Abstract
To introduce DNA into mitochondria efficiently, we fused adenoassociated virus capsid VP2 with a mitochondrial targeting sequence to carry the mitochondrial gene encoding the human NADH ubiquinone oxidoreductase subunit 4 (ND4). Expression of WT ND4 in cells with the G11778A mutation in ND4 led to restoration of defective ATP synthesis. Furthermore, with injection into the rodent eye, human ND4 DNA levels in mitochondria reached 80% of its mouse homolog. The construct expressed in most inner retinal neurons, and it also suppressed visual loss and optic atrophy induced by a mutant ND4 homolog. The adenoassociated virus cassette accommodates genes of up to ∼5 kb in length, thus providing a platform for introduction of almost any mitochondrial gene and perhaps even allowing insertion of DNA encompassing large deletions of mtDNA, some associated with aging, into the organelle of adults.
Mitochondrial dysfunction underlies a large number of diseases as well as aging (1–4). Many of these conditions arise from point mutations or deletions in the mitochondrial genome. Others can appear from mutations in nuclear genes that are required for mtDNA replication, oxidative phosphorylation (OXPHOS), and structure of the organelle (1, 5, 6). There is currently no effective remedy for any of these disorders, especially for those caused by mutations in the mitochondrial genome (5, 7–9). To date, more than 200 pathogenic mtDNA mutations have been identified that are associated with a large spectrum of clinical phenotypes, many of which are fatal (10, 11).
Most mutations associated with early blindness do not significantly alter life span. A connection between Leber’s hereditary optic neuropathy (LHON) and mtDNA was firmly established in 1988, when Wallace et al. (12) reported a homoplasmic nucleotide transition from guanosine to adenosine at position 11778 that resulted in an arginine-to-histidine substitution in ubiquinone oxidoreductase (NADH) subunit 4 (ND4) of complex I. Most LHON cases are associated with mutations in one of three mitochondrial genes that encode subunits of complex I of the mitochondrial respiratory chain (13, 14). This enzyme contains 7 subunits encoded by mtDNA that are intimately associated with the inner mitochondrial membrane and 35 subunits encoded by nuclear DNA and imported into the organelle (15). Approximately half of all patients with LHON have the G11778A mutation, making it an attractive target for mtDNA gene transfer.
The double membrane and autonomous gene expression system of mitochondria pose challenges to the functional complementation of mutant genes within the organelle. Although there are several reports describing DNA import into mitochondria by means of nucleic acid-protein conjugation, electroporation, or bacterial conjugation, there remain significant obstacles to correcting any mtDNA mutation in living animals: There is no delivery system for DNA into mitochondria, and there are no selectable markers for mitochondrial transformation (16–21). Still, there is some recent evidence for recombination between mtDNA molecules (22), which could enable gene replacement. Although a natural DNA mitochondrial import has been reported in isolated rat liver mitochondria (23) and in plants (24), exogenous DNA cannot be transfected into mitochondria by conventional techniques (25, 26). Therefore, the most important challenge at present is the development of procedures for introducing DNA into mitochondria, ideally in intact cells or living tissues.
In this report, we describe redirection of the adenoassociated virus (AAV) virion to mitochondria by addition of a mitochondrial targeting sequence (MTS) to the capsid to deliver the WT human ND4 gene in the mitochondrial genetic code for rescue of the defective respiration of LHON G11778A cybrids. We also report in vivo testing for the presence and expression of the human ND4 in the adult rodent visual system, where it prevented visual loss and atrophy of the optic nerve induced by the mutant ND4 allele.
Results
MTS-Modified AAV for Import of DNA into Mitochondria.
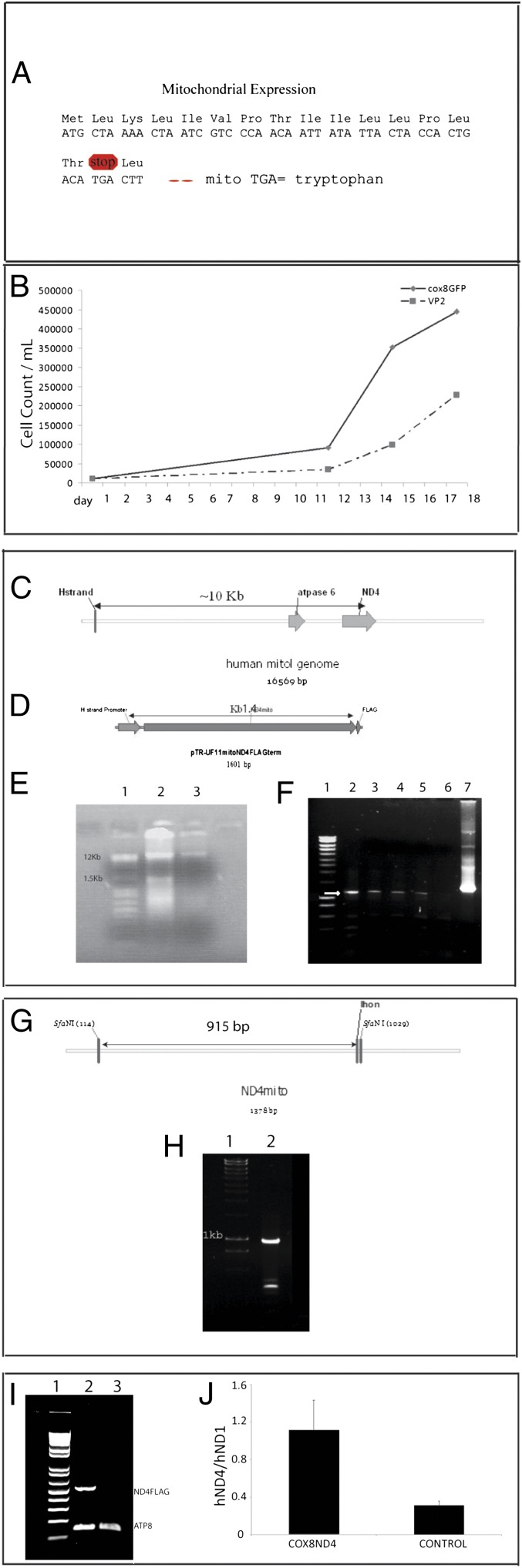
We asked whether the addition of an MTS to an AAV capsid protein could redirect import of AAV to mitochondria so as to deliver its DNA cargo, the ND4 subunit gene in this case, directly to the organelle. Mitochondrial ND4 can only be translated in the mitochondria and not on cytoplasmic ribosomes, because the TGA codon at amino acid 16, which encodes for tryptophan in the mitochondria, is a stop codon in the nuclear genetic code (Fig. 1A).
Fig. 1.
Nucleic acid analysis of infected LHON cell cultures. (A) Illustration shows that amino acids with their respective codons for human ND4 in the mitochondrial genetic code would result in a stop codon at amino acid 16 in the nuclear genetic code but not in the mitochondria, where this same TGA codon encodes for the amino acid tryptophan. For ND4 expression in the cytoplasm, the TGA codon must be changed to TGG, which encodes for tryptophan in the nucleus (allotopic expression). (B) Following 5 d of selection in restrictive media and 2 wk of growth in normal media, homoplasmic G11778A cybrids infected with WT ND4 packaged with COX8GFP VP2 exhibited better growth in standard glucose media compared with those infected with AAV VP2 lacking the MTS. (C) With a sense PCR primer flanking the H-strand promoter and antisense primer flanking the 3′end of ND4, the amplificant of endogenous mouse mtDNA would be greater than 10 kb. (D) In contrast, with our construct, amplification of the H-strand promoter adjacent to human ND4 would be ∼1.4 kb. (E) Agarose gel electrophoresis of mtDNA isolated from COX8GFP VP2 (lane 2) or VP2 transfected G11778A cells (lane 3) shows that in addition to endogenous mtDNA (16 kb), a smaller band likely representing our mitochondria-targeted HSP-ND4FLAG was only seen with COX8GFP VP2 transfection. Lane 1 is the molecular weight standards. (F) With an extension time of 2 min, PCR of mtDNA isolated from homoplasmic G11778A cells infected with ND4 delivered by COX8 VP2 revealed a 1.4-kb band (arrow) several weeks after growth in restrictive media (lanes 2–5). No PCR product was obtained from the VP2-delivered ND4 lacking the MTS (lane 6). PCR amplification of ND4 plasmid DNA is shown in lane 7. Lane 1 is the molecular weight standards. (G) Illustration of WT ND4 showing the two SfaN1 restriction sites that should cut twice, with the largest digestion product being 915 bp. With the second SfaN1 site lost in mutant G11778A ND4, a larger 1.2-kb fragment would be generated by digestion with SfaN1. (H) SfaN1 digestion of the above PCR-amplified HSP-ND4 DNA revealed a 915-bp band indicating the presence of two SfaN1 sites in the PCR product (lane 2). Lane 1 is the molecular weight marker. (I) RT-PCR of RNA isolated from mitochondrial pellets performed with the forward primer nested in ND4 and the reverse primer nested in the FLAG epitope revealed the expected 500-bp band only in LHON cybrids that received the COX8 VP2 AAV (lane 2) but not in RNA isolated from cybrids that received the VP2 construct lacking the MTS (lane 3). Expression of the endogenous ATP8 gene is seen in both samples (150-bp bands in lanes 2 and 3). Lane 1 is the molecular weight standards. (J) Bar plot of quantitative RT-PCR results comparing transcription of the human ND4 (hND4) introduced by the MTS AAV with the control shows an almost fourfold elevation. The ND4 mRNA level was indicated as a ratio to that of endogenous mitochondrial human ND1 gene (hND1).
AAV is a single-stranded (ss) DNA parvovirus with a 4.7-kb genome and a particle diameter of 21 nm. The AAV genome consists of two genes, REP and CAP, which encode the nonstructural REP proteins (REP78, REP68, REP52, and REP40) and the capsid proteins (VP1, VP2, and VP3), respectively. Flanking these two genes are the inverted terminal repeat (ITR) sequences that provide all the cis-acting sequence required for replication, packaging, and integration (27).
The VP2 capsid protein of AAV serotype 2 can tolerate large peptide insertions at the N terminus (28). We directed the AAV virion toward the mitochondria by addition of the 23-aa cytochrome oxidase subunit 8 (COX8) presequence fused in-frame to the N terminus of GFP into the AAV2 capsid ORF. The COX8GFP coding sequence was ligated into AAV capsid VP2 gene at a unique Eag1 site in residue 138 within the VP1-VP2 overlap. This insertion did not substantially reduce infectivity of the recombinant AAV (29), because we achieved relatively high titers [1.08 × 1011 vector genomes (VG)/mL] for this presumably mitochondria-targeted AAV. It should be noted that VP2 is not essential for the infectivity of AAV and that in the AAV capsid, only 3 of 60 subunits are VP2. To direct expression of exogenous WT human ND4 in the organelle, we linked a mitochondrial promoter, the heavy strand promoter (HSP), to the N terminus of ND4 cDNA to an AAV backbone (pTR-UF11) that contained AAV2 ITRs. For immunodetection, we linked a FLAG epitope to the C terminus of ND4.
MTS AAV Delivers WT Human ND4 DNA to Cultured G11778A Mutant Cells.
The resulting plasmid pTR-UF11-ND4FLAG packaged into the COX8 MTS AAV capsid was delivered to cybrid cell lines containing mitochondria with 100% mutated human G11778A ND4 (29). This same ND4 construct packaged in AAV without the MTS on VP2 was used as a control. Two days following infection, we selected for infected G11778A cybrid cells expressing the MTS-delivered WT ND4 using glucose-free galactose media. Cybrids without rescue die in this restrictive media (29, 30). Following 5 d of selection in restrictive media, cells were amplified for 2 wk in normal glucose media (Fig. 1B); the mitochondria were then isolated, and their DNA was extracted.
The mitochondrial genome is ∼16 kb (Fig. 1C). Our HSP-ND4FLAG construct is ∼1.5 kb (Fig. 1D). Agarose gel electrophoresis of the extracted mtDNA showed that in addition to the endogenous mtDNA (∼16 kb), a smaller band (∼1.5 kb), comparable to the size of HSP-ND4FLAG, was seen with COX8 AAV-delivered ND4 (Fig. 1E, gel lane 2) that was not observed in cells infected with AAV lacking the MTS (Fig. 1E, gel lane 3). This finding suggests that episomal HSP-ND4FLAG DNA was present in mitochondria of the MTS AAV-infected cybrids but was absent in mitochondria of cybrids infected with VP2 AAV without the MTS.
To test these DNA preparations for the WT ND4 allele delivered by the mitochondria-targeted AAV vector, we designed PCR primers flanking the H-strand promoter and flanking ND4. Using these primers, amplification of endogenous mtDNA would produce a gene product >10 kb (Fig. 1C). In contrast, our WT human ND4 with H-strand promoter should produce a 1.4-kb product (Fig. 1D). Using an extension time of 2 min to avoid amplifying the endogenous mtDNA, we tested for the imported WT ND4 in mitochondria. PCR amplification of the mtDNA, isolated from cells containing ND4 delivered with COX8 VP2, resulted in a product of 1.4 kb expected from the HSP adjacent to ND4, that is, the AAV-delivered gene (Fig. 1F, gel lanes 2–5). This product was not observed in cells infected with AAV lacking the MTS (Fig. 1F, gel lane 6).
This finding suggests that our modified AAV vector delivered the WT ND4 allele to the mitochondria. If so, a SfaNI digest on the PCR product should cut twice, with the largest digestion product being 915 bp (Fig. 1G), because there are two SfaNI sites in WT ND4. With the second site lost in mutant G11778A ND4, a 1.2-kb fragment would be seen (12). The SfaNI digest revealed a 915-bp band, indicating the presence of two SfaNI sites in the PCR product (Fig. 1H, gel lane 2). Thus, the WT ND4 allele was delivered to the mitochondria by the MTS-modified AAV.
Transcription of this template was tested next. RT-PCR of RNA isolated from mitochondrial pellets performed with the forward primer nested in ND4 and the reverse primer nested in the FLAG epitope revealed the expected 500-bp band only in cybrids that received the COX8 VP2 (Fig. 1I, gel lane 2) but not from RNA isolated from cybrids that received the VP2 AAV construct lacking the MTS (Fig. 1I, gel lane 3). Transcription of an endogenous mitochondrial gene (ATP8, 150-bp band) is apparent in both samples (Fig. 1I, gel lanes 2 and 3). Quantitative RT-PCR revealed that the levels of ND4 mRNA introduced by the MTS-targeted AAV were almost fourfold greater than the transcription of the same gene in control cells, which was indicated as a ratio to mRNA levels of the endogenous mitochondrial ND1 complex I subunit gene (Fig. 1J). These findings confirmed transcription of the MTS-delivered WT ND4FLAG allele in cultured cells.
MTS AAV Capsid Localizes Within Mitochondria, Where the Delivered WT Human ND4 Is Translated in Cultured Cells.
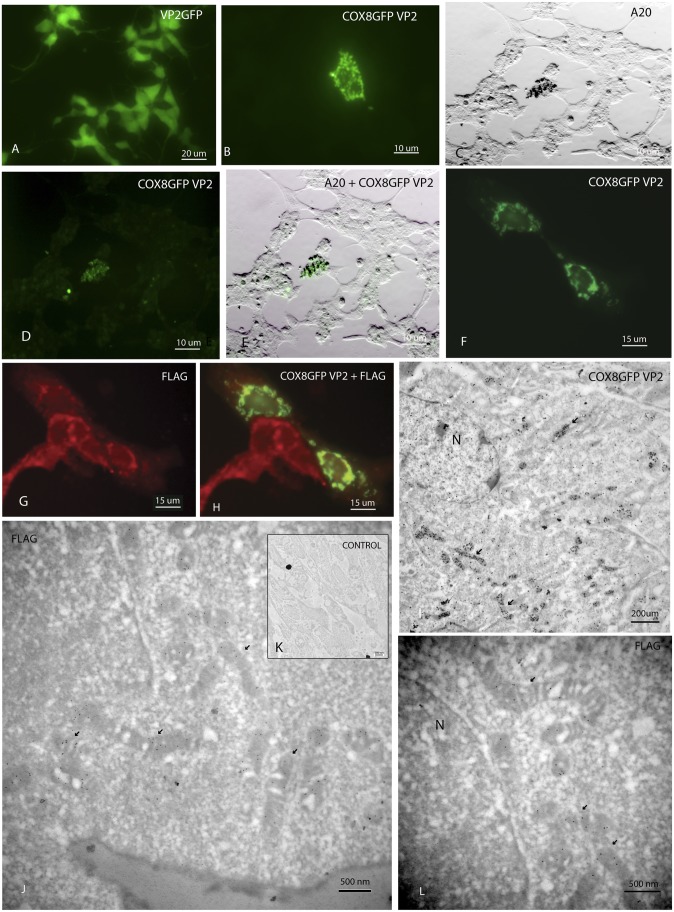
Fluorescence microscopy of cells infected with VP2 AAV containing GFP without the MTS revealed nuclear and cytoplasmic expression of GFP that did not associate with mitochondria (Fig. 2A). In contrast, cells infected with the COX8GFP VP2 MTS AAV revealed punctate and perinuclear expression of GFP (Fig. 2B) suggestive of mitochondrial localization. An antibody (A20) recognized fully assembled AAV virions (Fig. 2C) in a COX8GFP fluorescent cell (Fig. 2D) that was colabeled by silver-enhanced A20 immunogold (Fig. 2E). This finding suggested that the MTS AAV virion was targeted to the mitochondria.
Fig. 2.
Microscopy of infected cell cultures. (A) Fluorescence microscopy of 293T cells infected with VP2 AAV containing GFP without the MTS reveals nuclear and cytoplasmic expression of GFP. (B) Cells infected with the COX8GFP VP2 MTS AAV show punctate and perinuclear expression of GFP. A light micrograph (C) shows that the A20 antibody recognized fully assembled AAV virions in a COX8GFP fluorescent cell (D) colabeled by silver-enhanced A20 immunogold (E). A neuronal cell line, with large mitochondria containing G11778A mutated mtDNA, shows perinuclear COX8GFP (F) and perinuclear ND4FLAG (G). (H) Colocalization of COX8GFP and ND4FLAG is seen in two of these cells. (I) Transmission electron micrograph of cells infected with COX8GFP VP2 AAV containing the WT mitochondrial ND4 DNA reveals that silver-enhanced GFP immunogold was present in many mitochondria (arrows). GFP immunogold was also evident in the nucleus (N) of the cell to which the virus typically translocates. (J) In a COX8GFP VP2 AAV mitochondrial ND4-infected cell, FLAG-tagged immunogold (arrows) was clearly evident within mitochondria (arrows). (K) Control stained with only the secondary antibody, conjugated to immunogold, shows no background. (L) In another MTS AAV-infected cell, perinuclear mitochondria contained the ND4FLAG immunogold particles. N, nucleus.
Next, we looked for translation of the MTS-delivered WT human ND4 gene within the organelle. Because the TGA codon at amino acid 16 that encodes for mitochondrial tryptophan is a stop codon in the nucleus, mitochondrial ND4 cannot be synthesized outside the mitochondria (Fig. 1A). Three days following infection with the MTS AAV, a neuronal cell line (31) with large mitochondria containing G11778A-mutated mtDNA showed perinuclear COX8GFP capsid (Fig. 2F) and perinuclear ND4FLAG synthesis (Fig. 2G) with colocalization in two cells (Fig. 2H). The adjacent ND4FLAG-expressing cells without VP2 COX8GFP fluorescent capsid suggests that transfer of MTS-delivered ND4FLAG cDNA may proceed in adjacent dividing cells that were not infected with the nonreplicating fluorescent AAV viral particles. Expression of ND4FLAG suggests that the delivered WT ND4 was synthesized inside the organelle.
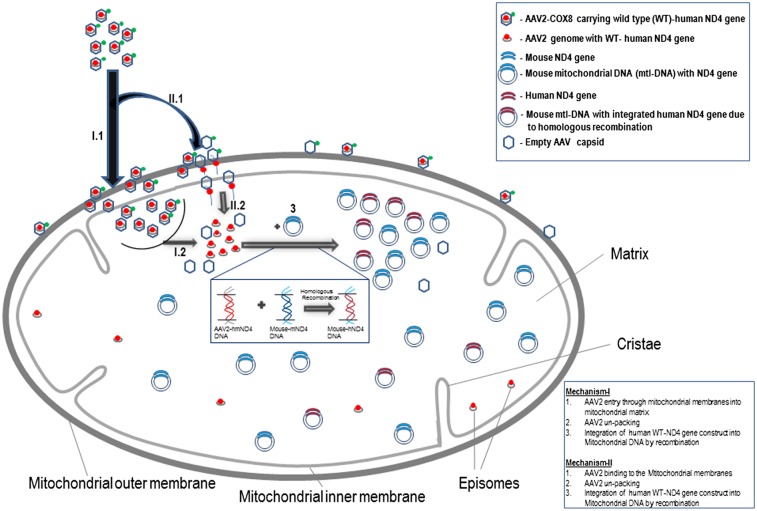
We wondered whether the MTS AAV virion entered into the mitochondria or whether DNA was released at the mitochondrial outer membrane, where the natural competence of the mitochondria to DNA would lead to internalization (23). Because fluorescence microscopy with a mitochondrial probe, such as MitoTracker Green, was unable to tell us whether the MTS AAV was stuck to the outer mitochondrial membrane or entered the mitochondria and to prove that the translation of ND4FLAG was confined to the mitochondria, we used transmission EM. In these COX8GFP VP2 AAV-infected cells, we found that GFP immunogold was located inside the mitochondria starting 1 d after infection (Fig. 2I). Two days after infection, FLAG-tagged immunogold was confined to the interior of mitochondria of cells infected with COX8GFP VP2 AAV containing the WT mitochondrial ND4FLAG gene (Fig. 2 J and L). A control stained with only the secondary goat antibody shows no background immunogold (Fig. 2K). Therefore, the COX8 MTS GFPVP2 AAV capsid protein entered mitochondria where the HSP-ND4FLAG DNA was likely released and the chimeric ND4FLAG protein was synthesized within the organelle.
Next, we looked for assembly of the AAV-delivered WT ND4 subunit into respiratory complex I. Using 1D blue native PAGE (BN/PAGE) loaded with a large amount of mitochondrial protein (250 μg), the ∼1,000-kDa complex I is evident in the blue gel for control (Fig. S1A, lane 2) and MTS AAV ND4FLAG-infected cells (Fig. S1A, lane 3). Immunoblotting of the transferred proteins stained for the nuclear-encoded NDUFA9 subunit of complex I shows NDUFA9 signal in both uninfected (Fig. S1A, lane 4) and infected (Fig. S1A, lane 5) mitochondrial pellets. Although a FLAG signal is not visualized in mitochondrial pellets of uninfected cells (Fig. S1A, lane 6), a FLAG signal is seen in MTS AAV-infected cell mitochondria reacted with the FLAG antibody (Fig. S1A, lane 7). Thus, the MTS AAV-transferred ND4FLAG assembled into complex I.
ND4 Restores Defective ATP Synthesis of MTS AAV-Infected LHON Cells.
If the WT ND4 is integrated into functional respiratory complexes, the defective respiration of LHON cells would be restored to normal levels. Because complex I is reduced only slightly in LHON cybrids and it is not significantly different from control cybrids with 100% WT ND4 mtDNA, complex I activity measurements were not a useful indicator for determining whether respiratory function was restored (29). Rather, to explore whether the imported ND4 was functional, we measured the end point of OXPHOS: the production of ATP using the complex I substrates malate and pyruvate. Relative to cells with WT ND4, ATP synthesis is reduced by approximately half in LHON cells (29). To perform this assay, we infected homoplasmic G11778A cybrid cells with the MTS ssAAV-delivered WT ND4 but got only a 48% increase in ATP synthesis relative to controls (P = 0.04).
To improve rescue, we turned to a self-complementary (sc) AAV vector. Because scAAV exhibits more efficient transduction than ssAAV (32) and contains both positive and negative strands of ND4FLAG DNA, we cloned the WT ND4FLAG from pTR-UF11mitoND4FLAG in a scAAV backbone still under the control of the HSP (sc-HSP-ND4FLAG). The sc-HSP-ND4FLAG plasmid packaged with COX8GFP VP2 AAV or VP2 without the MTS was used to infect G11778A cybrids. Following 5 d of selection in restrictive media, homoplasmic G11778A cybrids infected with ND4 packaged with scCOX8 VP2 or VP2 lacking the MTS, the GFP negative control or the allotopic p1-ND4 positive control was reseeded in 96-well plates and cultured for up to 5 d in either glucose-free galactose or glucose media. Given that cybrids with the mutant G11778A ND4 do not survive in restrictive media containing galactose (33), cell viability in culture was measured based on luciferin-luciferase–induced ATP luminescence indicated as a percentage of that of the corresponding cells grown in high-glucose media. Cells infected with ND4 packaged with the MTS-tagged COX8VP2 showed more ATP luminescence compared with packaging with VP2 lacking the MTS (VP2-ND4) or a control scAAV-GFP lacking WT human ND4 (Fig. S1B). The differences began to be significant on day 2 (P = 0.05) and became more so on day 5 (P = 0.001). The allotopic construct p1-ND4, previously shown to rescue these cells to levels statistically indistinguishable from WT cells with normal mtDNA, was used as a positive control for comparisons of rescue by the COX8 MTS-delivered ND4 gene (29).
Using the kinetic assay, we found the rate of ATP synthesis increased fourfold in response to the WT ND4 allele delivered by scCOX8 VP2-ND4 (50.5 nM ATP per minute per 106 cells) compared with scVP2-ND4 (12.5 nM ATP per minute per 106 cells) (P = 0.046). These in vitro results illustrate that exogenous WT ND4 rescues cultured cells from defective respiration attributable to the mutant ND4.
Stable Transmission of Human ND4 in Mice.
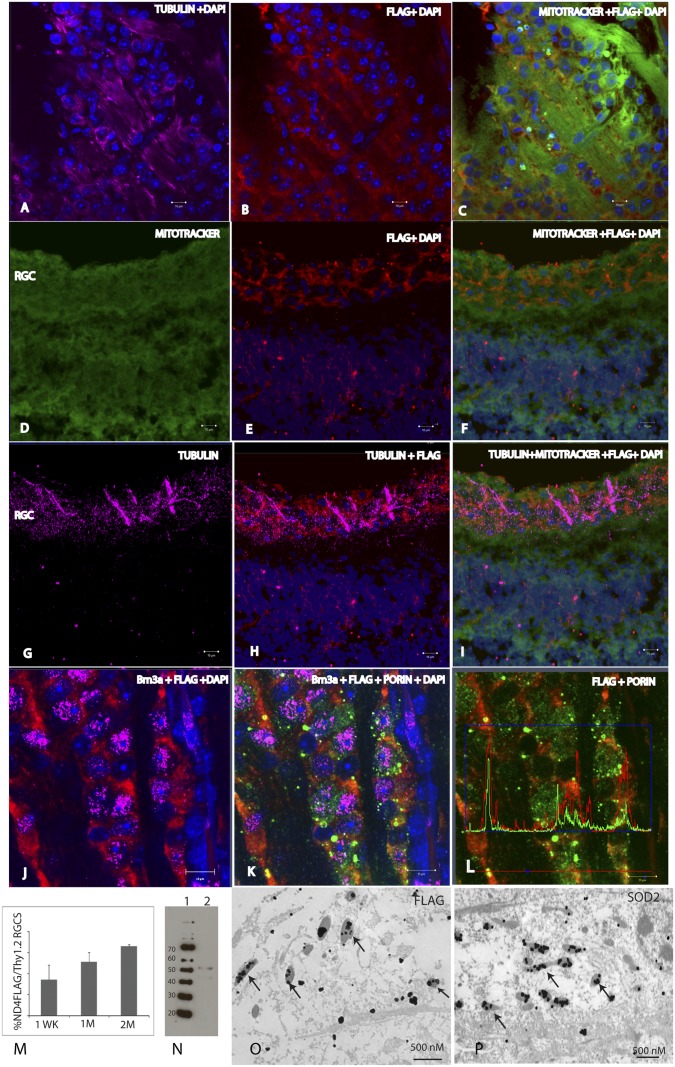
Next, we examined whether the mitochondria-targeted AAV would lead to the import of exogenous DNA in animals. The right eyes of mice received injections into the vitreous body of sc-HSP-ND4FLAG packaged by COX8 VP2 AAV or VP2 AAV lacking the MTS. Injections of scAAV-GFP into the left eyes served as controls. Mice were killed between 1 wk and 6 mo after intraocular injections. In retinal flat mounts stained with the III β-tubulin antibody to identify retinal ganglion cells (RGCs) (Fig. 3A), evidence of the expression of the ND4FLAG chimera was apparent (Fig. 3B). The ND4FLAG chimeric protein had a perinuclear distribution surrounding the nuclei of RGCs counterstained with DAPI. The ND4FLAG chimera colocalized with MitoTracker Green (Fig. 3C) in RGCs identified by III β-tubulin in Fig. 3A. In longitudinal retinal sections counterstained with MitoTracker Green (Fig. 3D), ND4FLAG was evident in cells of the RGC layer (Fig. 3E) and it colocalized with MitoTracker Green (Fig. 3F). RGCs labeled by the anti–III β-tubulin antibody (Fig. 3G) also expressed the ND4FLAG chimera (Fig. 3H), and they colocalized with MitoTracker Green (Fig. 3I). RGCs immunostained with Brn3a had a perinuclear distribution of ND4FLAG (Fig. 3J) that colocalized with mitochondrial porin (Fig. 3K), as also demonstrated by colocalization software analysis of fluorescent intensity (Fig. 3L). The fraction of ND4FLAG-expressing cells increased from almost 50% at 1 wk to ∼90% of Thy1.2-positive RGCs 1 to 2 mo after injection (Fig. 3M). ND4FLAG expression could be detected up to 6 mo after intraocular injection (Fig. 3N), the longest interval studied. Transmission EM revealed silver-enhanced ND4FLAG immunogold in optic nerve mitochondria (Fig. 3O) that were also labeled by an endogenous mitochondrial protein, manganese superoxide dismutase (MnSOD) (Fig. 3P). Therefore, MTS AAV infection resulted in expression of the ND4FLAG transgene in mitochondria of live animals.
Fig. 3.
Microscopy and immunoblotting of MTS AAV-infected retina and optic nerve. A retinal flat mount stained with the anti–III β-tubulin antibody (A) identified RGCs in which evidence of expression of the ND4FLAG chimera was apparent (B). The ND4FLAG chimera had a perinuclear distribution surrounding the nuclei of RGCs counterstained with DAPI. (C) ND4FLAG chimera colocalized with MitoTracker Green in RGCs identified by III β-tubulin as shown in A. (D) Longitudinal retinal sections were counterstained with MitoTracker Green. ND4FLAG was evident in cells of the RGC layer (E), and it colocalized with MitoTracker Green (F). RGCs labeled by the anti–III β-tubulin antibody (G) also expressed the ND4FLAG chimera (H) that colocalized with MitoTracker Green (I). In a flat-mount preparation, RGCs labeled by the Brn3a antibody had a perinuclear distribution of ND4FLAG (J) that colocalized with mitochondria immunolabeled by an antibody against porin (K). (L) ND4FLAG chimeric protein colocalized with porin. This finding was confirmed by software showing the peaks of FLAG immunofluorescence (red) colocalized with those of porin (green). (M) Bar plot shows that the number of ND4FLAG-expressing cells increased from almost half of Thy1.2-positive RGCs at 1 wk (1WK) to almost 90% at 1 mo (1M) to 2 mo (2M) after injection. (N) Immunoblotting of ND4FLAG protein in the retina could be detected up to 6 mo after intraocular injection (lane 2). Lane 1 is the molecular weight standards. Transmission EM of lightly fixed LR White resin-embedded optic nerves revealed silver-enhanced ND4FLAG immunogold (arrows) within optic nerve mitochondria (O) that were identified by mitochondrial SOD2 immunogold (arrows) (P).
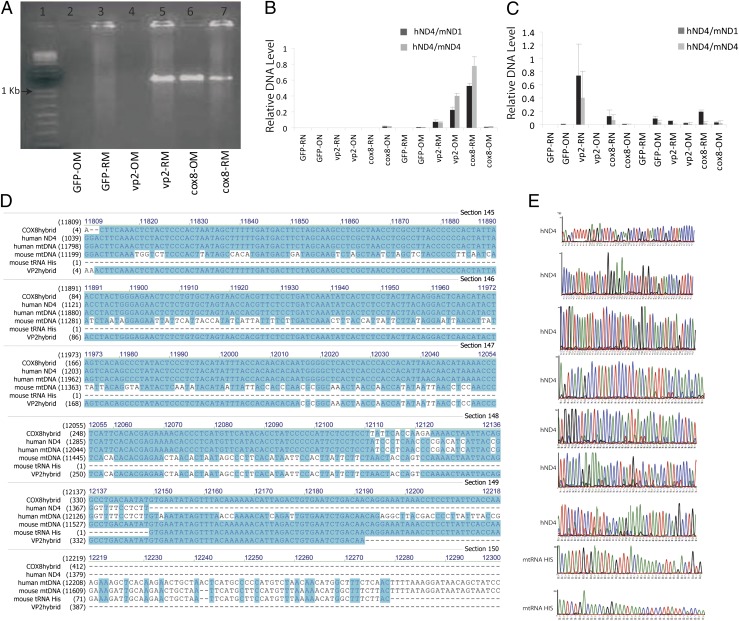
To determine if the mitochondria-associated virus transferred human ND4FLAG DNA, mtDNA and nuclear DNA were extracted from the retinas and optic nerves of mice 3 d and 9 d after intraocular viral injections. PCR for ND4FLAG gave the expected 1.4-kb band for the retinas and optic nerves of COX8-targeted ND4FLAG and in the retinas but not for the optic nerves of untargeted ND4FLAG (Fig. 4A). Quantitative PCR assay showed that in the retinal mitochondrial fraction (COX8-RM), human ND4 was ∼80% of its mouse homolog at 9 d after injection (Fig. 4B). Earlier, at day 3, human ND4 levels were much lower, except in the nuclear fraction of the untargeted AAV (Fig. 4C), where virus without modifications in VP2 normally delivers its DNA. The corresponding DNA sequences confirmed that PCR products were human ND4, further supporting that exogenous ND4 was imported into optic nerve mitochondria in mice by a mitochondria-targeted AAV. These findings indicate that the MTS-modified AAV is efficient at delivering its DNA cargo to mouse mitochondria in vivo.
Fig. 4.
Nucleic acid analysis of the infected rodent visual system. (A) Nine days after intraocular injections, PCR assay of mtDNA extracts probed for the human ND4FLAG gave the anticipated 1.4-kb band for both the retinas and optic nerves of COX8-targeted ND4FLAG. The untargeted VP2 was also positive for ND4FLAG in retinal mitochondrial (RM) extracts but was absent in optic nerve mitochondria (OM). (B) Quantitative PCR assay showed that in the retinal mitochondrial fraction (COX8-RM), human ND4 was ∼80% of its mouse homolog at 9 d postinjection. (C) Earlier, at day 3 postinjection, human ND4 levels in mice mitochondria were much lower. (D) Alignment of the DNA sequences obtained from COX8 (COX8hybrid)- or VP2 (VP2hybrid)-injected eyes with human or mouse ND4 and the sequencing chromatographs (COX8hybrid) (E) confirmed that the COX8 MTS VP2 delivered human ND4 in the infected murine optic nerve and retina. The DNA sequences after the 3′ end of the human ND4 (COX8hybrid) are homologous to the mouse tRNA histidine. The mouse tRNA histidine is just downstream of the murine ND4 in the mouse mitochondrial genome. cox8-OM, optic nerve mtDNA isolated from eyes injected with scAAV-mND4FLAG containing the VP2 cox8MTS; cox8-ON, nuclear DNA isolated from the optic nerves eyes injected with scAAV-mND4FLAG containing the VP2 cox8MTS; cox8-RM, retinal mtDNA isolated from eyes injected with scAAV-mND4FLAG containing the VP2 cox8MTS; cox8-RN, nuclear DNA isolated from the retinas of eyes injected with scAAV-mND4FLAG containing the VP2 cox8MTS; GFP-OM, optic nerve mtDNA isolated from AAV-GFP injected eyes; GFP-ON, nuclear DNA isolated from the optic nerves of AAV-GFP–injected eyes; GFP-RM, retinal mtDNA isolated from AAV-GFP injected eyes; GFP-RN, nuclear DNA isolated from the retinas of AAV-GFP–injected eyes; hND4, human ND4; mND1, mouse ND1; mND4, mouse ND4; vp2-OM, optic nerve mtDNA isolated from eyes injected with scAAV-mND4FLAG lacking the VP2 MTS; vp2-ON, nuclear DNA isolated from the optic nerves of eyes injected with scAAV-mND4FLAG lacking the VP2 MTS; vp2-RM, retinal mtDNA isolated from eyes injected with scAAV-mND4FLAG lacking the VP2 MTS; vp2-RN, nuclear DNA isolated from the retinas of eyes injected with scAAV-mND4FLAG lacking the VP2 MTS.
The increase in human ND4 over time suggests that the delivered DNA is being replicated rather than gradually released from AAV particles (in which case the level of human ND4 would be constant). Our HSP-ND4FLAG construct excluded mtDNA replication elements present within the D loop. Therefore, HSP-ND4 DNA would be expected to diminish over time rather than increase as we observed here. On the other hand, if the delivered viral DNA integrated into the DNA of the mouse mitochondrial genome, where replication elements are present in the D loop, this would explain our observation of increasing AAV-delivered ND4 with time.
Because the mouse ND4 and human ND4 are ∼23% dissimilar, we tested for homologous recombination by designing a forward primer flanking the human ND4 and three reverse primers flanking three different regions of mouse medina (P1, P2, and P3). With DNA isolated from COX8 or VP2 without an MTS-injected mouse retina as a template, PCR revealed the expected band of 500 bp from each pair of primers. The corresponding DNA sequence showed that four were human ND4 and two were human–mouse chimeras with human ND4 replacing mouse ND4 followed by mouse mtDNA starting from position 11,519. Alignment of the sequenced PCR products to the human or mouse mtDNA nucleotide sequences shown in Fig. 4D and examination of DNA sequencing chromatographs (Fig. 4E) revealed that COX8 MTS VP2 delivered human ND4 upstream of the mouse tRNA histidine, where human ND4 completely replaced murine ND4.
To investigate the possibility of homologous recombination of human ND4 replacing mouse endogenous ND4, we used Southern blots. Our human ND4 probe hybridized to DNA isolated from COX8 MTS-injected retinas failed to discriminate between COX8-, VP2-, and GFP-injected eyes (Fig. S2). Thus, we were unable to conclude that AAV-delivered human ND4 integrated into mouse mtDNA.
Human ND4 Expression in the Mouse Eye Does Not Cause Visual Loss.
Because human ND4 and mouse ND4 are approximately three-quarters homologous (Fig. 4D), we tested for an adverse consequence of human ND4 expression in the murine retina. Using the pattern electroretinogram (PERG), which is a sensitive measure of RGC function, we found no significant differences in amplitude relative to uninjected eyes of normal animals (Fig. 5 A and B). Thus, rodent visual function did not appear to be altered by the import and expression of human ND4 (34).
Fig. 5.
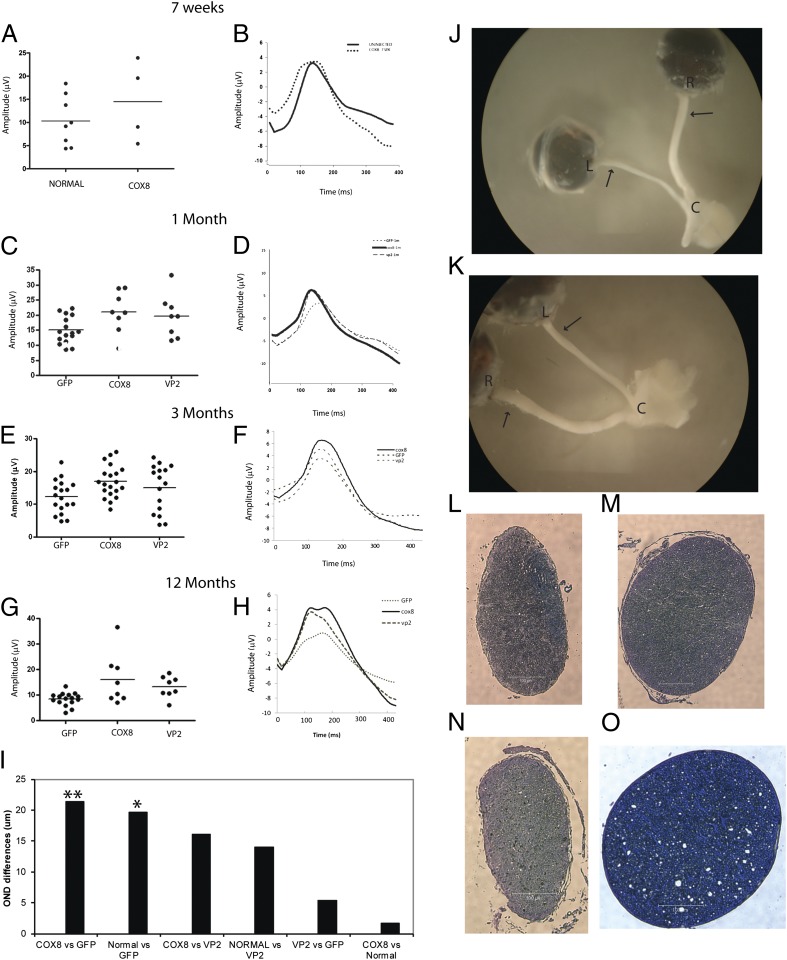
MTS AAV-mediated rescue of visual loss and optic atrophy of rodents. A scatterplot of amplitudes of individual animals (line = median value) (A) and averaged waveforms (B) of PERGs done 7 wk after intraocular injections of scAAV2/COX8 VP2 containing human ND4FLAG revealed no loss of amplitude relative to uninjected eyes of normal mice. One month after intraocular injections, a scatterplot of PERG amplitudes (C) and averaged waveforms (D) revealed that loss of visual function induced by an allotopic R340H mutant ND4 was prevented by ocular injection of the COX8-targeted human ND4 AAV. At this time point, PERG amplitude for the control left eyes injected with AAV-GFP and AAV containing mutant R340H ND4 decreased by 29% compared with eyes injected with COX8-delivered WT ND4 and also injected with R340H mutant ND4 AAV (P = 0.0151). Differences between the contralateral control eyes and eyes injected with VP2-delivered WT ND4 were not significant. In another series of experiments, both eyes of the same animal received injections of WT human ND4 delivered either by mitochondria-targeted COX8GFP VP2, VP2, or AAV-GFP. All eyes of these animals were also injected with R340H mutant ND4. Three months postinjection, a scatterplot (E) and averaged waveforms (F) show that the protective effect of COX8-delivered human ND4 in preventing loss of vision relative to the GFP control animals was highly significant (P = 0.0064). Relative to these controls, differences between mice that received the VP2-delivered human ND4 were not significant (P = 0.339). Twelve months after intravitreal injections, a scatterplot (G) and averaged waveforms (H) show that differences in PERG amplitude between eyes injected with AAV-GFP (and AAV containing mutant R340H ND4) relative to COX8-delivered WT ND4-injected eyes (that were also injected with R340H mutant ND4 AAV) were still highly significant (P = 0.0073). Differences between eyes injected with COX8-delivered WT ND4 AAV (that were also injected with R340H mutant ND4 AAV) relative to eyes injected with standard VP2 AAV containing the WT ND4 (that were also injected with R340H mutant ND4 AAV) were not significant. (I) Bar plot of postmortem optic nerve diameter differences shows that with both eyes injected with the mutant ND4, differences in optic nerve diameters between those treated with COX8-delivered WT ND4 relative to mock treatment with AAV-GFP were highly significant (**P = 0.0053). As an additional control, we injected AAV-GFP into the eyes of normal mice that received no other intraocular injections and were killed 1 y later. Comparisons of this normal group (injected only with AAV-GFP) with those that received the mutant R340H ND4 and mock treatment with AAV-GFP were also significant (*P = 0.047). Comparisons of the normal group with the COX8-treated eyes that received the mutant ND4 showed the least differences. (J and K) Gross specimens of two animals dissected 13 mo after intravitreal injections revealed significant thinning of the entire mock-treated left optic nerve from the globe to the optic chiasm in eyes injected with AAV-GFP and AAV containing mutant R340H ND4, whereas the opposite right eyes treated by injection of COX8-delivered WT ND4 (and also injected with R340H mutant ND4 AAV) were much thicker. Histology of cross-sections of the retrobulbar segment of optic nerve taken at the sites indicated by the arrows confirmed the atrophy of the untreated left optic nerves (L and N), in contrast to the preservation of optic nerve structure with treatment by COX8-delivered WT ND4 (M and O). (Scale bar, 100 μm.) C, optic chiasm; L, left eye; R, right eye.
MTS AAV Rescues Visual Dysfunction Caused by Mutated ND4.
Next, we wished to determine whether the delivered human ND4 would rescue visual function caused by mutated R340H ND4 (35, 36). For these experiments, we injected the right eyes of mice with WT human ND4 delivered by either VP2 or mitochondria-targeted COX8GFP VP2. The left eyes received GFP in a similar AAV backbone. Because there was no transgenic animal model with mutated ND4 mtDNA available, both eyes were injected, 2 d later, with allotopic R340H human mutant ND4 AAV, previously shown to cause visual loss in rodents (35, 36). One month after the double-AAV injections, electrophysiological function of mice was assessed with the PERG, which reflects visual capability. As a result of the mutant ND4 allele, the PERG amplitude for the control left eyes injected with both AAV-GFP and AAV containing mutant R340H ND4 decreased by 29% compared with eyes injected with COX8-delivered WT ND4 and also injected with R340H mutant ND4 AAV (P = 0.0151) (Fig. 5 C and D). The differences between the contralateral control eyes and eyes injected with VP2-delivered WT ND4 were not statistically significant. Therefore, only the MTS-targeted, AAV-delivered WT human ND4 prevented early visual loss induced by the mutant ND4.
MTS-AAV–Mediated Rescue of Mice Persists Long Term.
To support these results further, we performed another series of experiments. Rather than compare differences between treated and untreated eyes of the same animal, both eyes of the animal received the same treatment in these experiments: injection of WT human ND4 delivered either by mitochondria-targeted COX8GFP VP2, VP2, or AAV-GFP (without WT human ND4). Two days later, both eyes of all animals in all three groups were injected with allotopic R340H human mutant ND4 AAV. Three months after the double-intravitreal injections, the PERG amplitude of eyes injected with mitochondria-targeted ND4 was significantly increased compared with the GFP control animals (P = 0.0064) (Fig. 5 E and F). Six months after injection, the PERG amplitude in eyes injected with mitochondria-targeted ND4 was highly significantly increased compared with the GFP control animals (P = 0.0006). More than a year after the double-intravitreal injections, differences in PERG amplitude between eyes injected with AAV-GFP and eyes injected with COX8-delivered WT ND4 were still highly significant (P = 0.0073) (Fig. 5 G and H). These results suggest that MTS-targeted AAV delivery of relevant exogenous DNA to mitochondria ameliorates the functional consequences of the mutated ND4 homolog, causing visual loss for almost the entire life span of the laboratory mouse.
MTS-AAV Ameliorates Murine Optic Atrophy Induced by Mutated ND4.
Postmortem analysis performed 1 y after intravitreal injections confirmed that the MTS-targeted AAV prevented optic atrophy induced by the mutant R340H ND4. With both eyes injected with the mutant ND4, differences in optic nerve diameters between those treated with COX8-delivered WT ND4 relative to mock treatment with AAV-GFP were highly significant (P = 0.0053) (Fig. 5I). As an additional control, we injected AAV-GFP into the eyes of normal mice that received no other intraocular injections. Comparisons of this normal group (injected only with AAV-GFP) with those that received the mutant R340H ND4 and mock treatment with AAV-GFP were also significant (P = 0.047). Comparisons of the normal group with the COX8-treated eyes that received the mutant ND4 showed the least differences. Marked atrophy of the entire optic nerve from the globe to the optic chiasm was prominent in eyes injected with AAV-GFP and AAV containing mutant R340H ND4 (Fig. 5 J and K). In contrast, the contralateral optic nerves of eyes injected with COX8-delivered WT ND4 and also injected with R340H mutant ND4 AAV were preserved. Histological cross-sections of the retrobulbar segment of the optic nerve taken at the sites indicated by the arrows confirmed that the MTS-targeted vector (Fig. 5 M and O) prevented optic atrophy (Fig. 5 L and N), the hallmark of LHON, induced by the mutant R340H ND4.
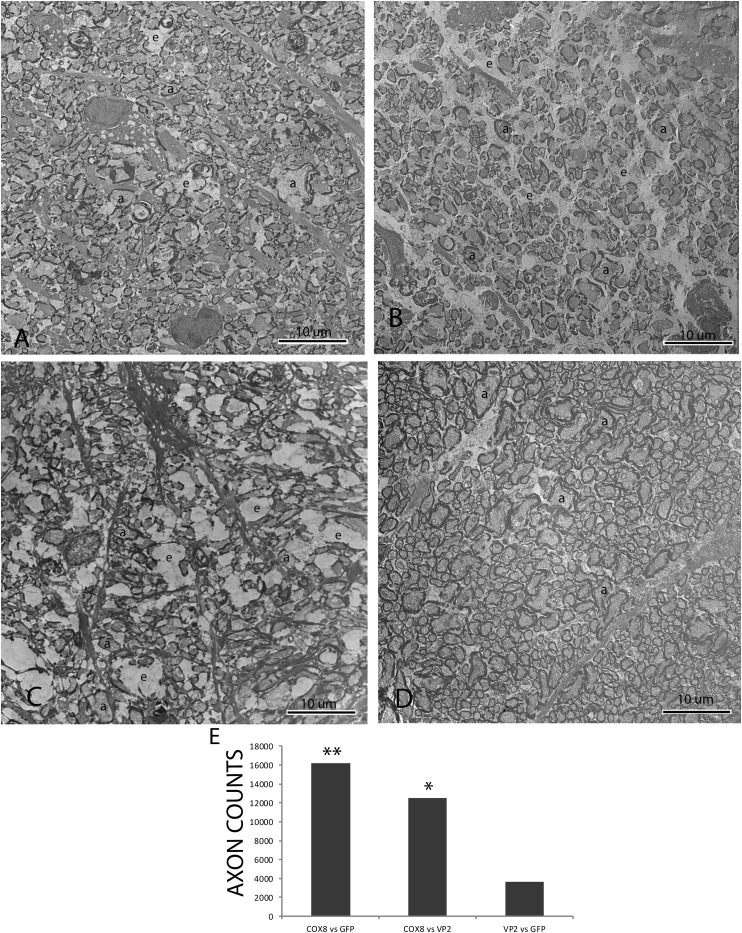
One year after intravitreal injections, transmission EM revealed that relative to the complement of axons seen with COX8 MTS scAAV-ND4 treatment (Fig. 6A), axonal density was markedly reduced with mock treatment with AAV-GFP (Fig. 6B) or with treatment with VP2 scAAV-ND4 lacking the MTS (Fig. 6C). The empty spaces between remaining axons where axons degenerated and were lost accounted for the cystic spaces seen by light microscopy. A normal optic nerve injected with only AAV-GFP a year earlier is shown for comparison (Fig. 6D). Differences in optic nerve axon counts between COX8 MTS scAAV-ND4–treated eyes relative to treatment with VP2 scAAV-ND4 or mock treatment with AAV-GFP were highly significant (P = 0.0178 and P = 0.0032, respectively) (Fig. 6E). Differences between VP2 scAAV-ND4 and AAV-GFP were not significant.
Fig. 6.
One year after intravitreal injections, a transmission electron micrograph shows that relative to the complement of axons seen with COX8 MTS scAAV-ND4 treatment (A), axonal density is markedly reduced with mock treatment with AAV-GFP (B) or with treatment with VP2 scAAV-ND4 lacking the MTS (C). The empty spaces between remaining axons where axons were lost accounted for the cystic spaces seen by light microscopy. (D) Micrograph of a normal optic nerve injected with only AAV-GFP a year earlier is shown for comparison. (E) Bar plot shows that differences in optic nerve axon counts between COX8 MTS scAAV-ND4–treated eyes relative to treatment with VP2 scAAV-ND4 or mock treatment with AAV-GFP were highly significant (*P = 0.0178; **P = 0.0032). a, axon; e, empty spaces where axons were lost.
Discussion
Our work shows that fusing an AAV capsid protein with an MTS makes an efficient delivery system for importing the functional homolog of a defective gene into the mitochondria of living cells in the mouse visual system. Studies by others have conjugated an MTS to exogenous proteins and small linear DNA, which enhanced their delivery to mitochondria (29, 37), but that strategy failed in cases of macromolecules and hydrophobic molecules, including mtDNA and mitochondrial proteins (38). Liposome-based carriers can import hybrid molecules into mitochondria of whole cells, but this approach suffers from low efficiency and high cytotoxicity (8, 25, 39). Mutated mtDNA can be removed from the germ line by transfer of the spindle–chromosomal complex from one egg to another that is enucleated and depleted of its mitochondria (40). A major limitation of this successful approach is that it cannot be used after birth and the technique has not yet been demonstrated to correct a mutated mtDNA phenotype.
In contrast, our MTS AAV approach resulted in expression of WT human ND4 that was translated in mitochondria, where it suppressed visual loss and optic atrophy induced by the mutant ND4 allele in rodents, thus suggesting that this approach may be useful in humans long after birth. Still, many questions remain to be answered. We are unsure of the exact mechanism of MTS AAV-mediated DNA entry into mitochondria. The presence of COX8GFP VP2 capsid detected by ultrastructural analysis of GFP suggests viral entry into the organelle where the ND4 DNA was deposited. Although homologous recombination has allowed insertion of foreign genes into the plant mitochondrial genome requiring only flanking regions of the insert to be homologous to the recipient mtDNA moiety (41), we were unable to confirm our PCR data suggesting in vivo homologous recombination of human ND4 into the mouse mitochondrial genome by Southern blot analysis. However, hybridization of the human probe to the endogenous mouse mitochondrial genome suggests the possibility that such an event may occur. Also, the increasing human ND4 DNA content with increasing time following ocular gene injections suggests replication of the MTS AAV-delivered human ND4. Because our HSP-ND4 construct lacked replication elements, replication of human ND4 would only occur if it had integrated into the mouse mitochondrial genome, where the replication elements present in the D-loop region would drive this event.
In summary, we were able to demonstrate expression of the MTS AAV-delivered ND4 with rescue of ATP synthesis in cultured cells as well as visual loss and optic atrophy in rodents for almost their entire laboratory life span. The AAV cassette accommodates genes up to ∼3.3 kb (scAAV) or 5 kb (ssAAV) in length, thus providing a platform for introduction of almost any mitochondrial gene, and perhaps even allowing insertion of large kilobase deletions associated with aging back into the adult vertebrae organelle. We look forward to providing this delivery system to other laboratories for confirmation of our results and for the benefit of the multitude of patients afflicted by disorders caused by mutated mtDNA, for whom there is currently no effective remedy.
Materials and Methods
Plasmids.
To construct the fusion gene containing the mitochondrial promoter, human WT ND4, and epitope tag, mtDNA was extracted from human cells. Using the high fidelity of pfu Turbo DNA polymerase (Stratagene), the 1.4-kb mitochondrial-encoded ND4 gene was cloned into the Topo TA cloning vector using a kit according to the manufacturer’s directions (Invitrogen). A final extension step using Taq DNA polymerase was also performed. The QuikChange in vitro mutagenesis kit (Stratagene) was used to add the FLAG epitope tag with appended AGA termination codon to the 3′ end of the ORF of the ND4 gene to obtain ND4FLAG. Base deletions and substitutions in the reading frame ND4FLAG were corrected using the QuikChange in vitro mutagenesis kit. The parent pTR-UF11 plasmid was digested with Xba1 and BamH1 to remove the GFP and neoR genes. The Topo plasmid containing the mitochondrial ND4FLAG was also digested with XbaI and BamHI for directional cloning into the similarly digested recipient pTR-UF11 plasmid. The ND4FLAG insert was then ligated into the recipient pTR-UF11 plasmid. This gave the desired plasmid, designated as pTR-UF11-mND4FLAG. Next, we removed the hybrid CMV enhancer and chicken β-actin promoter elements (used to drive gene expression in the nucleus) from pTR-UF11-ND4FLAG using Kpn1 and Xba1 digests. We purchased complementary oligonucleotides (Invitrogen) containing the mitochondrial HSP, sequence 5′-TAACCCCATACCCCGAACCAACCAAACCCCAAAGACAC-3′, with added linkers containing the Kpn1 restriction site at the 5′ end and Xba1 at the 3′ end of HSP. The annealed double-stranded HSP oligonucleotide was directionally ligated into the pTR-UF11-ND4FLAG to give plasmid pTR-UF11-HSP-ND4FLAG. The pTR-UF11 plasmid also contains flanking ITR sequences that provide all the cis-acting sequence required for replication, packaging, and integration of an ssAAV vector.
We then inserted HSP-ND4FLAG into a scAAV backbone. The parent SC-trs-smCBA-hGFP plasmid was digested with Kpn1 and BamH1 to remove the hybrid CMV enhancer and small chicken β-actin promoter elements as well as GFP. The insert containing HSP-ND4FLAG was digested out of pTR-UF11-HSP-ND4FLAG using restriction enzymes Kpn1 and BamH1 and it was then ligated into the recipient scAAV backbone generating the construct sc-HSP-ND4FLAG containing flanking ITRs.
To generate an AAV directed toward the mitochondria, we added an MTS to the ORF of VP2, one of the three capsid proteins that constitute an AAV virion. This MTS-targeted VP2 was generated by linking the 23-aa MTS presequence of COX8, 5′-ATGTCCGTCCTGACGCCGCTGCTGCTGCGGGGCTTGACAGGCTCGGCCCGGCGGCTCCCAGTGCCGCGCGCCAA-3′, in-frame to GFP. EagI linkers were added to the COX8GFP insert. The parent VP2 plasmid with an EagI site inserted at residue 138 within the VP1/VP2 overlap region was digested with EagI. The COX8GFP was ligated into the mutant VP2 parent plasmid. This created the plasmid designated COX8GFP VP2. Construction of the mutant VP2 parent plasmid and the GFP containing VP2 mutant plasmid without the MTS has been described previously (28).
AAVs.
The ssAAV2 pTR-UF11-HSP-ND4FLAG or scAAV2/sc-HSP-ND4FLAG, or a control scAAV2-GFP, was produced by the plasmid cotransfection method (42). The plasmids were amplified and purified by cesium chloride gradient centrifugation and then packaged with the VP2 COX8GFP or standard VP2 plus VP1, VP3, and helper plasmid PXX6 (3-fold excess) into AAV2 recombinant virus by transfection into human 293 cells with standard procedures. In brief, the crude iodixanol fractions were purified using the Pharmacia AKTA FPLC system, the virus was then eluted from the column with 215 mM NaCl (pH 8.0), and the recombinant AAV (rAAV) peak was collected. The rAAV-containing fractions were then concentrated and buffer-exchanged in Alcon balanced salt solution with 0.014% Tween 20, with a Biomax 100-K concentrator (Millipore). Virus was then titered for DNase-resistant viral genomes by real-time PCR relative to a standard. Finally, the purity of the virus was validated by silver-stained SDS/PAGE, assayed for sterility and lack of endotoxin, and then aliquoted and stored at −80 °C.
Cell Culture, Transfection, and Infection.
Human embryonal kidney cells (293T), homoplasmic G11778A cybrid cells, or neuronal G11778A NT-2 cells (with large mitochondria) were infected with AAV vectors or transfected with sc-HSP-ND4FLAG, PXX6 (3-fold excess), VP1, VP3, VP2, or VP2COX8GFP plasmid DNAs and cultured in complete high-glucose medium (DMEM) for 2 d. Cells were then transferred to galactose medium for selection at 5 d and recultured in complete high-glucose media until reaching the desired confluence to conduct other experiments. Cells were trypsinized and counted with an automated Coulter Z-100 particle counter.
Immunostaining.
Cells were cultured on plastic chamber slides and fixed in 4% paraformaldehyde. Injected mice were perfused with PBS followed by 4% paraformaldehyde. Whole retinas or longitudinal retinal sections were used for immunostaining. The following antibodies were used: Cy3-conjugated FLAG (1:100; Sigma), Porin (1:100; Abcam), Thy1.2 (1:1,500; Sigma), Brn3a (1:100; Santa Cruz Biotechnology), III β-tubulin (TUJI, 1:200; Covance), A20 (1:100; Sigma), MnSOD (1:100; Abcam), GFP (1:100; Sigma), NADH dehydrogenase subunit 4 (1:480; Abcam), and secondary antibodies that included anti-rat488, anti-mouse488, anti-mouseCy5, anti-rabbit 647, anti-rabbit Cy3 (1:600; Invitrogen) or anti-mouse IgG conjugated to immunogold that was enhanced with a kit according to the manufacturer’s specifications (Ted Pella, Inc.). Images were visualized with a Leica TCS SP5 confocal microscope. FLAG-positive cells and Thy1.2-immunopositive RGC counts were performed on the longitudinal sections. The total cell count was an average of the counting data performed on longitudinal sections of the retina per 40-mm2 area. The images were captured with a video camera mounted on a fluorescent microscope at a magnification of 400× for counting of the cells. Immunogold labeling was also evaluated by transmission EM.
BN/PAGE and Immunoblotting.
BN/PAGE was performed based on the method of Calvaruso et al. (43). We used the Invitrogen NativePAGE Gel system with minor modifications. Cell pellets (108) were washed twice with cold PBS and resuspended in 600 μL of cold mitochondrial extract buffer [0.25 mM sucrose, 20 mM Hepes, 10 mM KCl, 1.5 mM MgCl2, 1 mM EDTA (pH 7.4)]. After being homogenized, lysed cells were centrifuged at 4 °C at 750 × g for 3 min, followed by spins at 3,500 × g for 3 min and 5,000 × g for 3 min. Supernatant was collected and centrifuged for 30 min at 20,000 × g at 4 °C to collect mitochondria. The crude mitochondrial pellets were washed twice with cold PBS and resuspended in 100 μL of cold 1× NativePAGE sample buffer containing 2.5% digitonin (Invitrogen), 1% 2-mercaptoethanol (Bio-Rad), and 1 mM protease inhibitor (Invitrogen). After being mixed by pipetting up and down and by inversion, the mitochondrial pellet was sonicated three times for 15 s each time and kept on ice for 30 min before being centrifuged at 20,000 × g for 30 min at 4 °C. The supernatant was used for the NB/PAGE gel. Electrophoresis was performed according to the manufacturer’s specifications (Invitrogen). Gels were transferred to PVDF membranes (Immobilon; Millipore) using the semitransfer system (Bio-Rad), incubated in proteinase K (1 μg/μL; Qiagen) for 5–6 min, and then washed three times in PBS. For 1D Western blot, the transferred membrane was incubated in 8% acetic acid for 15 min, rinsed with deionized water, and air-dried. The dried membrane was rewetted with methanol and rinsed with deionized water before immunodetection. After blocking with 5% milk (Bio-Rad), the following antibodies were used: anti-FLAG (1:1,000; Sigma), anti-ND4 (1:200; Santa Cruz Biotechnology), and mouse monoclonal anti-NDUFA9 antibody (Mitosciences). Secondary probing with anti-mouse or anti-rabbit HRP-conjugated antibodies (1:5,000; Sigma) was performed for 60 min, followed by detection using electrochemiluminescence reagents (Amersham) and a FUJIFilm imaging system.
Supplementary Material
Acknowledgments
We thank Mabel Wilson for editing the manuscript and Profs. Douglas Anderson and Carlos Moraes for critically reviewing the manuscript. This study was supported by National Eye Institute (NEI) Grants R01 EY017141 (to J.G.), EY014801 (to V.P.), and P30-EY021721 to (W.W.H.), and in part from an unrestricted grant to Bascom Palmer Eye Institute from Research to Prevent Blindness, Inc.
Footnotes
Conflict of interest statement: W.W.H. and the University of Florida have a financial interest in the use of AAV therapies, and own equity in a company (AGTC Inc.) that might, in the future, commercialize some aspects of this work.
This article is a PNAS Direct Submission.
See Author Summary on page 7599 (volume 109, number 20).
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1119577109/-/DCSupplemental.
References
- 1.Wallace DC. Mitochondrial diseases in man and mouse. Science. 1999;283:1482–1488. doi: 10.1126/science.283.5407.1482. [DOI] [PubMed] [Google Scholar]
- 2.Wallace DC. The mitochondrial genome in human adaptive radiation and disease: On the road to therapeutics and performance enhancement. Gene. 2005;354:169–180. doi: 10.1016/j.gene.2005.05.001. [DOI] [PubMed] [Google Scholar]
- 3.Lane N. Mitochondrial disease: Powerhouse of disease. Nature. 2006;440:600–602. doi: 10.1038/440600a. [DOI] [PubMed] [Google Scholar]
- 4.Balaban RS, Nemoto S, Finkel T. Mitochondria, oxidants, and aging. Cell. 2005;120:483–495. doi: 10.1016/j.cell.2005.02.001. [DOI] [PubMed] [Google Scholar]
- 5.Koene S, Smeitink J. Mitochondrial medicine: Entering the era of treatment. J Intern Med. 2009;265:193–209. doi: 10.1111/j.1365-2796.2008.02058.x. [DOI] [PubMed] [Google Scholar]
- 6.Wallace DC, Fan W, Procaccio V. Mitochondrial energetics and therapeutics. Annu Rev Pathol. 2010;5:297–348. doi: 10.1146/annurev.pathol.4.110807.092314. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.DiMauro S, Andreu AL. Mutations in mtDNA: Are we scraping the bottom of the barrel? Brain Pathol. 2000;10:431–441. doi: 10.1111/j.1750-3639.2000.tb00275.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Kyriakouli DS, Boesch P, Taylor RW, Lightowlers RN. Progress and prospects: Gene therapy for mitochondrial DNA disease. Gene Ther. 2008;15:1017–1023. doi: 10.1038/gt.2008.91. [DOI] [PubMed] [Google Scholar]
- 9.Horvath R, Gorman G, Chinnery PF. How can we treat mitochondrial encephalomyopathies? Approaches to therapy. Neurotherapeutics. 2008;5:558–568. doi: 10.1016/j.nurt.2008.07.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Wallace DC, Fan W. The pathophysiology of mitochondrial disease as modeled in the mouse. Genes Dev. 2009;23:1714–1736. doi: 10.1101/gad.1784909. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Wang X, et al. The role of abnormal mitochondrial dynamics in the pathogenesis of Alzheimer’s disease. J Neurochem. 2009;109(Suppl 1):153–159. doi: 10.1111/j.1471-4159.2009.05867.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Wallace DC, et al. Mitochondrial DNA mutation associated with Leber’s hereditary optic neuropathy. Science. 1988;242:1427–1430. doi: 10.1126/science.3201231. [DOI] [PubMed] [Google Scholar]
- 13.Howell N. Leber hereditary optic neuropathy: Mitochondrial mutations and degeneration of the optic nerve. Vision Res. 1997;37:3495–3507. doi: 10.1016/S0042-6989(96)00167-8. [DOI] [PubMed] [Google Scholar]
- 14.Kogelnik AM, Lott MT, Brown MD, Navathe SB, Wallace DC. MITOMAP: A human mitochondrial genome database. Nucleic Acids Res. 1996;24:177–179. doi: 10.1093/nar/24.1.177. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Sazanov LA, Peak-Chew SY, Fearnley IM, Walker JE. Resolution of the membrane domain of bovine complex I into subcomplexes: Implications for the structural organization of the enzyme. Biochemistry. 2000;39:7229–7235. doi: 10.1021/bi000335t. [DOI] [PubMed] [Google Scholar]
- 16.Vestweber D, Schatz G. DNA-protein conjugates can enter mitochondria via the protein import pathway. Nature. 1989;338:170–172. doi: 10.1038/338170a0. [DOI] [PubMed] [Google Scholar]
- 17.Collombet JM, Wheeler VC, Vogel F, Coutelle C. Introduction of plasmid DNA into isolated mitochondria by electroporation. A novel approach toward gene correction for mitochondrial disorders. J Biol Chem. 1997;272:5342–5347. doi: 10.1074/jbc.272.8.5342. [DOI] [PubMed] [Google Scholar]
- 18.Yoon YG, Koob MD. Efficient cloning and engineering of entire mitochondrial genomes in Escherichia coli and transfer into transcriptionally active mitochondria. Nucleic Acids Res. 2003;31:1407–1415. doi: 10.1093/nar/gkg228. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Khan SM, Bennett JP., Jr Development of mitochondrial gene replacement therapy. J Bioenerg Biomembr. 2004;36:387–393. doi: 10.1023/B:JOBB.0000041773.20072.9e. [DOI] [PubMed] [Google Scholar]
- 20.Ibrahim N, et al. DNA delivery to mitochondria: Sequence specificity and energy enhancement. Pharm Res. 2011;28:2871–2882. doi: 10.1007/s11095-011-0516-4. [DOI] [PubMed] [Google Scholar]
- 21.Mileshina D, et al. Mitochondrial transfection for studying organellar DNA repair, genome maintenance and aging. Mech Ageing Dev. 2011;132:412–423. doi: 10.1016/j.mad.2011.05.002. [DOI] [PubMed] [Google Scholar]
- 22.Bacman SR, Williams SL, Moraes CT. Intra- and inter-molecular recombination of mitochondrial DNA after in vivo induction of multiple double-strand breaks. Nucleic Acids Res. 2009;37:4218–4226. doi: 10.1093/nar/gkp348. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Koulintchenko M, Temperley RJ, Mason PA, Dietrich A, Lightowlers RN. Natural competence of mammalian mitochondria allows the molecular investigation of mitochondrial gene expression. Hum Mol Genet. 2006;15:143–154. doi: 10.1093/hmg/ddi435. [DOI] [PubMed] [Google Scholar]
- 24.Koulintchenko M, Konstantinov Y, Dietrich A. Plant mitochondria actively import DNA via the permeability transition pore complex. EMBO J. 2003;22:1245–1254. doi: 10.1093/emboj/cdg128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Yamada Y, Akita H, Kogure K, Kamiya H, Harashima H. Mitochondrial drug delivery and mitochondrial disease therapy—An approach to liposome-based delivery targeted to mitochondria. Mitochondrion. 2007;7:63–71. doi: 10.1016/j.mito.2006.12.003. [DOI] [PubMed] [Google Scholar]
- 26.Vaidya B, Mishra N, Dube D, Tiwari S, Vyas SP. Targeted nucleic acid delivery to mitochondria. Curr Gene Ther. 2009;9:475–486. doi: 10.2174/156652309790031102. [DOI] [PubMed] [Google Scholar]
- 27.Samulski RJ, Chang LS, Shenk T. Helper-free stocks of recombinant adeno-associated viruses: Normal integration does not require viral gene expression. J Virol. 1989;63:3822–3828. doi: 10.1128/jvi.63.9.3822-3828.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Warrington KH, Jr, et al. Adeno-associated virus type 2 VP2 capsid protein is nonessential and can tolerate large peptide insertions at its N terminus. J Virol. 2004;78:6595–6609. doi: 10.1128/JVI.78.12.6595-6609.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Guy J, et al. Rescue of a mitochondrial deficiency causing Leber Hereditary Optic Neuropathy. Ann Neurol. 2002;52:534–542. doi: 10.1002/ana.10354. [DOI] [PubMed] [Google Scholar]
- 30.Manfredi G, et al. Rescue of a deficiency in ATP synthesis by transfer of MTATP6, a mitochondrial DNA-encoded gene, to the nucleus. Nat Genet. 2002;30:394–399. doi: 10.1038/ng851. [DOI] [PubMed] [Google Scholar]
- 31.Wong A, et al. Differentiation-specific effects of LHON mutations introduced into neuronal NT2 cells. Hum Mol Genet. 2002;11:431–438. doi: 10.1093/hmg/11.4.431. [DOI] [PubMed] [Google Scholar]
- 32.McCarty DM. Self-complementary AAV vectors; advances and applications. Mol Ther. 2008;16:1648–1656. doi: 10.1038/mt.2008.171. [DOI] [PubMed] [Google Scholar]
- 33.Zanna C, et al. Caspase-independent death of Leber’s hereditary optic neuropathy cybrids is driven by energetic failure and mediated by AIF and Endonuclease G. Apoptosis. 2005;10:997–1007. doi: 10.1007/s10495-005-0742-5. [DOI] [PubMed] [Google Scholar]
- 34.Guy J, et al. Efficiency and safety of AAV-mediated gene delivery of the human ND4 complex I subunit in the mouse visual system. Invest Ophthalmol Vis Sci. 2009;50:4205–4214. doi: 10.1167/iovs.08-3214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Qi X, Sun L, Lewin AS, Hauswirth WW, Guy J. The mutant human ND4 subunit of complex I induces optic neuropathy in the mouse. Invest Ophthalmol Vis Sci. 2007;48:1–10. doi: 10.1167/iovs.06-0789. [DOI] [PubMed] [Google Scholar]
- 36.Ellouze S, et al. Optimized allotopic expression of the human mitochondrial ND4 prevents blindness in a rat model of mitochondrial dysfunction. Am J Hum Genet. 2008;83:373–387. doi: 10.1016/j.ajhg.2008.08.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Flierl A, et al. Targeted delivery of DNA to the mitochondrial compartment via import sequence-conjugated peptide nucleic acid. Mol Ther. 2003;7:550–557. doi: 10.1016/s1525-0016(03)00037-6. [DOI] [PubMed] [Google Scholar]
- 38.Esaki M, Kanamori T, Nishikawa S, Endo T. Two distinct mechanisms drive protein translocation across the mitochondrial outer membrane in the late step of the cytochrome b(2) import pathway. Proc Natl Acad Sci USA. 1999;96:11770–11775. doi: 10.1073/pnas.96.21.11770. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Yasuzaki Y, Yamada Y, Harashima H. Mitochondrial matrix delivery using MITO-Porter, a liposome-based carrier that specifies fusion with mitochondrial membranes. Biochem Biophys Res Commun. 2010;397:181–186. doi: 10.1016/j.bbrc.2010.05.070. [DOI] [PubMed] [Google Scholar]
- 40.Tachibana M, et al. Mitochondrial gene replacement in primate offspring and embryonic stem cells. Nature. 2009;461:367–372. doi: 10.1038/nature08368. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Mileshina D, Koulintchenko M, Konstantinov Y, Dietrich A. Transfection of plant mitochondria and in organello gene integration. Nucleic Acids Res. 2011;39:e115. doi: 10.1093/nar/gkr517. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Hauswirth WW, et al. Production and purification of recombinant AAV vectors. In: Palczewski K, editor. Vertebrate Phototransduction and the Visual Cycle. Vol. 316. New York: Academic; 2000. pp. 743–761. [Google Scholar]
- 43.Calvaruso MA, Smeitink J, Nijtmans L. Electrophoresis techniques to investigate defects in oxidative phosphorylation. Methods. 2008;46:281–287. doi: 10.1016/j.ymeth.2008.09.023. [DOI] [PubMed] [Google Scholar]