Abstract
Constitutive activation of the NF-κB pathway is associated with diffuse large B-cell lymphoma (DLBCL) pathogenesis, but whether microRNA dysfunction can contribute to these events remains unclear. Starting from an integrative screening strategy, we uncovered that the negative NF-κB regulator TNFAIP3 is a direct target of miR-125a and miR-125b, which are commonly gained and/or overexpressed in DLBCL. Ectopic expression of these microRNAs in multiple cell models enhanced K63-linked ubiquitination of proximal signaling complexes and elevated NF-κB activity, leading to aberrant expression of its transcriptional targets and the development of a proproliferative and antiapoptotic phenotype in malignant B cells. Concordantly, genetic inhibition of miR-125a/miR-125b blunted NF-κB signals, whereas rescue assays and genetic modulation of a TNFAIP3-null model defined the essential role of the TNFAIP3 targeting on miR-125a/miR-125b-mediated lymphomagenesis. Importantly, miR-125a/mir-125b effects on TNFAIP3 expression and NF-κB activity were confirmed in a well-characterized cohort of primary DLBCLs. Our data delineate a unique epigenetic model for aberrant activation of the NF-κB pathway in cancer and provide a coherent mechanism for the role of these miRNAs in immune cell activation and hematopoiesis. Further, as miR-125b is a direct NF-κB transcriptional target, our results suggest the presence of a positive self-regulatory loop whereby termination of TNFAIP3 function by miR-125 could strengthen and prolong NF-κB activity.
The initiation, duration, and termination of NF-κB signals reflects the concerted action of positive and negative regulators (1). Part of this intricacy resides upstream to IκB kinase (IKK) activation, and involves the assembly and termination of signaling complexes. One of the best characterized molecules that impinge on this process is the ubiquitin editing enzyme TNFAIP3, which blocks the interactions between E3 ligases and E2 ubiquitin conjugating enzymes, thus inhibiting the “positive” K63-linked ubiquitination, while promoting K48-linked ubiquitination and proteasome-dependent degradation (2). Although much progress has been made recently in understanding the catalytic and noncatalytic activities of TNFAIP3 (3, 4), how TNFAIP3 activity is terminated, including the potential role of microRNAs (miRNAs) in this process, has been less studied.
Dysfunctional NF-κB activity contributes to various human conditions, including autoimmune and inflammatory disorders and cancer (5), suggesting that aberrant regulation of TNFAIP3 may contribute to these events. Indeed, genome-wide association studies have linked functional variants of this gene to autoimmune conditions (6) and in a subset of B-cell lymphomas loss-of-function mutations in TNFAIP3 have been reported (7–9). However, in a substantial fraction of lymphomas the molecular basis for the pathogenic constitutive activation of NF-κB remains uncertain (10).
Bidirectional interplays between the NF-κB pathway and miRNAs have been recently illustrated, albeit primarily in a nonmalignant setting (11, 12), thus indicating that dysfunction of these interactions could contribute to the development of NF-κB “addicted” tumors. We recently created an integrative map of the miRNA genome in diffuse large B-cell lymphoma (DLBCL) (13); in that study, copy number analysis revealed widespread gain and loss of chromosomal material targeting multiple miRNA loci, whereas expression measurements defined unique miRNA-driven DLBCL substructures. These data offered an entry point to the investigations reported here, in which we defined a role for miR-125a and miR-125b in suppressing TNFAIP3 and aberrantly activating the NF-κB pathway in DLBCL.
Results
MiR-125a and miR-125b Directly Inhibit TNFAIP3 Expression.
To uncover the putative interactions between DLBCL-relevant miRNAs and the NF-κB pathway, we performed an annotated screening strategy that built on a copy number/expression map of the miRNA genome that we reported earlier (13). In brief, we integrated this original dataset with target prediction algorithms (TargetScan) focusing on miRNAs expected to bind to NF-κB regulators known to play a role in DLBCL pathogenesis (7–9, 14–16). Next, we filtered the initial output to primarily retain miRNAs that (i) map to loci that we showed to be frequently disrupted in DLBCL (13), (ii) are expressed in normal or malignant mature B lymphocytes (13, 17), and (iii) displayed the highest target prediction scores. Among the potential interactions identified by this strategy (Fig. S1), the putative targeting of TNFAIP3 by miR-125a/b was particularly relevant because (i) TNFAIP3 is a tumor suppressor gene in lymphomas (7, 8); (ii) we earlier found miR-125a and miR-125b to be gained and/or overexpressed in 15–20% of DLBCLs (13) (Fig. S1), and the miR-125b1 locus is targeted by chromosomal translocations in both lymphoid and myeloid malignancies (18, 19); and (iii) miR-125a and miR-125b overexpression has been linked to dysfunctional hematopoiesis and aberrant immune cell responses, although a coherent mechanism to explain these effects remains unavailable (20–24).
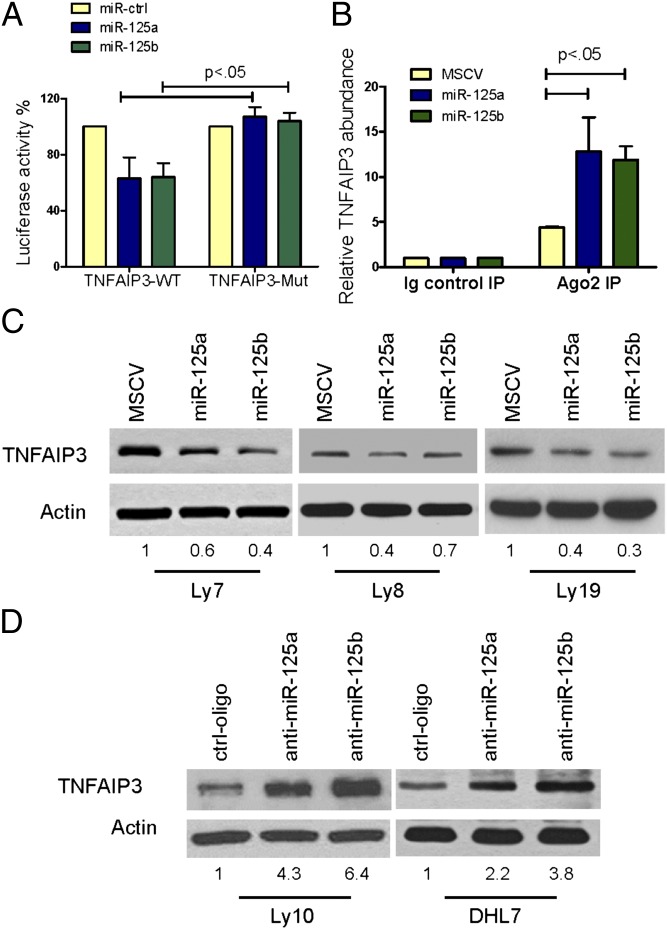
Putative miR-125a/b binding sites were found in the 3′-UTR and coding region of TNFAIP3 (Fig. S2A). Using reporter constructs containing these sites in wild-type (WT) or seed-sequence mutant (Mut) configuration (SI Materials and Methods), we confirmed that miR-125a and miR-125b directly bind to both TNFAIP3 sites (Fig. 1A and Fig. S2B). In addition, we immunoprecipitated Argonaute 2 (AGO2), a core component of RNA-induced silencing complex (RISC), as previously described (25), and demonstrated the binding of miR-125a/b to endogenous TNFAIP3 (Fig. 1B and Fig. S2C). Retrovirus-mediated stable expression of miR-125a or miR-125b in three independent DLBCL cell lines (Fig. S3A) confirmed that these interactions resulted in inhibition of TNFAIP3 protein expression (Fig. 1B), whereas antagomiRs specific to miR-125a and miR-125b elevated TNFAIP3 levels in additional cell models (Fig. 1C and Fig. S3B).
Fig. 1.
MiR-125a and miR-125b directly target TNFAIP3. (A) Luciferase constructs containing miR-125a/b binding sites in wild-type (WT) or mutant (Mut) configuration were cotransfected with miR-125a, miR-125b, or control oligonucleotides. MiR-125a and miR-125b inhibited luciferase activity in the TNFAIP3 WT but had no effect in the mutant construct (P < 0.05, Student’s t test). Data shown are mean ± SD of the ratio of luciferase activity (miR-125a or miR-125b vs. control oligo); experiments were performed in triplicate and repeated three times. (B) Lysates from Ly8 cells expressing MSCV, MSCV–miR-125a, or MSCV–miR-125b were immunoprecipitated with anti-Ago2 antibody or nonspecific IgG; RNA was isolated from the IP fractions and real-time RT-PCR used to demonstrate the significant enrichment for TNFAIP3 in the immunoprecipitates of miR-125a and miR-125b loaded RISC. Data shown are mean ± SD of TNFAIP3 relative abundance in the various Ago2-IPs. RISC-associated GAPDH was used for normalization. (C) Western blots depict TNFAIP3 inhibition in DLBCL cell lines stably expressing miR-125a and miR-125b. (D) TNFAIP3 expression is elevated in DLBCL cell lines transiently transfected with miR-125a– or miR-125b–specific antagomiRs. Densitometric quantification of TNFAIP3 expression, normalized by actin, is shown in C and D.
MiR-125a and miR-125b Expression Elevates NF-κB Activity in DLBCL.
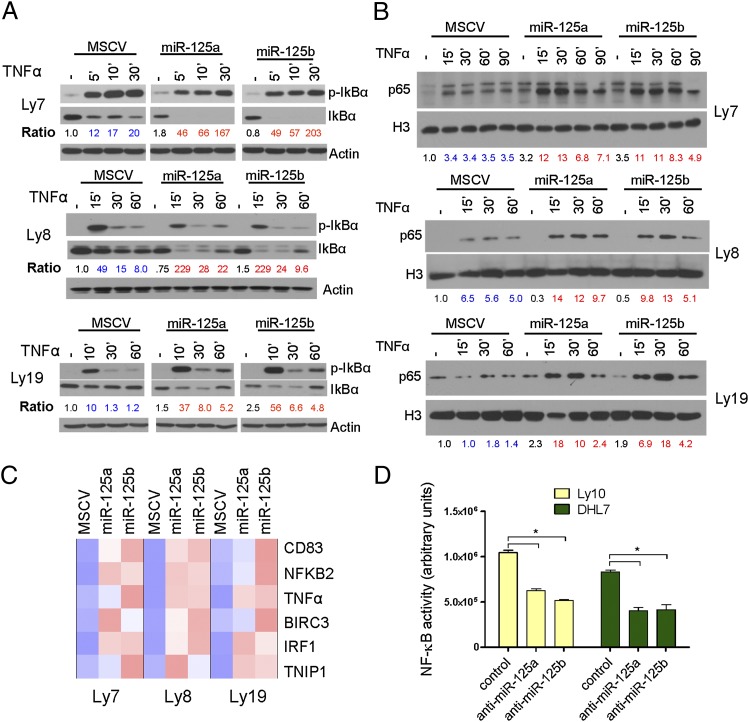
TNFAIP3 functions to terminate the NF-κB activation that follows cellular exposure to various pathogens and proinflammatory cytokines; these activities are well characterized upon several ligand/receptor pairings, including the TNFα/tumor necrosis factor receptor 1 (TNFR1) engagement (2–4). Thus, to test whether miR-125a and/or miR-125b expression enhanced NF-κB activity, we first used multiple independent cell models to measure the degree of IκBα phosphorylation/degradation following TNFα exposure; ectopic expression of these miRNAs led to a higher phosphorylation and sustained downmodulation of IκBα, which was accompanied by nuclear accumulation of p65 and higher expression of multiple NF-κB transcriptional targets (Fig. 2 A–C and Fig. S4A) (P < 0.05, Student’s t test). Concordantly, antagomiRs to miR-125a and miR-125b significantly decreased NF-κB activity (Fig. 2D) (P < 0.05, Student t test), in two additional DLBCL models with high basal levels of NF-κB signals (15, 26).
Fig. 2.
MiR-125a and miR-125b expression modulates NF-κB activity in DLBCL. (A) Immunoblot analyses of phospho-IκBα and IκBα after TNFα stimulation. Phosphorylation and degradation of IκBα are elevated in DLBCL cell lines expressing miR-125a or miR-125b. Ratios of p-IκBα/IκBα are listed in blue (control) and red fonts (miR-125a or miR-125b) and were normalized to time 0 of MSCV-only cells. (B) Ectopic expression of miR-125a and miR-125b increase accumulation of p65 in the nuclear fraction of DLBCL cell lines. Data shown in A and B were independently confirmed in two to three biological replicates. Densitometric quantification of p65 nuclear expression, normalized by H3, is shown. (C) Heat-map display of quantitative real-time RT-PCR measurements of six independent NF-κB transcriptional targets show significantly higher expression in miR-125a and miR-125b expressing cells (P < 0.05, Student’s t test). Data displayed are mean ± SD of the mRNA fold induction following exposure to TNFα (200 ng/mL for 120 min), normalized by MSCV-only cells. All assays were performed in triplicate and confirmed in three biological replicates. (D) AntagomiR-mediated inhibition of miR-125a or miR-125b expression in DLBCL cell lines with high basal NF-κB activity significantly inhibits this pathway, as determined by an ELISA-based measurement of p65 levels (*P < 0.05, Student’s t test). Data shown are mean ± SD of a representative assay performed in triplicate and independently confirmed in three biological replicates.
Inhibition of TNFAIP3 Is Essential for the miR-125a/miR-125b Activation of NF-κB.
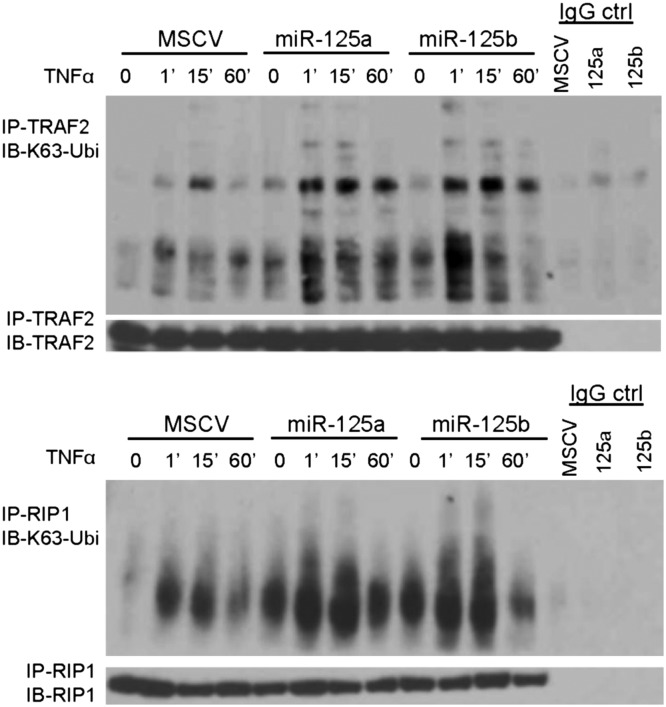
TNFAIP3 controls NF-κB signals in part by antagonizing the K63-linked ubiquitination of members of the activation complexes that assemble downstream of TNFR1 engagement (4). We investigated whether miR-125 expression can interfere in this process by immunoprecipitating TRAF2 and RIP1 following TNFα exposure. Immunoblotting with a K63-specific antibody showed that compared to isogenic controls containing an empty murine stem cell virus (MSCV) vector, in miR-125a– and miR-125b–expressing cells TRAF2 and RIP1 had much higher level of this ubiquitin modification (Fig. 3). Demonstration of the expected TNFα-mediated IκBα phosphorylation and degradation, and TRAF2 and RIP1 expression in input lysates is shown in Fig. S5. These data indicated that the miR-125a/b–mediated fine-tuning of TNFAIP3 expression can prominently down-modulate its activity.
Fig. 3.
miR-125a and miR-125b expression modulate K63-linkage ubiquitination. Immunoprecipitation of TRAF2 (Upper) or RIP1 (Lower) followed by immunoblotting for K63-linkage ubiquitin, in isogenic Ly8 cells stably expressing miR-125a, miR-125b, or empty vector show that these miRNAs enhance the K63 ubuiquitination that follows engagement of the TNFR1 by TNFα (100 ng/mL). Data were confirmed in four biological replicates. Immunoblotting with TRAF2 and RIP1 antibodies confirm equal amount of these proteins in the pulldowns, and IgG controls confirm the specificity of the IPs.
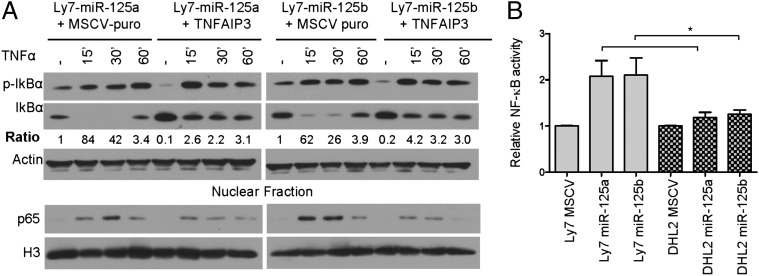
However, as each miRNA is predicted to target several genes (27), it became important to confirm that the elevation of NF-κB activity that followed miR-125a and miR-125b expression resulted primarily, if not exclusively, from the inhibition of TNFAIP3. To address this issue, we used a MSCV–puromycin retrovirus to stably reconstitute TNFAIP3 in our models of ectopic miR-125a and miR-125b expression. To evade miR-125a/b targeting, we created a FLAG-tagged TNFAIP3 construct that lacked 3′-UTR and contained synonymous mutations that disrupted the miR-125 binding site located in its coding region (SI Materials and Methods and Fig. S4B). Upon measuring IκBα phosphorylation/degradation, nuclear accumulation of p65, and the expression of NF-κB transcriptional target in these cells (Fig. 4A and Fig. S4B), we found that reconstitution of TNFAIP3 fully rescued the miR-125a/b–mediated activation of NF-κB. Next, to categorically establish that the targeting and inhibition of TNFAIP3 is essential for the activation of NF-κB in this model, we performed complementary assays in TNFAIP3-null cells. The DLBCL cell line DHL2 (a gift from Laura Pasqualucci, Columbia University, New York, NY) has biallelic truncating mutations in TNFAIP3 that result in a nonfunctional protein (7). We reasoned that if the primary mechanism for the miR-125a/b–mediated activation of NF-κB was indeed down-modulation of TNFAIP3, stable expression of these miRNAs would not influence NF-κB in these cells. Indeed, only a negligible elevation in NF-κB activity was detected in DHL2 cells ectopically expressing miR-125a/b, in marked contrast to the activation detected in TNFAIP3-competent B cells (P < 0.01, Student’s t test) (Fig. 4B and Fig. S4C). Together, these data show that TNFAIP3 targeting is indispensable for the miR-125a and miR-125b–mediated activation of NF-κB signals.
Fig. 4.
Targeting of TNFAIP3 is essential for the miR125a/b–mediated NF-κB activation in DLBCL. (A) Immunoblot analyses of p-IκBα and IκBα show that stable reconstitution of TNFAIP3 expression (Upper) rescued the effects of miR-125a and miR-125b in DLBCL cells; note the markedly lower ratio IκBα phosphorylation/degradation in TNFAIP3–FLAG-expressing cells in comparison with isogenic MSCV–puromycin-expressing cells. MiR-125a/miR-125b–expressing DLBCL cell lines reconstituted with TNFAIP3–FLAG also displayed a limited nuclear accumulation of p65 following TNFα stimulation (Lower). (B) ELISA-based measurement of p65 activity shows that stable expression of miR-125a and miR-125b in the TNFAIP3-null DHL2 cell line has minimal effect on NF-κB function, in statistically significant contrast to the effects of these miRNAs in a TNFAIP3-competent DLBCL cell line (Ly7) (*P < 0.05, Student’s t test). Data shown are mean ± SD (four independent data points) of NF-κB induction following exposure to TNFα (100 ng/mL), normalized by MSCV-only cells.
MiR-125 Mediated Targeting of TNFAIP3 Enhances DLBCL Aggressiveness.
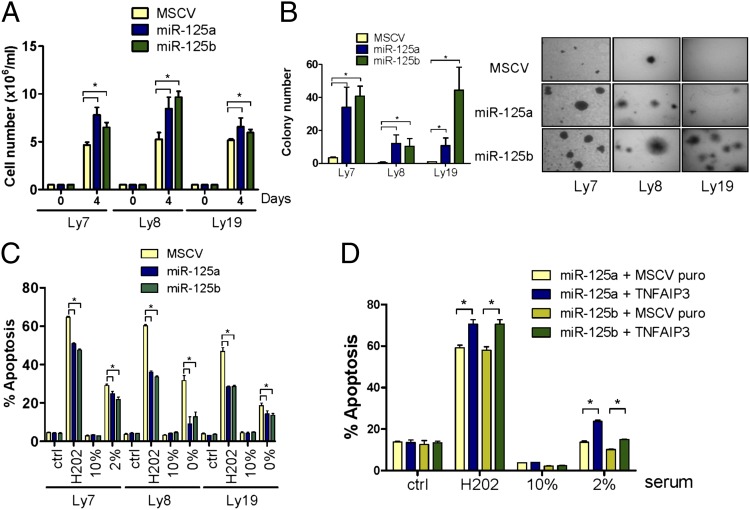
In a group of mature B-cell malignancies, deregulation of NF-κB signals is central to tumor pathogenesis (10). Our discovery that miR-125a and miR-125b targets TNFAIP3 suggested that overexpression of these miRNAs could elicit higher proliferative capacity and an antiapoptotic profile, features often associated with constitutive NF-κB activation in malignant cells (5, 10). In agreement with this prediction, stable expression of miR-125a or miR-125b significantly increased the growth rate of DLBCLs (Fig. 5A and Fig. S6A) (P < 0.05, Student’s t test). Ascertaining the relevance of TNFAIP3 down-regulation in this phenomenon, its reconstitution rescued this progrowth phenotype, whereas miR-125a and miR-125b overexpression did not increase the proliferation of the TNFAIP3-null DHL2 cell line (Fig. S6B). Further documenting the oncogenic properties of these miRNAs in DLBCL, we found a significantly elevated capacity for colony formation in soft agar in the miR-125a and miR-125b gain-of-function models (Fig. 5B), which was rescued by stable expression of the miR-125a/b–resistant TNFAIP3 construct (Fig. S6C).
Fig. 5.
MiR-125 targeting of TNFAIP3 enhances DLBCL aggressiveness. (A) DLBCL cell lines stably expressing miR-125a or miR-125b grow at significantly faster pace than their empty-vector isogenic counterparts. (B) Ectopic expression of miR-125a or miR-125b enhanced colony-formation capability in soft agar in all DLBCL models examined. (C) DLBCL cells expressing miR-125a or miR-125b became resistant to apoptosis induced by H202 and serum deprivation. Ctrl (control) and “10%” data indicate the basal apoptosis rate in cells exposed to vehicle or grown in media supplemented with 10% FBS, respectively. (D) Reconstitution of TNFAIP3 expression rescued DLBCLs from the miR-125a and miR-125b–induced antiapoptotic effects. All assays were performed in triplicate and repeated from two to four times; *P < 0.05, Student’s t test.
One fundamental property ascribed to aberrant NF-κB activation is restriction of apoptotic responses. Therefore, we tested whether overexpression of miR-125a and miR-125b could establish an antiapoptotic profile in DLBCL. To address this issue in the broadest possible manner, we used two general triggers of programed cell death, serum starvation and induction of reactive oxygen species. In all examined instances, miR-125a and miR-125b expression rendered DLBCL cells significantly more resistant to apoptosis than their isogenic counterparts (P < 0.05, Student’s t test) (Fig. 5C and Fig. S6D). Again, reconstitution of TNFAIP3 expression rescued the antiapoptotic properties derived from miR-125a/b expression, whereas stable expression of miR-125a/b in TNFAIP3-null cells did not change their apoptotic rate in response to H202 or serum starvation (Fig. 5D and Fig. S6E).
Expression of miR-125a/b Associates with TNFAIP3 Levels and Is Significantly Correlated with NF-κB Activity in Primary DLBCL.
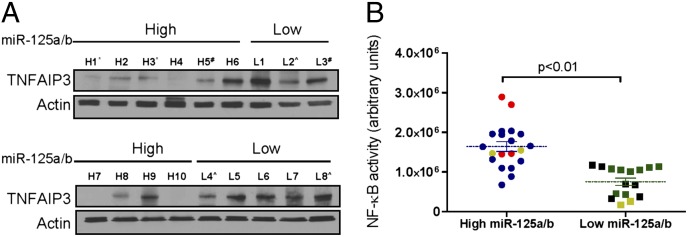
We sought to validate the relevance of our findings in primary human tissues, studying a small collection of well-characterized DLBCL biopsies (13). First, we used real-time stem-loop RT-PCR to define miR-125a and miR-125b levels and dichotomized this tumor cohort into samples with high (n = 10) or low (n = 8) miRNA expression (P < 0.05, Student’s t test; Fig. S7A). With Western blotting, we found a significantly lower expression of TNFAIP3 in primary DLBCLs expressing high levels of miR-125a and/or miR-125b than in tumors in the low miRNA expression category (Fig. 6A and Fig. S7B) (P < 0.01, Mann–Whitney test, normalized densitometric quantification). It should be noted that despite the coregulation of these miRNAs expression in primary DLBCL (r = 0.68, Pearson correlation coefficient), for the purpose of this analysis, higher levels of either miR-125a or miR-125b placed the tumor in the “high expression” category. Subsequently, using ELISA-based measurements, we detected a significantly elevated NF-κB activity in tumors with high miR-125a and/or miR-125b expression than in those expressing these miRNAs at lower levels (P < 0.01, Student’s t test) (Fig. 6B). Nonetheless, primary tumors are complex samples where a single genetic event (e.g., high expression of miR-125a/b) cannot be studied in isolation. Thus, to refine the relevance of our information, we used our previously generated (13) immunohistochemistry (IHC) data to classify these tumors as germinal center B cells (GCB) and non-GC (NGC) types, array-comparative genomic hybridization (CGH) analysis to define the copy number at the TNFAIP3, miR-125b1, miR-125b2, and miR-125a loci, and sequenced CARD11, MYD88, C79B, and TNFAIP3, genes that can be somatically mutated in DLBCL with high NF-κB activity.
Fig. 6.
MiR-125a/b expression in primary DLBCL modulates TNFAIP3 expression and NF-κB activity. (A) Western blot analysis showed a significant suppression in TNFAIP3 expression in tumors with high (H1–H10) vs. low miR-125a and/or miR-125b levels (L1–L8) (P < 0.01 Mann–Whitney, normalized densitometric values (also Fig. S7). *, samples with copy number gain at the miR-125b1 locus; #, samples with hemizygous loss of miR-125; ^, samples monoallelic mutation or loss of TNFAIP3. (B) ELISA-based quantification of p65 in protein lysates from primary DLBCLs showed significantly higher NF-κB activity (P < 0.01, Student’s t test) in tumors expressing high miR-125a and/or miR-125b levels and consequently low TNFAIP3; p65 measurements were performed in duplicate and all 36 values are included in the display. Data points labeled in red correspond to the samples with copy number gain at the miR-125b1 locus; those with loss of miR-125b are shown in gold; and the samples with monoallelic loss or mutation in TNFAIP3 are shown in black.
There was no statistically significant enrichment for GCB or NGC samples in either tumor category (i.e., high or low miR-125a/b expression) (P = 0.34, Fisher’s exact probability test) (Table S1). In addition, only 1 of 13 tumors analyzed showed a hemizygous deletion of the TNFAIP3 locus (tumor L8, which displayed low miR-125a/b expression and correspondingly high expression of TNFAIP3 (Fig. 6A and Table S1), suggesting that genomic loss at this locus is not a confounding variable in our analysis. Conversely, in 2 tumors with high and in 1 with low miR-125b expression, we identified copy number gain and loss of the miR-125b1 locus, respectively (Table S1) thus indicating a contribution of genomic abnormalities in the expression of miR-125b in DLBCL. Finally, we found five mutations in 4 of the 18 DLBCLs analyzed: two DLBCLs that belonged to the low miR-125a/b group, harbored heterozygous mutations in TNFAIP3 (splice-site truncating and nonsense), whereas two samples in the high miR-125 expression category had mutations in MYD88 and CD79B (1 tumor with both genes mutated, and the second with MYD88 mutation only) (Table S1). These findings suggest that high miR-125a/miR-125b expression and genetic loss of TNFAIP3 may be mutually exclusive events in this setting, but cosegregate with gain-of-function mutations in positive regulators of this pathway. However, given the relatively small numbers of primary DLBCLs analyzed in the present investigation, further studies will be needed to confirm the strength of these associations.
Discussion
We found that targeting of TNFAIP3 links miR-125a and miR-125b to NF-κB and lymphomagenesis. These data suggest a unique model for constitutive activation of the NF-κB pathway in DLBCL and provide a mechanism for the association of miR-125 expression with heightened activation of immune cells and dysfunctional hematopoiesis (20–24).
In earlier work, we had identified miR-125a and miR-125b among a small number of miRNAs that could efficiently subclassify DLBCLs on the basis of their differential expression (13). Further, in that DLBCL cohort miR-125b expression was correlated with the copy number status at the miR-125b1 locus (chromosome 11q24), with overexpression significantly associated with copy number gain (13), suggesting that miR-125 could play a role in DLBCL biology; the identification and validation of TNFAIP3 as direct target of miR-125a and miR-125b confirms this concept.
Stable expression of miR-125a/b in three well-characterized GCB DLBCL cell lines consistently amplified NF-κB function, whereas antagomiRs directed at miR-125a or miR-125b, via up-regulation of TNFAIP3 expression, significantly decreased NF-κB activity in two DLBCL cell lines with high miR-125 expression and oncogenic mutations in NF-κB regulators (Ly10, MYD88; DHL7, CARD11) (15, 26). These data are relevant at several levels: (i) they suggest that in tumors with proximal gain-of-function mutations (i.e., CARD11, MYD88, and CD79), the plasticity of the NF-κB pathway is preserved and elevating the expression/function of a physiologic negative regulator (e.g., TNFAIP3) can reset signal intensity; (ii) they indicate that overexpression of these miRNAs may be more frequent in mutant tumors, thus removing a physiologic break on NF-κB activation, as we preliminarily showed in our primary tumor cohort; and (iii) the results from the cell line Ly10, which also is hemizygous at the TNFAIP3 locus (7), suggest that a single copy of this gene is functionally relevant and that when the suppressive effects of mir-125a/b on the remaining allele are lifted it can curtail NF-κB signals. These considerations may all be instructive in the eventual design of therapeutic strategies to attack NF-κB in DLBCL.
The assembly of the signaling complexes that converge to activate IKK is regulated by distinct models of polyubiquitylation. TNFAIP3 plays a key role at this juncture by antagonizing the formation of the “pro-signal” K63-linked ubiquitin chains, and by promoting K48 ubiquitination, a mark for proteasome degradation (2). We showed that K63 ubiquitination of TRAF2 and RIP1, important components of the IKK activation complex downstream to TNFR1 engagement, were elevated in DLBCL expressing miR-125a/b, indicating that this miRNA-mediated fine-tuning of TNFAIP3 expression can prominently down-modulate its activity. Still, as each miRNA is predicted to target hundreds of transcripts, we resorted to two complementary strategies, TNFAIP3 reconstitution in a high miR-125a/b background and miR-125a/b stable expression in TNFAIP3-null cells, to conclusively demonstrate that the down-regulation of TNFAIP3 is essential for these miRNAs to enhance NF-κB activity. MiR-125a and miR-125b regulate other genes that are relevant for lymphoma biology. Particularly important is the recently reported targeting of BLIMP1 and IRF4 by miR-125b (which we now demonstrate to be also mediated by miR-125a; Fig. S8), transcription factors important in the differentiation of germinal center B cells into plasma cells (24, 28). Thus, together with our discovery of the TNFAIP3 targeting, miR-125b and miR125a is now shown to impact on two well-defined cooperative lymphomagenic processes: constitutive activation of NF-κB and blockade in B-cell differentiation. Indeed, to create a faithful murine model of human B-cell lymphoma, dual conditional mutant mice with enforced expression of constitutively active IKK2 and deletion of BLIMP1 were recently generated (29); conceivably this model can be recapitulated with conditional expression of a miR-125b or miR-125a in GC B cells.
Ectopic expression of miR-125a/b enhanced the proliferative capacity and defined an antiapoptotic profile in TNFAIP3-competent DLBCL cell lines, features associated with activation of NF-κB signals. These findings agree with the antiapoptotic behavior recently detected in a B-cell conditional TNFAIP3 knockout model (30). In these mice, homozygous or hemizygous deletion of TNFAIP3 led to the accumulation of GC B cells in association with resistance to programmed cell death. Importantly, detection of a clear phenotype in mice hypomorphic for TNFAIP3 further highlights the biological relevance of miR-125a/b–mediated fine-tuning of this protein levels, as miRNAs classically down-regulate but do not abolish target gene expression (31).
Recurrent chromosomal translocations targeting the miR-125b1 locus have been reported in myelodysplastic syndrome and acute myeloid and lymphoid leukemia, in most of these instances in association with strong miRNA up-regulation (18, 19). Our data in DLBCL reinforce the concept that elevated miR-125a/b expression plays a role in the pathogenesis of human hematological malignancies. The mechanism for overexpression of miR-125a and miR-125b in DLBCL is probably multiple and could include copy number gain at the miR-125b1 locus, as we reported earlier (13), and transcriptional up-regulation of the miRNA primary transcripts. In the latter, a previously reported functional NF-κB binding site within the promoter region of primary miR-125b1 is particularly relevant (12). This finding suggests that NF-κB could induce miR-125b1 transcription, which in turn may extend/strengthen these signals by suppressing TNFAIP3. We found that miR-125a and miR-125b expression is often coregulated in DLBCL. Thus, although the promoter region at the pri–miR-125a locus has not yet been identified, it is possible this putative self-regulatory loop be shared by both miRNAs.
Our investigation of a well-characterized primary tumor cohort confirmed that DLBCLs overexpressing miR-125a and/or miR-125b have a significantly lower TNFAIP3 expression and higher NF-κB activity. We did not find an association between these features and the GCB and non-GCB phenotypes, as defined by IHC analysis. This was somewhat surprising given the elevated NF-κB activity in the subset of tumors with high 125a/b expression. However, considering the relatively small number of cases included in the present study it is not possible at the moment to the precisely establish the correlation between miR-125a/b expression and subsets of DLBCL; a follow-up analysis of a large tumor set molecularly classified into GCB and activated B-cell–like (ABC) DLBCL should define this issue. Of interest, analysis of subpopulations of normal human mature B cells indicated that miR-125a/b levels are highest in centroblasts (17, 28, 32), a finding that may also impinge on the expression pattern of these miRNAs in distinct molecular subsets of DLBCL.
In our sequencing studies, we found that only cases with low miR-125a/b expression had heterozygous loss-of-function mutation in TNFAIP3, suggesting that high miRNA expression and genetic loss of TNFAIP3 may be mutually exclusive in DLBCL. Gain-of-function mutations in positive regulators of the NF-κB pathway may have a particularly ominous consequence to DLBCL pathogenesis when associated with loss of TNFAIP3, as was the case in the lymphomas in our series that contained mutations in MYD88 and CD79b. Nonetheless, in most instances miR-125a/b overexpression and elevated NF-κB occurred independently of these genetic alterations. Future investigations should define whether rarer mutations in NF-κB regulators are present in these cases, or whether the activation of NF-κB derives from microenvironmental factors that are epigenetically sustained by miR-125a/b–mediated suppression of TNFAIP3.
In this work, we identified and characterized miR-125a and miR-125b as regulators of TNFAIP3 expression and function, and consequently NF-κB activity. The fine-tuning of TNFAIP3 levels mediated by miR-125 expression had a striking impact on K-63 ubiquitination of TRAF2 and RIP1, IκBα degradation, p65 nuclear accumulation, and transcription of NF-κB target genes. These events defined an antiapoptotic profile compatible with constitutive NF-κB activation and enhanced the fitness of B-lymphoma cells. Together, our data provide a coherent mechanism for a miR-125 role in immune cell activation and oncogenesis, and point to a unique model for constitutive activation of the NF-κB pathway in DLBCL.
Materials and Methods
Cell Lines and Primary Tumors.
Human DLBCL cell lines were cultured as we previously described (33) and are detailed in SI Materials and Methods. The relevant features of the primary DLBCLs analyzed have been described (13) and are summarized in the Table S1.
Reporter Assays.
The two TNFAIP3 miR-125a/b binding sites (WT or mutant) were cloned in the 3′-UTR of the luciferase gene and these constructs, used to determine reporter activity, are detailed in SI Materials and Methods.
Modulation of miR-125a, miR-125a, and TNFAIP3 Expression in DLBCL Cell Lines.
Knockdown of miR-125a/b expression and generation of multiple DLBCL cell lines stably expressing these miRNAs, or TNFAIP3, is described in SI Materials and Methods.
Subcellular Fractionation and Immunoblotting.
Cytoplasmic and nuclear protein fractions were isolated from genetically modified DLBCL cell lines as we described (33) and are detailed in SI Materials and Methods.
Expression of NF-κB Target Genes and NF-κB Activity Assay.
Expression of CD83, BIRC3, NFKB2, TNF, IRF1, and TNIP1 was determined by quantitative real-time RT-PCR, in DLBCL cell lines stably expressing miR-125a, miR-125b, or empty MSCV vector, as described in SI Materials and Methods. DNA-binding activity of p65 was examined in protein extracts of DLBCL cell lines, as well as primary DLBCL biopsies, according to the manufacturer’s instructions (TransAM p65 kit; Active Motif).
Immunoprecipitations.
TRAF2 and RIP1 were immunoprecipitated from DLBCL cell lines genetically modified to express miR-125a/b and pulldowns immunoblotted to detect K63 ubiquitination. Immunoprecipitation of AGO2-containing RISC in a relevant DLBCL model was performed as described (25). Both assays are detailed in SI Materials and Methods.
Cell Growth, Soft Agar Colony-Forming Assays, and Apoptosis Rate.
Cell growth, soft agar colony-forming assays, and apoptosis rate, defined in multiple DLBCL cell lines genetically modified to express miR-125a and miR-125b, are described in detail in SI Materials and Methods. All assays were performed in triplicate and repeated three times.
Statistics.
The Mann–Whitney test was used to determine the significance of the difference in TNFAIP3 expression in primary DLBCL. For all other assays, the statistical analyses were performed with two-tailed Student’s t test. P < 0.05 was considered significant. Data analyses were performed in the Prism software (version 5.0; GraphPad) and Excel software (Microsoft).
Supplementary Material
Acknowledgments
We thank Patricia Dahia for suggestions throughout the execution of this project. This work was supported by Grant 5R01CA138747 (to R.C.T.A.), a Young Investigator Award from the Voelcker Fund (to R.C.T.A.), and Grant 2P30CA054174-17.
Footnotes
The authors declare no conflict of interest.
*This Direct Submission article had a prearranged editor.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1200081109/-/DCSupplemental.
References
- 1.Baltimore D. NF-κB is 25. Nat Immunol. 2011;12:683–685. doi: 10.1038/ni.2072. [DOI] [PubMed] [Google Scholar]
- 2.Hymowitz SG, Wertz IE. A20: From ubiquitin editing to tumour suppression. Nat Rev Cancer. 2010;10:332–341. doi: 10.1038/nrc2775. [DOI] [PubMed] [Google Scholar]
- 3.Skaug B, et al. Direct, noncatalytic mechanism of IKK inhibition by A20. Mol Cell. 2011;44:559–571. doi: 10.1016/j.molcel.2011.09.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Shembade N, Ma A, Harhaj EW. Inhibition of NF-kappaB signaling by A20 through disruption of ubiquitin enzyme complexes. Science. 2010;327:1135–1139. doi: 10.1126/science.1182364. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Ben-Neriah Y, Karin M. Inflammation meets cancer, with NF-κB as the matchmaker. Nat Immunol. 2011;12:715–723. doi: 10.1038/ni.2060. [DOI] [PubMed] [Google Scholar]
- 6.Adrianto I, et al. BIOLUPUS and GENLES Networks Association of a functional variant downstream of TNFAIP3 with systemic lupus erythematosus. Nat Genet. 2011;43:253–258. doi: 10.1038/ng.766. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Compagno M, et al. Mutations of multiple genes cause deregulation of NF-kappaB in diffuse large B-cell lymphoma. Nature. 2009;459:717–721. doi: 10.1038/nature07968. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Schmitz R, et al. TNFAIP3 (A20) is a tumor suppressor gene in Hodgkin lymphoma and primary mediastinal B cell lymphoma. J Exp Med. 2009;206:981–989. doi: 10.1084/jem.20090528. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Kato M, et al. Frequent inactivation of A20 in B-cell lymphomas. Nature. 2009;459:712–716. doi: 10.1038/nature07969. [DOI] [PubMed] [Google Scholar]
- 10.Staudt LM. Oncogenic activation of NF-kappaB. Cold Spring Harb Perspect Biol. 2010;2:a000109. doi: 10.1101/cshperspect.a000109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Li T, et al. MicroRNAs modulate the noncanonical transcription factor NF-kappaB pathway by regulating expression of the kinase IKKalpha during macrophage differentiation. Nat Immunol. 2010;11:799–805. doi: 10.1038/ni.1918. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Zhou R, et al. NF-kappaB p65-dependent transactivation of miRNA genes following Cryptosporidium parvum infection stimulates epithelial cell immune responses. PLoS Pathog. 2009;5:e1000681. doi: 10.1371/journal.ppat.1000681. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Li C, et al. Copy number abnormalities, MYC activity, and the genetic fingerprint of normal B cells mechanistically define the microRNA profile of diffuse large B-cell lymphoma. Blood. 2009;113:6681–6690. doi: 10.1182/blood-2009-01-202028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Davis RE, et al. Chronic active B-cell-receptor signalling in diffuse large B-cell lymphoma. Nature. 2010;463:88–92. doi: 10.1038/nature08638. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Ngo VN, et al. Oncogenically active MYD88 mutations in human lymphoma. Nature. 2011;470:115–119. doi: 10.1038/nature09671. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Lenz G, et al. Oncogenic CARD11 mutations in human diffuse large B cell lymphoma. Science. 2008;319:1676–1679. doi: 10.1126/science.1153629. [DOI] [PubMed] [Google Scholar]
- 17.Basso K, et al. Identification of the human mature B cell miRNome. Immunity. 2009;30:744–752. doi: 10.1016/j.immuni.2009.03.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Bousquet M, et al. Myeloid cell differentiation arrest by miR-125b-1 in myelodysplastic syndrome and acute myeloid leukemia with the t(2;11)(p21;q23) translocation. J Exp Med. 2008;205:2499–2506. doi: 10.1084/jem.20080285. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Chapiro E, et al. A new recurrent translocation t(11;14)(q24;q32) involving IGH@ and miR-125b-1 in B-cell progenitor acute lymphoblastic leukemia. Leukemia. 2010;24:1362–1364. doi: 10.1038/leu.2010.93. [DOI] [PubMed] [Google Scholar]
- 20.Bousquet M, Harris MH, Zhou B, Lodish HF. MicroRNA miR-125b causes leukemia. Proc Natl Acad Sci USA. 2010;107:21558–21563. doi: 10.1073/pnas.1016611107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Guo S, et al. MicroRNA miR-125a controls hematopoietic stem cell number. Proc Natl Acad Sci USA. 2010;107:14229–14234. doi: 10.1073/pnas.0913574107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.O’Connell RM, et al. MicroRNAs enriched in hematopoietic stem cells differentially regulate long-term hematopoietic output. Proc Natl Acad Sci USA. 2010;107:14235–14240. doi: 10.1073/pnas.1009798107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Ooi AG, et al. MicroRNA-125b expands hematopoietic stem cells and enriches for the lymphoid-balanced and lymphoid-biased subsets. Proc Natl Acad Sci USA. 2010;107:21505–21510. doi: 10.1073/pnas.1016218107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Chaudhuri AA, et al. MicroRNA-125b potentiates macrophage activation. J Immunol. 2011;187:5062–5068. doi: 10.4049/jimmunol.1102001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Glorian V, et al. HuR-dependent loading of miRNA RISC to the mRNA encoding the Ras-related small GTPase RhoB controls its translation during UV-induced apoptosis. Cell Death Differ. 2011;18:1692–1701. doi: 10.1038/cdd.2011.35. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Pasqualucci L, et al. Analysis of the coding genome of diffuse large B-cell lymphoma. Nat Genet. 2011;43:830–837. doi: 10.1038/ng.892. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Bartel DP. MicroRNAs: Target recognition and regulatory functions. Cell. 2009;136:215–233. doi: 10.1016/j.cell.2009.01.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Malumbres R, et al. Differentiation-stage-specific expression of microRNAs in B-lymphocytes and diffuse large B-cell lymphomas. Blood. 2008 doi: 10.1182/blood-2008-10-184077. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Calado DP, et al. Constitutive canonical NF-κB activation cooperates with disruption of BLIMP1 in the pathogenesis of activated B cell-like diffuse large cell lymphoma. Cancer Cell. 2010;18:580–589. doi: 10.1016/j.ccr.2010.11.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Tavares RM, et al. The ubiquitin modifying enzyme A20 restricts B cell survival and prevents autoimmunity. Immunity. 2010;33:181–191. doi: 10.1016/j.immuni.2010.07.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Selbach M, et al. Widespread changes in protein synthesis induced by microRNAs. Nature. 2008;455:58–63. doi: 10.1038/nature07228. [DOI] [PubMed] [Google Scholar]
- 32.Gururajan M, et al. MicroRNA 125b inhibition of B cell differentiation in germinal centers. Int Immunol. 2010;22:583–592. doi: 10.1093/intimm/dxq042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Rai D, Kim SW, McKeller MR, Dahia PL, Aguiar RC. Targeting of SMAD5 links microRNA-155 to the TGF-beta pathway and lymphomagenesis. Proc Natl Acad Sci USA. 2010;107:3111–3116. doi: 10.1073/pnas.0910667107. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.