. S3, run 1), whereas calibration P plus calibrations C, D, H, I, M, N, O, and P (subset NVND; SI Text and Table S4) gave much younger ages (Fig. S3, run 6).
A BEAST analysis that used lognormal prior distributions on the ages of calibration P and subset VND (and uniform priors for subset NVND) (run 7) generated age estimates younger than but largely overlapping those obtained with uniform priors for calibration A through P (run 1) (Fig. S3, Table 1, and Tables S3 and S4; see SI Text for a detailed comparison). In all nine BEAST analyses, effective sample sizes for each parameter were well above 200.
Ancestral Areas and Diversification Rate Changes.
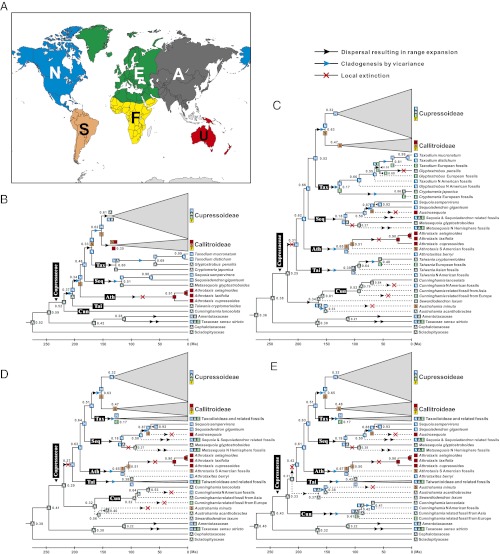
Likelihood AARs were implemented under the dispersal-extinction-cladogenesis model in LAGRANGE (49). We defined eight continent pairs (NS, NE, SF, NA, AE, FE, SU, NF; area codes are explained in Materials and Methods and are illustrated in Fig. 2A) and one continent group (NAE), which reflect continental connections known from plate tectonics (1, 3). The most likely scenarios (Fig. 2B) required 31 dispersal events, 21 vicariance events, and five local extinctions. Likelihood AAR for living Cupressaceae (Fig. 2B) suggested that the family originated in Asia and its early members expanded to North America from where Callitroideae and Athrotaxis entered Gondwanan South America. The integration of fossil Cupressaceae (Fig. 2 C–E, Fig. S4, and Table S5) in likelihood AAR resulted in a similar scenario but with two more incursions from Laurasian to Gondwanan continents (Austrosequoia and Austrohamia minuta). Reconstructions using alternative placements of certain fossil taxa (two Austrohamia species and Sewardiodendron laxum) affected only the ancestral areas inferred for the nodes closest to them (compare Fig. 2 B and C–E).
Fig. 2.
AARs for Cupressaceae. (A) The six areas (“N,” “S,” “E,” “F,” “A,” and “U”) used in the analyses (Left) and the modeled biogeographic processes (Right). (B) Likelihood reconstruction without fossil lineages. (C–E) Likelihood reconstructions that include fossil taxa in the tree and assume alternative placements of three early Cupressaceae fossils (see SI Text for details). The AARs with the highest likelihood are shown as colored boxes at each node. Single-area boxes indicate an ancestor confined to a single geographic area; combined boxes indicate an ancestor with a distribution encompassing two or more areas; two boxes separated by a space indicate the ancestral ranges inherited by each of the daughter lineages arising from the node. For each node with alternative reconstructions (within log2 likelihood units of the maximum), the relative probability of the global likelihood for the optimal reconstruction is given. Fossil lineages are shown by a dashed line, indicating their extinct status. Area codes are explained in SI Text.
Because the 122 species that we sequenced are not a random sample of the family’s 162 species but instead overrepresent deep nodes, we fitted birth/death diversification models to the maximum likelihood topology after statistical completion, assuming nonrandom species sampling (40). When the nonsequenced 40 species were added to the tree under a constant-rate birth-death model, assuming they originated during the past 10 million y, the best fit to the 1,000 simulated completed trees was a two-rate model with a decrease in diversification rate at 1.37 Ma. Of the alternative models [constant-rate pure-birth (CR-PB), constant-rate birth-death, logistic density dependence, and exponential density dependence], the CR-PB model provided the second-best fit to the completed trees.
Discussion
The dense taxon sampling and large amount of sequence data used here yielded a solidly supported phylogeny for the Cupressaceae (Fig. S1), which are monophyletic and sister to the Taxaceae sensu lato (i.e., Taxaceae sensu stricto plus Cephalotaxaceae and Amentotaxaceae) (31), as found previously (50, 51). The divergence of Cupressaceae from their sister lineage occurred >200 Ma (node 1 in Fig. 1 and Table 1; 209–282 Ma), while Pangea was still intact, matching fossil evidence of Cupressaceae in the Jurassic of Europe (Hughmillerites juddii) (35), Asia (Sewardiodendron laxum and Austrohamia acanthobractea) (32, 36), and South America (Austrohamia minuta) (34). Cupressaceae diversified into seven major lineages (subfamilies) during the Triassic and Jurassic (nodes 2–7 in Fig. 1, Fig. S2, Table 1, and Table S3), predating or coinciding with the separation of Gondwana and Laurasia (orange column in Fig. 1). Furthermore, AAR (with or without fossil taxa) yielded Asia as the ancestral area for the family (Fig. 2). Cunninghamioideae may have originated in Asia (Fig. 2), and the divergence of Taiwanioideae from their sister lineage (Fig. 2B) may match the separation of Asia from North America at ∼200 Ma (3); the three subfamilies, Athrotaxidoideae, Sequoioideae, and Taxodioideae (nodes 4–6 in Fig. 1) probably diverged from their sister lineage in North America (Fig. 2 C–E); the divergence of Callitroideae from Cupressoideae was dated to 124–183 Ma (node 7 in Fig. 1 and Table 1; mean: 153 Ma), an age range almost coinciding with the separation of Gondwana from Laurasia (Fig. 1) during the Late Jurassic (160–138 Ma) (1, 3). Living members of Cupressoideae occur mainly in former Laurasian continents (Fig. 1 Lower, B), whereas Callitroideae are endemic to fragments of Gondwana (30) (Fig. 1 Lower, B). African Cupressoideae apparently derived from a series of southward expansions during the middle and late Tertiary (Fig. 1). Unambiguous fossils (with reproductive organs) of Cupressoideae are known only from former Laurasia and those of Callitroideae from Gondwana (33). Overland connections between Laurasian and Gondwanan continents were severed from the Late Jurassic until the middle Tertiary, when India connected with Eurasia, followed by the subsequent reconnection of Africa to Eurasia and South America with North America (1, 3). It is clear from our results that the divergence between Cupressoideae and Callitroideae correlates with the break-up of Pangea (Fig. 1) and most likely was caused by it, as shown in the likelihood AARs (Fig. 2B).
In the remaining five subfamilies, we further inferred three intercontinental disjunctions between the Northern and Southern Hemispheres (Fig. 2 and Fig. S4). The most recent involves the extinct Austrosequoia and its extant sister lineage, comprising Sequoia and Sequoiadendron (Fig. 2 C–E and Fig. S4). Austrosequoia dispersed from North America (via South America) to Australia around 94–100 Ma, as judged from the mid-Cretaceous (Cenomanian) fossil remains in Australia (33, 52). The second disjunction involves Athrotaxis and its putative sister species Athrotaxites berryi (53) (Fig. S5I). Athrotaxis currently is endemic to Australia (30) but is known from fossils in Argentina (Athrotaxis ungeri) (54), and Athrotaxites berryi is known from the Aptian (ca. 111–126 Ma) (48) of North America (53). The Gondwanan taxon Athrotaxis probably originated from a southward expansion from North America, as suggested by likelihood AARs (Fig. 2 C–E). The third inferred intercontinental disjunction involves the extinct Austrohamia, with one species (Austrohamia minuta) from the Jurassic of southern Argentina (34) and the other (Austrohamia acanthobractea) from the late Jurassic of northern China (36). The South American A. minuta might have arrived there following dispersal from the Laurasian North America, as suggested by likelihood AAR (Fig. 2 C–E). These three instances of intercontinental disjunctions all involve north-to-south expansion. We found no instance of range expansion from the Southern to the Northern Hemisphere but detected a clear signal of dispersal or overland expansion among the Southern Gondwanan continents themselves (Fig. S4).
Previous studies of gymnosperm radiations mostly have inferred Oligocene-age crown groups (14–26), and a recent meta-analysis found a median crown age for gymnosperm genera of 32 Ma, younger than that found for angiosperm genera (25). Our dating of those genera with more than one species in the Cupressaceae similarly suggests relatively recent diversifications (Fig. S2B). The young ages of most living gymnosperm clades probably reflect rediversification following extinction. In Cupressaceae, the evidence for widespread extinction and range shrinkage is particularly strong (as visualized in Fig. S6). For example, Cunninghamioideae (Fig. S6A) and Taiwanioideae (55) were widely distributed in the Northern Hemisphere during the Cretaceous but now are restricted to Asia. Sequoioideae and Taxodioideae were widespread in the Northern Hemisphere in the Cretaceous and Early Tertiary, with Sequoioideae also found in Australia, but now are reduced to two species each in southern North America and one and two species, respectively, in East Asia (Fig. 2 C–E and Fig. S6 B and C). Athrotaxidoideae were present in both North and South America during the Cretaceous (53, 54) but today consist of three species in Australia (Fig. 2 C–E). Finally, the genera Austrocedrus (56), Calocedrus (57), Chamaecyparis (58), Fitzroya (59), Papuacedrus (60), and Tetraclinis (61) all had wider distributions in the past. The ice ages of the past 2 million years further contributed to population extinction and reductions in species range, as inferred from our diversification modeling, which indicated a downturn in Cupressaceae diversification rates at 1.37 Ma. Relatively few Cupressaceae lineages have adapted to the strongly seasonal and semiarid habitats that became more widespread with the global cooling during the Oligocene/Miocene (30). Those that did, such as Juniperus, experienced a diversification burst during the Miocene (19).
Besides throwing light onto the diversification of a Triassic/Jurassic Pangean spermatophyte lineage, our findings confirm and illustrate the power of incorporating fossils directly into AARs, rather than using them only for molecular-clock calibration (62, 63). Specifically, it was the incorporation in the AARs of up to 29 fossil taxa (groups) (Fig. S4 and Table S5), most from areas where the respective lineage no longer occurs, that provided insights about range changes, but with the direction of range expansion (predominantly north-to-south) being inferred less from the fossil record than the molecular topology.
Materials and Methods
Plant Material, DNA Isolation and Sequencing, and Sequence Alignment.
Table S1 lists all plant materials used in this study, with species name and author, geographic provenance, herbarium voucher and deposition, and GenBank accession numbers. A total of 290 sequences were newly produced. Phylogenetic and dating analyses were conducted on two datasets. The 56-taxon dataset comprised 35 ingroup species, 21 outgroups, and 10,472 aligned nucleotides from 10 DNA regions (see below). The 144-taxon dataset comprised 122 ingroup species, 22 outgroups, and 7,171 aligned nucleotides from six plastid DNA regions. The ingroup species represented all 32 Cupressaceae genera; outgroups represented the other conifer families, Cycas, Ginkgo, and a basal angiosperm for rooting purposes. We sequenced the mitochondrial regions atpA and cox1, the nuclear regions 18S and 26S, and the plastid regions matK, rbcL, psbB, petB-D, rps4, and trnL-F (for primer sequences, see refs. 19 and 64). For DNA extraction, PCR, and sequencing procedures, see Mao et al. (19). The sequences produced were aligned using ClustalX version 1.83 (65), followed by manual adjustments in Mega4 (66).
Phylogenetic Analyses.
Phylogenetic relationships were reconstructed using parsimony, Bayesian, and maximum likelihood inference. Parsimony analyses relied on PAUP version 4.10b (67) and the University of Oslo Bioportal (http://www.bioportal.uio.no) (68) using heuristic searching, starting trees obtained via stepwise addition, tree-bisection-reconnection branch swapping, steepest descent, and the MulTrees and Collapse options in effect, with no upper limit for the number of trees held in memory; support values for all nodes (on a 50% majority rule bootstrap tree) were calculated with the same settings as above for 1,000 replicates; 10 searches with random taxon additions were conducted for each replicate, and the strict consensus tree of all shortest trees were saved. Bayesian analysis relied on MrBayes version 3.1.2 (69) and the GTR+I+G model as suggested by MrModeltest version 2.3 (70). We used the default of one cold and three heated Markov chain Monte Carlo chains, starting from random initial trees, and chains were run for 6,000,000 generations, sampling every 200th. The default options in MrBayes were used for chain heating and mixing. We discarded a burn-in of the first 2,000,000 generations and used 20,000 trees from the posterior distribution to obtain a majority rule consensus tree. Maximum likelihood analyses relied on Garli version 1.0 (71) with the GTR+I+G substitution model, starting from random trees and using 5,000,000 generations per search; 30 independent searches were performed, and the best tree was saved.
Separate phylogenetic analyses of the nuclear, plastid, and mitochondrial datasets did not yield statistically supported (>75% likelihood bootstrap support) topological contradictions (data available upon request). Therefore we combined the three data partitions in the 56-taxon dataset.
Molecular-Clock Models and Calibration.
A likelihood ratio test in PAUP 4.10b (67, 72) suggested that the 56-taxon and 144-taxon datasets reject a strict molecular clock (P < 0.01), and we therefore used relaxed molecular-clock approaches: Bayesian coestimation of branch lengths and topology with uncorrelated lognormally distributed rates in BEAST 1.5.3 (43), Bayesian estimation with an input phylogeny in MULTIDIVTIME (46), and penalized likelihood rate smoothing in R8S (44, 45). In each case, genetic distances were transformed into absolute time (in million years) by using 16 fossil calibration points, of which 12 were within Cupressaceae (Fig. S2 and Table S4). Fossils were assigned to the stem of their most closely related lineages; Table S4 lists the morphological features used for each fossil taxonomic assignment. For BEAST analyses, we used uniform prior distributions for minimum constraint (calibration points A–O), with the younger bound set by the youngest date of the respective fossil and the older bound set to 366.8 Ma (maximum constraint for calibration point P place near the root) (Table S4). Calibration point P was restricted to fall between 306.2 and 366.8 Ma (Table S4). We tested the effects of other time intervals at calibration point P, calibrations with different subsets of fossils, and different distribution prior for calibrations by carrying out eight additional BEAST runs (SI Text and Fig. S3). BEAST analyses were run on the Cyberinfrastructure for Phylogenetic Research (CIPRES) Science Gateway (http://www.phylo.org/portal2) (73). For MULTIDIVTIME and penalized likelihood, constraints were as in BEAST, except that these programs do not allow specific prior distributions on fossil constraints.
Ancestral Area Reconstructions (AAR).
AAR relied on the likelihood dispersal-extinction-cladogenesis approach implemented in LAGRANGE (49). The matrix of migration probabilities among continents in LAGRANGE (SI Text and Table S6) allowed dispersal between six operational geographic areas: E, Europe, north Africa, and northern Arabia; A, Asia; N, North America, Caribbean, and Central America; S, South America; F, south to middle Africa and southern Arabia; and U, Australia, New Guinea, New Caledonia, and New Zealand (see Fig. 2A). Boundaries between A, E, and F were defined to minimize the number of species that fell in two areas. The northern boundary between A and E was defined by the Ural Mountains, which is the conventional boundary between European Russia and Asian Russia. The boundary between E and F is the Tropic of Cancer, which runs along the middle of a broad belt of very low precipitation (<100 mm y−1) stretching across all of North Africa and most of Arabia (74); this belt of low precipitation is a significant biogeographic barrier for Cupressaceae (30).
For details on the selection of fossils, the inference of calibration fossils' phylogenetic position, cross validation of calibration fossils (Fig. S7), Ancestral Area Reconstructions, and diversification modeling, see SI Text.
Supplementary Material
Acknowledgments
We thank Damon Little, Elena Conti, Richard Abbott, the editor, and three anonymous reviewers for their constructive suggestions. The University of Oslo Bioportal, CIPRES Science Gateway, and the Willi Hennig Society provided computation and software resources. This research was supported by Ministry of Science and Technology of China Grant 2012CB114504 and 2010DFB63500, National Natural Science Foundation of China Grants 30725004 and 31100488, and International Joint Program (“111” project) of China. K.M. was supported by the China Scholarship Council for 1 y study at the University of Edinburgh. The Royal Botanic Garden Edinburgh is supported by the Scottish Government’s Rural and Environment Science and Analytical Services Division.
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission. C.C.D. is a guest editor invited by the Editorial Board.
Data deposition: The sequences reported in this paper have been deposited in the GenBank database (JF725702–JF725991). GenBank accession numbers and provenance of sequenced samples are provided in Table S1.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1114319109/-/DCSupplemental.
References
- 1.Scotese CR. Atlas of Earth History. Arlington, TX: Department of Geology, University of Texas; 2001. [Google Scholar]
- 2.Rogers JJW, Santosh M. Supercontinents in earth history. Gondwana Res. 2003;6(3):357–368. [Google Scholar]
- 3.Smith AG, Smith DG, Funnell BM. Atlas of Cenozoic and Mesozoic Coastlines. Cambridge, UK: Cambridge Univ Press; 2004. [Google Scholar]
- 4.Whiteside JH, Grogan DS, Olsen PE, Kent DV. Climatically driven biogeographic provinces of Late Triassic tropical Pangea. Proc Natl Acad Sci USA. 2011;108:8972–8977. doi: 10.1073/pnas.1102473108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Springer MS, Murphy WJ, Eizirik E, O’Brien SJ. Placental mammal diversification and the Cretaceous-Tertiary boundary. Proc Natl Acad Sci USA. 2003;100:1056–1061. doi: 10.1073/pnas.0334222100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Roelants K, Bossuyt F. Archaeobatrachian paraphyly and Pangaean diversification of crown-group frogs. Syst Biol. 2005;54:111–126. doi: 10.1080/10635150590905894. [DOI] [PubMed] [Google Scholar]
- 7.San Mauro D, Vences M, Alcobendas M, Zardoya R, Meyer A. Initial diversification of living amphibians predated the breakup of Pangaea. Am Nat. 2005;165:590–599. doi: 10.1086/429523. [DOI] [PubMed] [Google Scholar]
- 8.Wildman DE, et al. Genomics, biogeography, and the diversification of placental mammals. Proc Natl Acad Sci USA. 2007;104:14395–14400. doi: 10.1073/pnas.0704342104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Gamble T, Bauer AM, Greenbaum E, Jackman TR. Evidence for Gondwanan vicariance in an ancient clade of gecko lizards. J Biogeogr. 2008;35(1):88–104. [Google Scholar]
- 10.Roelants K, Haas A, Bossuyt F. Anuran radiations and the evolution of tadpole morphospace. Proc Natl Acad Sci USA. 2011;108:8731–8736. doi: 10.1073/pnas.1100633108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Crane PR. Vegetational consequences of the angiosperm diversification. In: Friis EM, Chaloner WC, Crane PR, editors. The Origin of Angiosperms and Their Biological Consequences. Cambridge, UK: Cambridge Univ Press; 1987. pp. 107–144. [Google Scholar]
- 12.Lupia R, Lidgard S, Crane PR. Comparing palynological abundance and diversity: Implications for biotic replacement during the Cretaceous angiosperm radiation. Paleobiology. 1999;25(3):305–340. [Google Scholar]
- 13.Wagstaff SJ. Evolution and biogeography of the austral genus Phyllocladus (Podocarpaceae) J Biogeogr. 2004;31:1569–1577. [Google Scholar]
- 14.Won H, Renner SS. Dating dispersal and radiation in the gymnosperm Gnetum (Gnetales)—clock calibration when outgroup relationships are uncertain. Syst Biol. 2006;55:610–622. doi: 10.1080/10635150600812619. [DOI] [PubMed] [Google Scholar]
- 15.Qiao CY, Ran JH, Li Y, Wang XQ. Phylogeny and biogeography of Cedrus (Pinaceae) inferred from sequences of seven paternal chloroplast and maternal mitochondrial DNA regions. Ann Bot (Lond) 2007;100:573–580. doi: 10.1093/aob/mcm134. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Knapp M, Mudaliar R, Havell D, Wagstaff SJ, Lockhart PJ. The drowning of New Zealand and the problem of Agathis. Syst Biol. 2007;56:862–870. doi: 10.1080/10635150701636412. [DOI] [PubMed] [Google Scholar]
- 17.Biffin E, Hill RS, Lowe AJ. Did Kauri (Agathis: Araucariaceae) really survive the Oligocene drowning of New Zealand? Syst Biol. 2010;59:594–602. doi: 10.1093/sysbio/syq030. [DOI] [PubMed] [Google Scholar]
- 18.Ickert-Bond SM, Rydin C, Renner SS. A fossil-calibrated relaxed clock for Ephedra indicates an Oligocene age for the divergence of Asian and New World clades and Miocene dispersal into South America. J Syst Evol. 2009;47(5):444–456. [Google Scholar]
- 19.Mao K, Hao G, Liu J, Adams RP, Milne RI. Diversification and biogeography of Juniperus (Cupressaceae): Variable diversification rates and multiple intercontinental dispersals. New Phytol. 2010;188:254–272. doi: 10.1111/j.1469-8137.2010.03351.x. [DOI] [PubMed] [Google Scholar]
- 20.Wei XX, Yang ZY, Li Y, Wang XQ. Molecular phylogeny and biogeography of Pseudotsuga (Pinaceae): Insights into the floristic relationship between Taiwan and its adjacent areas. Mol Phylogenet Evol. 2010;55:776–785. doi: 10.1016/j.ympev.2010.03.007. [DOI] [PubMed] [Google Scholar]
- 21.Biffin E, Brodribb TJ, Hill RS, Thomas P, Lowe AJ. Leaf evolution in Southern Hemisphere conifers tracks the angiosperm ecological radiation. Proc Biol Sci. 2012;279:341–348. doi: 10.1098/rspb.2011.0559. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Willyard A, Syring J, Gernandt DS, Liston A, Cronn R. Fossil calibration of molecular divergence infers a moderate mutation rate and recent radiations for pinus. Mol Biol Evol. 2007;24:90–101. doi: 10.1093/molbev/msl131. [DOI] [PubMed] [Google Scholar]
- 23.Gernandt DS, et al. Use of simultaneous analyses to guide fossil-based calibrations of pinaceae phylogeny. Int J Plant Sci. 2008;169:1086–1099. [Google Scholar]
- 24.Treutlein J, Wink M. Molecular phylogeny of cycads inferred from rbcL sequences. Naturwissenschaften. 2002;89(5):221–225. doi: 10.1007/s00114-002-0308-0. [DOI] [PubMed] [Google Scholar]
- 25.Crisp MD, Cook LG. Cenozoic extinctions account for the low diversity of extant gymnosperms compared with angiosperms. New Phytol. 2011;192:997–1009. doi: 10.1111/j.1469-8137.2011.03862.x. [DOI] [PubMed] [Google Scholar]
- 26.Nagalingum NS, et al. Recent synchronous radiation of a living fossil. Science. 2011;334:796–799. doi: 10.1126/science.1209926. [DOI] [PubMed] [Google Scholar]
- 27.Eckenwalder JE. Re-evaluation of Cupressaceae and Taxodiaceae: A proposed merger. Madrono. 1976;23(5):237–256. [Google Scholar]
- 28.Brunsfeld SJ, et al. Phylogenetic relationships among the genera of Taxodiaceae and Cupressaceae: Evidence from rbcL sequences. Syst Bot. 1994;19(2):253–262. [Google Scholar]
- 29.Gadek PA, Alpers DL, Heslewood MM, Quinn CJ. Relationships within Cupressaceae sensu lato: A combined morphological and molecular approach. Am J Bot. 2000;87:1044–1057. [PubMed] [Google Scholar]
- 30.Farjon A. A Monograph of Cupressaceae and Sciadopitys. London: Royal Botanic Gardens Kew; 2005. pp. 228–400. [Google Scholar]
- 31.Christenhusz MJM, et al. A new classification and linear sequence of extant gymnosperms. Phytotaxa. 2011;19:55–70. [Google Scholar]
- 32.Yao XL, Zhou ZY, Zhang BL. Reconstruction of the Jurassic conifer Sewardiodendron laxum (Taxodiaceae) Am J Bot. 1998;85:1289–1300. [PubMed] [Google Scholar]
- 33.Stockey RA, Kvaček J, Hill RS, Rothwell GW, Kvaček Z. In: Fossil Record of Cupressaceae s. lat. A Monograph of Cupressaceae and Sciadopitys. Farjon A, editor. London: Royal Botanic Gardens Kew; 2005. pp. 54–68. [Google Scholar]
- 34.Escapa I, Cúneo R, Axsmith B. A new genus of the Cupressaceae (sensu lato) from the Jurassic of Patagonia: Implications for conifer megasporangiate cone homologies. Rev Palaeobot Palynol. 2008;151:110–122. [Google Scholar]
- 35.Rothwell GW, Stockey RA, Mapes G, Hilton J. Structure and relationships of the Jurassic conifer seed cone Hughmillerites juddii gen. et comb. nov.: Implications for the origin and evolution of Cupressaceae. Rev Palaeobot Palynol. 2011;164:45–59. [Google Scholar]
- 36.Zhang JW, D'Rozario A, Wang LJ, Li Y, Yao JX. A new species of the extinct genus Austrohamia (Cupressaceae s.l) in the Daohugou Jurassic flora of China and its phytogeographical implications. J Syst Evol. 2012;50(1):72–82. [Google Scholar]
- 37.Doyle JA, Donoghue MJ. Phylogenies and angiosperm diversification. Paleobiology. 1993;19(2):141–167. [Google Scholar]
- 38.Ho SY, Phillips MJ. Accounting for calibration uncertainty in phylogenetic estimation of evolutionary divergence times. Syst Biol. 2009;58:367–380. doi: 10.1093/sysbio/syp035. [DOI] [PubMed] [Google Scholar]
- 39.Clarke JT, Warnock RC, Donoghue PC. Establishing a time-scale for plant evolution. New Phytol. 2011;192:266–301. doi: 10.1111/j.1469-8137.2011.03794.x. [DOI] [PubMed] [Google Scholar]
- 40.Cusimano N, Stadler T, Renner SS. A new method for handling missing species in diversification analysis applicable to randomly or non-randomly sampled phylogenies. Syst Biol. 2012 doi: 10.1093/sysbio/sys031. [DOI] [PubMed] [Google Scholar]
- 41.Pybus OG, Harvey PH. Testing macro-evolutionary models using incomplete molecular phylogenies. Proc Biol Sci. 2000;267:2267–2272. doi: 10.1098/rspb.2000.1278. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Cusimano N, Renner SS. Slowdowns in diversification rates from real phylogenies may not be real. Syst Biol. 2010;59:458–464. doi: 10.1093/sysbio/syq032. [DOI] [PubMed] [Google Scholar]
- 43.Drummond AJ, Rambaut A. BEAST: Bayesian evolutionary analysis by sampling trees. BMC Evol Biol. 2007;7:214. doi: 10.1186/1471-2148-7-214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Sanderson MJ. Estimating absolute rates of molecular evolution and divergence times: A penalized likelihood approach. Mol Biol Evol. 2002;19(1):101–109. doi: 10.1093/oxfordjournals.molbev.a003974. [DOI] [PubMed] [Google Scholar]
- 45.Sanderson MJ. 2003. R8S: Analysis of rates (“r8s”) of evolution (and other stuff), version1.60. Available at: http://ginger.ucdavis.edu/r8s.
- 46.Thorne JL, Kishino H. Divergence time and evolutionary rate estimation with multilocus data. Syst Biol. 2002;51:689–702. doi: 10.1080/10635150290102456. [DOI] [PubMed] [Google Scholar]
- 47.Linder HP, Hardy CR, Rutschmann F. Taxon sampling effects in molecular clock dating: An example from the African Restionaceae. Mol Phylogenet Evol. 2005;35:569–582. doi: 10.1016/j.ympev.2004.12.006. [DOI] [PubMed] [Google Scholar]
- 48.Ogg JG. 2010. International Stratigraphic Chart (International Commission on Stratigraphy, West Lafayette, IN). Available at: https://www.seegrid.csiro.au/wiki/pub/CGIModel/GeologicTime/StratChart2010.jpg.
- 49.Ree RH, Smith SA. Maximum likelihood inference of geographic range evolution by dispersal, local extinction, and cladogenesis. Syst Biol. 2008;57:4–14. doi: 10.1080/10635150701883881. [DOI] [PubMed] [Google Scholar]
- 50.Cheng YC, Nicolson RG, Tripp K, Chaw SM. Phylogeny of Taxaceae and Cephalotaxaceae genera inferred from chloroplast matK gene and nuclear rDNA ITS region. Mol Phylogenet Evol. 2000;14:353–365. doi: 10.1006/mpev.1999.0710. [DOI] [PubMed] [Google Scholar]
- 51.Rai HS, Reeves PA, Peakall R, Olmstead RG, Graham SW. Inference of higher-order conifer relationships from a multi-locus plastid data set. Botany. 2008;86:658–669. [Google Scholar]
- 52.Peters MD, Christophel DC. Austrosequoia wintonensis, a new taxodiaceous cone from Queensland, Australia. Can J Bot. 1978;56:3119–3128. [Google Scholar]
- 53.Miller CN, Jr, LaPasha CA. Structure and affinities of Athrotaxites berryi Bell, an Early Cretaceous conifer. Am J Bot. 1983;70:772–779. [Google Scholar]
- 54.Del Fueyo GM, Archangelsky S, Llorens M, Cuneo R. Coniferous ovulate cones from the Lower Cretaceous of Santa Cruz Province, Argentina. Int J Plant Sci. 2008;169:799–813. [Google Scholar]
- 55.Lepage BA. Earliest occurrence of Taiwania (Cupressaceae) from the Early Cretaceous of Alaska: Evolution, biogeography, and paleoecology. Proc Acad Nat Sci Philadelphia. 2009;158(1):129–158. [Google Scholar]
- 56.Paull R, Hill RS. Oligocene Austrocedrus from Tasmania (Australia): Comparisons with Austrocedrus chilensis. Int J Plant Sci. 2008;169(2):315–330. [Google Scholar]
- 57.Shi G, Zhou Z, Xie Z. A new Oligocene Calocedrus from South China and its implications for transpacific floristic exchanges. Am J Bot. 2012;99:108–120. doi: 10.3732/ajb.1100331. [DOI] [PubMed] [Google Scholar]
- 58.Liu YS, Mohr BAR, Basinger JF. Historical biogeography of the genus Chamaecyparis (Cupressaceae, Coniferales) based on its fossil record. Palaeobio Palaeoenv. 2009;89:203–209. [Google Scholar]
- 59.Paull R, Hill RS. Early Oligocene Callitris and Fitzroya (Cupressaceae) from Tasmania. Am J Bot. 2010;97:809–820. doi: 10.3732/ajb.0900374. [DOI] [PubMed] [Google Scholar]
- 60.Wilf P, et al. Papuacedrus (Cupressaceae) in Eocene Patagonia: A new fossil link to Australasian rainforests. Am J Bot. 2009;96:2031–2047. doi: 10.3732/ajb.0900085. [DOI] [PubMed] [Google Scholar]
- 61.Kvaček Z, Manchester SR, Schorn HE. Cones, seeds, and foliage of Tetraclinis Salicornioides (Cupressaceae) from the Oligocene and Miocene of Western North America: A geographic extension of the European Tertiary species. Int J Plant Sci. 2000;161:331–344. doi: 10.1086/314245. [DOI] [PubMed] [Google Scholar]
- 62.Clayton JW, Soltis PS, Soltis DE. Recent long-distance dispersal overshadows ancient biogeographical patterns in a pantropical angiosperm family (Simaroubaceae, Sapindales) Syst Biol. 2009;58:395–410. doi: 10.1093/sysbio/syp041. [DOI] [PubMed] [Google Scholar]
- 63.Crisp MD, Trewick SA, Cook LG. Hypothesis testing in biogeography. Trends Ecol Evol. 2011;26:66–72. doi: 10.1016/j.tree.2010.11.005. [DOI] [PubMed] [Google Scholar]
- 64.Liu N, et al. Phylogenetic relationships and divergence times of the family Araucariaceae based on the DNA sequences of eight genes. Chin Sci Bull. 2009;54:2648–2655. [Google Scholar]
- 65.Thompson JD, Gibson TJ, Plewniak F, Jeanmougin F, Higgins DG. The CLUSTAL_X windows interface: Flexible strategies for multiple sequence alignment aided by quality analysis tools. Nucleic Acids Res. 1997;25:4876–4882. doi: 10.1093/nar/25.24.4876. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Tamura K, Dudley J, Nei M, Kumar S. MEGA4: Molecular Evolutionary Genetics Analysis (MEGA) software version 4.0. Mol Biol Evol. 2007;24:1596–1599. doi: 10.1093/molbev/msm092. [DOI] [PubMed] [Google Scholar]
- 67.Swofford DL. PAUP*: Phylogenetic Analyses Using Parsimony (*and Other Methods), Version 4. Sunderland, MA: Sinauer Associates; 2002. [Google Scholar]
- 68.Kumar S, et al. AIR: A batch-oriented web program package for construction of supermatrices ready for phylogenomic analyses. BMC Bioinformatics. 2009;10:357. doi: 10.1186/1471-2105-10-357. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Huelsenbeck JP, Ronquist F. MRBAYES: Bayesian inference of phylogenetic trees. Bioinformatics. 2001;17:754–755. doi: 10.1093/bioinformatics/17.8.754. [DOI] [PubMed] [Google Scholar]
- 70.Nylander JA. 2004. MrModeltest v2 (Program distributed by the author, Evolutionary Biology Centre, Uppsala University, Uppsala, Sweden). Available at: http://www.abc.se/znylander/mrmodeltest2/mrmodeltest2.html.
- 71.Zwickl DJ. Austin, TX: University of Texas; 2006. Genetic algorithm approaches for the phylogenetic analysis of large biological sequence datasets under the maximum likelihood criterion. Ph.D. dissertation. [Google Scholar]
- 72.Felsenstein J. Evolutionary trees from DNA sequences: A maximum likelihood approach. J Mol Evol. 1981;17:368–376. doi: 10.1007/BF01734359. [DOI] [PubMed] [Google Scholar]
- 73.Miller MA, Pfeiffer W, Schwartz T. Proceedings of the Gateway Computing Environments Workshop (GCE) New Orleans. New York: Institute of Electrical and Electronics Engineers (IEEE); 2010. Creating the CIPRES Science Gateway for inference of large phylogenetic trees; pp. 1–8. [Google Scholar]
- 74.Geelan P, Lewis H. Times Atlas of the World (Comprehensive Edition) 9th Ed. London: Times Books; 1992. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.