Abstract
X-ray crystallography and small-angle X-ray scattering (SAXS) in solution have been used to show that a mutant aspartate transcarbamoylase exists in an intermediate quaternary structure between the canonical T and R structures. Additionally, the SAXS data indicate a pH-dependent structural alteration consistent with either a pH-induced conformational change or a pH-induced alteration in the T to R equilibrium. These data indicate that this mutant is not a model for the R state, as has been proposed, but rather represents the enzyme trapped along the path of the allosteric transition between the T and R states.
Keywords: allostery, cooperativity, intermediate structure, structural stabilization
Although many aspects of allosteric regulation and cooperativity have been established for Escherichia coli aspartate transcarbamoylase (ATCase), the enzyme that catalyzes the committed step in pyrimidine nucleotide biosynthesis, many of the details of how the active sites change from low-activity, low-affinity to high-activity, high-affinity during the T to R transition have not been delineated (1). The major limitation has been that stabilization of the enzyme in the R state requires the presence of active site ligands, thus making interpretation of the structural rearrangements upon the T to R transition, in the absence of ligands, difficult. Newell and Schachman (2) have concluded, from sedimentation velocity experiments, that the quaternary structures of ATCase with the catalytic chain mutation K164E and the catalytic chain double-mutation K164E and E239K exist in the R state, irrespective of the presence of active site ligands. The enzymatic properties of the double mutant (K164E/E239K ATCase) include a lack of homotropic cooperativity and an inability to be activated by ATP or inhibited by CTP: properties coincident of an enzyme that cannot transition between the two allosteric states (2). More recently, Velyvis et al. (3) demonstrated that the partially labeled K164E/E239K ATCase exhibited certain chemical shifts in solution NMR studies that they conclude are characteristic of the R state of the enzyme. These same chemical shifts were also observed when the wild-type enzyme was bound with the bisubstrate analog N-phosphonacetyl-l-aspartate (PALA) (3), which is known to stabilize fully the R state of the enzyme (4). Thus, a crystal structure of K164E/E239K ATCase should provide us a means to obtain the structural details of an R-state active site in the absence of ligands. However, here we report the X-ray crystal structure and the solution small-angle X-ray scattering (SAXS) data for the K164E/E239K ATCase, which clearly demonstrate that this double-mutant enzyme is not in the R-quaternary structure. Instead this mutant enzyme is in a unique intermediate state on the path between the T and R structural states.
Results and Discussion
Crystal Structure of K164E/E239K ATCase Is Different from the Wild-Type R State Structure.
The K164E/E239K ATCase was purified and subsequently crystallized in the absence of ligands as described in Methods. The crystals were determined to be in the P212121 space group with unit cell dimensions of a = 124.1 Å, b = 144.8 Å, and c = 203.4 Å with α = β = γ = 90°. A previously determined structure of R-state ATCase in the presence of the PALA has also been solved in the P212121 space group (5). However, the unit cell dimensions (a = 125.5 Å, b = 153.5 Å, c = 185.7 Å with α = β = γ = 90°) vary significantly from those determined for the K164E/E239K ATCase. In the crystal of the PALA–ATCase complex the molecular threefold axis is parallel to the crystallographic b-axis (5). The almost 9 Å shorter b-axis of the K164E/E239K ATCase crystals was the first indication that the structure of this double-mutant enzyme may not be an R-state structure.
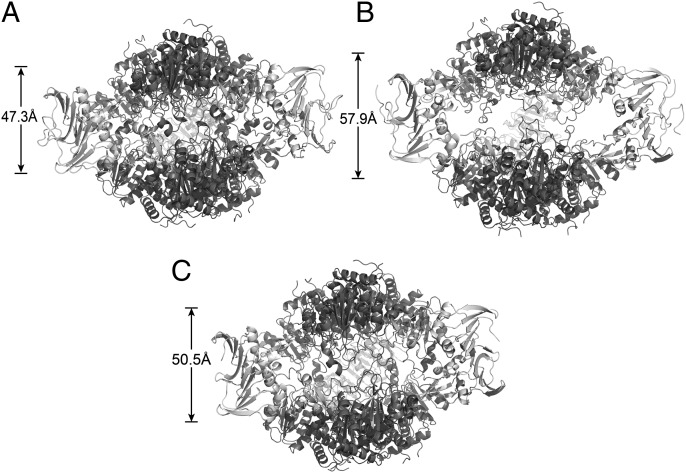
To quantitatively compare the quaternary conformation of the K164E/E239K structure with the canonical ATCase T and R structures, the vertical separation (6) between the upper and lower catalytic subunits, planar angle between allosteric domains of the regulatory dimer, and the rotation about the threefold axis (Fig. 1) of the K164E/E239K structure were determined and compared with the corresponding values for the published T and R structures (Fig. 2). The vertical separation for the K164E/E239K structure was 50.5 Å, compared with 47.3 Å and 57.9 Å for the T state [Protein Data Bank (PDB) ID 1ZA1 used throughout] and R state (PDB ID 1D09 used throughout), respectively. The average planar angle between allosteric domains of the regulatory dimer in the K164E/E239K structure was 153°, which compares to 152.0° and 155.8° for the T and R structures, respectively, of the wild-type enzyme. For the wild-type enzyme the relative rotation around the molecular threefold axis between the T and R structures is 12°, whereas the rotation from the wild-type T structure to the K164E/E239K structure is only 4.8°. The vertical separation, the angle between the allosteric domains of the regulatory dimer, and the rotation of the catalytic subunits about the threefold axis all indicate that the K164E/E239K structure is not globally in either the wild-type T or R structure.
Fig. 1.
ATCase holoenzyme showing the threefold and twofold axes. One-third of the molecule (c1-r1-r6-c6) is represented as a ribbon diagram. The other two-thirds of the molecule is shown without secondary structure. Planar angles between the CP and ASP domains of the catalytic chain (PA1) and the allosteric domains of the r1–r6 regulatory chains (PA2) are also shown. Insets: The c1 and r1–r6 chains with the corresponding PA1 and PA2 are shown in an orientation to better visualize the planar angle between the domains as well as the centers of gravity of these domains. Also indicated is the vertical separation between the centers of gravity of an upper and lower catalytic chain. This figure was produced using MOLSCRIPT (32) and RASTER3D (33).
Fig. 2.
Comparison of the structures of (A) wild-type ATCase in the T state (PDB ID 1ZA1), (B) wild-type ATCase in the R state (PDB ID 1D09), and (C) the K164E/E239K mutant ATCase (PDB ID 4E2F). The vertical separations between the center of masses of the upper and lower catalytic trimers are indicated. The catalytic chains are dark and the regulatory chains light in color. This figure was drawn with PyMol (34).
The analysis of the global conformation of the K164E/E239K structure by means of X-ray crystallography does not agree with the previously reported data as determined by sedimentation velocity (2). Therefore, we used the program HydroPRO (7) to calculate the radius of gyration (Rg) on the basis of the wild-type and K164E/E239K structures. These values have some uncertainty because the highly mobile portions of the structures are not reported in the structural data. The Rg values of the wild-type T and R states were calculated to be 46.4 and 49.1 Å, respectively, which compares well with the reported values of 46.6 ± 0.3 Å and 49.3 ± 0.2 Å, respectively (8). The calculated Rg for the K164E/E239K structure was 47.2 Å, a value closer to the T than the R state of the wild-type enzyme.
K164E/E239K ATCase Exists in a Structural State Different from the Wild-Type R State in Solution.
For the wild-type enzyme, the percent change in the sedimentation coefficient between the T- and R-state structures has been reported to be −3.8% (2). The corresponding percent difference calculated between the K164E/E239K and wild-type R-state structures was −0.4% (2). These data suggest that the structure of K164E/E239K enzyme may be different in solution than it is in the crystal. Therefore, we used SAXS to obtain additional structural data on the K164E/E239K ATCase in solution (9, 10).
SAXS data were recorded using Beamline 4-2 at the Stanford Synchrotron Research Laboratory at pH values 8.3, 7.5, and 7.0. In addition to recording SAXS data for the K164E/E239K enzyme, data were also recorded for the wild-type ATCase in the absence of ligands (T-state control) and in the presence of PALA (R-state control). The SAXS patterns (Fig. 3), plotted as normalized Kratky Plots (11), for the T and R states of the wild-type enzyme are dramatically different. The SAXS pattern for the K164E/E239K enzyme at pH 8.3 (Fig. 3A) is intermediate between the T and R states observed for the wild-type enzyme. This clear difference in the scattering patterns between the T and R states and the K164E/E239K enzyme can also be visualized when the data are plotted as I(q) vs. I (Fig. S1A) and as log I(q) vs. I (Fig. S2A). The intermediate curve observed for the K164E/E239K enzyme can be explained in at least two ways. First, the K164E/E239K enzyme exists in a new structural state that is different from the wild-type T or R states; or second, in solution the K164E/E239K enzyme is a mixture of two or more structures. The second explanation is consistent with the K164E/E239K enzyme having a T to R equilibrium shifted from the T state-dominated equilibrium observed for the wild-type enzyme.
Fig. 3.
SAXS of the wild-type and the K164E/E239K enzymes at (A) pH 8.3, (B) pH 7.5, and (C) pH 7.0. At each pH the wild-type enzyme in the absence of ligands is shown in black and in the presence of PALA in blue. The K164E/E239K enzyme in the absence of ligands is shown in red and in the presence of PALA in orange. The data are plotted as normalized Kratky Plots (11). The green dashed line indicates the best fit of a combination of the wild-type T pattern and the K164E/E239K PALA R pattern to the K164E/E239K pattern in the absence of ligands. Each scattering curve corresponds to 140 data points connected by linear line segments.
If the SAXS pattern of the K164E/E239K enzyme is a mixture of wild-type T and R structures, it should be possible to generate the SAXS pattern of the double-mutant enzyme by a linear combination of the wild-type T- and R-state patterns. The dotted line in Fig. 3 A–C corresponds to the best fit of the SAXS pattern of the K164E/E239K enzyme using the wild-type T and R curves. At each pH, the best-fit curve does not match that observed for the K164E/E239K enzyme. In particular, the position and amplitude of the first subsidiary minimum is not predicted correctly. The inability to deconvolute the curve into T and R components suggests that in solution the K164E/E239K enzyme is neither the T nor R quaternary structure of the wild-type enzyme or a mixture thereof.
As opposed to the wild-type enzyme in the absence or presence of PALA, the SAXS pattern for the K164E/E239K enzyme is pH dependent (compare Fig. 3 A–C). Thus, by altering the pH either the quaternary structure of the K164E/E239K enzyme is shifting, or the population of molecules in the R state is increasing relative to the T state. According to these SAXS data, at no pH between 7.0 and 8.3 does the unliganded K164E/E239K enzyme exist in a structure that would be considered the same as the R structure of the wild-type enzyme. As the pH is varied from 8.3 to 7.0 the SAXS pattern of the unliganded K164E/E239K enzyme shifts toward the wild-type R structure. This shift in structure could be modeled on the basis of the SAXS pattern of the K164E/E239K enzyme at pH 8.3 in the presence and absence of PALA, suggesting that this shift in the SAXS pattern was due to a change in the relative population of the two structures in solution. The SAXS data for the wild-type and the K164E/E239K enzymes are also plotted as scattering intensity and the log of the scattering intensity vs. the momentum transfer (q) (Figs. S1 and S2).
The Rg values can be determined directly from the SAXS data using the Guinier plot (Table S1). The values for the wild-type enzyme, 48.1 ± 0.1 Å and 51.9 ± 0.1 Å for the T and R states, respectively, are similar to the previously determined Rg values in solution of 47.3 ± 0.2 Å and 49.9 ± 0.2 Å for the T and R states (12). Most strikingly, we found that in solution the K164E/E239K enzyme has a slightly greater Rg than the PALA liganded wild-type or mutant enzymes. At the two other pHs tested this trend was the same: the Rg was greater (0.5–0.6 Å) for the unliganded mutant before the addition of PALA and conversion to the R state. This discrepancy between the Rg as determined in solution by SAXS and the crystal structure suggest that the ground state structure of the mutant enzyme is much more flexible than that of the wild-type; however, whatever the explanation may be the crystal structure and SAXS data both support the conclusion that the mutant without ligands is not in an R-state structure.
These results suggest that the previously reported findings that the K164E/E239K ATCase is in the R state may need to be reevaluated (2, 3). The evidence presented here clearly demonstrates that the quaternary structure of the K164E/E239K ATCase is not identical to the wild-type ATCase–PALA complex, the typical structure used to define the R state. The sedimentation velocity experiments on the K164E/E239K ATCase, which indicated R state-like sedimentation values, were performed at 20 °C and pH 7.0 (2), a temperature and pH that may perturb the enzyme toward a more R-like structure. The SAXS data in Fig. 3 show that lowering the pH from 8.3 to 7.0 causes a shift in the structure toward the R state. The solution NMR report by Velyvis et al. (3) also asserts the K164E/E239K ATCase is in the R state and concludes that the Monod, Wyman, and Changeux model (13) can fully account for the allosteric properties of ATCase. The experiments performed by Velyvis et al. (3) were carried out at 37 °C and pH 7.5. At this pH value and 25 °C, the K164E/E239K ATCase is not in the canonical R-state structure (Fig. 3B). Although the higher temperature of the NMR experiments may cause a shift in the structure toward the R state, the temperature dependence of SAXS data has been determined for the wild-type and the D236A enzyme. The wild-type enzyme showed no change in structure by SAXS through the temperature range of 4° to 55 °C, whereas the D236A ATCase exhibited a large shift toward the T state between 4 and 30 °C, with a minor shift back toward the R state between 30 °C and 45 °C (14). These data suggest that under the NMR conditions (37 °C) the K164E/E239K ATCase, which has properties similar to those of D236A ATCase, would not be shifted toward the R state. This study underscores the need for extreme caution when making inferences about the allosteric mechanism of a wild-type protein when working with a model system, which in this case is the double-mutant enzyme.
K164E/E239K ATCase Is in an Intermediate State During the Allosteric Transition from the T to the R State.
Both the X-ray crystallographic and SAXS data indicate that the structure of the K164E/E239K enzyme in the absence of ligands is not in the R structure, but rather it is an intermediate structure between the canonical T and R structures. Instead of providing a model for the R state, the K164E/E239K enzyme gives us structural information on the pathway of the T- to R-state transition of ATCase. When wild-type ATCase undergoes the allosteric transition to the R state, the enzyme expands 10.6 Å and the upper catalytic trimer rotates 12° relative to the lower trimer along the threefold axis. Compared with the wild-type T-state structure the K164E/E239K enzyme only expands 2.8 Å and the upper catalytic trimer rotates only 4.4° relative to the lower trimer (Fig. 4A). Previous studies revealed that the planar angle between the allosteric domains of the regulatory dimer changes almost linearly with changes in the size of the molecule (6), which increases from 152.0° to 155.8° as the enzyme transitions from the T to the R state. The fact that in the K164E/E239K structure this planar angle is only 153° suggests that the size of the double mutant is between the size of the wild-type T- and R-state enzymes. As seen in Fig. 4A, the percent change in the vertical separation, the angle between the allosteric domains of the regulatory dimer, the rotation about the threefold axis, and the radius of gyration observed in the K164E/E239K structure are all approximately one-third of the difference in the parameters observed between the T and R states of the wild-type enzyme. These data suggest that the K164E/E239K enzyme exists in an intermediate structure shifted approximately one-third toward the wild-type R structure from the T structure.
Fig. 4.
(A) Percent change in quaternary structural properties of the K164E/E239K ATCase. As reference, the wild-type enzyme characteristics go from 0 (T state) to 100% (R state). (B) T structure of one catalytic chain of wild-type ATCase (PDB ID 1ZA1). (C) R structure of one catalytic chain of wild-type ATCase (PDB ID 1D09). In B and C the diameter of the tube corresponds to the main chain rmsd between the wild-type and the K164E/E239K structures. The portions of the tube highlighted in black correspond to those with rmsd >1.5 Å.
Not only is the quaternary conformation of the K164E/E239K structure intermediate between the canonical ATCase T and R structures, but also the tertiary structure of the individual chains are intermediate as well. As seen in Fig. 4 B and C, the rmsd of the Cα in the catalytic chain between the wild-type and the K164E/E239K structures is reflected in the diameter of the tube. The catalytic chain of the K164E/E239K structure has less rmsd to that of the T-state structure than the R-state structure. Thus, the tertiary structure of the double mutant represents an intermediate state closer to the T state.
Another prominent feature of the T to R transition is the closure of the two domains of each catalytic chain to rearrange the active site residues so as to push the substrates toward one another in the bimolecular reaction. However, the active site of the K164E/E239K ATCase more closely resembles the T-state than the R-state active site (Fig. 5).
Fig. 5.
Stereo view of the K164E/E239K ATCase 2 Fo–Fc electron density map of the active site region contoured at 1.3 σ. Overlaid onto the electron density are the side chains known to be involved in substrate binding and catalysis. Side chains are shown for the K164E/E239K (cyan carbons), wild-type T-state (magenta carbons), and wild-type R-state (orange carbons) ATCases. The three structures were overlaid on the basis of the α-carbon positions of the active site residues.
A comparison of the active site region of the K164E/E239K ATCase with the T and R states of the wild-type enzyme is shown with electron density in Fig. 5. Key differences include significant alterations in the positions of Arg54 and Arg167, which are two key residues involved in catalysis. Both Arg54 and Arg167 swing out of the active site in the K164E/E239K ATCase structure. These alterations in side chain position may be responsible for the reduced catalytic activity and increased [Asp]0.5 of the K164E/E239K compared with the wild-type enzyme (2).
In addition to the K164E/E239K enzyme reported here, SAXS experiments have shown that the E239Q and D236A enzymes also exhibit patterns intermediate between the T and R patterns of the wild-type enzyme (8, 14). These mutations break the T-state interchain c1–c4 interactions between Glu239 and both Lys164 and Tyr165 or the c1–r4 interaction between Asp236c and Lys143r. For ATCase to transition from the T- to the R-quaternary structure, these interchain interactions must break (Fig. 6 A and C), although for the wild-type enzyme in the R state, c1 intrachain interactions are formed between Glu239 to both Lys164 and Tyr165 (Fig. 6C). In the case of the K164E/E239K enzyme, the possible salt link between Glu164 and Lys239 does not form (average distance between Glu164 Oɛ and Lys239 Nζ is 4.78 ± 0.7 Å). Furthermore, the salt-link between Asp236 (c1/c4) and Lys143 (r4/r1) observed in the T-state structure (Fig. 6A) is broken in the K164E/E239K enzyme, with an average distance between the Asp236 Oδ and the Lys143 Nζ of 8.17 ± 3.5 Å (Fig. 6B).
Fig. 6.
The c1–c4, c1–r4, and c4–r1 interfaces. (A) T-state residue positions and interactions of the wild-type enzyme (PDB ID 1ZA1), (B) the same residue positions in the K164E/E239K ATCase reported here, and (C) the R-state residue positions and interactions of the wild-type enzyme (PDB ID 1D09). Shown are the interactions involving Glu239 [wild-type T state (A) and R state (C) or Lys239 in the K164E/E239K structure (B)]. Also shown are the interactions between Asp236 of the catalytic chains and Lys143 of the regulatory chains. For clarity Tyr165 is not labeled.
Most interestingly, the scattering curve of the K164E/E239K ATCase at pH 8.3 is comparable to the intermediate curve of the structure at the end of the early fast-rising phase of the time-resolved stop-flow X-ray scattering of ATCase upon mixing with substrates carbamoyl phosphate and l-aspartate (15). The elongation of the enzyme is a necessary part of the allosteric transition, which requires the breaking of the T-state intersubunit interactions involving both Glu239 and Asp236; thus the K164E/E239K structure, lacking these interactions, is a unique structure along the path of the quaternary structural transition from the T to the R state. As such, these findings suggest that there may be transiently stable intermediate structure(s) that the protein passes through during the allosteric transition.
Methods
Enzyme Preparation.
The mutations K164E/E239K were introduced to the catalytic chains of ATCase on plasmid pEK152 (16) by site-specific mutagenesis using the QuikChange Site-Directed Mutagenesis Kit (Stratagene) to produce plasmid pEK695. The double mutant was isolated according to the procedure described by Nowlan and Kantrowitz (17), from overproducing E. coli strain EK1104 [F− ara, thi, Δ(pro-lac), ΔpyrB, pyrF±, rpsL], containing the plasmid pEK695. The enzyme was purified by isoelectric precipitation (17), ion exchange, and hydrophobic interaction chromatography. For the ion-exchange purification, a Q-Sepharose Fast Flow (GE Healthcare) column (11 cm × 2.5 cm) was used. The protein was eluted using a linear gradient from 0.05 M Tris·acetate, 2 mM 2-mercaptoethanol, pH 8.3 (Low Q buffer) to Low Q buffer plus 0.5 M sodium chloride. For the hydrophobic interaction chromatography, a Phenyl Sepharose High Performance (GE Healthcare) column (8.5 cm × 2 cm) was used. After the ion-exchange chromatography, ATCase-containing fractions were dialyzed into Low Q and brought to 20% saturation with ammonium sulfate and loaded onto the Phenyl Sepharose column. The protein was eluted using a linear gradient from Low Q buffer plus 20% saturating ammonium sulfate to Low Q buffer. The homogeneity of the protein was checked by SDS/PAGE (18), nondenaturing PAGE (19, 20), and by size-exclusion chromatography (Fig. S3). The concentration of purified double-mutant enzyme was determined by the Bio-Rad version of Bradford’s dye-binding assay using wild-type ATCase as the standard (21).
Crystallization and Data Collection.
A sample of 10 mg/mL purified K164E/E239K ATCase was sent to the Hauptman-Woodward Institute for high-throughput screening of crystallization conditions (22). The final crystallization condition was developed on the basis of one of the screening results. The mutant enzyme was crystallized by the hanging-drop vapor diffusion method. Two microliters of enzyme (10 mg/mL) in 50 mM Tris·acetate (pH 8.3) was mixed with 2 μL of crystallization buffer [16% (wt/vol) PEG 4000, 0.04 M Na2MoO4·2H2O, 0.04 M N-cyclohexyl-3-aminopropanesulfonic acid, and 30 mM Tris·acetate (pH 8.75)] and equilibrated over a reservoir of 1.0 mL of crystallization buffer at 20 °C. Bar-shaped crystals grew to average dimensions of 0.3 × 0.3 × 0.4 mm in approximately 2 wk.
Crystals were transferred into a freezing solution containing 20% (vol/vol) ethylene glycol in crystallization buffer for 1 min before freezing in liquid N2. Data were collected in the Boston College Crystallography Facility. X-rays were generated using a Rigaku MicroMax-007HF rotating-anode generator operating at 40 kV and 30 mA, and data collected using a Rigaku R-axis IV++ detector. The diffraction data were integrated, scaled, and averaged using d*TREK (23).
Structure Determination.
Attempts to solve the structure of the K164E/E239K ATCase by molecular replacement succeeded using the T-state ATCase structure (PDB ID 1ZA1) rather than the R-state PALA-ATCase structure in the same space group (PDB ID 1Q95). First, one catalytic trimer (c3) of the double mutant was found by using the c3 of the T-state structure (PDB ID 1ZA1) in MOLREP (24) as implemented in CCP4 (25). MOLREP was also able to locate the second c3. MOLREP was then used to locate the three regulatory dimers (r2) by fixing the coordinates of the two c3 subunits. Combining the two c3 and three r2 subunits positioned by MOLREP the initial model of the holoenzyme (c6r6) in the asymmetric unit was established. Refinement of the structure was performed using PHENIX (26). The 80’s loops (residues 77–84) and 240’s loops (residues 231–245) of all of the catalytic chains were rebuilt into the refined structure using the Autobuild protocol in PHENIX (26). The loop positions were further checked using composite omit maps, calculated using RESOLVE (27) within PHENIX, and displayed in COOT (28, 29) averaging over the sixfold noncrystallography symmetry. The 80’s and 240’s loops of the catalytic chains had elevated b-factors, which were higher than in the T- or R-state structures of the wild-type enzyme. The higher b-factors are most likely due to loss of a number of loop-stabilizing interactions. Even though the density of these loops was weak and the b-factors high, the atomic coordinates have been included for completeness. Water molecules were added to the structure using PHENIX (26) on the basis of the Fo–Fc map at or above the 3.0σ level. After completion of the refinement, the final Rwork and Rfree were 0.214 and 0.274, respectively. The model was checked for errors using PROCHECK (30). The detailed statistics of data collection and refinement are given in Table 1. Residues 1–9 in the regulatory chains were omitted from the deposited structure owing to extremely weak electron density. Coordinates for the K164E/E239K ATCase structure have been deposited with the PDB (PDB ID 4E2F).
Table 1.
Data collection and refinement statistics for the K164E/E239K ATCase structure
| Data collection | |
| Space group | P212121 |
| Cell dimensions | |
| a, b, c (Å) | 124.1, 144.8, 203.4 |
| α, β, γ (°) | 90, 90, 90 |
| Resolution (Å) | 29.89–2.80 (2.90–2.80) |
| Rsym (%) | 0.079 (0.473) |
| Average (I/σ) | 8.9 (2.2) |
| Completeness (%) | 92.4 (92.4) |
| Redundancy | 3.33 (3.35) |
| Refinement | |
| Resolution (Å) | 29.89–2.80 |
| Reflections | 83,904 |
| Rwork/Rfree | 0.214/0.274 |
| No. of atoms | |
| Protein | 21,258 |
| Waters | 506 |
| rmsd | |
| Bond lengths (Å) | 0.014 |
| Angles (°) | 1.51 |
| Mean B value (Å2) | 81.5 |
Values in parentheses are for the highest resolution shell.
Small-Angle X-Ray Scattering.
The SAXS experiments were performed at Beamline 4-2 at the Stanford Synchrotron Radiation Laboratory (3.0 Gev, 50–100 Ma). The experimental setup and procedures were performed as described by Sakash et al. (31). For experiments at pH 8.3 the buffer used was 0.05 M Tris·acetate and 10 mM 2-mercaptoethanol, whereas for pH 7.0 and pH 7.5 the buffer used was 0.05 M Hepes buffer, 50 mM KCl, and 20 mM 2-mercaptoethanol.
Supplementary Material
Acknowledgments
This work was supported by National Institutes of Health (NIH) Grant GM026237 (to E.R.K) and Grant CHE-0923264 from the National Science Foundation. The Stanford Synchrotron Radiation Laboratory (SSRL) is operated by the Department of Energy, Office of Basic Energy Sciences. The SSRL Structural Biology Resource is supported by NIH, National Center for Research Resources (P41RR01209) and by the Department of Energy, Office of Biological and Environmental Research.
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
Data deposition: The atomic coordinates and structure factors have been deposited in the Protein Data Bank, www.pdb.org (PDB ID code 4E2F).
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1119683109/-/DCSupplemental.
References
- 1.Lipscomb WN. Aspartate transcarbamylase from Escherichia coli: Activity and regulation. Adv Enzymol Relat Areas Mol Biol. 1994;68:67–151. doi: 10.1002/9780470123140.ch3. [DOI] [PubMed] [Google Scholar]
- 2.Newell JO, Schachman HK. Amino acid substitutions which stabilize aspartate transcarbamoylase in the R state disrupt both homotropic and heterotropic effects. Biophys Chem. 1990;37:183–196. doi: 10.1016/0301-4622(90)88018-n. [DOI] [PubMed] [Google Scholar]
- 3.Velyvis A, Yang YR, Schachman HK, Kay LE. A solution NMR study showing that active site ligands and nucleotides directly perturb the allosteric equilibrium in aspartate transcarbamoylase. Proc Natl Acad Sci USA. 2007;104:8815–8820. doi: 10.1073/pnas.0703347104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Moody MF, Vachette P, Foote AM. Changes in the x-ray solution scattering of aspartate transcarbamylase following the allosteric transition. J Mol Biol. 1979;133:517–532. doi: 10.1016/0022-2836(79)90405-4. [DOI] [PubMed] [Google Scholar]
- 5.Huang J, Lipscomb WN. Aspartate transcarbamylase (ATCase) of Escherichia coli: A new crystalline R-state bound to PALA, or to product analogues citrate and phosphate. Biochemistry. 2004;43:6415–6421. doi: 10.1021/bi030213b. [DOI] [PubMed] [Google Scholar]
- 6.Stieglitz K, Stec B, Baker DP, Kantrowitz ER. Monitoring the transition from the T to the R state in E. coli aspartate transcarbamoylase by X-ray crystallography: Crystal structures of the E50A mutant enzyme in four distinct allosteric states. J Mol Biol. 2004;341:853–868. doi: 10.1016/j.jmb.2004.06.002. [DOI] [PubMed] [Google Scholar]
- 7.García De La Torre J, Huertas ML, Carrasco B. Calculation of hydrodynamic properties of globular proteins from their atomic-level structure. Biophys J. 2000;78:719–730. doi: 10.1016/S0006-3495(00)76630-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Tauc P, Vachette P, Middleton SA, Kantrowitz ER. Structural consequences of the replacement of Glu239 by Gln in the catalytic chain of Escherichia coli aspartate transcarbamylase. J Mol Biol. 1990;214:327–335. doi: 10.1016/0022-2836(90)90164-H. [DOI] [PubMed] [Google Scholar]
- 9.Putnam CD, Hammel M, Hura GL, Tainer JA. X-ray solution scattering (SAXS) combined with crystallography and computation: Defining accurate macromolecular structures, conformations and assemblies in solution. Q Rev Biophys. 2007;40:191–285. doi: 10.1017/S0033583507004635. [DOI] [PubMed] [Google Scholar]
- 10.Rambo RP, Tainer JA. Bridging the solution divide: Comprehensive structural analyses of dynamic RNA, DNA, and protein assemblies by small-angle X-ray scattering. Curr Opin Struct Biol. 2010;20:128–137. doi: 10.1016/j.sbi.2009.12.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Glatter O, Kratky O. Small Angle X-Ray Scattering. London: Academic Press; 1982. [Google Scholar]
- 12.Fetler L, Vachette P. The allosteric activator Mg-ATP modifies the quaternary structure of the R-state of Escherichia coli aspartate transcarbamylase without altering the T<—>R equilibrium. J Mol Biol. 2001;309:817–832. doi: 10.1006/jmbi.2001.4681. [DOI] [PubMed] [Google Scholar]
- 13.Monod J, Wyman J, Changeux JP. On the nature of allosteric transitions: A plausible model. J Mol Biol. 1965;12:88–118. doi: 10.1016/s0022-2836(65)80285-6. [DOI] [PubMed] [Google Scholar]
- 14.Fetler L, Kantrowitz ER, Vachette P. Direct observation in solution of a preexisting structural equilibrium for a mutant of the allosteric aspartate transcarbamoylase. Proc Natl Acad Sci USA. 2007;104:495–500. doi: 10.1073/pnas.0607641104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Tsuruta H, et al. Structural kinetics of the allosteric transition of aspartate transcarbamylase produced by physiological substrates. FEBS Lett. 1990;263:66–68. doi: 10.1016/0014-5793(90)80706-o. [DOI] [PubMed] [Google Scholar]
- 16.Baker DP, Kantrowitz ER. The conserved residues glutamate-37, aspartate-100, and arginine-269 are important for the structural stabilization of Escherichia coli aspartate transcarbamoylase. Biochemistry. 1993;32:10150–10158. doi: 10.1021/bi00089a034. [DOI] [PubMed] [Google Scholar]
- 17.Nowlan SF, Kantrowitz ER. Superproduction and rapid purification of Escherichia coli aspartate transcarbamylase and its catalytic subunit under extreme derepression of the pyrimidine pathway. J Biol Chem. 1985;260:14712–14716. [PubMed] [Google Scholar]
- 18.Laemmli UK. Cleavage of structural proteins during the assembly of the head of bacteriophage T4. Nature. 1970;227:680–685. doi: 10.1038/227680a0. [DOI] [PubMed] [Google Scholar]
- 19.Ornstein L. Disc electrophoresis. I. Background and theory. Ann N Y Acad Sci. 1964;121:321–349. doi: 10.1111/j.1749-6632.1964.tb14207.x. [DOI] [PubMed] [Google Scholar]
- 20.Davis BJ. Disc electrophoresis. II. Method and application to human serum proteins. Ann N Y Acad Sci. 1964;121:404–427. doi: 10.1111/j.1749-6632.1964.tb14213.x. [DOI] [PubMed] [Google Scholar]
- 21.Bradford MM. A rapid and sensitive method for the quantitation of microgram quantities of protein utilizing the principle of protein-dye binding. Anal Biochem. 1976;72:248–254. doi: 10.1016/0003-2697(76)90527-3. [DOI] [PubMed] [Google Scholar]
- 22.Luft JR, et al. A deliberate approach to screening for initial crystallization conditions of biological macromolecules. J Struct Biol. 2003;142:170–179. doi: 10.1016/s1047-8477(03)00048-0. [DOI] [PubMed] [Google Scholar]
- 23.Pflugrath JW. The finer things in X-ray diffraction data collection. Acta Crystallogr D Biol Crystallogr. 1999;55:1718–1725. doi: 10.1107/s090744499900935x. [DOI] [PubMed] [Google Scholar]
- 24.Vagin A, Teplyakov A. MOLREP: An automated program for molecular replacement. J Appl Cryst. 1997;30:1022–1025. [Google Scholar]
- 25.Winn MD, et al. Overview of the CCP4 suite and current developments. Acta Crystallogr D Biol Crystallogr. 2011;67:235–242. doi: 10.1107/S0907444910045749. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Adams PD, et al. PHENIX: A comprehensive Python-based system for macromolecular structure solution. Acta Crystallogr D Biol Crystallogr. 2010;66:213–221. doi: 10.1107/S0907444909052925. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Terwilliger TC. Automated main-chain model building by template matching and iterative fragment extension. Acta Crystallogr D Biol Crystallogr. 2003;59:38–44. doi: 10.1107/S0907444902018036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Emsley P, Cowtan K. Coot: Model-building tools for molecular graphics. Acta Crystallogr D Biol Crystallogr. 2004;60:2126–2132. doi: 10.1107/S0907444904019158. [DOI] [PubMed] [Google Scholar]
- 29.Emsley P, Lohkamp B, Scott WG, Cowtan K. Features and development of Coot. Acta Crystallogr D Biol Crystallogr. 2010;66:486–501. doi: 10.1107/S0907444910007493. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Laskowski RA, MacArthur MW, Moss DS, Thornton JM. PROCHECK: A program to check the stereochemical quality of protein structures. J Appl Cryst. 1993;26:283–291. [Google Scholar]
- 31.Sakash JB, Kantrowitz ER. The contribution of individual interchain interactions to the stabilization of the T and R states of Escherichia coli aspartate transcarbamoylase. J Biol Chem. 2000;275:28701–28707. doi: 10.1074/jbc.M005079200. [DOI] [PubMed] [Google Scholar]
- 32.Kraulis PJ. MOLSCRIPT: A program to produce both detailed and schematic plots of protein structures. J Appl Cryst. 1991;24:946–950. [Google Scholar]
- 33.Merritt EA, Bacon DJ. Raster3D: Photorealistic molecular graphics. Methods Enzymol. 1997;277:505–524. doi: 10.1016/s0076-6879(97)77028-9. [DOI] [PubMed] [Google Scholar]
- 34.DeLano WL. The PyMol Molecular Graphics System. San Carlos, CA: DeLano Scientific; 2002. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.