Abstract
Herein we provide a concise review of the state of methylation research as it pertains to clinical oral cancerous and precancerous tissues. We provide context for ongoing research efforts in this field and describe technologies that are presently being applied to analyze clinical specimens. We also discuss the various recurrent methylation changes that have been reported for oral malignancy (including those genes frequently silenced by promoter methylation and the small RNAs with activity modulated by methylation changes) and describe surrogate disease markers identified via epigenetic analysis of saliva and blood specimens from patients with oral cancer.
1. Background
Oral cancer remains a major global killer [1]. Its entrenched poor survival rates make essential the ongoing molecular evaluation of oral cancers and precancers for the purpose of uncovering new tools for detecting and treating this disease. As with most solid epithelial tumors, oral cancers develop through a series of histopathological stages; hyperplasia leads to various degrees of dysplasia, which are followed by carcinoma in situ and finally invasive disease stages. The accumulation of various genetic and epigenetic alterations is understood to drive this progression paradigm. Herein we will discuss the role of DNA methylation in clinical oral tumorigenesis, focusing specifically on oral squamous cell carcinomas (OSCCs).
DNA methylation occurs most frequently at cytosine residues of CpG dinucleotides in gene promoter regions, and much less frequently, within a gene [2]. CpG islands (CpG-rich regions spanning >500 bp with >55% GC content) exist in approximately 60–70% of promoters in the human genome [2, 3]. Methylation in the promoter region of a given gene can serve to decrease expression of that gene. This is thought to occur by either physically inhibiting the binding of proteins essential for transcription, or by recruiting proteins that have transcription repressive properties [4]. This reversible process helps govern gene expression activity in individual cells and is commonly disrupted in cancer, where gene silencing via methylation in particular can contribute to Knudsonian two-hit disruption of tumor suppressor genes [5].
Further, global hypomethylation of genes is understood to serve as a mechanism of oncogene activation, providing another avenue for methylation changes to contribute to tumorigenesis. In addition to being identified as an early event in tumorigenesis for many epithelial cancers, aberrant methylation has also been identified in dysplasia—and tumor—adjacent “normal” tissues, indicating additional complexity at this level of epigenetic dysregulation.
2. Techniques and Methodologies for Evaluating Methylation Changes
Myriad technologies exist for detecting DNA methylation alterations and most of these have already been applied in an oral cancer context. The most common technique is methylation-specific PCR (MSP). This approach involves bisulfate conversion of unmethylated cytosines within CpG islands of the genome, with methylated cytosines unchanged by this conversion. In MSP, following design of primers for both methylated and unmethylated sequences of a specific locus, converted DNA is amplified and then separated by gel electrophoresis. Differential analysis of resultant MSP products—say for tumor versus patient-matched normal tissues—reveals changes in methylation status. MSP is particularly useful because it has sufficient sensitivity to detect one methylated cytosine in 1000 and the primers used are also highly specific (i.e., have a low false positive rate). MSP is also a relatively quick and affordable technique, though its limitations include the reality that it is more qualitative than quantitative and the fact that it is generally unhelpful for the assessment of genome-wide methylation changes.
Other techniques for evaluating methylation changes are built on the principles underpinning MSP. MethyLight—which integrates sodium bisulfite conversion and quantitative fluorescence PCR—offers a sensitive, highly specific, and rapid means for assessing methylation status for a particular locus [6]. This technique involves use of primers with a fluorescent 5′ reporter dye (typically FAM) and a 3′ quencher dye. During amplification, Taq DNA polymerase cleaves the probe and releases the reporter dye. The resultant fluorescence is quantified by laser in associated equipment and will be proportional to the number of methylated cytosines at a given locus. This approach allows a more precise quantitation of methylation status compared to MSP, which can only be measured qualitatively. Additionally, MethyLight has been shown to be more sensitive than MSP by a factor of ten (detecting even one methylated cytosine in 10,000) [6].
Sequence alterations resulting from bisulfite conversion can remove cut sites recognized by specific restriction enzymes. For example, if methylation has been lost, the cut site for BstUI may be lost via conversion of a CGCG sequence to TGTG. COBRA (combined bisulfite restriction analysis) leverages this fact to allow quantification of methylation in PCR products generated following bisulfite conversion [7]. Ultimately, the amount of total methylation per locus corresponds to the degree to which the strands are cut by the restriction enzyme. The strength of this technique lies in its quantitative results, ease of use, and compatibility with paraffin embedded samples. However, COBRA does have its limitations, as it is a locus specific technique and thus unhelpful for a genome wide analysis.
Pyrosequencing is a technique that is quantitative, easy to use, and accurate [8]. It utilizes a sequence-by-synthesis method that monitors the bioluminometric signal that results when a pyrophosphate group is released during DNA synthesis. This technique can assess the methylation status of genes by bisulphate conversion, PCR, and subsequent comparison of the ratio of T and Cs in the samples.
A recurring disadvantage to the above techniques is the fact that they do not easily facilitate global analyses of methylation changes; they are all locus-specific. Several techniques for global evaluation of changes in the methylome have been developed to address this issue. Currently, the most common approach for genome-wide analysis involves methylation-specific microarrays. A range of arrays are available, but most are based on one of three general principles to sample processing: (MSRE) methylation-sensitive restriction enzyme digests, methylation-specific immunoprecipitation, or sodium bisulfite conversion [9].
Regarding enzyme digestion approaches, sample processing with endonucleases specific to methylation sites (e.g., HpA1I) was one of the first methods of array-based methylation profiling [10]. There are several different types of arrays that use this approach. For example, in differential methylation hybridization arrays, DNA is digested by MSREs. This pool of DNA fragments is labeled with one fluorescent dye, and a pool of the same sample with no digestion is labeled with second dye. They are concurrently hybridized on an array and relative signal intensities are measured to determine methylation status at a given locus [10]. This method of methylation profiling can interrogate everything from hundreds of different CpG islands to the entire genome. These approaches all have relatively high sensitivity; however, their utility for analyzing disease tissues can be variable as DNA sample input requirements can be quite high, requiring approximately 2 μg of tissue, and clinical lesions—particularly premalignant ones—can be quite small (thus limiting the amount of specimen available for analysis). Additionally, these methylation arrays can be prone to false positives, have a low throughput as only several samples can be processed in parallel, and analysis is limited to those sequences that possess recognition sites for the restriction enzyme associated with a given platform [9].
Immunoprecipitation of either methylated DNA or chromatin, known as MeDIP and ChIP, respectively, has also been used to facilitate global analyses of methylation alterations [11, 12]. These methods use antibodies to pull down material of interest and subsequently subject it to microarray analysis. This method has the ability to detect methylation status at a genome-wide level, does not require specific primers, and avoids the sequence bias introduced by restriction enzyme methods [9]. However, this technique can also have lower throughput than other approaches and has low sensitivity to methylated areas that occur in CpG poor regions. Additionally, instead of microarray analysis of pulled down material, ChIP-Seq combines immunoprecipitation with sequencing technology to elucidate protein-DNA interactions on a genome-wide scale [13].
A third common approach to methylation-specific microarrays involves use of the sodium bisulfite conversion methods described above. Illumina arrays offer one platform based on this approach [14]. These arrays are automated in use and allow analysis of hundreds of samples simultaneously. They are also capable of delineating methylation status for thousands of CpG sites (if not the entire genome), allow determination of the methylation status of specific CpG sites, and have relatively low sample input requirements (thus more readily facilitating analysis of low yield clinical tissues). That said, this technique can introduce bias via incomplete bisulfite conversion reactions and PCR.
Lastly, a technique that is rapidly becoming increasingly common is whole genome bisulfite sequencing. Due to improvements in protocols, increase of throughput, and the rapid decline of sequencing cost, this method is robust and highly sensitive. In brief, this protocol involves bisulfite conversion, PCR amplification, and sequencing of products to determine methylation status to a single base pair resolution [15]. Historically, this method was problematic due to processing errors of bisulfite conversion, degradation of DNA due to long processing times, and the need for a relatively large amount of sample [16]. However, improvements on protocols have greatly increased efficiency of this technique, which has great promise for the profiling of methylation status.
3. Genes Evincing Methylation Changes in Oral Squamous Cell Carcinomas
As specific genes are implicated as critical drivers of malignant phenotypes, they become attractive candidates for evaluation as targets of novel therapeutics or as biomarkers for guiding patient management decisions. Recurrent methylation-mediated alterations of several genes have been reported for all invasive epithelial cancer types, including oral tumors. It is understood that in general cancer cells exhibit increased global hypomethylation with specific regions of hypermethylation. However, it is currently not known how specific genes are targeted. The major etiological factors for oral squamous cell carcinoma (OSCC) are smoking, alcohol use, and (to a lesser extent) Human Papillomavirus (HPV) infection [17]. How these factors influence DNA methylation in oral tumorigenesis has not been evaluated. However, smoking has been shown in other cancer types to activate DNA methyltransferase 1 (DNMT1) which catalyzes DNA methylation [18, 19]. A summary of specific genes implicated in oral cancers and understood to be governed by methylation changes follows below.
CDKN2A, mapping to chromosome 9p21.3, produces two major proteins: p16(INK4), which is a cyclin-dependent kinase inhibitor, and p14(ARF), which binds the p53-stabilizing protein MDM2 and is involved in cell cycle control. Deletion at this locus is regularly reported to be one of the earliest events in oral cancer initiation and progression [20]. Hypermethylation of the CDKN2A promoter region has been extensively evaluated in oral cancers with the frequency of hypermethylation being reported as anywhere from 28% to 86% [21, 22]. Aberrant methylation of this locus in noncancer controls has not been detected [23–25]. A panel of cell lines was investigated for homozygous deletion, hypermethylation, and point mutations at p16, with results indicating that the first two alteration types were the more common modes of p16 disruption in OSCC [26].
In a specific cohort of betel chewing individuals with oral cancer, methylation of p16 was detected in 63% of OSCCs and 67% of verrucous carcinomas [27]. In a panel of individuals of Indian descent, methylation of p16 was detected in 23% of OSCC cases [24]. In general, data do suggest that differences in patient ethnicity, etiological factors, and tissue type (since the OSCC category actually spans different tissues) can influence the molecular alterations detected for disease.
Several groups have correlated methylation of p16INK4A/p14ARF with various clinical features for oral cancers, though results have varied. In one study, it was observed that people with p16 promoter methylation had a lower mean age, a higher risk of lymph node invasion in young patients, a higher risk of distant metastasis in older patients, and shortened disease free survival in older patients [28]. Other work showed that p16INK4A methylation was associated with increased likelihood of disease recurrence, whereas p14ARF is associated with lower recurrence rates [29]. Concurrent promoter hypermethylation of p16 and p14 correlated significantly with tumor size and lymph node metastasis and with later stage of OSCC in one study [30], while a separate study found methylation of p14ARF alone correlated with a good prognosis for patients [31]. Larger scale trials are needed to fine tune how methylation status of p16 and p14 promoters may best be applied to manage clinical decisions.
P16 promoter methylation has been assessed for squamous cell carcinomas of the tongue as well as in margin tissues that remain in patients following surgical resections. As expected, tumors showed a high frequency of p16 promoter hypermethylation (86.8%) [32]. Regarding tissues at surgical margins—which were all histologically-characterized as disease-free—43.3% exhibited p16 promoter hypermethylation [32]. Significantly, those cases with margin tissues harboring p16 hypermethylation had a 6.3-fold increased risk for local recurrence.
Similarly, a separate group assessed p16 promoter hypermethylation status in OSCC tumors, associated normal tissues, and a panel of healthy controls [33]. They found no methylation of p16 in the healthy control group whereas p16 methylation in OSCC-associated normal tissues was detected for 27.3% of cases (and in all of those cases, concurrent p16 hypermethylation was also detected for matched tumors). In this study, clinical features and habitual factors did not correlate with methylation status. Another study did not report prognostic significance for p16 methylation, again showing that there is lingering ambiguity regarding whether p16 hypermethylation will have utility as a clinical biomarker [34].
E-cadherin (CDH1) plays a critical role in cell adhesion processes and is known to significantly influence epithelial tissue architecture [35]. With respect to malignancy, it is known to function as a suppressor of invasion and metastasis formation and has previously been reported as undergoing hypermethylation-mediated silencing in several cancer types [35]. Previous studies of tongue squamous cell carcinoma revealed that downregulation of CDH1 expression via promoter hypermethylation was significantly associated with poorer rates of disease-free survival [36]. Independent reports have confirmed association of epigenetically-silenced CDH1 expression and poorer overall survival for oral cancer (also demonstrating that CDH1 promoter hypermethylation is associated specifically with poorer survival for node-positive cases and individuals with stage III disease) [34]. Other groups, on the other hand, while reporting associations between CDH1 promoter hypermethylation and oral cancers (when compared to normal oral mucosa), have failed to detect significant associations between these same clinical parameters and this epigenetic event [28]. Independent evaluation based on a large OSCC patient cohort is needed to more accurately determine the significance of the methylation status of CDH1 vis-à-vis clinical outcomes (something that can also be said for other genes reported as epigenetically dysregulated in oral tumors).
O 6 -methylguanine-DNA methyltransferase (MGMT) is a DNA repair gene that protects from toxicity and mutations that occur by alkylating agents through the removal of O6-guanine DNA adducts. CpG island hypermethylation of the MGMT promoter region results in gene silencing, with loss of MGMT repair capacity thought to drive cancer progression via the emergence of genomic instability. Decreased expression of MGMT via epigenetic silencing has been reported for many cancer types and loss of its expression can be tied to greater sensitivity to alkylating chemotherapeutic agents [37]. Epigenetic silencing of MGMT has been associated with OSCCs where tobacco exposure and betel quid chewing are suspected etiological factors [38–40]. MGMT promoter hypermethylation has also been associated with poorer outcomes for oral cancer, including a greater likelihood of nodal metastases, tumor recurrence, and decreased survival [41, 42]. MGMT promoter hypermethylation has also been associated with poorer outcomes for oral cancer, including a greater likelihood of nodal metastases, tumor recurrence, and decreased survival [32, 33]. Reduced MGMT expression has also been associated with these parameters in head and neck squamous cell carcinomas generally, and OSCCs specifically [41, 43]. Ongoing or elevated MGMT expression has been associated with resistance to alkylating agents in multiple cancer types including gliomas, astrocytomas, and melanomas [44, 45]. While alkylating agents such as ifosfamide and cyclophosphamide have been applied to manage various stages of oral and other head and neck malignancy, we have not found any reports to date regarding the role of MGMT silencing in modulating response to these compounds in these cancer types [46, 47]. Existing data from other cancer types provide a strong rationale for pursuing such studies.
Death-associated protein kinase (DAPK) encodes a serine/threonine kinase that is required for apoptosis induced by IFN-γ [48]. Loss of its expression via promoter hypermethylation has been associated with formation of metastases and advanced disease stages in multiple cancer types, including head and neck cancers [38, 48, 49]. Regarding OSCCs, DAPK hypermethylation has been reported as associated with increased likelihood of lymph node involvement, though these results have not always attained statistical significance [34, 50]. Interestingly, other groups have not reported associations between these clinical features and hypermethylation of DAPK (or other genes discussed here) [42, 51]. Again, this may be a function of the tissue heterogeneity that exists within the oral squamous cell carcinoma category. Detection of DAPK promoter hypermethylation at resection margins of oral tumors has been significantly associated with decreased overall survival, suggesting that it may have utility as a biomarker for guiding patient follow-up strategies [52]. As with MGMT, DAPK hypermethylation has also been associated with oral tumors where tobacco consumption is a suspected etiological factor [39].
The TGFβ superfamily transcription factor, runt-related transcription factor 3 (RUNX3), functions as a tumor suppressor gene and is involved in mediating apoptotic processes [53]. Promoter hypermethylation-mediated silencing of RUNX3 has been reported for many cancer types [53]. Recently, hypermethylation of the RUNX3 promoter region was found to be significantly associated with the presence of lymph node metastases and tumor stage in tongue carcinomas [52]. Other groups have reported that reduced RUNX3 expression or promoter hypermethylation is associated not only with progression in oral cancers, but also with disease recurrence and poorer prognoses [52]. The fact that other groups have not found significant associations between RUNX3 promoter hypermethylation and patient outcomes suggests that the role of epigenetic silencing of this gene in oral cancers bears further scrutiny [54]. The emerging role of RUNX3-mediated perturbation of the canonical Wnt signaling pathway in oral cancers also needs to be further evaluated.
3.1. Wnt Pathway Genes
Dysregulation of the canonical Wnt signaling pathway—via disrupted function of genes such as adenomatous polyposis coli (APC), AXIN1, β-catenin (CTNNB1), and secreted frizzled-related proteins (SFRPs)—has been noted for a variety of cancer types, including oral malignancies [55–58]. Briefly, canonical Wnt signaling involves stabilizing CTNNB1. When the canonical Wnt pathway is inactive, CTNNB1 exists in a phosphorylated form and is marked for degradation, with this phosphorylation mediated by a protein complex that incorporates Glycogen Synthase Kinase 3β (GSK3β), APC, and AXIN proteins. When Wnt binds to receptors encoded by Frizzled (Fz) genes, Dishevelled (Dsh) is activated and, consequently, GSK3β is inhibited. This in turn causes CTNNB1 dephosphorylation, which stabilizes the molecule and allows it to accumulate in the cell nucleus, where it can induce TCF/LEF-mediated transcription of several target genes. Regarding oncogenic processes, the downstream effects of CTNNB1 activation via Wnt signaling include enhanced cell proliferation and antiapoptotic activity.
Given their role as Wnt antagonists, SFRPs function as tumor suppressors. Marsit et al., reporting on head and neck cancers in general, described methylation differences for SFRP genes based on alcohol consumption, smoking behaviors, and HPV16 status [59]. More specifically, multiple reports involving analysis of both oral cancer cell lines and clinical OSCC cases have reported that promoter hypermethylation for SFRPs is associated with disease [60, 61]. Interestingly, the findings in these studies for SFRP1 have been conflicting, with both hypermethylation and demethylation of this gene reported [60–62].
APC also functions as a tumor suppressor gene and has also been reported as downregulated in oral tumors. Disruption of APC function in OSCC has been attributed to loss of heterozygosity (LOH alterations), mutations, and epigenetic alterations [63]. Regarding the latter, in vitro studies demonstrated that treatment with demethylating agent 5-aza-2′-deoxycytidine resulted in restoration of APC expression in oral cancer cells, supporting the functional importance of epigenetic silencing for this gene [64]. The drug received FDA approval for the treatment of myelodysplastic syndrome and is currently in clinical trials testing its utility in the treatment of several solid tumors. Increased promoter methylation for APC has been reported in clinical oral cancer tissues and some data do suggest a relationship between APC methylation status and development of lymph node metastases when it is analyzed in concert with the promoter methylation status of CDH1 [34].
As described above, activation of CTNNB1 is a critical consequence of canonical Wnt pathway signaling—and a variety of oncogenic processes can be turned on by this activity [56]. Elevated expression and greater nuclear localization of CTNNB1 have been reported for oral cancer and a multitude of other cancer types [56, 65]. While recent work has shown CTNNB1 immunostaining levels to be significantly associated with lymph node status, survival outcomes, and different invasive stages for oral cancers, data regarding the association of CTNNB1 methylation status for this disease have not [66]. Data from other cancer types suggest that activating mutations of CTNNB1 that prevent its downregulation may be a more common event than methylation [67].
WIF1 functions as a tumor suppressor that inhibits Wnt signaling through direct interaction with Wnt proteins, its activation leading to cell cycle arrest [56]. It has been described as downregulated in several cancer types [56]. Promoter hypermethylation-mediated downregulation of WIF1 in oral cancers has been reported by multiple groups, so there is evidence to suggest that epigenetic alteration of this gene can contribute to invasive disease phenotypes [52, 61]. More recently, a group studying tongue cancers found no significant associations between WIF1 promoter hypermethylation and lymph node metastasis formation, tumor stage, or overall survival [52]. In a separate study, this same group found no significant association between WIF1 promoter hypermethylation status in tissues from histologically negative resection margins and oral cancer recurrence [52]. Results so far suggest that WIF1 alterations may only have clinical utility as oral cancer biomarkers where used for diagnosis of invasive disease.
Figure 1 places the genes discussed in this section in their proper context in the canonical Wnt signaling cascade.
Figure 1.
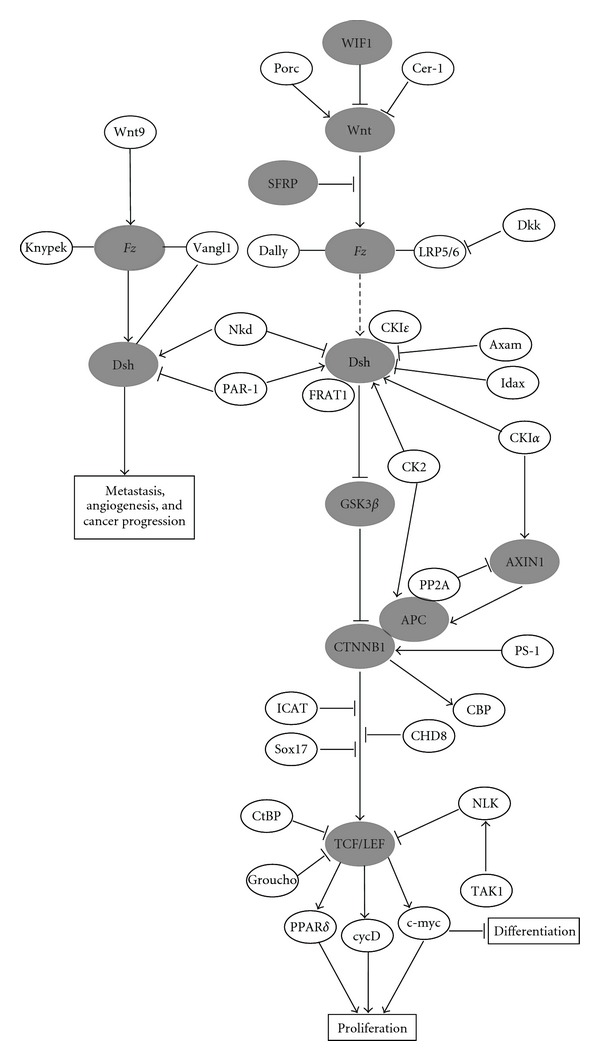
An overview of the canonical WNT signaling pathway. Genes highlighted in this paper as implicated in OSCC are shaded grey. Gene interactions are shown as activating (arrows) or inhibiting (blocked arrows). Boxes indicate processes that will ultimately be influenced by WNT signaling. Solid lines represent direct interactions and dashed lines indicate indirect effects. Lines connecting genes represent binding or association interactions. Complexes are represented by ovals that overlap.
4. Methylation Changes Governing Behavior of Noncoding RNAs in Oral Cancers
Noncoding RNAs, particularly microRNAs (miRNAs), have been shown to play a role in many biological processes and cellular pathways—and their behavior has been shown to be modulated by methylation changes [68, 69]. MiRNAs are 19–24 nucleotide transcripts that regulate mRNA expression by binding to and subsequently silencing the mRNA target [70–72]. MiRNAs can act as oncogenes or tumor suppressors and have been identified as dysregulated in several cancer types, including oral cancer [73–77]. As is the case with protein coding genes, there are several mechanisms that can lead to abnormal miRNA expression, including hypermethylation of CpG island promoters [78–82]. Using a functional-based screen in two OSCC cell lines, Uesugi et al. profiled tumor suppressive miRNAs that are silenced by hypermethylation [76]. They identified 110 miRNAs that exhibited inhibitory properties and, when compared to additional cell lines and tumor tissue, they found that miR-218 and miR-585 were frequently silenced by DNA hypermethylation [76].
Separate work has also shown that silencing of four tumor suppressive miRNAs (miR-34b, miR-137, miR-193a, and miR-203) can be mediated by aberrant methylation in OSCC cells [68]. This same work reported downregulation of miRNA expression through tumor-specific hypermethylation as being more frequent for miR-137 and miR-193a than for miR-34b and miR-203 when analyzed in primary tumors and paired normal oral mucosa. A separate group investigated the association between promoter methylation of miR-137 and both overall survival and disease-free survival, as well as with various prognostic factors [83]. This study found an association between methylation at the miR-137 promoter and poorer overall survival, though no associations were observed with disease-free survival or any of the other evaluated prognostic factors. MiR-137 has been reported as a negative regulator of CDK6/CCND1-mediated cell cycle progression [84], hence promoter methylation might serve as a means of inactivating tumor suppressive miR-137 function. Langevin et al. suggest that their inability to detect associations between miR-137 and other prognostic features in their study may have been a product of factors such as limited sample sizes and insufficiently long followup, proposing that stronger associations between miR-137 and oral cancer outcomes may exist [83]. Very recent work has suggested that epigenetically-mediated miR-137 expression may differ in oral cancer cells depending on whether they are part of a stem-like subpopulation [85]. The argument for further evaluation of miR-137 in the context of oral cancer outcomes is bolstered by evidence from other cancer types that also suggest a tumor suppressive biological role for this miRNA and an association with poorer disease outcomes [86–88]. Though only a few reports have evaluated the impact of methylation changes on small RNA behaviors in oral cancers and precancers, these early data do suggest a role for these molecules in oral malignant processes and provide a strong rationale for further studies in this area.
5. Altered Methylation States in Oral Premalignant Lesions and Cancer Progression
Premalignant lesions in the oral cavity are readily detectable owing to the accessibility of the organ site. That said, one factor that contributes to the poor prognosis of OSCC is the current inability to determine which premalignant lesions will ultimately progress to invasive disease; the clinical standard of care is “watchful waiting” since histopathological review is presently incapable of delineating progression risk and intervention with all early lesions would lead to overtreatment that is costly both in terms of dollars and patient quality of life. Since earlier stage lesions are smaller in size and more readily treatable, biomarkers delineating the progression likelihood for a given dysplasia case represent a key means for improving patient outcomes.
Several studies have indicated that deletion and silencing of loci mapping to chromosome arms 9p and 3p are common events in the early development of OSCC [89, 90]. Thus, studies investigating the role methylation changes in oral premalignant lesion (OPL) behavior have generally investigated genes mapping within these regions.
More specifically, several studies have investigated the impact of methylation changes p14 and p16—both of which, as described above, are encoded by CDKN2A and map to chromosome 9p21.3—in OPLs. A much higher degree of p16 promoter hypermethylation has been reported relative to p14 promoter hypermethylation in patients with histologically confirmed severe dysplasias (57.5% and 3.8% resp.) [91]. Another group reported that p16 hypermethylation in OPLs was associated with a greater risk for progression to invasive disease; patients with confirmed OPLs (as determined by WHO criteria and independent review by two pathologists) that harbored hypermethylated p16 promoters were approximately two and a half times more likely to develop OSCC than patients with OPLs that did not exhibit p16 promoter hypermethylation [92]. Interestingly, another study reported higher rates of promoter methylation for neighboring p15 relative to p16 across multiple stages of histopathologically confirmed oral dysplasia (50% versus 18%) [93]. This same study also reported a much higher frequency of p14 promoter methylation; though the fact that analyzed tissues came from a population of betel quid chewers—where the suspected disease etiology was therefore different than other populations (where tobacco is the predominant etiological factor)—may have impacted these results.
Another study assessed whether hypermethylated genes previously implicated in oral dysplasias could be used as markers for progression likelihood [94]. In this instance, longitudinal follow-up data made this evaluation possible since progression status for each patient with clinically confirmed disease was known. Interestingly, the results of this work suggested a much lower incidence of promoter methylation at previously identified candidate genes than in the studies discussed above. The authors in this instance posited that use of pyrosequencing instead of MSP may explain the discrepancy, the former technique being less prone to false positives. This study did conclude that p16 could be a predictor of progression, though its specificity came in at a fairly low value (57%).
Though wealth of clinical data associated with canonical genomic alterations at chromosome 9p and 3p makes these regions attractive for locus-specific analysis of methylation alterations, other loci are also worthy of further scrutiny for the utility in predicting disease progression. 14-3-3-σ has also found to be recurrently methylated in histologically confirmed oral dysplasias and has been associated with coincident methylation at p16, making it an attractive candidate for further evaluation in an oral cancer progression context [95]. Another group observed that RECK, a gene that functions to inhibit angiogenesis, invasion, and metastasis, has a potential to be a biomarker as it is recurrently found to be hypermethylated in the normal mucosa adjacent to the tumor [96]. Ultimately, whole methylome analyses of a large panel of well-annotated OPLs with extensive followup are needed to uncover disease-relevant biomarkers that will impact disease management and oral cancer survival rates.
6. Global Methylome Changes and the Role of Hypomethylation in Oral Malignancy
In addition to investigating the methylation status of specific genes, some studies have begun analyzing the oral cancer methylome as a whole. Early evidence suggests that the distribution of methylation at various gene promoters across a series of lesions of progressing grade from a single individual can follow a specific pattern. Further, the CpG island methylation phenotype (CIMP) has been observed in several cancer types and is based on the observation that those tumors exhibiting aberrant methylation of one gene are more likely to have other sites of aberrant methylation [97, 98]. To determine whether the CIMP phenomenon was detectable in OSCC, the methylation status of ten genes was evaluated in a large panel of oral tumors [99]. This analysis revealed a cluster of tumors with a greater degree of promoter methylation than would be predicted by chance alone. These cases were identified as “CIMP +ve” and results suggest (1) that these tumors exhibited a less aggressive tumor biology than other oral cancers and (2) that these cases were characterized by a greater host inflammatory response (a finding which the authors note is consistent with findings in other cancer types). Another group has reported observing CIMP in head and neck cancers, though it is clear additional studies are needed to elucidate the mechanisms driving this phenotype [100].
While the majority of literature has focused on promoter hypermethylation silencing of tumor suppressor genes as a critical mechanism driving oral tumorigenesis, DNA hypomethylation is also understood to contribute to development of many epithelial cancers by facilitating activation of candidate oncogenes [101]. More specifically, it has been proposed that DNA hypomethylation contributes to tumorigenesis by two potential mechanisms. The first represents another global level of methylation dysregulation, where highly methylated repetitive elements (such as long interspersed nuclear element-1 LINE-1 and Alu sequences) are demethylated, resulting in increased chromosomal instability that leads to irregular mitoses that, in turn, drive the emergence of further mutations that can activate oncogenes [97, 102, 103]. The second is more specific, where inadvertent demethylation in a local context activates oncogenes that are normally methylation-silenced in the human genome [97, 102, 103].
With respect to oral cancer specifically, little work has been done in clinical tissues. Analysis of in vitro OSCC models of interleukin-mediated chronic inflammation have indicated that chronic inflammation can drive both global hypomethylation of LINE-1 sequences as well as specific CpG methylation changes [104]. More precisely, this model showed that IL-6 specifically was inducing these changes, with methylation alterations also found to be associated with downstream changes in gene expression. The suggestion that inflammatory responses can play a critical role in mediating cancer-causing methylation changes is of particular interest given that the oral cavity, which experiences regular exposure to carcinogens, is frequently under inflammatory stress. Another group investigated the role of hypomethylation in a murine model of oral cancer which used DNMT1 hypomorphic alleles to reduce genomic methylation as lingual and esophageal carcinogenesis was induced by 4-nitroquinoline 1-oxide [105]. This group reported that reduction of DNA methylation levels led to the suppression of tumor formation in certain cell types.
A separate group detected global LINE-1 hypomethylation in oral rinses obtained from OSCC patients (relative to healthy controls) [106]. This is touched on further in the following section. The relative scarcity of studies addressing either global trends in the oral cancer methylome or the role of specific instances of hypomethylation in oral tumorigenesis points to a need to develop these research streams more fully.
7. Methylation Changes as Surrogate Markers for Oral Malignancy
7.1. Saliva
The use of saliva for the diagnosis or establishing prognosis of OSCC patients is a promising screen, as it is both noninvasive and inexpensive. The potential of this tool has been studied by utilizing a genome-wide methylation array to profile the methylation status of 13 OSCC patients before and after surgery, as well as ten normal samples [107]. This work identified 34 genes of interest, including p16, and proposed panels ranging from 4–7 genes that are likely to have the optimal sensitivity and specificity for clinical applications (the range of specificity being 62–77% and sensitivity being 83–100%). This group also used saliva samples taken from the same patients at both the pre- and postoperative stages, eliminating issues arising from using saliva samples from unmatched normal subjects as a control.
Another group analyzed the methylation status of 11 genes by MSP for a panel of primary tumors and saliva samples from 90 patients with HNSCC [108]. They found several genes that were frequently methylated in the paired saliva and tissue samples including previously-implicated candidates such as p16, MGMT, DAPK, and RASSF1. Additionally, they were able to accurately detect malignant cells in saliva several months before tumor recurrence was otherwise detected in patients, indicating that saliva-based assays may be a very useful tool for effective followup of oral cancer patients.
As referenced above, a separate study of oral rinses identified LINE-1 hypomethylation as being more prevalent in rinses obtained from OSCC patients when compared to healthy controls [106]. While the authors did not establish any statistically significant associations between their rinse results and disease stage, histological grade, lesions site, or carcinogen exposures (e.g., tobacco smoke), the utility of this noninvasive approach for differential diagnosis of oral malignancy does suggest it is worth evaluating in independent patient cohorts.
To assess the potential of saliva as a screen for premalignant lesions, another group collected saliva samples from patients that had leukoplakia and were at risk for the development of OSCC [109]. They assessed the methylation status of p16, p14, and MGMT from these specimens using MSP and observed a relatively high frequency of hypermethylation of MGMT and p16 relative to levels typically found in OSCC patients. However, followup had not been done on the patients, so whether or not the hypermethylation of p16 and MGMT was unique to patients at risk for progressing into OSCC is not known.
7.2. Serum
In addition to saliva rinses, methylation changes detected in DNA isolated from serum from peripheral blood also hold potential as a diagnostic or prognostic tools for detection and management of disease. This approach has previously been used in a variety of other cancers such as lung, liver, and colorectal [110–112]. As with studies involving saliva, the methylation status of p16 has been the predominant focus of this work in oral cancer.
One group found that parallel evaluation of promoter methylation status of multiple genes (including p16) improved the sensitivity and specificity of disease discrimination [113]. Another group similarly analyzed the promoter methylation status of multiple genes in serum samples obtained from OSCC cases and found that in a majority of cases, at least one of the four assayed genes was detectable as hypermethylated [49]. It has been reported that when a gene is found to be hypermethylated within a tumor, DNA from a matched peripheral blood sample will harbor similar methylation changes at a frequency ranging from 31 and 54.5% [49, 114, 115].
Evaluation of methylation alterations in samples obtained by minimally invasive procedures such as collection of saliva or blood remains a very attractive avenue for the discovery of biomarkers for management of clinical oral malignancy. Expanded collection efforts and evaluation of these biological materials should be a component of all molecular investigations of oral cancers and precancers.
8. Conclusion
Interrogation of methylation changes in clinical oral cancers and precancers has revealed multiple recurrent alterations at genes and chromosomal loci that are associated with oncogenic processes in this and other cancer types. Some analyses have indicated strong associations between these changes and specific patient outcomes, including the likelihood of progressing from premalignant to invasive disease and posttreatment recurrence. Evaluation of blood and saliva samples, that are easily accessible, has also generated data that methylation changes detected in these specimens might have utility as surrogate biomarkers for managing disease. Given that oral cancers and precancers represent a significant global health challenge, these more affordably processed specimen types should be of particular research interest going forward if the field is serious about developing tools with utility worldwide. We conclude that changes in DNA methylation from clinical tissues do show early promise as markers to improve management for this cancer type, which has had survival rates that have remained stubbornly low over recent decades. We ultimately feel that the true utility of methylation markers for management of oral malignancy will only be realized when analyses are expanded to much larger patient cohorts. This reasoning holds whether the methylation status is being evaluated for an individual locus or by the most current and robust platform for whole methylome analysis. We also feel that a much stronger research emphasis on developing new methylation markers for managing smaller, more treatable premalignant lesions would address a currently underserved path for improving oral cancer outcomes.
Conflicts of Interest
None declared.
Acknowledgments
The authors acknowledge financial support from Pacific Otolaryngology Foundation, Rotary Hearing Foundation, and CIHR (201003MOP-221786-CPT-CAAA-136662).
References
- 1.Ferlay J, Shin HR, Bray F, Forman D, Mathers C, Parkin DM. Estimates of worldwide burden of cancer in 2008: GLOBOCAN 2008. International Journal of Cancer. 2010;127(12):2893–2917. doi: 10.1002/ijc.25516. [DOI] [PubMed] [Google Scholar]
- 2.Takai D, Jones PA. Comprehensive analysis of CpG islands in human chromosomes 21 and 22. Proceedings of the National Academy of Sciences of the United States of America. 2002;99(6):3740–3745. doi: 10.1073/pnas.052410099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Illingworth RS, Bird AP. CpG islands—“a rough guide”. FEBS Letters. 2009;583(11):1713–1720. doi: 10.1016/j.febslet.2009.04.012. [DOI] [PubMed] [Google Scholar]
- 4.Bird A. DNA methylation patterns and epigenetic memory. Genes and Development. 2002;16(1):6–21. doi: 10.1101/gad.947102. [DOI] [PubMed] [Google Scholar]
- 5.Knudson AG., Jr. Mutation and cancer: statistical study of retinoblastoma. Proceedings of the National Academy of Sciences of the United States of America. 1971;68(4):820–823. doi: 10.1073/pnas.68.4.820. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Eads CA, Danenberg KD, Kawakami K, et al. MethyLight: a high-throughput assay to measure DNA methylation. Nucleic Acids Research. 2000;28(8):p. E32. doi: 10.1093/nar/28.8.e32. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Xiong Z, Laird PW. COBRA: a sensitive and quantitative DNA methylation assay. Nucleic Acids Research. 1997;25(12):2532–2534. doi: 10.1093/nar/25.12.2532. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Tost J, Gut IG. DNA methylation analysis by pyrosequencing. Nature Protocols. 2007;2(9):2265–2275. doi: 10.1038/nprot.2007.314. [DOI] [PubMed] [Google Scholar]
- 9.Bibikova M, Fan J-B. Genome-wide DNA methylation profiling. Wiley Interdisciplinary Reviews: Systems Biology and Medicine. 2010;2(2):210–223. doi: 10.1002/wsbm.35. [DOI] [PubMed] [Google Scholar]
- 10.Huang THM, Perry MR, Laux DE. Methylation profiling of CpG islands in human breast cancer cells. Human Molecular Genetics. 1999;8(3):459–470. doi: 10.1093/hmg/8.3.459. [DOI] [PubMed] [Google Scholar]
- 11.Weber M, Davies JJ, Wittig D, et al. Chromosome-wide and promoter-specific analyses identify sites of differential DNA methylation in normal and transformed human cells. Nature Genetics. 2005;37(8):853–862. doi: 10.1038/ng1598. [DOI] [PubMed] [Google Scholar]
- 12.Wu J, Smith LT, Plass C, Huang THM. ChIP-chip comes of age for genome-wide functional analysis. Cancer Research. 2006;66(14):6899–6902. doi: 10.1158/0008-5472.CAN-06-0276. [DOI] [PubMed] [Google Scholar]
- 13.Park PJ. ChIP-seq: advantages and challenges of a maturing technology. Nature Reviews Genetics. 2009;10(10):669–680. doi: 10.1038/nrg2641. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Bibikova M, Le J, Barnes B, et al. Genome-wide DNA methylation profiling using Infinium assay. Epigenomics. 2009;1(1):177–200. doi: 10.2217/epi.09.14. [DOI] [PubMed] [Google Scholar]
- 15.Frommer M, McDonald LE, Millar DS, et al. A genomic sequencing protocol that yields a positive display of 5-methylcytosine residues in individual DNA strands. Proceedings of the National Academy of Sciences of the United States of America. 1992;89(5):1827–1831. doi: 10.1073/pnas.89.5.1827. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Ehrich M, Zoll S, Sur S, van den Boom D. A new method for accurate assessment of DNA quality after bisulfite treatment. Nucleic Acids Research. 2007;35(5, article e29) doi: 10.1093/nar/gkl1134. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Silva SDD, Ferlito A, Takes RP, et al. Advances and applications of oral cancer basic research. Oral Oncology. 2011;47(9):783–791. doi: 10.1016/j.oraloncology.2011.07.004. [DOI] [PubMed] [Google Scholar]
- 18.Lin RK, Hsieh YS, Lin P, et al. The tobacco-specific carcinogen NNK induces DNA methyltransferase 1 accumulation and tumor suppressor gene hypermethylation in mice and lung cancer patients. Journal of Clinical Investigation. 2010;120(2):521–532. doi: 10.1172/JCI40706. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Belinsky SA, Klinge DM, Stidley CA, et al. Inhibition of DNA Methylation and Histone Deacetylation Prevents Murine Lung Cancer. Cancer Research. 2003;63(21):7089–7093. [PubMed] [Google Scholar]
- 20.Papadimitrakopoulou V, Izzo J, Lippman SM, et al. Frequent inactivation of p16(INK4α) in oral premalignant lesions. Oncogene. 1997;14(15):1799–1803. doi: 10.1038/sj.onc.1201010. [DOI] [PubMed] [Google Scholar]
- 21.Nakahara Y, Shintani S, Mihara M, Ueyama Y, Matsumura T. High frequency of homozygous deletion and methylation of p16INK4A gene in oral squamous cell carcinomas. Cancer Letters. 2001;163(2):221–228. doi: 10.1016/s0304-3835(00)00699-6. [DOI] [PubMed] [Google Scholar]
- 22.Akanuma D, Uzawa N, Yoshida MA, Negishi A, Amagasa T, Ikeuchi T. Inactivation patterns of the p16 (INK4a) gene in oral squamous cell carcinoma cell lines. Oral Oncology. 1999;35(5):476–483. doi: 10.1016/s1368-8375(99)00020-2. [DOI] [PubMed] [Google Scholar]
- 23.Cody DT, II, Huang Y, Darby CJ, Johnson GK, Domann FE. Differential DNA methylation of the p16 INK4A/CDKN2A promoter in human oral cancer cells and normal human oral keratinocytes. Oral Oncology. 1999;35(5):516–522. doi: 10.1016/s1368-8375(99)00026-3. [DOI] [PubMed] [Google Scholar]
- 24.Viswanathan M, Tsuchida N, Shanmugam G. Promoter hypermethylation profile of tumor-associated genes p16, p15, hMLH1, MGMT and E-cadherin in oral squamous cell carcinoma. International Journal of Cancer. 2003;105(1):41–46. doi: 10.1002/ijc.11028. [DOI] [PubMed] [Google Scholar]
- 25.Shaw RJ, Liloglou T, Rogers SN, et al. Promoter methylation of P16, RARβ, E-cadherin, cyclin A1 and cytoglobin in oral cancer: quantitative evaluation using pyrosequencing. British Journal of Cancer. 2006;94(4):561–568. doi: 10.1038/sj.bjc.6602972. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Lee JK, Kim MJ, Hong SP, Hong SD. Inactivation patterns of p16/INK4A in oral squamous cell carcinomas. Experimental and Molecular Medicine. 2004;36(2):165–171. doi: 10.1038/emm.2004.23. [DOI] [PubMed] [Google Scholar]
- 27.Tran TN, Liu Y, Takagi M, Yamaguchi A, Fujii H. Frequent promoter hypermethylation of RASSF1A and p16INK4a and infrequent allelic loss other than 9p21 in betel-associated oral carcinoma in a Vietnamese non-smoking/non-drinking female population. Journal of Oral Pathology and Medicine. 2005;34(3):150–156. doi: 10.1111/j.1600-0714.2004.00292.x. [DOI] [PubMed] [Google Scholar]
- 28.Su PF, Huang WL, Wu HT, Wu CH, Liu TY, Kao SY. P16INK4A promoter hypermethylation is associated with invasiveness and prognosis of oral squamous cell carcinoma in an age-dependent manner. Oral Oncology. 2010;46(10):734–739. doi: 10.1016/j.oraloncology.2010.07.002. [DOI] [PubMed] [Google Scholar]
- 29.Sailasree R, Abhilash A, Sathyan KM, Nalinakumari KR, Thomas S, Kannan S. Differential roles of p16INK4A and p14ARF genes in prognosis of oral carcinoma. Cancer Epidemiology, Biomarkers & Prevention. 2008;17(2):414–420. doi: 10.1158/1055-9965.EPI-07-0284. [DOI] [PubMed] [Google Scholar]
- 30.Ishida E, Nakamura M, Ikuta M, et al. Promotor hypermethylation of p14ARF is a key alteration for progression of oral squamous cell carcinoma. Oral Oncology. 2005;41(6):614–622. doi: 10.1016/j.oraloncology.2005.02.003. [DOI] [PubMed] [Google Scholar]
- 31.Ogi K, Toyota M, Ohe-Toyota M, et al. Aberrant methylation of multiple genes and clinicopathological features in oral squamous cell carcinoma. Clinical Cancer Research. 2002;8(10):3164–3171. [PubMed] [Google Scholar]
- 32.Sinha P, Bahadur S, Thakar A, et al. Significance of promoter hypermethylation of p16 gene for margin assessment in carcinoma tongue. Head & Neck. 2009;31(11):1423–1430. doi: 10.1002/hed.21122. [DOI] [PubMed] [Google Scholar]
- 33.Kato K, Hara A, Kuno T, et al. Aberrant promoter hypermethylation of p16 and MGMT genes in oral squamous cell carcinomas and the surrounding normal mucosa. Journal of Cancer Research and Clinical Oncology. 2006;132(11):735–743. doi: 10.1007/s00432-006-0122-8. [DOI] [PubMed] [Google Scholar]
- 34.Šupić G, Kozomara R, Branković-Magić M, Jović N, Magić Z. Gene hypermethylation in tumor tissue of advanced oral squamous cell carcinoma patients. Oral Oncology. 2009;45(12):1051–1057. doi: 10.1016/j.oraloncology.2009.07.007. [DOI] [PubMed] [Google Scholar]
- 35.Berx G, van Roy F. Involvement of members of the cadherin superfamily in cancer. Cold Spring Harbor Perspectives in Biology. 2009;1(6):p. a003129. doi: 10.1101/cshperspect.a003129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Chang HW, Chow V, Lam KY, Wei WI, WingYuen AP. Loss of E-cadherin expression resulting from promoter hypermethylation in oral tongue carcinoma and its prognostic significance. Cancer. 2002;94(2):386–392. doi: 10.1002/cncr.10211. [DOI] [PubMed] [Google Scholar]
- 37.Esteller M. Aberrant DNA methylation as a cancer-inducing mechanism. Annual Review of Pharmacology and Toxicology. 2005;45:629–656. doi: 10.1146/annurev.pharmtox.45.120403.095832. [DOI] [PubMed] [Google Scholar]
- 38.Rosas SLB, Koch W, Da Costa Carvalho MDG, et al. Promoter hypermethylation patterns of p16, O6-methylguanine-DNA-methyltransferase, and death-associated protein kinase in tumors and saliva of Head & Neck cancer patients. Cancer Research. 2001;61(3):939–942. [PubMed] [Google Scholar]
- 39.Kulkarni V, Saranath D. Concurrent hypermethylation of multiple regulatory genes in chewing tobacco associated oral squamous cell carcinomas and adjacent normal tissues. Oral Oncology. 2004;40(2):145–153. doi: 10.1016/s1368-8375(03)00143-x. [DOI] [PubMed] [Google Scholar]
- 40.Huang S-H, Lee H-S, Mar K, Ji D-D, Huang M-S, Hsia K-T. Loss expression of O6-methylguanine DNA methyltransferase by promoter hypermethylation and its relationship to betel quid chewing in oral squamous cell carcinoma. Oral Surgery, Oral Medicine, Oral Pathology, Oral Radiology and Endodontology. 2010;109(6):883–889. doi: 10.1016/j.tripleo.2009.12.019. [DOI] [PubMed] [Google Scholar]
- 41.Zuo C, Ai L, Ratliff P, et al. O6-methylguanine-DNA methyltransferase gene: epigenetic silencing and prognostic value in Head & Neck squamous cell carcinoma. Cancer Epidemiology Biomarkers and Prevention. 2004;13(6):967–975. [PubMed] [Google Scholar]
- 42.Paluszczak J, Misiak P, Wierzbicka M, Woźniak A, Baer-Dubowska W. Frequent hypermethylation of DAPK, RARbeta, MGMT, RASSF1A and FHIT in laryngeal squamous cell carcinomas and adjacent normal mucosa. Oral Oncology. 2011;47(2):104–107. doi: 10.1016/j.oraloncology.2010.11.006. [DOI] [PubMed] [Google Scholar]
- 43.Sawhney M, Rohatgi N, Kaur J, et al. MGMT expression in oral precancerous and cancerous lesions: correlation with progression, nodal metastasis and poor prognosis. Oral Oncology. 2007;43(5):515–522. doi: 10.1016/j.oraloncology.2006.05.007. [DOI] [PubMed] [Google Scholar]
- 44.Jiang G, Wei Z-P, Pei D-S, Xin Y, Liu Y-Q, Zheng J-N. A novel approach to overcome temozolomide resistance in glioma and melanoma: inactivation of MGMT by gene therapy. Biochemical and Biophysical Research Communications. 2011;406(3):311–314. doi: 10.1016/j.bbrc.2011.02.042. [DOI] [PubMed] [Google Scholar]
- 45.Sharma S, Salehi F, Scheithauer BW, Rotondo F, Syro LV, Kovacs K. Role of MGMT in tumor development, progression, diagnosis, treatment and prognosis. Anticancer Research. 2009;29(10):3759–3768. [PubMed] [Google Scholar]
- 46.Airoldi M, Cortesina G, Giordano C, Pedani F, Bumma C. Ifosfamide in the treatment of Head & Neck cancer. Oncology. 2003;65, supplement 2(2):37–43. doi: 10.1159/000073356. [DOI] [PubMed] [Google Scholar]
- 47.Kies MS, Boatright DH, Li G, et al. Phase II trial of induction chemotherapy followed by surgery for squamous cell carcinoma of the oral tongue in young adults. doi: 10.1002/hed.21906. Head & Neck. In press. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Michie AM, McCaig AM, Nakagawa R, Vukovic M. Death-associated protein kinase (DAPK) and signal transduction: regulation in cancer. FEBS Journal. 2010;277(1):74–80. doi: 10.1111/j.1742-4658.2009.07414.x. [DOI] [PubMed] [Google Scholar]
- 49.Sanchez-Cespedes M, Esteller M, Wu L, et al. Gene promoter hypermethylation in tumors and serum of Head & Neck cancer patients. Cancer Research. 2000;60(4):892–895. [PubMed] [Google Scholar]
- 50.Kong WJ, Zhang S, Guo C, Zhang S, Wang Y, Zhang D. Methylation-associated silencing of death-associated protein kinase gene in laryngeal squamous cell cancer. Laryngoscope. 2005;115(8):1395–1401. doi: 10.1097/01.MLG.0000166708.23673.3A. [DOI] [PubMed] [Google Scholar]
- 51.Dikshit RP, Gillio-Tos A, Brennan P, et al. Hypermethylation, risk factors, clinical characteristics, and survival in 235 patients with laryngeal and hypopharyngeal cancers. Cancer. 2007;110(8):1745–1751. doi: 10.1002/cncr.22975. [DOI] [PubMed] [Google Scholar]
- 52.Supic G, Kozomara R, Jovic N, Zeljic K, Magic Z. Prognostic significance of tumor-related genes hypermethylation detected in cancer-free surgical margins of oral squamous cell carcinomas. Oral Oncology. 2011;47(8):702–708. doi: 10.1016/j.oraloncology.2011.05.014. [DOI] [PubMed] [Google Scholar]
- 53.Kudo Y, Tsunematsu T, Takata T. Oncogenic role of RUNX3 in Head & Neck cancer. Journal of Cellular Biochemistry. 2011;112(2):387–393. doi: 10.1002/jcb.22967. [DOI] [PubMed] [Google Scholar]
- 54.Gao F, Huang C, Lin M, et al. Frequent inactivation of RUNX3 by promoter hypermethylation and protein mislocalization in oral squamous cell carcinomas. Journal of Cancer Research and Clinical Oncology. 2009;135(5):739–747. doi: 10.1007/s00432-008-0508-x. [DOI] [PubMed] [Google Scholar]
- 55.Reya T, Clevers H. Wnt signalling in stem cells and cancer. Nature. 2005;434(7035):843–850. doi: 10.1038/nature03319. [DOI] [PubMed] [Google Scholar]
- 56.Barker N, Clevers H. Mining the Wnt pathway for cancer therapeutics. Nature Reviews Drug Discovery. 2006;5(12):997–1014. doi: 10.1038/nrd2154. [DOI] [PubMed] [Google Scholar]
- 57.Fodde R, Brabletz T. Wnt/β-catenin signaling in cancer stemness and malignant behavior. Current Opinion in Cell Biology. 2007;19(2):150–158. doi: 10.1016/j.ceb.2007.02.007. [DOI] [PubMed] [Google Scholar]
- 58.Molinolo AA, Amornphimoltham P, Squarize CH, Castilho RM, Patel V, Gutkind JS. Dysregulated molecular networks in Head & Neck carcinogenesis. Oral Oncology. 2009;45(4-5):324–334. doi: 10.1016/j.oraloncology.2008.07.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Marsit CJ, McClean MD, Furniss CS, Kelsey KT. Epigenetic inactivation of the SFRP genes is associated with drinking, smoking and HPV in Head & Neck squamous cell carcinoma. International Journal of Cancer. 2006;119(8):1761–1766. doi: 10.1002/ijc.22051. [DOI] [PubMed] [Google Scholar]
- 60.Sogabe Y, Suzuki H, Toyota M, et al. Epigenetic inactivation of SFRP genes in oral squamous cell carcinoma. International Journal of Oncology. 2008;32(6):1253–1261. doi: 10.3892/ijo_32_6_1253. [DOI] [PubMed] [Google Scholar]
- 61.Pannone G, Bufo P, Santoro A, et al. WNT pathway in oral cancer: epigenetic inactivation of WNT-inhibitors. Oncology Reports. 2010;24(4):1035–1041. doi: 10.3892/or.2010.1035. [DOI] [PubMed] [Google Scholar]
- 62.Lee CH, Hung YJ, Lin CY, Hung PH, Hung HW, Shieh YS. Loss of SFRP1 expression is associated with aberrant β-catenin distribution and tumor progression in mucoepidermoid carcinoma of salivary glands. Annals of Surgical Oncology. 2010;17(8):2237–2246. doi: 10.1245/s10434-010-0961-z. [DOI] [PubMed] [Google Scholar]
- 63.Pérez-Sayáns M, Suárez-Peñaranda JM, Herranz-Carnero M, et al. The role of the adenomatous polyposis coli (APC) in oral squamous cell carcinoma. Oral Oncology. 2012;48(1):56–60. doi: 10.1016/j.oraloncology.2011.09.001. [DOI] [PubMed] [Google Scholar]
- 64.Uesugi H, Uzawa K, Kawasaki K, et al. Status of reduced expression and hypermethylation of the APC tumor suppressor gene in human oral squamous cell carcinoma. International Journal of Molecular Medicine. 2005;15(4):597–602. [PubMed] [Google Scholar]
- 65.Fujii M, Katase N, Lefeuvre M, et al. Dickkopf (Dkk)-3 and β-catenin expressions increased in the transition from normal oral mucosal to oral squamous cell carcinoma. Journal of Molecular Histology. 2011;42(6):499–504. doi: 10.1007/s10735-011-9357-z. [DOI] [PubMed] [Google Scholar]
- 66.Ravindran G, Devaraj H. Aberrant expression of β-catenin and its association with ΔNp63, Notch-1, and clinicopathological factors in oral squamous cell carcinoma. doi: 10.1007/s00784-011-0605-0. Clinical Oral Investigations. In press. [DOI] [PubMed] [Google Scholar]
- 67.Ying Y, Tao Q. Epigenetic disruption of the WNT/β-catenin signaling pathway in human cancers. Epigenetics. 2009;4(5):34–39. doi: 10.4161/epi.4.5.9371. [DOI] [PubMed] [Google Scholar]
- 68.Lopez-Serra P, M. Esteller. DNA methylation-associated silencing of tumor-suppressor microRNAs in cancer. Oncogene. 2012;31(13):1609–1622. doi: 10.1038/onc.2011.354. Article ID 10.1038/onc.2011.354. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Lujambio A, Portela A, Liz J, et al. CpG island hypermethylation-associated silencing of non-coding RNAs transcribed from ultraconserved regions in human cancer. Oncogene. 2010;29(48):6390–6401. doi: 10.1038/onc.2010.361. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Miranda KC, Huynh T, Tay Y, et al. A pattern-based method for the identification of microRNA binding sites and their corresponding heteroduplexes. Cell. 2006;126(6):1203–1217. doi: 10.1016/j.cell.2006.07.031. [DOI] [PubMed] [Google Scholar]
- 71.Iwama H, Masaki T, Kuriyama S. Abundance of microRNA target motifs in the 3′-UTRs of 20 527 human genes. FEBS Letters. 2007;581(9):1805–1810. doi: 10.1016/j.febslet.2007.03.066. [DOI] [PubMed] [Google Scholar]
- 72.Duursma AM, Kedde M, Schrier M, Le Sage C, Agami R. miR-148 targets human DNMT3b protein coding region. RNA. 2008;14(5):872–877. doi: 10.1261/rna.972008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Esquela-Kerscher A, Slack FJ. Oncomirs—microRNAs with a role in cancer. Nature Reviews Cancer. 2006;6(4):259–269. doi: 10.1038/nrc1840. [DOI] [PubMed] [Google Scholar]
- 74.Hammond SM. MicroRNAs as tumor suppressors. Nature Genetics. 2007;39(5):582–583. doi: 10.1038/ng0507-582. [DOI] [PubMed] [Google Scholar]
- 75.Lujambio A, Esteller M. How epigenetics can explain human metastasis: a new role for microRNAs. Cell Cycle. 2009;8(3):377–382. doi: 10.4161/cc.8.3.7526. [DOI] [PubMed] [Google Scholar]
- 76.Uesugi A, Kozaki K-I, Tsuruta T, et al. The tumor suppressive microRNA miR-218 targets the mTOR component rictor and inhibits AKT phosphorylation in oral cancer. Cancer Research. 2011;71(17):5765–5778. doi: 10.1158/0008-5472.CAN-11-0368. [DOI] [PubMed] [Google Scholar]
- 77.Zheng J, Xue H, Wang T, et al. miR-21 downregulates the tumor suppressor P12 CDK2AP1and stimulates cell proliferation and invasion. Journal of Cellular Biochemistry. 2011;112(3):872–880. doi: 10.1002/jcb.22995. [DOI] [PubMed] [Google Scholar]
- 78.Saito Y, Liang G, Egger G, et al. Specific activation of microRNA-127 with downregulation of the proto-oncogene BCL6 by chromatin-modifying drugs in human cancer cells. Cancer Cell. 2006;9(6):435–443. doi: 10.1016/j.ccr.2006.04.020. [DOI] [PubMed] [Google Scholar]
- 79.Han L, Witmer PD, Casey E, Valle D, Sukumar S. DNA methylation regulates microRNA expression. Cancer Biology and Therapy. 2007;6(8):1284–1288. doi: 10.4161/cbt.6.8.4486. [DOI] [PubMed] [Google Scholar]
- 80.Lujambio A, Ropero S, Ballestar E, et al. Genetic unmasking of an epigenetically silenced microRNA in human cancer cells. Cancer Research. 2007;67(4):1424–1429. doi: 10.1158/0008-5472.CAN-06-4218. [DOI] [PubMed] [Google Scholar]
- 81.Toyota M, Suzuki H, Sasaki Y, et al. Epigenetic silencing of microRNA-34b/c and B-cell translocation gene 4 is associated with CpG island methylation in colorectal cancer. Cancer Research. 2008;68(11):4123–4132. doi: 10.1158/0008-5472.CAN-08-0325. [DOI] [PubMed] [Google Scholar]
- 82.Lujambio A, Calin GA, Villanueva A, et al. A microRNA DNA methylation signature for human cancer metastasis. Proceedings of the National Academy of Sciences of the United States of America. 2008;105(36):13556–13561. doi: 10.1073/pnas.0803055105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Langevin SM, Stone RA, Bunker CH, et al. MicroRNA-137 promoter methylation is associated with poorer overall survival in patients with squamous cell carcinoma of the head and neck. Cancer. 2011;117(7):1454–1462. doi: 10.1002/cncr.25689. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Kozaki KI, Imoto I, Mogi S, Omura K, Inazawa J. Exploration of tumor-suppressive microRNAs silenced by DNA hypermethylation in oral cancer. Cancer Research. 2008;68(7):2094–2105. doi: 10.1158/0008-5472.CAN-07-5194. [DOI] [PubMed] [Google Scholar]
- 85.Wiklund ED, Gao S, Hulf T, et al. MicroRNA alterations and associated aberrant DNA methylation patterns across multiple sample types in oral squamous cell carcinoma. PLoS ONE. 2011;6(11) doi: 10.1371/journal.pone.0027840. Article ID e27840. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Zhi F, Chen X, Wang S, et al. The use of hsa-miR-21, hsa-miR-181b and hsa-miR-106a as prognostic indicators of astrocytoma. European Journal of Cancer. 2010;46(9):1640–1649. doi: 10.1016/j.ejca.2010.02.003. [DOI] [PubMed] [Google Scholar]
- 87.Silber J, Lim DA, Petritsch C, et al. miR-124 and miR-137 inhibit proliferation of glioblastoma multiforme cells and induce differentiation of brain tumor stem cells. BMC Medicine. 2008;6, article 14 doi: 10.1186/1741-7015-6-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Bemis LT, Chen R, Amato CM, et al. MicroRNA-137 targets microphthalmia-associated transcription factor in melanoma cell lines. Cancer Research. 2008;68(5):1362–1368. doi: 10.1158/0008-5472.CAN-07-2912. [DOI] [PubMed] [Google Scholar]
- 89.Rosin MP, Lam WL, Poh C, et al. 3p14 and 9p21 loss is a simple tool for predicting second oral malignancy at previously treated oral cancer sites. Cancer Research. 2002;62(22):6447–6450. [PubMed] [Google Scholar]
- 90.Mao L, Lee JS, Fan YH, et al. Frequent microsatellite alterations at chromosomes 9p21 and 3p14 in oral premalignant lesions and their value in cancer risk assessment. Nature Medicine. 1996;2(6):682–685. doi: 10.1038/nm0696-682. [DOI] [PubMed] [Google Scholar]
- 91.Kresty LA, Mallery SR, Knobloch TJ, et al. Alterations of p16INK4a and p14ARF in patients with severe oral epithelial dysplasia. Cancer Research. 2002;62(18):5295–5300. [PubMed] [Google Scholar]
- 92.Cao J, Zhou J, Gao Y, et al. Methylation of p16 CpG island associated with malignant progression of oral epithelial dysplasia: a prospective cohort study. Clinical Cancer Research. 2009;15(16):5178–5183. doi: 10.1158/1078-0432.CCR-09-0580. [DOI] [PubMed] [Google Scholar]
- 93.Takeshima M, Saitoh M, Kusano K, et al. High frequency of hypermethylation of p14, p15 and p16 in oral pre-cancerous lesions associated with betel-quid chewing in Sri Lanka. Journal of Oral Pathology and Medicine. 2008;37(8):475–479. doi: 10.1111/j.1600-0714.2008.00644.x. [DOI] [PubMed] [Google Scholar]
- 94.Hall GL, Shaw RJ, Field EA, et al. p16 promoter methylation is a potential predictor of malignant transformation in oral epithelial dysplasia. Cancer Epidemiology Biomarkers and Prevention. 2008;17(8):2174–2179. doi: 10.1158/1055-9965.EPI-07-2867. [DOI] [PubMed] [Google Scholar]
- 95.Gasco M, Bell AK, Heath V, et al. Epigenetic inactivation of 14-3-3 σ in oral carcinoma: association with p16INK4a silencing and human papillomavirus negativity. Cancer Research. 2002;62(7):2072–2076. [PubMed] [Google Scholar]
- 96.Long NK, Kato K, Yamashita T, et al. Hypermethylation of the RECK gene predicts poor prognosis in oral squamous cell carcinomas. Oral Oncology. 2008;44(11):1052–1058. doi: 10.1016/j.oraloncology.2008.02.004. [DOI] [PubMed] [Google Scholar]
- 97.Toyota M, Ahuja N, Ohe-Toyota M, Herman JG, Baylin SB, Issa JPJ. CpG island methylator phenotype in colorectal cancer. Proceedings of the National Academy of Sciences of the United States of America. 1999;96(15):8681–8686. doi: 10.1073/pnas.96.15.8681. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Toyota M, Ahuja N, Suzuki H, et al. Aberrant methylation in gastric cancer associated with the CpG island methylator phenotype. Cancer Research. 1999;59(21):5438–5442. [PubMed] [Google Scholar]
- 99.Shaw RJ, Hall GL, Lowe D, et al. CpG island methylation phenotype (CIMP) in oral cancer: associated with a marked inflammatory response and less aggressive tumour biology. Oral Oncology. 2007;43(9):878–886. doi: 10.1016/j.oraloncology.2006.10.006. [DOI] [PubMed] [Google Scholar]
- 100.Marsit CJ, Houseman EA, Christensen BC, et al. Examination of a CpG island methylator phenotype and implications of methylation profiles in solid tumors. Cancer Research. 2006;66(21):10621–10629. doi: 10.1158/0008-5472.CAN-06-1687. [DOI] [PubMed] [Google Scholar]
- 101.Sun W, Liu Y, Glazer CA, et al. TKTL1 is activated by promoter hypomethylation and contributes to Head & Neck squamous cell carcinoma carcinogenesis through increased aerobic glycolysis and HIF1α stabilization. Clinical Cancer Research. 2010;16(3):857–866. doi: 10.1158/1078-0432.CCR-09-2604. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Yamada Y, Jackson-Grusby L, Linhart H, et al. Opposing effects of DNA hypomethylation on intestinal and liver carcinogenesis. Proceedings of the National Academy of Sciences of the United States of America. 2005;102(38):13580–13585. doi: 10.1073/pnas.0506612102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Laird PW, Jackson-Grusby L, Fazeli A, et al. Suppression of intestinal neoplasia by DNA hypomethylation. Cell. 1995;81(2):197–205. doi: 10.1016/0092-8674(95)90329-1. [DOI] [PubMed] [Google Scholar]
- 104.Gasche JA, Hoffmann J, Boland CR, Goel A. Interleukin-6 promotes tumorigenesis by altering DNA methylation in oral cancer cells. International Journal of Cancer. 2011;129(5):1053–1063. doi: 10.1002/ijc.25764. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Baba S, Yamada Y, Hatano Y, et al. Global DNA hypomethylation suppresses squamous carcinogenesis in the tongue and esophagus. Cancer Science. 2009;100(7):1186–1191. doi: 10.1111/j.1349-7006.2009.01171.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Subbalekha K, Pimkhaokham A, Pavasant P, et al. Detection of LINE-1s hypomethylation in oral rinses of oral squamous cell carcinoma patients. Oral Oncology. 2009;45(2):184–191. doi: 10.1016/j.oraloncology.2008.05.002. [DOI] [PubMed] [Google Scholar]
- 107.Viet CT, Schmidt BL. Methylation array analysis of preoperative and postoperative saliva DNA in oral cancer patients. Cancer Epidemiology Biomarkers and Prevention. 2008;17(12):3603–3611. doi: 10.1158/1055-9965.EPI-08-0507. [DOI] [PubMed] [Google Scholar]
- 108.Righini CA, De Fraipont F, Timsit JF, et al. Tumor-specific methylation in saliva: a promising biomarker for early detection of Head & Neck cancer recurrence. Clinical Cancer Research. 2007;13(4):1179–1185. doi: 10.1158/1078-0432.CCR-06-2027. [DOI] [PubMed] [Google Scholar]
- 109.López M, Aguirre JM, Cuevas N, et al. Gene promoter hypermethylation in oral rinses of leukoplakia patients—a diagnostic and/or prognostic tool? European Journal of Cancer. 2003;39(16):2306–2309. doi: 10.1016/s0959-8049(03)00550-1. [DOI] [PubMed] [Google Scholar]
- 110.Esteller M, Sanchez-Cespedes M, Resell R, Sidransky D, Baylin SB, Herman JG. Detection of aberrant promoter hypermethylation of tumor suppressor genes in serum DNA from non-small cell lung cancer patients. Cancer Research. 1999;59(1):67–70. [PubMed] [Google Scholar]
- 111.Wong IHN, Lo YMD, Zhang J, et al. Detection of aberrant p16 methylation in the plasma and serum of liver cancer patients. Cancer Research. 1999;59(1):71–73. [PubMed] [Google Scholar]
- 112.Hibi K, Robinson CR, Booker S, et al. Molecular detection of genetic alterations in the serum of colorectal cancer patients. Cancer Research. 1998;58(7):1405–1407. [PubMed] [Google Scholar]
- 113.Carvalho AL, Jeronimo C, Kim MM, et al. Evaluation of promoter hypermethylation detection in body fluids as a screening/diagnosis tool for Head & Neck squamous cell carcinoma. Clinical Cancer Research. 2008;14(1):97–107. doi: 10.1158/1078-0432.CCR-07-0722. [DOI] [PubMed] [Google Scholar]
- 114.Nakahara Y, Shintani S, Mihara M, Hino S, Hamakawa H. Detection of p16 promoter methylation in the serum of oral cancer patients. International Journal of Oral and Maxillofacial Surgery. 2006;35(4):362–365. doi: 10.1016/j.ijom.2005.08.005. [DOI] [PubMed] [Google Scholar]
- 115.Hibi K, Taguchi M, Nakayama H, et al. Molecular detection of p16 promoter methylation in the serum of patients with esophageal squamous cell carcinoma. Clinical Cancer Research. 2001;7(10):3135–3138. [PubMed] [Google Scholar]


