Abstract
Ion channels operate in intact tissues as part of large macromolecular complexes that can include cytoskeletal proteins, scaffolding proteins, signaling molecules, and a litany of other molecules. The proteins that make up these complexes can influence the trafficking, localization, and biophysical properties of the channel. TRIP8b (tetratricopetide repeat-containing Rab8b-interacting protein) is a recently discovered accessory subunit of hyperpolarization-activated cyclic nucleotide-gated (HCN) channels that contributes to the substantial dendritic localization of HCN channels in many types of neurons. TRIP8b interacts with the carboxyl-terminal region of HCN channels and regulates their cell-surface expression level and cyclic nucleotide dependence. Here we examine the molecular determinants of TRIP8b binding to HCN2 channels. Using a single-molecule fluorescence bleaching method, we found that TRIP8b and HCN2 form an obligate 4:4 complex in intact channels. Fluorescence-detection size-exclusion chromatography and fluorescence anisotropy allowed us to confirm that two different domains in the carboxyl-terminal portion of TRIP8b—the tetratricopepide repeat region and the TRIP8b conserved region—interact with two different regions of the HCN carboxyl-terminal region: the carboxyl-terminal three amino acids (SNL) and the cyclic nucleotide-binding domain, respectively. And finally, using X-ray crystallography, we determined the atomic structure of the tetratricopepide region of TRIP8b in complex with a peptide of the carboxy-terminus of HCN2. Together, these experiments begin to uncover the mechanism for TRIP8b binding and regulation of HCN channels.
Keywords: beta subunit, single molecule, protein–protein interaction
Over the past two decades, it has become apparent that in living tissues ion channels do not function in isolation, but instead exist as macromolecular complexes that include auxiliary subunits, signaling molecules, scaffolding proteins, and cytoskeletal proteins (1–3). These accessory proteins associate with channels and alter their gating, expression levels, localization, and response to signals such as phosphorylation or internal calcium. Until recently, little was known about which proteins may associate with and modulate hyperpolarization-activated cyclic nucleotide-gated (HCN) channels under physiological conditions. HCN channels are members of the superfamily of voltage-gated channels that contain four subunits around a centrally located pore. Unlike most of the members of the family, however, HCN channels open in response to hyperpolarizing voltages (4, 5). In addition to their unique voltage dependence, activation of HCN channels is facilitated by direct binding of cAMP to a cyclic nucleotide-binding domain (CNBD) in the carboxyl-terminal region of each subunit. The CNBD is connected to the pore region via a gating ring formed from the C-linker of each subunit. The binding of cAMP to the CNBD causes a conformational change in the C-linker and potentiation of the pore opening, which leads to a speeding of channel activation and a depolarizing shift in the voltage dependence of activation (6).
In pyramidal neurons, HCN channels display a striking subcellular localization best seen in cortical pyramidal neurons and hippocampal CA1 pyramidal neurons (7, 8). HCN channels are expressed in a gradient in the dendrites of these neurons showing increasing density with increasing distance from the soma. Differences in surface expression level as high as 60-fold between the soma and distal dendrites have been seen in hippocampal pyramidal cells (7). This gradient of expression of HCN channels has been suggested to contribute to the distance-independent temporal summation of excitatory postsynaptic potentials (8), to modulate integrative properties of neurons (9), and to impact the dendritic control of synaptic plasticity (10).
Until recently, the mechanism by which HCN channels achieve their diversity of expression and function was unknown. In 2004, Santoro and colleagues (11) identified an accessory subunit for HCN channels called TRIP8b (tetratricopetide repeat-containing Rab8b-interacting protein), which has recently been shown to target HCN1 and HCN2 channels to the membrane of the distal dendrites of CA1 pyramidal neurons (12, 13). TRIP8b is a cytoplasmic protein primarily found in neurons that interacts with the carboxyl-terminal tripeptide of HCN channels (SNL in HCN1, -2, and -4; ANM in HCN3) and regulates their surface expression levels (11, 14, 15). In addition, TRIP8b dramatically reduces the cAMP dependence of HCN channel gating through a second interaction site on HCN channels involving the CNBD (15–17). Yeast two-hybrid and coimmunoprecipitation experiments demonstrated that the tetratricopetide repeat (TPR) region of TRIP8b interacts with the carboxyl-terminal SNL sequence and a conserved stretch of amino acids amino-terminal to the TPR region likely interacts with the CNBD of HCN channels (16, 17). The two binding sites have been shown to have distinct effects on channel function. Whereas the interaction with the C-terminal HCN tripeptide contributes to TRIP8b effects on channel trafficking to and from the plasma membrane, interaction with the CNBD contributes to TRIP8b-mediated effects on trafficking and is uniquely responsible for TRIP8b-mediated regulation of HCN channel gating (16, 17).
Here, by using a single-molecule bleaching method, we show that TRIP8b and HCN2 form an obligate 4:4 complex in intact channels in the membrane. Using size-exclusion chromatography and fluorescence anisotropy, we confirm that TRIP8b forms a bipartite interaction with the carboxyl-terminal region of HCN2 channels, making a stable interaction with both the carboxyl-terminal SNL sequence and the C-linker/CNBD. Finally, with X-ray crystallography we elucidate the molecular mechanism of binding of the carboxyl-terminal SNL sequence to TRIP8b. Our structure shows that the carboxyl-terminal tripeptide (SNL) of HCN2 channels interacts with TRIP8b via contacts made with a series of TPR domains in the carboxyl-terminal portion of TRIP8b. These results extend our understanding of the structure of the TRIP8b:HCN complex to include intact channels and atomic resolution.
Results
EGFP-Tagged TRIP8b-1a Isoform Modulates HCN2 Channels in Oocytes.
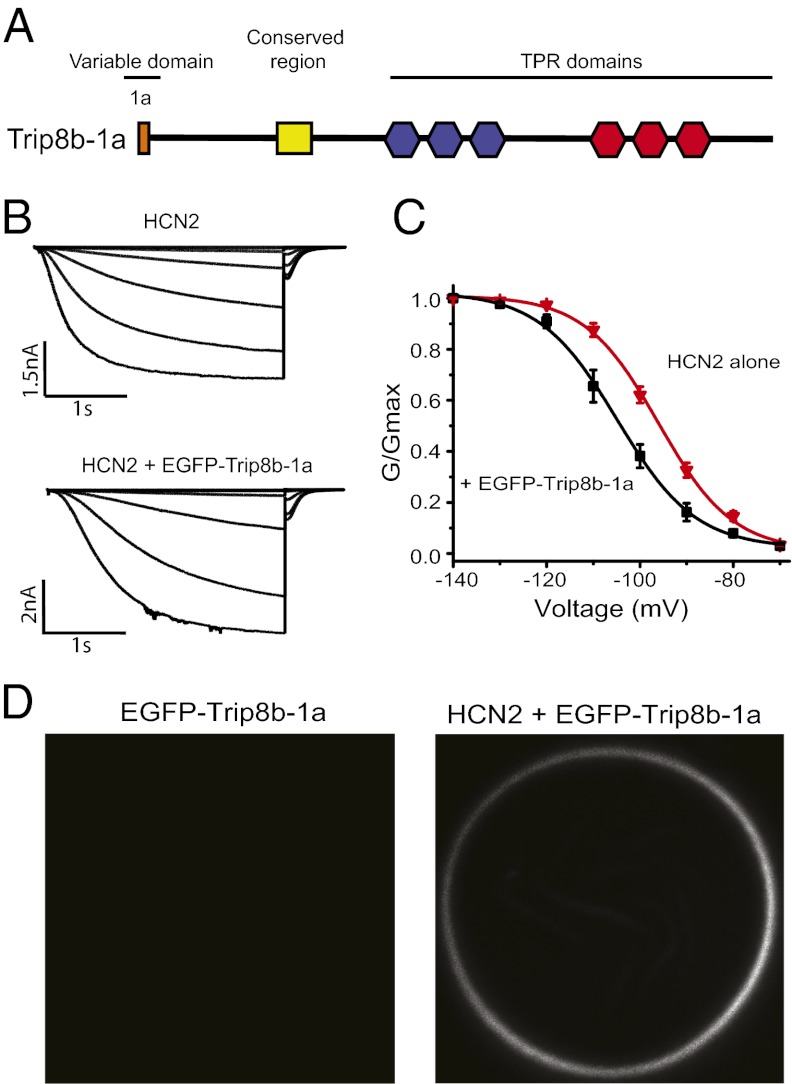
TRIP8b (also called PEX5R and PEX5L) is a heavily alternatively spliced protein with the amino-terminal portion varying among isoforms and the carboxyl-terminal portion constant across isoforms (14, 15). These splice variants are able to up- and down-regulate HCN channel surface expression in an isoform-dependent manner (14, 15). The amino-terminal portion includes five alternatively spliced exons, and the carboxyl-terminal portion includes six TPR repeats as well as a domain, termed the TRIP8b conserved domain, that is conserved in all orthologs of TRIP8b and the human peroxisomal import protein peroxin-5 (PEX5) (Fig. 1A and Fig. S1). For this study, we have used isoform TRIP8b-1a (Fig. 1A and Fig. S1).
Fig. 1.
EGFP-tagged TRIP8b-1a bound to HCN2 channels regulates their voltage dependence. (A) Cartoon showing the isoform TRIP8b-1a used in this study. (B) Current families from cell-attached patches on oocytes that expressed HCN2 alone and HCN2 with EGFP-TRIP8b-1a. (C) Conductance-voltage relationship for HCN2 channels and HCN2 channels with EGFP-TRIP8b-1a measured in the cell-attached patch configuration. (D) Confocal microscopy images of oocytes expressing EGFP-TRIP8b-1a alone and EGFP-TRIP8b-1a coexpressed with HCN2 channels.
To verify that an EGFP-tagged version of TRIP8b-1a was able to associate with and regulate HCN2, EGFP was fused to the amino-terminal end of TRIP8b-1a (EGFP-TRIP8b-1a) and then coexpressed with HCN2 in Xenopus laevis oocytes. We monitored both the change in the voltage dependence of activation of the HCN2 channels and the fluorescence associated with the membrane. In cell-attached patches, EGFP-TRIP8b-1a produced a significant shift in the voltage dependence of HCN2 channel activation in the hyperpolarizing direction (HCN2: V1/2 = −96.1 ± 0.94 mV, n = 4; HCN2 + EGFP-TRIP8b-1a: V1/2 = −105.4 ± 2.14 mV, n = 4) (Fig. 1C). In addition, currents from oocytes coinjected with EGFP-TRIP8b-1a activated more slowly than currents from oocytes injected with HCN2 alone (Fig. 1B). These results are consistent with the previously reported reduction of the cAMP dependence of HCN family channels produced by all TRIP8b isoforms tested (14, 15, 18). cAMP normally produces a 15- to 20-mV depolarizing shift in the activation of HCN2 channels and speeds up the rate of activation (6). As there is substantial cAMP inside the oocyte, the effects of TRIP8b on HCN2 currents are thought to be largely an indirect effect on channel activation resulting from a reduction or elimination of the cyclic nucleotide effect (Fig. 1C, black). In addition, confocal microscopy showed that oocytes injected with EGFP-TRIP8b-1a alone displayed little or no fluorescence associated with the membrane and that coexpression with HCN2 localizes the EGFP-TRIP8b-1a signal to the membrane (Fig. 1D). Together, these results indicate that the EGFP-tagged TRIP8b-1a is able to bind to and modulate HCN channels in the membrane.
TRIP8b and HCN2 Form a 4:4 Complex in Intact Channel Complexes.
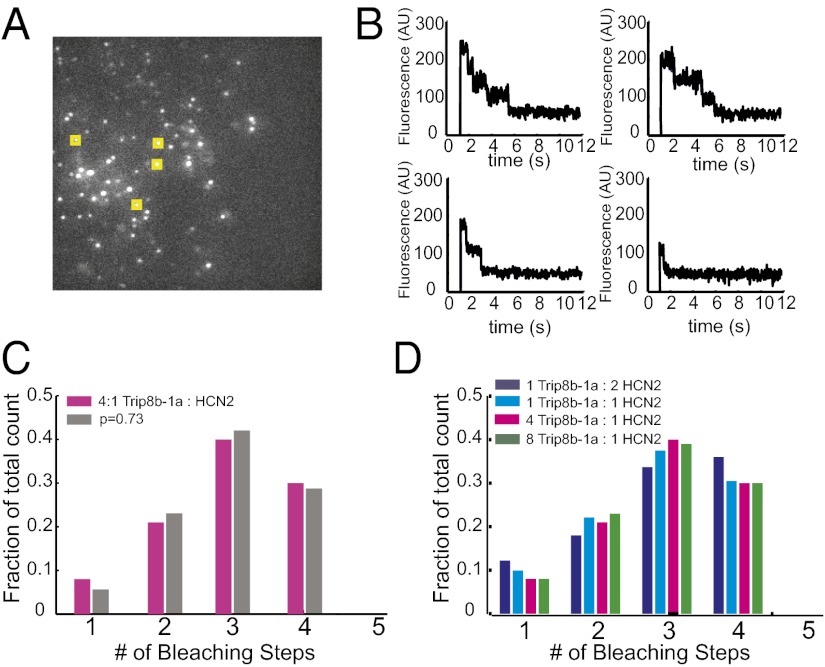
To begin to understand how TRIP8b associates with HCN2 channels, we sought to determine the stoichiometry of the complex using a single-molecule bleaching method developed by Ulbrich and Isacoff (19). To do this, we coexpressed EGFP-TRIP8b-1a with HCN2 channels in X. laevis oocytes at a reduced expression level. The membranes of the oocytes were then imaged using total internal reflection (TIRF) microscopy to record from single-channel complexes in the membrane while minimizing the autofluorescence from inside the oocyte (Fig. 2A). In response to illumination at 488 nm, the EGFP molecules bleached in a stepwise fashion and were then counted to determine their number, and therefore the number of TRIP8b-1a subunits, in the channel complex.
Fig. 2.
Single-molecule bleaching revealed a 4:4 TRIP8b/HCN2 stoichiometry. (A) TIRF microscopy was used to image the membranes of oocytes. The image shows an average of five frames of a representative movie of EGFP-TRIP8b-1a coexpressed with HCN2. Yellow boxes represent a sample of selected spots for analysis. (B) Examples of bleaching steps for EGFP-TRIP8b-1a associated with HCN2 channels. Each graph shows a spot that bleaches in a different number of steps from 1 to 4. (C) Distribution of bleaching steps for an RNA injection ratio of 4:1 EGFP-TRIP8b-1a:HCN2. Magenta bars show the fraction of spots that bleached in a given number of steps. Data were obtained from 508 spots. The gray bars show the fit of a binomial distribution with n = 4 and P = 0.73. (D) Distribution of bleaching steps from various injection ratios of EGFP-TRIP8b-1a and HCN2 RNA. These data were obtained from 242, 289, and 277 spots for the 8:1, 2:1, and 1:2 ratios, respectively.
We first injected RNA for EGFP-TRIP8b-1a and HCN2 at a ratio of 4:1. Only stationary, diffraction-limited fluorescent spots were included in the analysis (Fig. 2A, yellow squares). The fluorescence signal was then plotted as a function of time, and bleaching steps were counted (Fig. 2B). Many of the spots bleach in four discrete steps, which is consistent with the idea that there are four TRIP8b-1a subunits in the channel complex (Fig. 2B, Top Left, and C). The large number of spots that bleach in one, two, or three steps most likely resulted from two sources. First, a significant fraction of the EGFP molecules are not fluorescent or are proteolytically cleaved at the start of the experiment, leading to an underestimate of subunit number (19). Second, the bleaching time for EGFP is fast enough that occasionally a single step could result from the near simultaneous bleaching of two EGFP molecules, again leading to an underestimate of the number of TRIP8b molecules present. On the basis of an exponential distribution of bleaching times and assuming that we would not be able to resolve two steps within 100 ms of one another, simultaneous bleaching of two EGFP molecules was estimated to occur less than 3.1% of the time. To correct for the nonfluorescent EGFP molecules, the distribution of the number of steps was fitted with different binomial distributions assuming two to five EGFP-TRIP8b-1a subunits and a probability, P, that the EGFP is fluorescent. The data fit well with a binomial with four EGFP-TRIP8b-1a subunits and a value of P = 0.73, similar to that recorded for other channels (Fig. 2C) (19, 20). These results indicate that there are four EGFP-TRIP8b-1a subunits in the channel complex and that the TRIP8b-1a:HCN2 stoichiometry is 4:4.
To determine if the stoichiometry of this complex depends on the amount of TRIP8b present in the cell, we varied the amount of TRIP8b-1a present by injecting varying ratios of TRIP8b-1a:HCN2 RNA into the oocytes. The distribution of the number of EGFP-TRIP8b-1a subunits was not significantly different, having RNA ratios ranging from 1:2–8:1 TRIP8b-1a:HCN2 (Fig. 2D). Even in the case where less TRIP8b-1a than HCN2 RNA was injected, the distribution was still consistent with four TRIP8b-1a subunits supporting the hypothesis that TRIP8b forms an obligate tetramer in the channel complex. At very low concentrations of TRIP8b (less than a 1:2 ratio of TRIP8b:HCN2), consistently measuring spots became prohibitively difficult. Despite this limitation, the fact that all ratios tested pointed toward a 4:4 stoichiometry suggests that TRIP8b can associate only with HCN channels as a 4:4 complex.
HCN2 Interacts with TRIP8b in Solution.
Previously, it has been shown, using yeast two-hybrid and coimmunoprecipitation experiments, that TRIP8b interacts directly with the carboxyl-terminal region of HCN channels (11, 14–17). To determine if TRIP8b forms a stable interaction with HCN2 in solution, we used fluorescence-detection size-exclusion chromatography (FSEC) (21). For each protein, amino-terminal or carboxyl-terminal fusions to EGFP were constructed and expressed in bacteria. After cell lysis, the cleared lysates were run on a size-exclusion chromatography system fitted with an in-line fluorescence detector where the elution of the EGFP-fusion protein could be detected in the crude cell lysate with fluorescence.
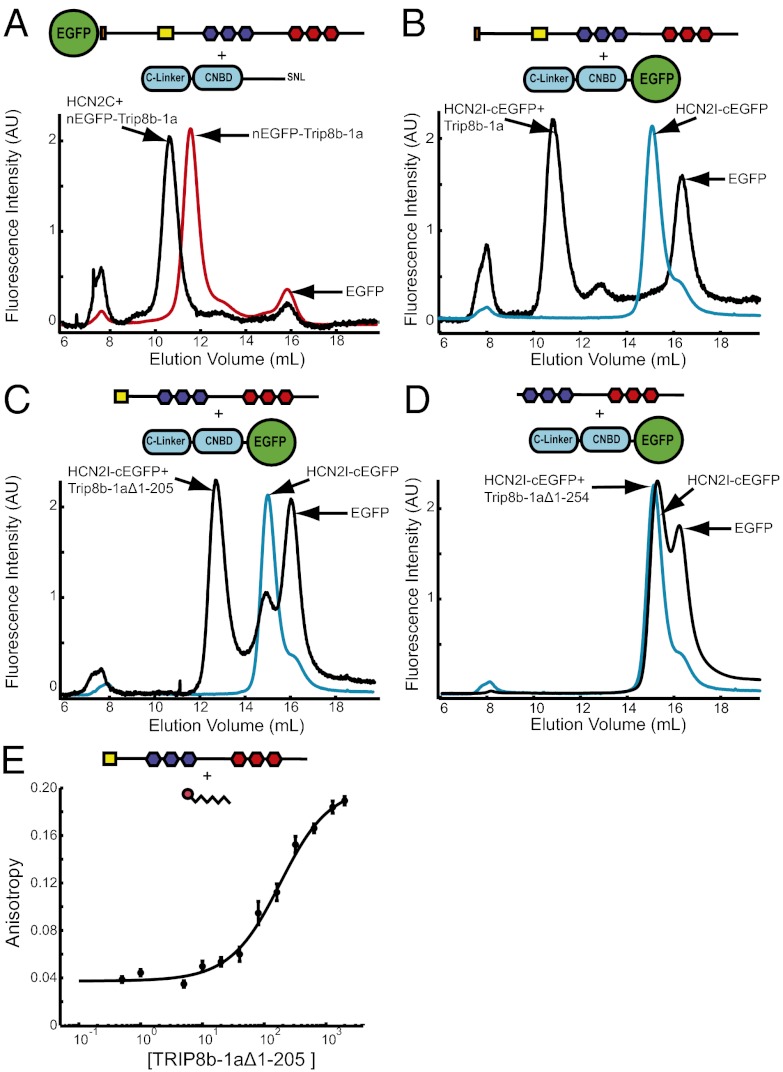
To examine the interaction between TRIP8b-1a and the HCN2 carboxyl-terminal region (residues 443–863, termed HCN2C), we used the amino-terminal EGFP fusion of TRIP8b-1a (TRIP8b residues 1–567). EGFP-TRIP8b-1a alone ran as a single monodispersed peak with an elution volume of 11.6 mL (Fig. 3A, red). A small second peak eluting at about 16 mL corresponded to free EGFP generated from proteolytic cleavage of the fusion protein. However, coexpression of EGFP-TRIP8b-1a with HCN2C caused a significant decrease in the elution volume of the EGFP-TRIP8b-1a to 10.6 mL, which is indicative of a stable interaction between TRIP8b-1a and the HCN2C fragment (Fig. 3A, black). Similar results were also observed with purified TRIP8b-1a without the EGFP tag (Fig. S2A). Although exact sizes and stoichiometries of the molecules cannot be accurately determined from FSEC, these results confirm and extend the previous findings that TRIP8b interacts directly with the HCN carboxyl-terminal region (11, 14–17).
Fig. 3.
TRIP8b-1a forms a bipartite interaction with HCN2. Each panel contains a cartoon showing the protein constructs used and the location of the fluorescent tag. (A) FSEC results of the crude lysates containing TRIP8b-1a (red) and TRIP8b-1a coexpressed with HCN2C (black). (B) FSEC results of the crude lysates containing HCN2I (cyan) and HCN2I coexpressed with TRIP8b-1a (black). (C) FSEC results of crude lysates containing HCN2I (cyan) and HCN2I coexpressed with TRIP8b-1aΔ1–205 (black). (D) FSEC results from crude lysates containing HCN2I (cyan) and HCN2 coexpressed with TRIP8b-1aΔ1–254 (black). (E) Fluorescence anisotropy of the TAMRA-SNL peptide plotted versus the total concentration of TRIP8b-1aΔ1–205. The data were fit with a Kd of 155 ± 31 nM (equation in Materials and Methods).
The TRIP8b protein was originally identified in a yeast two-hybrid screen using the distal carboxyl-terminal region of HCN as bait, and TRIP8b was found to bind directly to the carboxyl-terminal three amino acids of HCN (SNL) (15). Subsequently, yeast two-hybrid and coimmunoprecipitation experiments revealed that TRIP8b also binds the C-linker/CNBD of HCN channels (15–17). To determine if the TRIP8b interaction with only the C-linker/CNBD of HCN is also stable in solution, we used a smaller piece of HCN2 (residues 443–640) that contains only the C-linker and CNBD (HCN2I). A carboxyl-terminal EGFP fusion of HCN2I (HCN2I-EGFP) alone elutes as a single elution peak at 15.2 mL (with a shoulder at 16 mL, corresponding to EGFP) (Fig. 3B, cyan). However, coexpression of HCN2I-EGFP with TRIP8b-1a caused a dramatic decrease in the elution volume to 10.85 mL, indicative of a stable interaction between HCN2I-EGFP and TRIP8b-1a (Fig. 3B, black). Again, similar results were also observed with purified HCN2I without the EGFP tag (Fig. S2B). This truncated HCN2I construct is missing the carboxyl-terminal SNL sequence of HCN2, and yet it retains the ability to stably bind TRIP8b-1a, suggesting that the C-linker/CNBD region is sufficient to bind TRIP8b.
To localize the HCN-binding site on TRIP8b, we constructed a truncated version of TRIP8b containing only the TPR domains and most of the TRIP8b conserved region (TRIP8b-1aΔ1–205, residues 206–567) (Fig. S1). FSEC experiments showed that TRIP8b-1aΔ1–205 was also able to stably interact with HCN2I-EGFP in solution, showing that the interaction with the C-linker/CNBD does not require the amino-terminal portion of TRIP8b (Fig. 3C). However, a further truncation of TRIP8b-1a that removes the TRIP8b conserved domain (TRIP8b-1aΔ1–254, residues 255–567) did not interact with HCN2I (Fig. 3D). These results confirm and extend previous findings that the interaction with the C-linker/CNBD involves the TRIP8b conserved domain.
To show that the SNL sequence is sufficient to bind to Trip8b, we measured the anisotropy of a peptide of the last seven amino acids of HCN2 (SNL peptide) with a covalently attached carboxy tetramethyl-rhodamine (TAMRA) on the N terminus (22). To measure the binding affinity between the SNL peptide and Trip8b-1aΔ1–205, we measured the anisotropy of 50 nM TAMRA-SNL peptide with increasing amounts of TRIP8b-1aΔ1–205. Anisotropy was then plotted versus total concentration of TRIP8b-1aΔ1–205, and the data were fit with a single binding isotherm (Materials and Methods). This analysis revealed that the SNL peptide is sufficient for binding to Trip8bΔ1–205 with an affinity of 155 ± 31 nM. Taken together, these results indicate that TRIP8b forms a bipartite interaction with the HCN2 carboxyl-terminal region, forming a stable interaction with both the CNBD (or C-linker) and the carboxyl-terminal SNL sequence.
X-Ray Crystal Structure of TRIP8b in Complex with Carboxyl-Terminal Residues of HCN2.
To determine the molecular mechanism for TRIP8b binding to the carboxyl-terminal SNL sequence of HCN, we determined the cocrystal structure of TRIP8b-1aΔ1–205 in complex with the SNL peptide (without the TAMRA tag). The TRIP8b-1aΔ1–205 protein was expressed in bacteria and purified, and the SNL peptide was added at 1 mM before crystallization. Cocrystals of the TRIP8b-1aΔ1–205 and SNL peptide grew under vapor diffusion in space group P41212 with a single molecule in the asymmetric unit and diffracted to 3.0 Å (Table S1). The structure of the TRIP8b-1aΔ1–205+SNL peptide was solved with molecular replacement using the structure of PEX5 as a search probe (23).
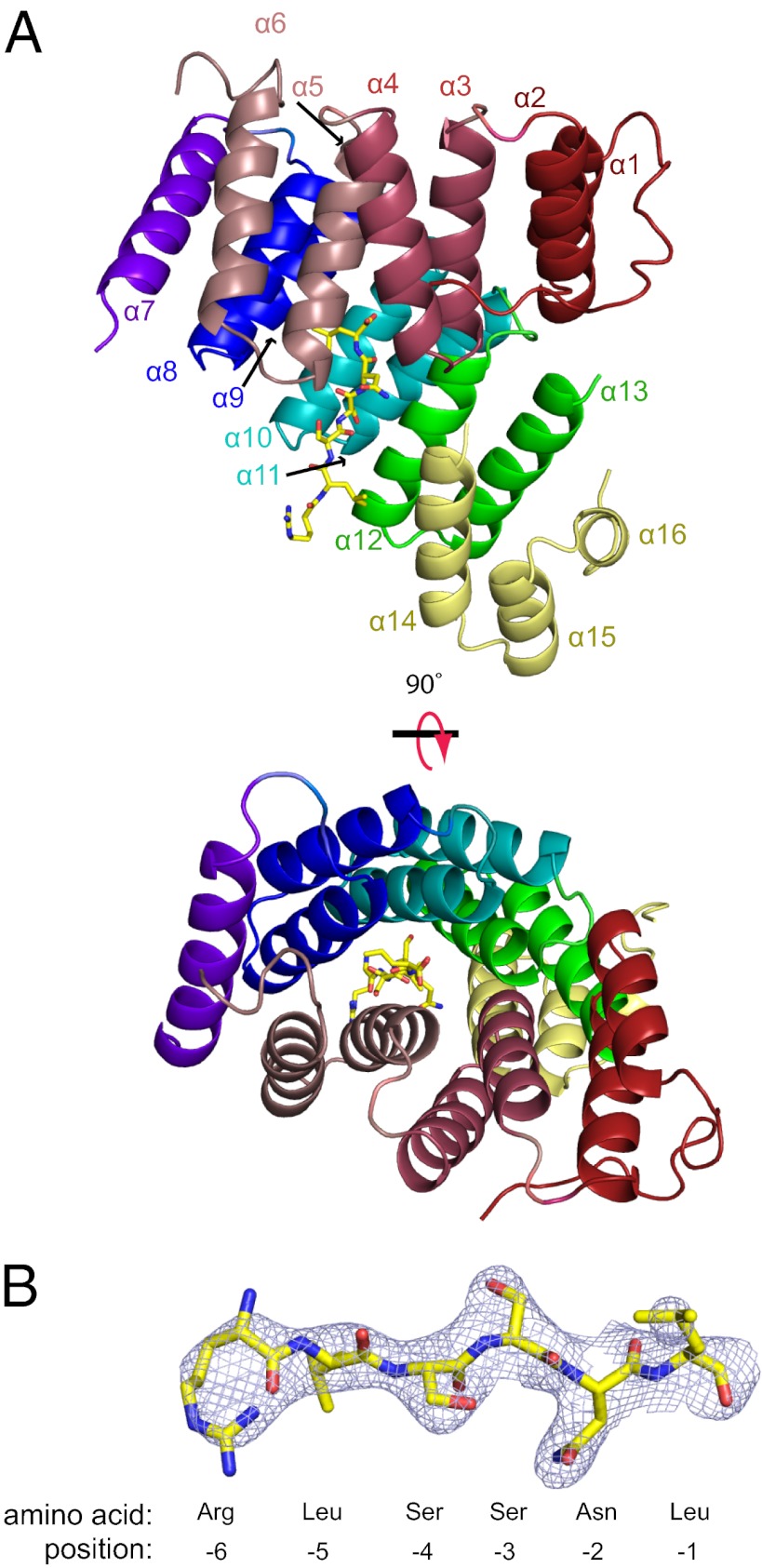
The overall structure of TRIP8b-1aΔ1–205 contained 16 α-helices (Fig. 4A). Helices α1–α6 and α8–α13 composed two clusters of TPRs separated by an α-helical hinge region containing helix α7 flanked by two disordered loops. Each of the two clusters had three TPR domains, each of which was made up of two antiparallel α-helices separated by a turn. Helix α7, in the hinge region, was a single 16-residue α-helix that did not adopt the typical TPR fold. At the carboxyl-terminal end of the protein, after the TPRs, there was also a small three-helix bundle (Fig. 4A, yellow) whose functional role in TRIP8b remains unknown. No electron density was observed for the first 49 amino acids containing the TRIP8b conserved region or in the segment between α6 and α7.
Fig. 4.
Crystal structure of TRIP8b-1aΔ1–205 in complex with a peptide of the last seven amino acids of HCN2. (A) Ribbon representation of TRIP8b-1aΔ1–205 shown from two different perspectives with the SNL peptide shown in stick representation. Each TPR is colored differently (TRP1, red; TPR2, dark pink; TRP3, light pink; TPR4, blue; TPR5, cyan; TPR6, green; hinge region helix, purple; three helix bundle, yellow). (B) Fo-Fc electron density “omit” map contoured at 1σ for the SNL peptide. In addition, the numbering system for the peptide is shown below.
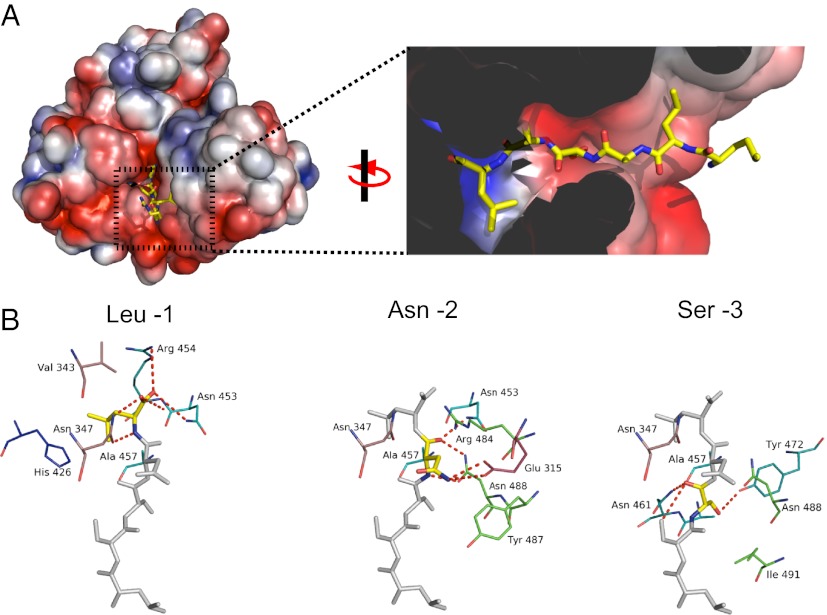
The carboxyl-terminal tripeptide sequence of HCN2 bound directly to the TPR domains, and electron density for residues −1 to −6 of the SNL peptide was clearly visible (Fig. 4B and Fig. S3B). The SNL peptide was situated in a strikingly narrow passage at the cleft between the two clusters of TPRs (Fig. 5A). Important contacts were made between the carboxyl-terminal three amino acids, SNL, and TPRs 2, 3, 5, and 6 (Fig. 4A, 5). The terminal carboxylate sat in a positively charged pocket of TRIP8b and formed hydrogen bonds with the side chains of Asn-347, Asn-453, Arg-454, and Arg-484 (Fig. 5 A and B and Fig. S3A). The side chain of the terminal leucine (−1) was buried in a hydrophobic pocket of TRIP8b composed of Val-343, Thr-346, Phe-438, and Thr-458, as well as the Cβ and Cγ of Arg-454 (Fig. 5A and Fig. S3). The asparagine at the −2 position was recognized by TRIP8b in part by hydrogen bonding of the NH2 to Glu-315. In addition, the side chains of Asn-453 and Asn-488 were hydrogen-bonded with the backbone carbonyl group and NH group of the asparagine at the −2 position of the SNL peptide. The side chain of the serine at the −3 position formed a hydrogen bond with Tyr-472 of TRIP8b, and the main chain carbonyl and NH groups formed hydrogen bonds with the side chain of Asn-461. In addition, the leucine at position −5 was surrounded by hydrophobic residues Ile-491 and Ile-494 (Fig. 5A). SNL peptide residue −6 showed no appreciable interactions with TRIP8b.
Fig. 5.
Binding site for the SNL peptide on TRIP8b. (A) Molecular surface representation of TRIP8b-1aΔ1–205 colored according to the calculated electrostatic potential (−3 to 3 kTe). Close-up of the binding pocket from A with the SNL peptide in stick representation. The surface display was clipped to show the pocket where HCN2 binds. In addition, two residues (Asn-347 and Cys-355) were removed from the figure for clarity. (B) Interactions with TRIP8b broken down by residue on the SNL peptide. The colors of the carbons on TRIP8b match the color scheme of the ribbon representation in Fig. 4A. Polar contacts are shown as dashed red lines.
Discussion
The results in this paper extend our understanding of the structure of the TRIP8b:HCN complex to include intact channels and atomic resolution. Previous studies have shown that TRIP8b interacts with HCN channels at two sites (11, 14–17). The first is the carboxyl-terminal tripeptide of HCN channels, which is SNL for HCN1, -2, and -4 and ANM for HCN3, and the second is the C-linker/CNBD. In addition, binding assays using truncated pieces of TRIP8b have shown that the TPR domains of TRIP8b likely interact with the carboxyl-terminal tripeptide of HCN channels whereas the TRIP8b conserved domain binds to the C-linker/CNBD (15–17). Our X-ray crystallography data show the molecular detail of the interaction with the carboxyl-terminal tripeptide, and our fluorescence and biochemical data begin to suggest a picture of what the complex looks like in vivo.
The overall structure of TRIP8b in complex with the SNL peptide is very similar to the structure of PEX5 in complex with PTS1 (Fig. S4A) (23). PEX5 is a cytosolic receptor for proteins carrying a peroxisomal targeting signal type 1 (PTS1) and is involved in translocating proteins across the peroxisomal membrane (24). PEX5 has significant sequence similarity in the TPR domains and conserved domain with TRIP8b (56% identity, Fig. S1), and the root-mean-square deviations in the α-carbons between the two proteins is 1.38A. Although the role of the conserved domain in PEX5 is not known, PEX5 binds its cargo, PTS1, to the TPR domains with a similar mechanism to TRIP8b (Fig. S4B). Although PEX5 also recognizes a carboxyl-terminal tripeptide sequence in its target protein, the target sequence in PTS1 is SKL instead of SNL in HCN2 channels (25). A study from Gatto et al. examined the sequence requirements for PEX5 recognition of the tripeptide and found that mutation of the lysine to asparagine reduced the affinity of the interaction 28-fold (26). Surprisingly, the structures of the binding pockets of PEX5 and TRIP8b are very similar, and many of the residues involved are identical (Fig. S4B). The details of the interaction at the −2 position, however, are different and may explain the discrimination between terminal tripeptides. In PEX5, the positively charged N of the Lys −2 side chain contacts the side-chain carboxylates of Glu-398 and Glu-430 via a single water molecule (Fig. S4B). In TRIP8b, the residue at the position equivalent to Glu-430 is Thr-341, which is not positioned to coordinate the water molecule seen in the PEX5 structure. In the absence of those water-mediated interactions, the N on the side chain of the asparagine at −2 in HCN2 channels forms a contact with a glutamate at position 308 in TRIP8b (Fig. S4B).
Zolles and colleagues showed that the effect of TRIP8b was concentration-dependent (18). As they injected increasing amounts of TRIP8b into the oocyte, the cAMP dependence of HCN2 became more and more reduced, saturating with a ninefold excess of TRIP8b RNA over HCN2 RNA. This TRIP8b concentration dependence could result from two possible mechanisms: (i) HCN channels could bind varying numbers of TRIP8b molecules, resulting in a variable regulation of individual HCN channels; or (ii) TRIP8b could always be associating with HCN channels as a tetramer, and the titratable effect could reflect the fact that not all channels have TRIP8b bound. The single-molecule data in our experiments strongly favor the second mechanism. Despite a wide range of TRIP8b/HCN2 ratios injected into the oocytes, the bleaching step counts consistently showed a 4:4 ratio. These data suggest that TRIP8b forms an obligate tetramer with the intact HCN2 channel. It remains to be seen precisely how TRIP8b interacts with the C-linker/CNBD of HCN channels and how that interaction reduces the cyclic nucleotide dependence of these channels.
Materials and Methods
Molecular Biology.
For experiments in Xenopus oocytes, mouse HCN2 and human TRIP8b-1a were each subcloned into the oocyte high-expression vector pGEMHE (27). For the EGFP-TRIP8b-1a construct, EGFP was fused in frame with TRIP8b-1a before the initial methionine on the N terminus of TRIP8b. For the biochemistry experiments, HCN2#443–863 (HCN2C), TRIP8b-1a, TRIP8b-1aΔ1–205, and TRIP8b-1aΔ1–254 were each subcloned into pETM11 vectors. For EGFP-TRIP8b-1a and HCN2I-EGFP (HCN2#443–640), the constructs were subcloned into pNGFP_BC and pCGFP_BC, respectively (kindly provided by E. Gouaux, Vollum Institute, Portland, OR). For experiments measuring the interaction between TRIP8b and HCN2, vectors were cotransformed into BL21 (DE3) cells and then grown in the presence of both kanamycin and carbenicillin. All constructs were confirmed with fluorescence-based automated sequencing.
Electrophysiology.
X. laevis oocytes were defolliculated and injected with cRNA as previously described (28). Recordings were made in the cell-attached patch configuration using an Axopatch 200A patch-clamp amplifier (Axon Instruments). Data were acquired with PULSE acquisition software (HEKA Elektronik). Patch pipettes were pulled from borosilicate glass and had resistances of 0.25–1 MΩ after fire polishing. The solutions for HCN2 recordings were as follows: pipette and bath solution, 130 mM KCl, 3 mM Hepes, and 0.2 mM EDTA, pH 7.2.
Confocal Microscopy.
Fluorescence images in whole oocytes were collected on a Zeiss LSM 710 confocal microscope using a 20× air objective. Excitation of the fluorophore was done with a 488-nm laser, and emission was measured between 502 and 551 nm.
Protein Purification and Biochemistry.
TRIP8b-1a, TRIP8b-1aΔ1–205, and TRIP8b-1aΔ1–254 were each screened for expression and solubility using FSEC as previously described (21). Briefly, 5 mL of bacterial cultures was induced, spun down, and resuspended in 800 μL of 150 mM KCl and 30 mM Hepes (pH 7.5) containing 2.5 μg/mL DNase and complete protease inhibitor tablets (Roche). The cells were lysed with a probe sonicator, the lysate was cleared by centrifugation, and 100 μL of the supernatant was loaded on a Superdex 200 10/300 GL column (GE Healthcare) mounted on an FPLC system with a fluorescence detector set for detection of GFP fluorescence.
For crystallography experiments, 2-L bacterial cultures of TRIP8b-1aΔ1–205 in the pETM11 vector were grown at 37 °C in LB to midlog phase and induced with 1 mM isopropyl-β-d-thiogalactopyranoside overnight at 18 °C. The cultures were spun down and resuspended in 150 mM KCl and 30 mM Hepes (pH 7.5) containing 1 mM 4-(2-aminoethyl) benzenesulfonyl fluoride hydrochloride, 2.5 μg/mL DNase, and complete protease inhibitor tablets (Roche). Cells were lysed with an EmulsiFlex C-3 homogenizer (Avestin), and the lysate was cleared by centrifugation at 131,000 × g for 45 min at 4 °C. The proteins were then purified on a Ni2+ affinity resin column (HisTrap HP, GE Healthcare). The octahistidine tag was removed by Tobacco Etch Virus protease cleavage, and the proteins were further purified on an anion exchange column (HiTrap Q HP, GE Healthcare) and concentrated to about 31 mg/mL.
X-Ray Crystallography.
Crystals were grown using the sitting drop, vapor diffusion method at 20 °C. The SNL peptide of the last seven amino acids of the HCN2 channel sequence (SRSSNL) was synthesized by American Peptide Company with an acetylation at the amino-terminal end and added to the protein at a concentration of 1 mM. The final concentration of TRIP8b-1aΔ1–205 after addition to the peptide solution was 0.5 mM. Crystals of TRIP8b-1aΔ1–205 grew using a 3:1 mixture of protein and reservoir solution containing 91 mM trisodium citrate, 91 mM MES, and 3.63 M NaCl at pH 6.0. Crystals developed within 8 d with space group P41212 that diffracted to a resolution of 3 Å. To serve as a cryoprotectant, 20% (vol/vol) glycerol was added to the reservoir solution before rapid freezing. Data were collected at 110° K on beamline 8.2.1 at the Advanced Light Source (Lawrence Berkeley National Laboratory). Integration, scaling, and merging of the diffraction data were done with the Mosflm program (29). The structures were solved by molecular replacement using the programs Phaser (30) and Phenix (31). The structure of human PEX5 (1FCH), with the PTS1 peptide removed from the structure, was used as a search probe. Structure refinement and validation were performed using the Phenix (31) software suite and the program Coot (32) using 2Fo–Fc “omit” maps (Table S1). The electrostatic surface potential was calculated using the Adaptive Poisson–Boltzmann Solver software (33). Additional real-space refinement was carried out in Rosetta, which helped reduce model bias (34).
Fluorescence Anisotropy.
Fluorescence anisotropy was recorded using the Fluorolog 3 spectrophotometer (HORIBA, Jobin Yvon). SNL peptide, synthesized by Biomatik, contained a TAMRA attached to the last seven amino acids of HCN2 (SRSSNL). A 50-nM TAMRA-SNL peptide was placed in a quartz cuvette, and anisotropy was measured with increasing concentrations of TRIP8b-1aΔ1–205. Anisotropy experiments with 540 nm excitation and 580 nm emission were performed as previously described (22). To estimate binding affinity, plots of the anisotropy versus total TRIP8b-1aΔ1–205 concentration were fit using the following first-order reaction scheme:
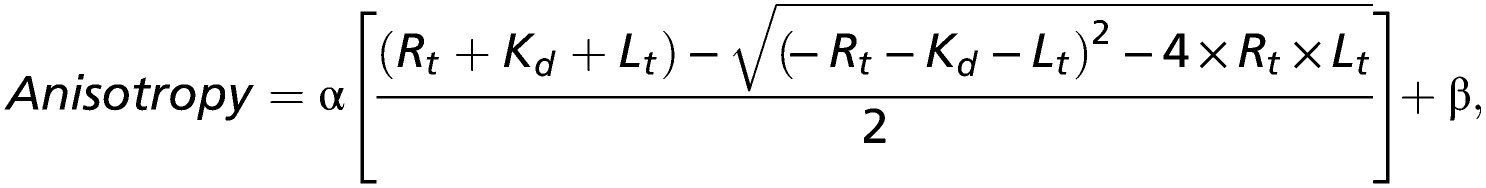 |
where R, L, and RL are concentrations of the free receptor and ligand and receptor-ligand complex, respectively. Rt and Lt are total receptor and ligand concentrations, Kd is the ligand-binding affinity, and α and β are a scaling factor and an offset factor, respectively.
Subunit Counting Using Single-Molecule Bleaching Method.
Oocytes were injected with varying ratios of TRIP8b and HCN2 mRNA. Oocytes were then kept under gentle agitation for 16–24 h at 18 °C before use. Before imaging, the oocytes were treated at room temperature in a solution containing 200 mM N-methyl-D-glucamine, 2 mM KCl, 1 mM MgCl2, 10 mM Hepes, 10 mM EGTA, 1 mg/mL hylauranidase, and 1.5 U/mL of neuraminidase for ∼20 min. The oocytes were then devitellinized manually using forceps and placed on a high refractive index coverslip with the animal pole (dark side) facing the objective.
TIRF movies were acquired using a Nikon TE2000-E microscope with a high numerical aperture objective (100×, 1.49 N.A.; Nikon) and the Evolve 512 EMCCD camera (Photometrics). Oocytes were illuminated with a 488-nm argon laser from Spectra Physics. An image stack of 800–1,200 frames was acquired at 30–50 Hz. The first five frames after opening of the laser shutter were averaged, and the background was subtracted using the rolling-ball method in Image J (National Institutes of Health). The image was then lowpass-filtered with a 2-pixel cutoff, and thresholding was applied to find connected regions of pixels that were above threshold. A region of interest (ROI) of 6 × 6 pixels was placed around the center of the spot. Spots smaller than 3 pixels and larger than 15 pixels were discarded manually. Finally, we measured the summed fluorescence intensity inside the 6 × 6 ROI and plotted the data versus time using programs written in MATLAB (MathWorks). Bleaching steps were then determined with an edge detector program written in MATLAB (Center for Computer Integrated Systems for Microscopy and Manipulation, available at http://www.cs.unc.edu/∼nanowork/cismm/download/) and then confirmed manually to catch mistakes and discard traces with erratic fluorescence signals. Approximately ∼20–60% of the data for each optical patch were discarded due to an erratic fluorescence signal.
Statistical Analysis.
Data were plotted as mean ± SEM. Student’s test was used to determine significance at P < 0.05.
Supplementary Material
Acknowledgments
X-ray diffraction data sets were collected at Advanced Light Source, and we thank the beamline personnel for their assistance. We also thank Michael Puljung and Yoni Haitin for critical reading of the manuscript and members of the W.N.Z. and D.M.C. laboratories for their technical assistance and helpful discussions. This work was supported by the Howard Hughes Medical Institute and National Institute of Neurological Disorders and Stroke Grants NS074053 (to W.N.Z.), NS074545 (to J.R.B.), NS064757 (to A.S.L.), and NS055995 and NS059934 (to D.M.C.). The Advanced Light Source is supported by the Director, Office of Science, Office of Basic Energy Sciences, of the Department of Energy under Contract DE-AC02-05CH11231.
Footnotes
The authors declare no conflict of interest.
*This Direct Submission article had a prearranged editor.
Data deposition: The structure factors reported in this paper have been deposited in the Protein Data Bank, www.pdb.org [PDB ID code 4EQF (TRIP8b-1aΔ1-205)].
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1201997109/-/DCSupplemental.
References
- 1.Buraei Z, Yang J. The ß subunit of voltage-gated Ca2+ channels. Physiol Rev. 2010;90:1461–1506. doi: 10.1152/physrev.00057.2009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Pongs O, Schwarz JR. Ancillary subunits associated with voltage-dependent K+ channels. Physiol Rev. 2010;90:755–796. doi: 10.1152/physrev.00020.2009. [DOI] [PubMed] [Google Scholar]
- 3.Trudeau MC, Zagotta WN. Calcium/calmodulin modulation of olfactory and rod cyclic nucleotide-gated ion channels. J Biol Chem. 2003;278:18705–18708. doi: 10.1074/jbc.R300001200. [DOI] [PubMed] [Google Scholar]
- 4.Craven KB, Zagotta WN. CNG and HCN channels: Two peas, one pod. Annu Rev Physiol. 2006;68:375–401. doi: 10.1146/annurev.physiol.68.040104.134728. [DOI] [PubMed] [Google Scholar]
- 5.Robinson RB, Siegelbaum SA. Hyperpolarization-activated cation currents: From molecules to physiological function. Annu Rev Physiol. 2003;65:453–480. doi: 10.1146/annurev.physiol.65.092101.142734. [DOI] [PubMed] [Google Scholar]
- 6.Wainger BJ, DeGennaro M, Santoro B, Siegelbaum SA, Tibbs GR. Molecular mechanism of cAMP modulation of HCN pacemaker channels. Nature. 2001;411:805–810. doi: 10.1038/35081088. [DOI] [PubMed] [Google Scholar]
- 7.Lörincz A, Notomi T, Tamás G, Shigemoto R, Nusser Z. Polarized and compartment-dependent distribution of HCN1 in pyramidal cell dendrites. Nat Neurosci. 2002;5:1185–1193. doi: 10.1038/nn962. [DOI] [PubMed] [Google Scholar]
- 8.Magee JC. Dendritic lh normalizes temporal summation in hippocampal CA1 neurons. Nat Neurosci. 1999;2:508–514. doi: 10.1038/9158. [DOI] [PubMed] [Google Scholar]
- 9.Williams SR, Stuart GJ. Site independence of EPSP time course is mediated by dendritic I(h) in neocortical pyramidal neurons. J Neurophysiol. 2000;83:3177–3182. doi: 10.1152/jn.2000.83.5.3177. [DOI] [PubMed] [Google Scholar]
- 10.Nolan MF, et al. A behavioral role for dendritic integration: HCN1 channels constrain spatial memory and plasticity at inputs to distal dendrites of CA1 pyramidal neurons. Cell. 2004;119:719–732. doi: 10.1016/j.cell.2004.11.020. [DOI] [PubMed] [Google Scholar]
- 11.Santoro B, Wainger BJ, Siegelbaum SA. Regulation of HCN channel surface expression by a novel C-terminal protein-protein interaction. J Neurosci. 2004;24:10750–10762. doi: 10.1523/JNEUROSCI.3300-04.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Piskorowski R, Santoro B, Siegelbaum SA. TRIP8b splice forms act in concert to regulate the localization and expression of HCN1 channels in CA1 pyramidal neurons. Neuron. 2011;70:495–509. doi: 10.1016/j.neuron.2011.03.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Lewis AS, et al. Deletion of the hyperpolarization-activated cyclic nucleotide-gated channel auxiliary subunit TRIP8b impairs hippocampal Ih localization and function and promotes antidepressant behavior in mice. J Neurosci. 2011;31:7424–7440. doi: 10.1523/JNEUROSCI.0936-11.2011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Santoro B, et al. TRIP8b splice variants form a family of auxiliary subunits that regulate gating and trafficking of HCN channels in the brain. Neuron. 2009;62:802–813. doi: 10.1016/j.neuron.2009.05.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Lewis AS, et al. Alternatively spliced isoforms of TRIP8b differentially control h channel trafficking and function. J Neurosci. 2009;29:6250–6265. doi: 10.1523/JNEUROSCI.0856-09.2009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Santoro B, et al. TRIP8b regulates HCN1 channel trafficking and gating through two distinct C-terminal interaction sites. J Neurosci. 2011;31:4074–4086. doi: 10.1523/JNEUROSCI.5707-10.2011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Han Y, et al. Trafficking and gating of hyperpolarization-activated cyclic nucleotide-gated channels are regulated by interaction with tetratricopeptide repeat-containing Rab8b-interacting protein (TRIP8b) and cyclic AMP at distinct sites. J Biol Chem. 2011;286:20823–20834. doi: 10.1074/jbc.M111.236125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Zolles G, et al. Association with the auxiliary subunit PEX5R/Trip8b controls responsiveness of HCN channels to cAMP and adrenergic stimulation. Neuron. 2009;62:814–825. doi: 10.1016/j.neuron.2009.05.008. [DOI] [PubMed] [Google Scholar]
- 19.Ulbrich MH, Isacoff EY. Subunit counting in membrane-bound proteins. Nat Methods. 2007;4:319–321. doi: 10.1038/NMETH1024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Ji W, et al. Functional stoichiometry of the unitary calcium-release-activated calcium channel. Proc Natl Acad Sci USA. 2008;105:13668–13673. doi: 10.1073/pnas.0806499105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Kawate T, Gouaux E. Fluorescence-detection size-exclusion chromatography for precrystallization screening of integral membrane proteins. Structure. 2006;14:673–681. doi: 10.1016/j.str.2006.01.013. [DOI] [PubMed] [Google Scholar]
- 22.Rossi AM, Taylor CW. Analysis of protein-ligand interactions by fluorescence polarization. Nat Protoc. 2011;6:365–387. doi: 10.1038/nprot.2011.305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Gatto GJ, Jr, Geisbrecht BV, Gould SJ, Berg JM. Peroxisomal targeting signal-1 recognition by the TPR domains of human PEX5. Nat Struct Biol. 2000;7:1091–1095. doi: 10.1038/81930. [DOI] [PubMed] [Google Scholar]
- 24.Williams CP, Stanley WA. Peroxin 5: A cycling receptor for protein translocation into peroxisomes. Int J Biochem Cell Biol. 2010;42:1771–1774. doi: 10.1016/j.biocel.2010.07.004. [DOI] [PubMed] [Google Scholar]
- 25.Gould SJ, Keller GA, Hosken N, Wilkinson J, Subramani S. A conserved tripeptide sorts proteins to peroxisomes. J Cell Biol. 1989;108:1657–1664. doi: 10.1083/jcb.108.5.1657. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Gatto GJ, Jr, et al. Correlating structure and affinity for PEX5:PTS1 complexes. Biochemistry. 2003;42:1660–1666. doi: 10.1021/bi027034z. [DOI] [PubMed] [Google Scholar]
- 27.Liman ER, Tytgat J, Hess P. Subunit stoichiometry of a mammalian K+ channel determined by construction of multimeric cDNAs. Neuron. 1992;9:861–871. doi: 10.1016/0896-6273(92)90239-a. [DOI] [PubMed] [Google Scholar]
- 28.Zagotta WN, Hoshi T, Aldrich RW. Gating of single Shaker potassium channels in Drosophila muscle and in Xenopus oocytes injected with Shaker mRNA. Proc Natl Acad Sci USA. 1989;86:7243–7247. doi: 10.1073/pnas.86.18.7243. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Collaborative Computational Project, Number 4 (1994) The CCP4 suite: programs for protein crystallography. Acta Crystallogr D Biol Crystallogr 50: 760–763. [DOI] [PubMed]
- 30.McCoy AJ. Solving structures of protein complexes by molecular replacement with Phaser. Acta Crystallogr D Biol Crystallogr. 2007;63:32–41. doi: 10.1107/S0907444906045975. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Adams PD, et al. PHENIX: A comprehensive Python-based system for macromolecular structure solution. Acta Crystallogr D Biol Crystallogr. 2010;66:213–221. doi: 10.1107/S0907444909052925. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Emsley P, Cowtan K. Coot: Model-building tools for molecular graphics. Acta Crystallogr D Biol Crystallogr. 2004;60:2126–2132. doi: 10.1107/S0907444904019158. [DOI] [PubMed] [Google Scholar]
- 33.Baker NA, Sept D, Joseph S, Holst MJ, McCammon JA. Electrostatics of nanosystems: Application to microtubules and the ribosome. Proc Natl Acad Sci USA. 2001;98:10037–10041. doi: 10.1073/pnas.181342398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.DiMaio F, et al. Improved molecular replacement by density- and energy-guided protein structure optimization. Nature. 2011;473:540–543. doi: 10.1038/nature09964. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.