Abstract
Despite structural knowledge of broadly neutralizing monoclonal antibodies (NMAbs) complexed to HIV-1 gp120 and gp41 envelope glycoproteins, virus inactivation mechanisms have been difficult to prove, in part because neutralization assays are complex and were previously not understood. Concordant with recent evidence that HIV-1 titers are determined by a race between entry of cell-attached virions and competing inactivation processes, we show that NMAb 2G12, which binds to gp120 N-glycans with α (1, 2)-linked mannose termini and inhibits replication after passive transfer into patients, neutralizes by slowing entry of adsorbed virions. Accordingly, apparent neutralization is attenuated when a kinetically competing virus inactivation pathway is blocked. Moreover, removing 2G12 from media causes its dissociation from virions coupled to accelerated entry and restored infectivity, demonstrating the reversibility of neutralization. A difference between 2G12 dissociation and infectivity recovery rates implies that the inhibited complexes at virus–cell junctions contain several 2G12’s that must dissociate before entry commences. Quantitative microscopy of 2G12 binding and dissociation from single virions and studies using a split CCR5 coreceptor suggest that 2G12 competitively inhibits interactions between gp120’s V3 loop and the tyrosine sulfate-containing CCR5 amino terminus, thereby reducing assembly of complexes that catalyze entry. These results reveal a unique reversible kinetic mechanism for neutralization by an antibody that binds near a critical V3 region in the glycan shield of gp120.
Few monoclonal antibodies broadly neutralize diverse HIV-1 isolates (NMAbs), and these have been extensively studied because they reveal conserved vulnerabilities in the viral envelope glycoproteins gp120 and gp41 (1, 2). Consequently, many previous studies used genetic and structural approaches to identify NMAb target epitopes and to enhance their presentations in vaccines (1, 3, 4). Although those investigations provided critical evidence about HIV-1 entry and about epitope shielding by glycans, variable loops, and conformational masking, neutralization mechanism(s) have been difficult to prove. For example, several NMAbs alter conformations of purified gp120 and/or inhibit its interactions with CD4 or coreceptors (5, 6), but it is unclear how or whether those effects contribute to neutralization. Indeed, it has been proposed that neutralization might ultimately require secondary processes such as NAb cross-linking, prevention of adsorption, or gp120 shedding (7–10). A central cause for uncertainty occurs because the infectivity assays required to identify and analyze neutralization have been poorly understood and give discrepant results using identical HIV-1s and NAbs (11–13). Consequently, it has become evident that elucidation of NMAb mechanisms requires improved understanding of the factors that influence infectivity assays (11–13).
HIV-1 envelope glycoproteins are trimers containing gp120 surface subunits that bind to CD4 and then to a coreceptor (usually CCR5 or CXCR4) and a gp41 transmembrane subunit that has a metastable conformation in native virions (14, 15). After CD4 binding, V3 loop regions of gp120 reduce their constraining hold on gp41 and move toward the trimer apex where they interact with coreceptors, thereby playing a pivotal role in controlling HIV-1 entry rates (16–19). These and additional conformational changes in gp120 enable gp41 to refold by a multistep process that fuses the viral and cellular membranes. The cell surface complexes that mediate membrane fusion (fusion complexes, FCs) are believed to contain several gp120–gp41 trimers, CD4s, and coreceptors (Fig. S1 and SI Discussion).
The most widely used neutralization assay, which has been recommended as a worldwide standard (13) and detects NAbs that contribute to host–virus coevolution in vivo (3, 20), uses TZM-bl(JC.53) cells that were derived from our HeLa-CD4/CCR5 cell clone JC.53 (21) by transfection with LTR–luciferase and LTR–β-galactosidase reporter plasmids (22). These reporters provide a facile means to monitor infections but have no effect on efficiencies of infection by diverse HIV-1 isolates or sensitivities to NAbs (23). This standard assay is done in the presence of DEAE-dextran, a polycation that increases virion adsorption and enhances HIV-1 titers ∼20-fold (20, 22, 24). We prefer to stain and count infected foci in JC.53 cultures using antibodies to HIV-1, which allows us to compare titers in cell clones having distinct amounts of wild-type or mutant CD4 and/or CCR5 (18, 21, 25). This provides information about entry inhibitor mechanisms and factors that limit entry of diverse HIV-1 isolates.
By systematically analyzing this assay system we found that HIV-1 infectivities depend on kinetics of adsorbed virion entry (24, 25). Many factors that reduce titers including limiting CD4 and/or coreceptor concentrations or using mutant coreceptors or coreceptor antagonists slow entry (25). Conversely, adaptive mutations centered in the V3 region of gp120 overcome these limitations and accelerate entry (18, 25). Recently, we found that the enhancement of titers caused by pretreating cultures with polycations results from the stabilization of virion adsorption, thus preventing a previously unknown competing process of rapid desorption and providing a longer opportunity for infection (24). These analyses demonstrate that titers in JC.53 cultures are limited by kinetic competition between entry and processes causing virus inactivation. Similar lines of evidence including the critical importance of V3, infectivity increases caused by adherence factors (26, 27), and inhibitions by coreceptor antagonists that slow entry, suggest that kinetic competition also limits HIV-1 replication in peripheral blood mononuclear cells (PBMCs) and other cultures. Incorporation of adhesion factors such as ICAM-1 into HIV-1 virions reduces neutralization by some NAbs in PBMC assays (28).
On the basis of this evidence, we tested the possibility that NAbs might function by slowing HIV-1 entry. We initially used NMAb 2G12, which binds to N-glycans with α (1, 2)-linked mannose termini at positions N295, N332, and N392 near the V3 loop in the “glycan shield” region of gp120 (1, 29, 30), because 2G12 strongly inhibits HIV-1 and SHIV in vivo (1, 31, 32). We report that 2G12 functions by this kinetic mechanism. Although another NMAb (b12) functions differently, kinetic competition also limits its potency.
Results
2G12 and Other Tested NAbs Neutralize Cell-Attached Virions.
It was reported that many NAbs including b12 and 2G12 neutralize by inhibiting HIV-1 adsorption (7, 10), a complex process that is influenced by many cell surface components other than CD4 and coreceptors (27). Those studies incubated HIV-1 with cells at 37 °C for 30–120 min before rinsing the cells extensively. However, we recently found that nearly all virions that adsorb onto cells in those assay conditions dissociate within ∼30 min (24), before CD4- and coreceptor-dependent entry steps induce gp41 conformational changes that stably anchor the virions (14). Consequently, the previous results could have been caused indirectly by inhibiting these entry steps, thereby enabling more dissociation. To test that possibility, we stabilized adsorption by pretreating cultures with DEAE-dextran before incubating with culture media containing HIV-1JRCSF for 30 min at 37 °C and then counting virions (24). Control virions and virions that were preincubated with 2G12 for 60 min at 37 °C bound equally onto cells (Table 1). Moreover, 2G12 efficiently neutralized the stably adsorbed virions, indicating that it inhibits a postadsorption process. Similar results occurred with b12, 2F5, and a pooled patient IgG that neutralized HIV-1SF162 (Table 1). When polycations were absent, control and 2G12-containing virions also adsorbed equally, implying that 2G12 did not alter the competing attachment or dissociation rates in those conditions (Table 1).
Table 1.
Neutralizing antibodies inhibit HIV-1 at a postadsorption step
| Antibody | Virions/cell (SEM) | Titer, FFU/0.1 mL (SD) | |
| 2G12 | − | 7.8 ± 0.47 | 1,700 ± 94.0 |
| + | 7.6 ± 0.48 | 21 ± 4.8 | |
| 2G12 | − | 2.4 ± 0.24 | 160 ± 29 |
| (− polycations) | + | 2.3 ± 0.22 | 0 ± 0 |
| b12 | − | 11.0 ± 0.54 | 760 ± 36.0 |
| + | 11.0 ± 0.63 | 73 ± 22.0 | |
| 2F5 | − | 5.9 ± 0.30 | 2,800 ± 320 |
| + | 6.2 ± 0.28 | 540 ± 200 | |
| HIV-IG | − | 2.1 ± 0.16 | 3,900 ± 630 |
| + | 1.1 ± 0.12 | 92 ± 14 |
HIV-1JRCSF was incubated in culture media in the presence (+) or absence (−) of NMAbs 2G12 (100 μg/mL), b12 (15 μg/mL), or 2F5 (100 μg/mL) for 1 h at 37 °C before adding onto JC.53 cultures and incubating an additional 30 min at 37 °C, after which cell-attached virions were fixed, permeabilized, and stained for Gag p24. The HIV-IG pool (50 μg/mL) from chronically infected patients was assayed identically using HIV-1SF162, which is efficiently neutralized by that antibody. The results for each NAb were obtained in independently performed experiments.
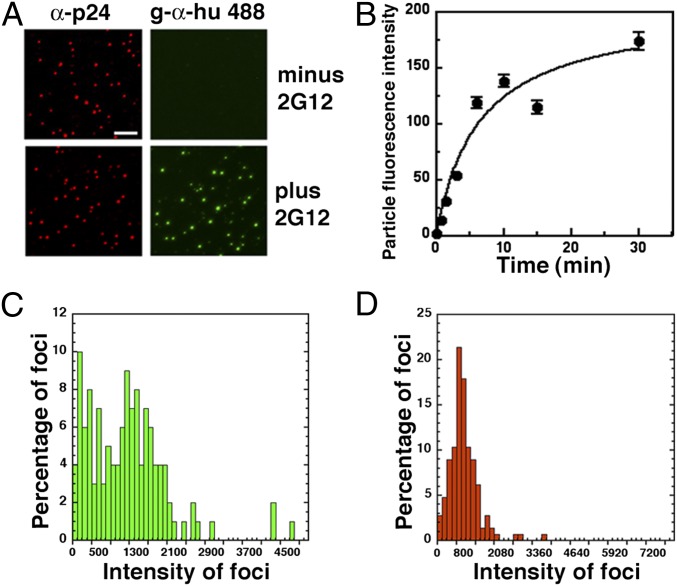
Immunofluorescence microscopy confirmed that 2G12 bound extensively onto cell-free HIV-1JRCSF at 37 °C (Fig. 1A). The preincubated virions were spinoculated onto DEAE-dextran–coated coverslips before fixation and immunostaining for p24 Gag and 2G12. Numbers of p24-containing fluorescent foci/μm2 were unaffected by 2G12 (20 ± 1.6 and 19 ± 1.0 virions/400 μm2 ± 2G12, respectively), suggesting that 2G12 had not caused cross-linking. Quantitative analyses of fluorescent intensities indicated that 2G12 bound rapidly and in diverse quantities onto HIV-1 (Fig. 1 B and C), confirming that virions contain different amounts of gp120 (33). Amounts of p24 per virion were more uniform (Fig. 1D).
Fig. 1.
Characterization of 2G12 binding to cell-free HIV-1JRCSF. (A) Virions incubated ± 50 μg/mL 2G12 for 1 h at 37 °C, were spinoculated onto DEAE-dextran coated coverslips, fixed, permeabilized, and then immunostained for Gag p24 and 2G12. Gag p24 was visualized using an anti-p24 MAb followed by goat antimouse Alexa-594, whereas 2G12 binding was detected using goat antihuman Alexa-488. (Scale bar, 5 μm.) (B) Kinetics of 2G12 virion binding. HIV-1 samples bound onto coverslips were incubated with 50 μg/mL 2G12 for different times at 37 °C, fixed, and stained for 2G12. Fluorescence intensities were averaged from at least 500 virions per time point. Error bars ± SEM. (C) Virion distributions of 2G12 and (D) Gag p24 quantities were generated using the binning function in Kaleidagraph.
2G12 Neutralizes by Slowing Entry.
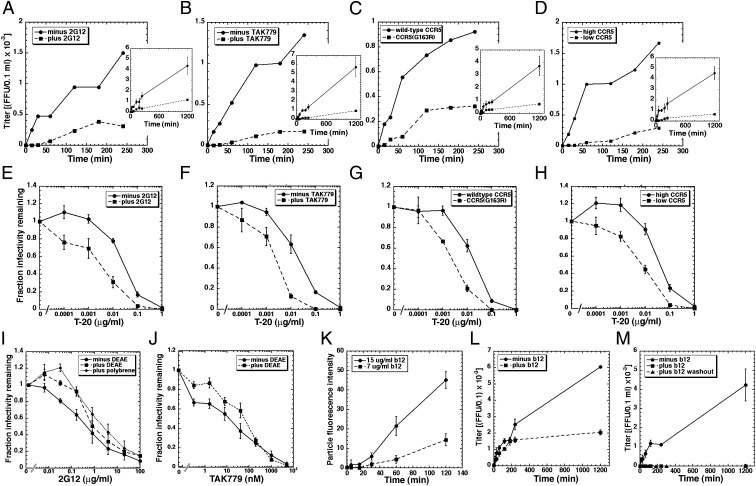
To analyze entry kinetics, we spinoculated HIV-1JRCSF at 4 °C onto cultures containing polycations, and after removing unadsorbed virions we warmed cultures to 37 °C. Entry was terminated at different times by adding an excess of the CCR5 antagonist TAK779. As reported previously (24), control virions entered in two phases, with a small burst within 1–2 h and the majority entering at a slower sustained rate for ∼20 h (Fig. 2A). When polycations are absent, slow virions nearly all dissociate and entry consequently appears to approach completion within ∼2 h (24). Thus, by stabilizing adsorption, we more accurately analyze entry kinetics of the entire virion cohort. Importantly, virions preincubated at 37 °C with 2G12 for 1 h before spinoculation commence entry only after a ∼40- to 50-min lag and their ensuing entry is also slowed compared with control virions (Fig. 2A). Identical slowing occurred with HIV-1SF162 (Fig. S2). Although 2G12 has no effect on numbers of adsorbed virions (Table 1), it reduces their rate and efficiency of entry.
Fig. 2.
Kinetic effects of NMAbs 2G12 and b12 on HIV-1 entry into cells. Unless otherwise noted, virions were preincubated with NMAbs for 1 h at 37 °C before spinoculating them onto cells at 4 °C and then warming to 37 °C in the same NMAb concentrations. (A) JC.53 cells ± 50 μg/mL 2G12. (B) JC.53 cells ± 250 nM TAK779. (C) Cells expressing low-affinity CCR5(G163R) compared with cells expressing a similar amount of wild-type CCR5. (D) JC.53 cells compared with JC.10 cells that express a low amount of wild-type CCR5. Insets show entry kinetics plotted to 1,200 min, whereas the larger graphs plot initial kinetics to 240 min. Single representative experiments (of at least three in total) are shown. Error bars are ± SD. (E–H) These same factors increase sensitivities to different concentrations of T-20. (I and J) Inactivations of HIV-1JRCSF as a function of 2G12 (I) or TAK779 (J) concentrations in cultures containing or lacking polycations DEAE-dextran or polybrene. Data in E–H, and I and J are averages of three independent experiments. Error bars are ± SEM. (K) Kinetics of b12 virion binding. Error bars ± SEM. (L) Entry kinetics of HIV-1 ± 15 μg/mL b12. (M) HIV-1 infection kinetics in which virions were preincubated ± 15 μg/mL b12 for 16 h at 37 °C. In one set of cultures, b12 was washed out after 60-min incubation with target JC.53 cells. Error bars are ± SD. Conditions in which b12 was present continuously (black squares) or was washed away (black triangles) yielded zero titers, indicating complete and irreversible inhibition by b12.
Long lags preceding entry followed by slowed steady-state entry rates occur whenever assembly of fusion-competent virus–CD4–CCR5 complexes is inhibited, for example by lowering functional CCR5 concentrations with partially inhibiting concentrations of TAK779 (Fig. 2B) or by using cells expressing CCR5(G163R), which has a reduced affinity for gp120–CD4 complexes (34) (Fig. 2C) or cells with low CCR5 concentrations (Fig. 2D). These similarities suggest that 2G12 inhibits assembly and lowers the steady-state concentration of FCs. Consistent with this interpretation and evidence that FCs mediate gp41 refolding (18, 25, 35), 2G12 and these other factors increase enfuvirtide (T-20) sensitivities (Fig. 2 E–H), suggesting that they prolong the cell surface lifetime of the T-20–susceptible gp41 conformational intermediate (SI Discussion).
Apparent neutralization by 2G12 is ∼10 times greater in the absence of polycations than in their presence (Fig. 2I). Other factors that slow entry including TAK779 are similarly less inhibiting when virions are stably adsorbed and have a longer time to infect (Fig. 2J) (24). Moreover, neither 2G12 (Table 1) nor TAK779 alter rates of HIV-1 adsorption or desorption in JC.53 cultures. By eliminating the kinetically competing process of virus dissociation, polycations enable slowed virions to infect more efficiently (24), suggesting that slowing entry is responsible for 2G12 neutralization.
2G12 Binds and Neutralizes Reversibly.
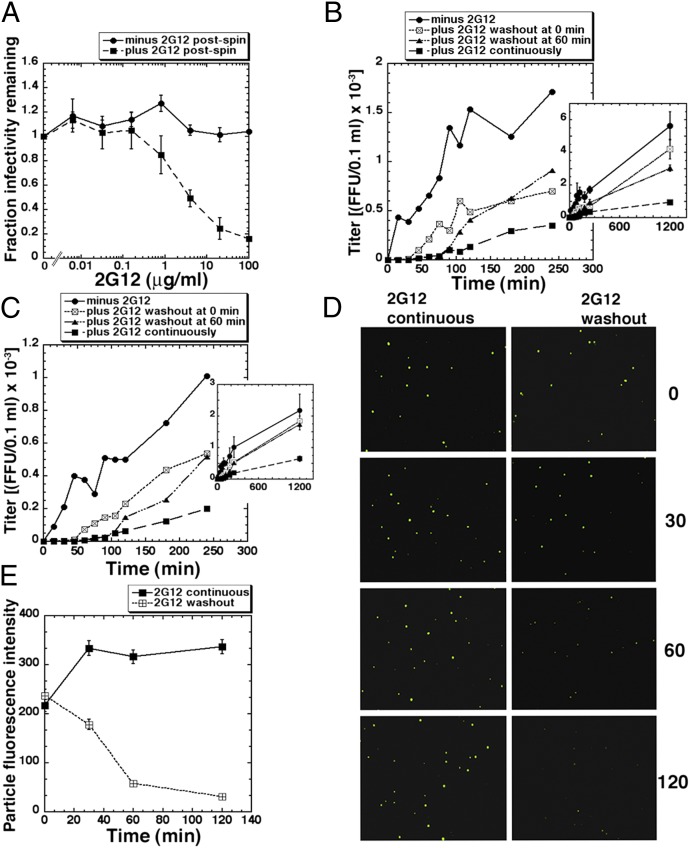
The fact that virions preincubated with 2G12 for 1 h enter JC.53 cells after a long lag, whereas control virions begin entry immediately after cultures are warmed to 37 °C, suggests that the 2G12-containing virions were all inhibited at the beginning of the kinetic assay (Fig. 2). However, in agreement with recent independent results (9), when 2G12 was removed from media at the onset of warming, neutralization was eliminated (Fig. 3A). Accordingly, virions preincubated with 2G12 for 1 h before spinoculating onto cells and warming to 37 °C entered cells slowly after a lag when the 2G12 concentration was maintained (Fig. 3B). In contrast, replacing 2G12-containing medium with control medium at the onset of warming greatly shortened the lag phase and induced accelerated entry and recovered infectivity. By 20 h postwarming, the titers in 2G12 and control cultures were not significantly different. Similar results occurred when 2G12 was removed after cultures had been at 37 °C for 1 h (Fig. 3B) and when virions were preincubated with 2G12 for 16 h before adsorbing onto cells (Fig. 3C). That 2G12 rapidly dissociates following its removal from media was confirmed by immunofluorescence microscopy (Fig. 3D) and by analyzing virion fluorescent intensities (Fig. 3E). These results substantiate the close relationship between 2G12 binding, neutralization, and entry slowdown.
Fig. 3.
Neutralization by 2G12 is reversible. (A) HIV-1JRCSF samples were preincubated with different concentrations of 2G12 for 1 h at 37 °C before spinoculating onto DEAE-treated JC.53 cultures at 4 °C. Cultures were warmed to 37 °C in fresh media in either the absence or presence of the same 2G12 concentrations (n = 3, error bars ± SEM). (B and C) HIV-1JRCSF preincubated for 1 h (B) or 16 h (C) at 37 °C ± 50 μg/mL 2G12 was spinoculated onto DEAE-dextran–containing JC.53 cells at 4 °C. After rinsing with fresh media containing the same concentrations of 2G12, entry kinetics were assayed. However, after 0 min or 60 min at 37 °C, 2G12 was washed out of some cultures. Single representative experiments (of three in total) are shown. Insets show data collected until 1,200 min (error bars ± SD). (D) 2G12 immunostaining of cell-free virions adsorbed onto DEAE-dextran–coated coverslips. Virions preincubated with 50 μg/mL 2G12 for 1 h at 37 °C were spinoculated at 4 °C and then fixed either immediately (0 min) or incubated at 37 °C for 30, 60, or 120 min either in the presence (Left) or absence (Right) of 2G12 before fixation. Representative CytoVision-derived images are shown. (E) Quantitation of 2G12 washout. Each data point represents the averaged fluorescence intensity from at least 500 virions. Error bars ± SEM.
Importantly, when 2G12 is removed from media, its dissociation begins immediately with first order kinetics (e.g., Fig. 3E), whereas infectivity recovers more slowly (Fig. 3 B and C). This delay is not caused by a slow rate of FC formation after 2G12 has dissociated, because entry begins rapidly in control JC.53 cultures (Figs. 2 and 3). We infer that the inhibited complexes at the virus–cell junctions contain several 2G12s that must dissociate before entry begins.
2G12 Inhibits CD4 and CCR5 Binding and Blocks a Coreceptor-Dependent Step in Entry.
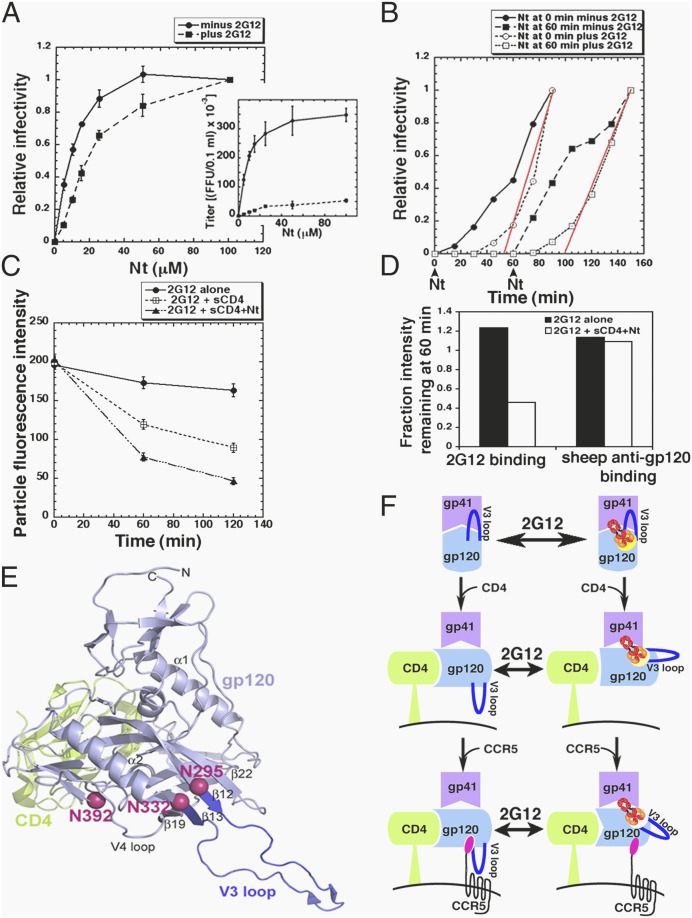
To learn how 2G12 inhibits FC assembly, we used HeLa-CD4/CCR5(Δ18) cells containing an amino terminally deleted coreceptor in conjunction with the adapted virus HIV-1JRCSF-Ad that efficiently infects these cells only when the tyrosine sulfate-containing amino terminal peptide Nt is present (18, 36). 2G12 increased the Nt concentration required for entry approximately threefold (Fig. 4A), consistent with evidence that it inhibits gp120–CCR5 interactions (6). Moreover, 2G12 delayed entry by ∼40 min regardless of whether Nt was added at the onset of warming or was withheld until cultures had been at 37 °C for 1 h (Fig. 4B). Thus, virions cannot progress past the 2G12 blockage until CCR5 is reconstituted, demonstrating that 2G12 interferes with a coreceptor-dependent entry step.
Fig. 4.
2G12 inhibits a coreceptor-dependent entry step. (A) HeLa-CD4/CCR5(Δ18) cells were infected in the presence of different concentrations of Nt with HIV-1JRCSF-Ad that had been pretreated for 1 h at 37 °C ± 4 μg/mL 2G12. Titers were normalized relative to maximum titers at 100 μM Nt (n = 3, error bars are ± SEM). (Inset) Nonnormalized data from a representative experiment (error bars are ± SD). (B) HIV-1JRCSF-Ad samples preincubated ± 2G12 as above were spinoculated at 4 °C onto HeLa-CD4/CCR5(Δ18) cultures that were then warmed to 37 °C. Nt (100 μM) was added at 0 or 60 min after warming, and entry kinetics were measured. To analyze the 2G12-dependent lag phases, titers for initial time points were normalized relative to titers obtained after 90 min exposures to Nt, by which time the lag phases were over and entry was progressing at a steady rate. Lines (red) were extrapolated through the linear data points to the time axis to graphically estimate 2G12-dependent lag times, which were 47 min for Nt added at 0 and 39 min when Nt was withheld for 60 min. A representative experiment of three is shown. Final titers after 20 h ± 2G12 were 2.2 × 104 and 2.3 x 105 for Nt added at 0 min and were 2.4 × 104 FFU/0.1 mL and 2.2 × 105 FFU/0.1 mL for Nt added at 60 min. (C) HIV-1JRCSF-Ad, pretreated for 1 h with 4 μg/mL 2G12, was spinoculated onto coverslips and incubated with ± 5 μg/mL sCD4 in the presence or absence of 100 μM Nt and in the continuous presence of 2G12 for 0, 60, or 120 min at 37 °C. The virions were then fixed, immunostained for 2G12, and virion fluorescence intensities were measured. Each data point represents at least 500 virions. A representative experiment of three is shown. (D) Gp120 shedding control. Virions that had been incubated with 2G12 ± sCD4 + Nt as in C were fixed after 60 min and 2G12 binding was detected by immunostaining, whereas virion-associated gp120 was detected using sheep anti-gp120. (E) Crystal structure [Protein Data Bank (PDB) ID 2B4C] of gp120 (light blue) bound with sCD4 (green) including N-glycans implicated in forming the 2G12 epitope (red spheres) and the V3 loop (dark blue). (Upper) Viral membrane; (Lower) cell membrane. (F) Proposed model of HIV-1 neutralization in which the NMAb 2G12 and CD4/CCR5 reciprocally compete for gp120 binding. Shown is one of three subunits of the HIV-1 glycoprotein trimer [gp120 (light blue), gp41 (purple)], which is oriented vertically with the target cell membrane (black curve) below. (Left, Top) In the absence of CD4 the V3 loop (dark blue) helps to stabilize gp120 association with gp41. (Left, Middle and Bottom) Upon CD4 and CCR5 binding sequential conformational transitions occur, including rotation of V3 toward the target cell/trimer apex and creation of a CCR5 amino terminal sulfotyrosine binding pocket at the V3 base [CD4 (green), CCR5 (black with magenta amino terminus), sulfotyrosine binding pocket (white outline surrounding the CCR5 amino terminus)]. (Right Top) The 2G12 epitope includes N-glycans located near the V3 base (bright yellow patch). (Right Middle and Bottom) HIV-1 with bound 2G12 resists conformational changes induced by CD4 and CCR5 binding, thereby reducing affinities of these receptors for gp120. Conversely, dissociation of 2G12 increases CD4 and CCR5 affinities, which reduces 2G12 affinity. (Decreases in 2G12 affinity are represented by a pale yellow patch for CD4 binding and white for CD4/CCR5). We believe the model system depicted is in equilibrium and that CD4 and CCR5 binding are reversible. However, for simplification, we have portrayed unidirectional CD4 and CCR5 binding.
Although 2G12 interferes with a CCR5-dependent entry step (Figs. 2E and 4B), conceivably it could also inhibit an earlier CD4-dependent process. To examine this, we spinoculated HIV-1JRCSF-Ad that had been preincubated for 1 h at 37 °C with a highly inhibiting 2G12 concentration onto coverslips in the continuous presence of 2G12. We then added sCD4 alone or sCD4 + Nt into some of the media and subsequently measured amounts of 2G12 on the virions. This analysis relied on our evidence that 2G12 binds reversibly and inhibits assembly of FCs containing CD4 and CCR5. Consequently, we anticipated that sCD4 and/or sCD4 + Nt would reciprocally displace 2G12, as confirmed in Fig. 4C. 2G12 dissociation was not caused by gp120 shedding (Fig. 4D and Fig. S3).
NMab b12 Slowly Inactivates HIV-1.
In striking contrast to 2G12, b12 binds HIV-1 slowly after a lag, suggestive of an avidity maturation process (Fig. 2K) and correspondingly only slowly terminates entry of cell-attached virions after a substantial delay (Fig. 2L). In contrast, virions that had been preincubated with the same b12 concentration for 16 h before spinoculating onto the cells were completely and irreversibly inactivated (Fig. 2M).
Discussion
2G12, which depends most critically on N-glycans with α (1, 2)-linked mannose termini at positions N295, N332, and N392 in the glycan shield of gp120JRCSF, has an unusual “crossover” domain-swapped heavy chain organization that pulls its arms together, thereby enabling it to bind several N-glycans on single gp120s with low entropic penalty (29, 30), consistent with our evidence that it does not cross-link virions (Fig. 1). Using a unique single virion immunofluorescence assay, we found that 2G12 binds reversibly to virions (Fig. 3) and that 2G12 binding and FC assembly are reciprocally competitive (Fig. 4C). According to that interpretation, 2G12 that is preadsorbed onto virions inhibits interactions with CD4 and CCR5, whereas the competitive FC assembly process that begins when JC.53 cells with adsorbed virions are warmed to 37 °C displaces some 2G12 and enables entry to commence after a lag required for this competition to reach equilibrium (Figs. 2–4). 2G12 competitively inhibits HIV-1JRCSF interactions with both CD4 and CCR5 (Fig. 4C), slows gp41 refolding (Fig. 2E), and blocks a coreceptor-dependent entry step (Fig. 4B).
These results are compatible with two hypotheses regarding 2G12 function. First, by holding or moving the target N-glycans, 2G12 might resist CD4 and CCR5 induced changes in their relative positions, and this allosteric constraint would reciprocally reduce CD4 and CCR5 affinities. Conversely, whenever preadsorbed 2G12 dissociated FC assembly could progress and the resulting shift in N-glycan positions would reciprocally reduce 2G12 affinity (Fig. 4C). Consistent with this constraint model, the CDRs in 2G12 complexes with oligosaccharides are ∼35 Å apart (29), whereas the bases of the targeted N-glycans in CD4–gp120 complexes are ∼7–24 Å apart (Fig. 4E). Another interpretation, diagrammed in Fig. 4F, derives from the fact that N295 and N332 occur directly adjacent to the disulfide bond on opposite borders of the V3 loop. This is a critical fulcrum for gp120 function because CD4 weakens V3 constraints on gp41 (19) and induces V3 movement toward the apex of the gp120 trimer (16, 17, 19) where it binds to the CCR5 amino terminus (37). By inhibiting these concerted processes, 2G12 would weaken CD4 and CCR5 affinities and retard entry. We favor this model because it relies entirely on prior evidence. Additionally, V3 mutations alter sensitivity to 2G12 (38). Moreover, residues 295 and 332 at the V3 border are also targets for autologous NAbs in newly infected individuals (3, 39), and terminal mannoses in glycans at N295, N301, and N332 plus nearby V3 amino acids in gp120JRCSF interact with NMAbs PGT127 and PGT128 from elite neutralizers (2, 4). Interestingly, these PGT NMAbs do not have domain swapped heavy chains and they apparently neutralize irreversibly by a slow process that may involve gp120 cross-linking (2). These apparent differences between the 2G12 and PGT NMAb neutralization mechanisms are intriguing because their epitopes overlap.
These results support other evidence that titers in TZM-bl(JC.53) cells (24, 25) and probably in other cultures (26–28) are determined by a race between entry and competing processes (24, 25). Indeed, 2G12 slows virions without inactivating them, yet it greatly reduces their titers. In contrast, b12 (Fig. 2 K–M), PGT, and MPER NMAbs neutralize HIV-1 irreversibly by slow processes that may require Env cross-linking and/or gp120 shedding (2, 9). As verified by our kinetic analyses of b12 (Fig. 2 L and M), slow neutralization mechanisms leave a window of opportunity for virus escape. Whereas previous studies using passively transferred NMAbs suggest that 2G12 may be more inhibitory than MPER NMAbs in vivo (32, 40), both b12 and 2G12 are highly protective (31, 40, 41). Further studies will be needed to learn whether information concerning NAb neutralization mechanisms and kinetics can be used to advance the HIV-1 vaccine effort.
Materials and Methods
Methods to isolate and grow HeLa-CD4/CCR5 cells including JC.53 cells were previously described (18, 21). The molecular clone pYK-JRCSF and HIV-1SF162 isolate were obtained from the National Institutes of Health AIDS Research and Reference Reagent Program, Division of AIDS, NIAID, NIH: pYK-JRCSF from Dr. Irvin SY Chen and Dr. Yoshio Koyanagi; HIV-1SF162 from Dr. Jay Levy. Virus stocks were prepared as described (21, 24). Details of methods used to characterize NMAb binding to HIV-1 and kinetic effects of NMAbs on HIV-1 entry, and methods to assess reversibility of 2G12 neutralization and to identify the entry steps inhibited by 2G12 are described in SI Materials and Methods.
Supplementary Material
Acknowledgments
We thank S. Kozak and L. Schwanemann for technical assistance, Chayne Piscitelli for initial help with structural modeling and for helpful suggestions, and Matthew Thayer and Leslie Smith for use of their CytoVision system. This work was supported by Grant R01 CA67358 from the National Institutes of Health.
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1109728109/-/DCSupplemental.
References
- 1.Pantophlet R, Burton DR. GP120: Target for neutralizing HIV-1 antibodies. Annu Rev Immunol. 2006;24:739–769. doi: 10.1146/annurev.immunol.24.021605.090557. [DOI] [PubMed] [Google Scholar]
- 2.Pejchal R, et al. A potent and broad neutralizing antibody recognizes and penetrates the HIV glycan shield. Science. 2011;334:1097–1103. doi: 10.1126/science.1213256. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Tang H, et al. Epitopes immediately below the base of the V3 loop of gp120 as targets for the initial autologous neutralizing antibody response in two HIV-1 subtype B-infected individuals. J Virol. 2011;85:9286–9299. doi: 10.1128/JVI.02286-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Walker LM, et al. Protocol G Principal Investigators Broad neutralization coverage of HIV by multiple highly potent antibodies. Nature. 2011;477:466–470. doi: 10.1038/nature10373. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Brower ET, Schön A, Klein JC, Freire E. Binding thermodynamics of the N-terminal peptide of the CCR5 coreceptor to HIV-1 envelope glycoprotein gp120. Biochemistry. 2009;48:779–785. doi: 10.1021/bi8021476. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Trkola A, et al. CD4-dependent, antibody-sensitive interactions between HIV-1 and its co-receptor CCR-5. Nature. 1996;384:184–187. doi: 10.1038/384184a0. [DOI] [PubMed] [Google Scholar]
- 7.Beirnaert E, De Zutter S, Janssens W, van der Groen G. Potent broad cross-neutralizing sera inhibit attachment of primary HIV-1 isolates (groups M and O) to peripheral blood mononuclear cells. Virology. 2001;281:305–314. doi: 10.1006/viro.2000.0802. [DOI] [PubMed] [Google Scholar]
- 8.Klasse PJ, Sattentau QJ. Occupancy and mechanism in antibody-mediated neutralization of animal viruses. J Gen Virol. 2002;83:2091–2108. doi: 10.1099/0022-1317-83-9-2091. [DOI] [PubMed] [Google Scholar]
- 9.Ruprecht CR, et al. MPER-specific antibodies induce gp120 shedding and irreversibly neutralize HIV-1. J Exp Med. 2011;208:439–454. doi: 10.1084/jem.20101907. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Ugolini S, et al. Inhibition of virus attachment to CD4+ target cells is a major mechanism of T cell line-adapted HIV-1 neutralization. J Exp Med. 1997;186:1287–1298. doi: 10.1084/jem.186.8.1287. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Fenyö EM, et al. International network for comparison of HIV neutralization assays: The NeutNet report. PLoS ONE. 2009;4:e4505. doi: 10.1371/journal.pone.0004505. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Mann AM, et al. HIV sensitivity to neutralization is determined by target and virus producer cell properties. AIDS. 2009;23:1659–1667. doi: 10.1097/QAD.0b013e32832e9408. [DOI] [PubMed] [Google Scholar]
- 13.Polonis VR, et al. Recent advances in the characterization of HIV-1 neutralization assays for standardized evaluation of the antibody response to infection and vaccination. Virology. 2008;375:315–320. doi: 10.1016/j.virol.2008.02.007. [DOI] [PubMed] [Google Scholar]
- 14.Eckert DM, Kim PS. Mechanisms of viral membrane fusion and its inhibition. Annu Rev Biochem. 2001;70:777–810. doi: 10.1146/annurev.biochem.70.1.777. [DOI] [PubMed] [Google Scholar]
- 15.Pancera M, et al. Structure of HIV-1 gp120 with gp41-interactive region reveals layered envelope architecture and basis of conformational mobility. Proc Natl Acad Sci USA. 2010;107:1166–1171. doi: 10.1073/pnas.0911004107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Liu J, Bartesaghi A, Borgnia MJ, Sapiro G, Subramaniam S. Molecular architecture of native HIV-1 gp120 trimers. Nature. 2008;455:109–113. doi: 10.1038/nature07159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Moscoso CG, et al. Quaternary structures of HIV Env immunogen exhibit conformational vicissitudes and interface diminution elicited by ligand binding. Proc Natl Acad Sci USA. 2011;108:6091–6096. doi: 10.1073/pnas.1016113108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Platt EJ, Durnin JP, Shinde U, Kabat D. An allosteric rheostat in HIV-1 gp120 reduces CCR5 stoichiometry required for membrane fusion and overcomes diverse entry limitations. J Mol Biol. 2007;374:64–79. doi: 10.1016/j.jmb.2007.09.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Xiang SH, et al. A V3 loop-dependent gp120 element disrupted by CD4 binding stabilizes the human immunodeficiency virus envelope glycoprotein trimer. J Virol. 2010;84:3147–3161. doi: 10.1128/JVI.02587-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Wei X, et al. Antibody neutralization and escape by HIV-1. Nature. 2003;422:307–312. doi: 10.1038/nature01470. [DOI] [PubMed] [Google Scholar]
- 21.Platt EJ, Wehrly K, Kuhmann SE, Chesebro B, Kabat D. Effects of CCR5 and CD4 cell surface concentrations on infections by macrophagetropic isolates of human immunodeficiency virus type 1. J Virol. 1998;72:2855–2864. doi: 10.1128/jvi.72.4.2855-2864.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Wei X, et al. Emergence of resistant human immunodeficiency virus type 1 in patients receiving fusion inhibitor (T-20) monotherapy. Antimicrob Agents Chemother. 2002;46:1896–1905. doi: 10.1128/AAC.46.6.1896-1905.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Platt EJ, Bilska M, Kozak SL, Kabat D, Montefiori DC. Evidence that ecotropic murine leukemia virus contamination in TZM-bl cells does not affect the outcome of neutralizing antibody assays with human immunodeficiency virus type 1. J Virol. 2009;83:8289–8292. doi: 10.1128/JVI.00709-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Platt EJ, Kozak SL, Durnin JP, Hope TJ, Kabat D. Rapid dissociation of HIV-1 from cultured cells severely limits infectivity assays, causes the inactivation ascribed to entry inhibitors, and masks the inherently high level of infectivity of virions. J Virol. 2010;84:3106–3110. doi: 10.1128/JVI.01958-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Platt EJ, Durnin JP, Kabat D. Kinetic factors control efficiencies of cell entry, efficacies of entry inhibitors, and mechanisms of adaptation of human immunodeficiency virus. J Virol. 2005;79:4347–4356. doi: 10.1128/JVI.79.7.4347-4356.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Losier M, Fortin JF, Cantin R, Bergeron MG, Tremblay MJL. Virion-bound ICAM-1 and activated LFA-1: A combination of factors conferring resistance to neutralization by sera from human immunodeficiency virus type 1-infected individuals independently of the disease status and phase. Clin Immunol. 2003;108:111–118. doi: 10.1016/s1521-6616(03)00093-7. [DOI] [PubMed] [Google Scholar]
- 27.Ugolini S, Mondor I, Sattentau QJ. HIV-1 attachment: Another look. Trends Microbiol. 1999;7:144–149. doi: 10.1016/s0966-842x(99)01474-2. [DOI] [PubMed] [Google Scholar]
- 28.Rizzuto CD, Sodroski JG. Contribution of virion ICAM-1 to human immunodeficiency virus infectivity and sensitivity to neutralization. J Virol. 1997;71:4847–4851. doi: 10.1128/jvi.71.6.4847-4851.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Calarese DA, et al. Antibody domain exchange is an immunological solution to carbohydrate cluster recognition. Science. 2003;300:2065–2071. doi: 10.1126/science.1083182. [DOI] [PubMed] [Google Scholar]
- 30.Scanlan CN, et al. The broadly neutralizing anti-human immunodeficiency virus type 1 antibody 2G12 recognizes a cluster of alpha1—>2 mannose residues on the outer face of gp120. J Virol. 2002;76:7306–7321. doi: 10.1128/JVI.76.14.7306-7321.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Hessell AJ, et al. Broadly neutralizing human anti-HIV antibody 2G12 is effective in protection against mucosal SHIV challenge even at low serum neutralizing titers. PLoS Pathog. 2009;5:e1000433. doi: 10.1371/journal.ppat.1000433. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Trkola A, et al. Delay of HIV-1 rebound after cessation of antiretroviral therapy through passive transfer of human neutralizing antibodies. Nat Med. 2005;11:615–622. doi: 10.1038/nm1244. [DOI] [PubMed] [Google Scholar]
- 33.Poignard P, et al. Heterogeneity of envelope molecules expressed on primary human immunodeficiency virus type 1 particles as probed by the binding of neutralizing and nonneutralizing antibodies. J Virol. 2003;77:353–365. doi: 10.1128/JVI.77.1.353-365.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Siciliano SJ, et al. A critical site in the core of the CCR5 chemokine receptor required for binding and infectivity of human immunodeficiency virus type 1. J Biol Chem. 1999;274:1905–1913. doi: 10.1074/jbc.274.4.1905. [DOI] [PubMed] [Google Scholar]
- 35.Reeves JD, et al. Sensitivity of HIV-1 to entry inhibitors correlates with envelope/coreceptor affinity, receptor density, and fusion kinetics. Proc Natl Acad Sci USA. 2002;99:16249–16254. doi: 10.1073/pnas.252469399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Farzan M, et al. Tyrosine-sulfated peptides functionally reconstitute a CCR5 variant lacking a critical amino-terminal region. J Biol Chem. 2002;277:40397–40402. doi: 10.1074/jbc.M206784200. [DOI] [PubMed] [Google Scholar]
- 37.Huang CC, et al. Structures of the CCR5 N terminus and of a tyrosine-sulfated antibody with HIV-1 gp120 and CD4. Science. 2007;317:1930–1934. doi: 10.1126/science.1145373. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Chaillon A, et al. The V1V2 domain and an N-linked glycosylation site in the V3 loop of the HIV-1 envelope glycoprotein modulate neutralization sensitivity to the human broadly neutralizing antibody 2G12. J Virol. 2011;85:3642–3648. doi: 10.1128/JVI.02424-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Walker LM, et al. Rapid development of glycan-specific, broad, and potent anti-HIV-1 gp120 neutralizing antibodies in an R5 SIV/HIV chimeric virus infected macaque. Proc Natl Acad Sci USA. 2011;108:20125–20129. doi: 10.1073/pnas.1117531108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Balazs AB, et al. Antibody-based protection against HIV infection by vectored immunoprophylaxis. Nature. 2012;481:81–84. doi: 10.1038/nature10660. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Parren PW, et al. Antibody protects macaques against vaginal challenge with a pathogenic R5 simian/human immunodeficiency virus at serum levels giving complete neutralization in vitro. J Virol. 2001;75:8340–8347. doi: 10.1128/JVI.75.17.8340-8347.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.