Abstract
Tardive dyskinesia (TD) is a debilitating, unpredictable and often irreversible side effect resulting from chronic treatment with typical antipsychotic agents such as haloperidol. TD is characterized by repetitive, involuntary, purposeless movements primarily of the orofacial region. In order to investigate genetic susceptibility to TD, we used a validated mouse model for a systems genetics analysis geared toward detecting genetic predictors of TD in human patients. Phenotypic data from 27 inbred strains chronically treated with haloperidol and phenotyped for vacuous chewing movements were subject to a comprehensive genomic analysis involving 426,493 SNPs, 4,047 CNVs, brain gene expression, along with gene network and bioinformatic analysis. Our results identified ~50 genes that we expect to have high prior probabilities for association with haloperidol-induced TD, most of which have never been tested for association with human TD. Among our top candidates were genes regulating the development of brain motor control regions (Zic4, Nkx6-1), glutamate receptors (Grin1, Grin2a), and an indirect target of haloperidol (Drd1a) that has not been as well studied as the direct target, Drd2.
Keywords: pharmacogenetic, adverse drug reaction, QTL, haloperidol, mouse
Introduction
First-generation or “typical” antipsychotics (prototype haloperidol) can cause a number of motor side effects that are collectively termed extrapyramidal syndromes (EPS) (Dayalu and Chou 2008; Hsin-tung E and Simpson 2000). Of all patients who initiate treatment, ~40% experience restlessness, involuntary spasms or muscular rigidity in the first few weeks and these symptoms are alleviated to varying degrees by anticholinergic agents (Simpson 1970). Of all patients who sustain long-term treatment (> 3 months), ~35% develop the EPS syndrome tardive dyskinesia (TD) (Dayalu and Chou 2008; Hsin-tung E and Simpson 2000). TD is characterized by repetitive, involuntary and purposeless movements, primarily of the orofacial region (e.g., chewing movements and tongue protrusion) (Crane 1968). Of all individuals who develop TD, it is irreversible in ~50% of cases (Soares-Weiser and Fernandez 2007b) and there is currently no validated and widely accepted treatment (Tandon et al. 2008). Therefore, the physician cannot predict whether a patient will develop TD and, without efficacious treatments, a large number of patients are left with a disfiguring condition.
While familial occurrence of tardive dyskinesia has been observed in a few small studies (Muller et al. 2001; O’Callaghan et al. 1990; Yassa and Ananth 1981), there are no heritability estimates for susceptibility to TD in humans. Despite lack of known heritability, a number of candidate genes have been tested for associations with TD. The results are generally inconsistent. The most encouraging data are for DRD3, HTR2A, HTR2C, and CYP2D6 with positive meta-analyses for each (Bakker et al. 2006; Lerer et al. 2005; Patsopoulos et al. 2005; Reynolds et al. 2005). Negative results have been obtained for DRD2, DRD4, COMT, MAOA, MAOB, and enzymes related to oxidative stress (Herken et al. 2003; Kaiser et al. 2002; Lai et al. 2005; Lee et al. 2007; Matsumoto et al. 2004). Many of these studies suffered from the limitations of examining just one gene at a time, and usually only a single genetic variant, and suboptimal power due to small sample sizes (n<500). We have completed a genome-wide association study (GWAS) of TD by analyzing 492,900 single nucleotide polymorphisms (SNPs) in 214 TD cases and 524 controls with schizophrenia, and no association exceeded chance expectations (Aberg et al. 2010).
Because of limited progress with human pharmacogenomic studies, we explored the potential of a complementary mouse-then-human experimental paradigm (Harrill et al. 2009; Rusyn et al. 2010). We exposed diverse inbred mouse strains to human-like steady-state drug concentrations and measured outcomes of relevance to TD. All strains were previously genotyped using a dense single-nucleotide polymorphism (SNP) chip (Yang et al. 2009b), allowing genetic mapping in silico. Human orthologs of genomic regions strongly implicated in mouse can then be used to reduce the genetic search space in humans to determine whether the association replicates across species.
After chronic treatment with typical antipsychotics, rodents show purposeless mouth openings in the vertical plane (vacuous chewing movements, VCMs)(Waddington et al. 1983). VCMs are a phenotypically and pharmacologically valid animal model of TD that has been used for decades by behavioral pharmacologists (Turrone et al. 2003; Turrone et al. 2002). A large body of research has shown that haloperidol-induced VCM closely mimic nearly every characteristic of human TD (Soares-Weiser and Fernandez 2007a). In a recent study from our laboratory (Crowley et al. 2010), we exposed 27 genetically inbred mouse strains to standardized doses of haloperidol for 120 days in order to calculate heritability and to identify optimal phenotypes for genetic association mapping. This study yielded five critical pieces of information. First, we showed that it is possible to deliver human-like steady state concentrations of haloperidol to diverse mouse strains in a reliable manner with implantable drug pellets. Second, we demonstrated that haloperidol plasma concentrations are highly variable between inbred strains with heritability estimates of ~0.7 and are not influenced by potential confounders such as the dose implanted or body mass. Third, we observed marked behavioral changes across multiple domains. Four measures of activity in the open field, rigidity on an inclined screen (a measure of EPS), and four measures of orofacial movement all exhibited, on average, marked changes following haloperidol exposure. Crucially, these measures were independent of haloperidol plasma level and strain was again the major predictor of phenotypic variation. Fourth, we observed that the behavioral domains we assessed were not discrete constructs but rather loaded onto two factors (Table S1). One factor loaded primarily on antipsychotic-induced changes in open field activity (“OFA”), while the other loaded primarily on haloperidol-induced orofacial movements (“Orofacial”). Finally, we found high heritabilities for haloperidol-induced effects on VCMs, activity in the open field, and EPS. Heritabilities for each of these phenotypes exceeded 75%, and heritability of the factors OFA and Orofacial were ~0.9, after incorporation of the longitudinal nature of the design (Crowley et al. 2010).
In the current study, we perform genetic association mapping of the highly heritable factors from Crowley et al. (“OFA” and “Orofacial”) (Crowley et al. 2010), haloperidol levels and a composite measure of EPS. A comprehensive genomic analysis approach was taken, including tests of association with genome-wide SNPs, biological pathways, copy number variants (CNVs) and gene expression from the brains of the animals tested.
Methods And Materials
Phenotypic data
The collection of the phenotypic data for this study is described in detail in Crowley et al. (Crowley et al. 2010). For GWAS mapping, we chose four phenotypes from Crowley et al.: 1) a principal component loading primarily on antipsychotic-induced changes in open field activity (“OFA”), 2) a principal component loading primarily on antipsychotic-induced changes in orofacial movements (“Orofacial”), 3) the log10 transformation of plasma haloperidol levels 30 days after drug pellet implantation (“HAL30”) and 4) a basic linear unbiased predictor of changes in inclined screen rigidity across 120 days of drug treatment (“EPS”), a commonly used measure of extrapyramidal symptoms in the literature (Chipkin et al. 1988).
Animals
All testing procedures were conducted in strict compliance with the Guide for the Care and Use of Laboratory Animals and approved by the Institutional Animal Care and Use Committee of the University of North Carolina. Male mice (aged 8-10 weeks at the start of testing) from 27 inbred strains (N = 5-9 mice/strain) were obtained from the Jackson Laboratory (Bar Harbor, ME) through the Mouse Phenome Project (Bogue and Grubb 2004). A total of 22 classical and 5 wild-derived strains were examined (Table S2). Animals were maintained on a 12h light:12h dark schedule with lights on at 0700. The housing room was maintained at 20-24°C with 40-50% relative humidity. Mice were housed in standard 20 cm × 30 cm ventilated polycarbonate cages with laboratory grade Bed-O-Cob bedding. Water and Purina ProLab IsoPro 3000 were available ad libitum. All mice were group-housed (maximum of five per cage) except that BALB/cByJ, CAST/EiJ, and SJL/J mice were separated due to fighting after 7, 10 and 13 weeks of housing, respectively. All phenotypes were measured on days 0, 30, 60, 90 and 120 relative to drug treatment (day 1).
Antipsychotic exposure
Slow release haloperidol pellets (3.0 mg/kg/day; Innovative Research of America; Sarasota, FL) (Fleischmann et al. 2002) designed for 60 days of continuous release were implanted subcutaneously with a trocar under two minutes of isoflurane anesthesia. Blood plasma was collected via tail nick for drug concentration assays after 30, 60, 90 and 120 days of exposure to haloperidol. Human-like steady-state concentrations of haloperidol (3.75-19 ng/ml) (Hsin-Tung and Simpson 2000) were achieved in 98% of mice. Figure S1 shows haloperidol plasma levels for each strain across the duration of this study.
Scoring VCMs
High-resolution digital videotapes of orofacial behavior were made by modifying the method of Tomiyama et al. (Tomiyama et al. 2001) (Figure S2). Each mouse was placed in a restrictor device for 25 minutes, and the final 15 minutes were scored for orofacial movement phenotypes: tongue protrusions, overt chewing movements, subtle chewing movements and jaw tremors.
Open field activity
Extrapyramidal side effects may appear as general motor deficits in mice. Therefore, spontaneous locomotor activity in the open field (Crawley 1985) was measured for 1 hour using an automated apparatus (Accuscan Instruments, Columbus, OH). Four phenotypes were extracted from these activity data: total distance traveled (cm), vertical activity, stereotypy, and time spent in the central region of the chamber (percent of total time; central 20cm × 20cm).
Extrapyramidal side effects (EPS)
The inclined screen test (Barnes et al. 1990) was used as an index of Parkinsonian rigidity and sedation. Mice were placed on a wire mesh screen inclined at 45° and the latency to move all four paws was recorded (maximum of 300 seconds).
Phenotypic statistical analysis
Linear mixed effects models (Crowley et al. 2010) were used to decompose phenotype variances for the calculation of heritability and to assess the significance of covariate fixed effects (R 2.6.0 and Stata 9.2). Heritability was calculated using intra-class correlation coefficients. The heritabilities of the over-time trajectories in haloperidol-induced movement disorder phenotypes were assessed using an extension of the mixed model for behavioral genetic analysis (Goldstein 1995). We applied factor analysis to examine the factor structure of the mouse-specific response trajectories (MPlus 5.21) (Joreskog 1969; Muthén and Muthén 2003; Van Prooijen and Van Der Kloot 2001) and individual response phenotype trajectories were decomposed into strain- and mouse-level components, with heritabilities calculated as the ratio of strain-level variance to strain-level + mouse-level variance (Figure S3).
SNP genotypes
All 27 strains were genotyped at the Jackson Laboratory using the Affymetrix Mouse Diversity Array (Yang et al. 2009a) which contains 623,124 SNPs. Prior to association mapping, we removed singletons, heterozygous or missing genotypes, and highly variable probes suggestive of variation within the probe sequence (Yang et al. 2009a). A total of 426,493 SNPs remained for association analysis following quality control.
Association mapping
Due to the population substructure among commercially-available inbred strains, it is critically important to avoid false associations owing to population stratification. Therefore, a three-step process was used for genome-wide association mapping in an effort to reduce the effect of population stratification and increase confidence in mapped loci. First, we used EMMA (Efficient Mixed Model Association) (Kang et al. 2008) to assess evidence of association between each SNP and phenotype. EMMA implements a linear mixed model to account for population structure and genetic relatedness among strains by estimating the pair-wise relatedness between all individuals and fitting these to the phenotype vector. We controlled Type I multiple testing error using permutation by shuffling the strain label while keeping the genotype vectors intact (1,000 permutations). The percentiles of the minimum p-value per permutation were used to determine genome-wide significance thresholds adjusting for multiple testing. Thresholds were 3.4E-8, 1.6E-6, 8.7E-13, and 9.5E-6 for OFA, Orofacial, EPS, and HAL30, respectively. As a check, EMMA was applied after removing the five wild-derived strains as population stratification artifacts are more likely with these genetically divergent strains (Kang et al. 2008). Second, we used TreeQA (Pan et al. 2009), to examine the reproducibility of EMMA results. TreeQA is a quantitative genome-wide association mapping algorithm which uses local phylogenies constructed in genomic regions exhibiting no evidence of historical recombination. Finally, to confirm the robustness of allelic effects, we performed a univariate test (Wilcoxin ranked-sum) using single SNP genotypes as predictors of the primary phenotypic values.
Pathway analysis
We used Ingenuity Pathway Analysis (v6.0) which contains curated biological interactions and functional annotation. As input, we selected the top 1% of genes in each phenotype ranked by minimum p-value. The p-value cutoffs for the top 1% genes were 0.00057, 0.000274, 8.04E-7, and 0.000321 for OFA, Orofacial, EPS, and HAL30.
Bioinformatic analysis
First, we selected all SNPs with EMMA p ≤ 0.0001. Second, we annotated each SNP using UCSC’s KnownGene (Fujita et al. 2011; Hsu et al. 2006), mouse QTL data (Blake et al. 2011), OMIM (McKusick 2007), The Jackson Laboratory’s Mouse Phenotype Database (Blake et al. 2011) and The Sanger Institute Mouse SNPs database (www.sanger.ac.uk/resources/mouse/genomes). We extracted information from the first three databases when the position of each SNP maps within the interval of start and end position of each entry. For the QTL database, we chose a window of ± 5 Mb, since QTL in standard crosses are not mapped to high resolution. The Sanger SNPs database has full genome sequence for 13 of the 27 strains we studied.
CNV analysis
The same Mouse Diversity Array .CEL files that were used to genotype SNPs were used for CNV calling. We analyzed hybridization intensity data from 622,995 SNPs and 597,225 exon probe sets using PennCNV (Wang et al. 2007) to generate an initial set of CNVs calls. We then applied a multi-step quality control procedure (remove CNVs that overlap array gaps, remove small, low confidence, or sparse CNVs) to derive the most confident call set (a total of 4,047 CNV regions were predicted across 26 strains). We do not currently know the level of false negative and false positive CNV calls in this dataset, but are currently using an independent methodology to examine the validity of these calls. This CNV dataset will be the focus of a future publication. We used binary CNV genotypes for genome-wide association with EMMA. Finally, we checked for the presence of CNVs in regions with EMMA SNPs with p < 0.0001 for each phenotype.
Gene expression analysis
The animals used for gene expression are the same ones described in detail in Crowley et al. (Crowley et al. 2010), in which each animal was treated with haloperidol for 120 days and behaviorally phenotyped. At the completion of drug treatment, whole brain was collected from 92 animals (25 strains, average of 4 brains/strain, Table S2) and total RNA was extracted using an automated Maxwell 16 Instrument (Promega; Madison, WI). All samples were processed according to manufacturer’s instructions and hybridized to an Affymetrix Mouse Gene 1.1 ST 96-Array Plate (Affymetrix; Santa Clara, CA). Before analysis, we removed probes containing a known genetic variant from Sanger Institute re-sequencing. We used the RMA method for background adjustment, quantile normalization, and to estimate target and probe effects. Using the probe-set summarized data, we ran a simple linear regression model to test whether changes of expression levels were associated with phenotype. For pathway analysis, we used SAFE (Barry et al. 2005), a two-stage, permutation-based method that accounts for the unknown correlation among genes. Finally, we calculated association between SNP genotype and gene expression levels for EMMA SNPs with p<0.0001. We collected all probe sets within 1Mb of these SNPs, stratified gene expression values by SNP genotype and tested for a significant expression difference between the two genotypes using t-test. For each phenotype, we calculated adjusted p-values using false discovery rate.
Results
Phenotypic data
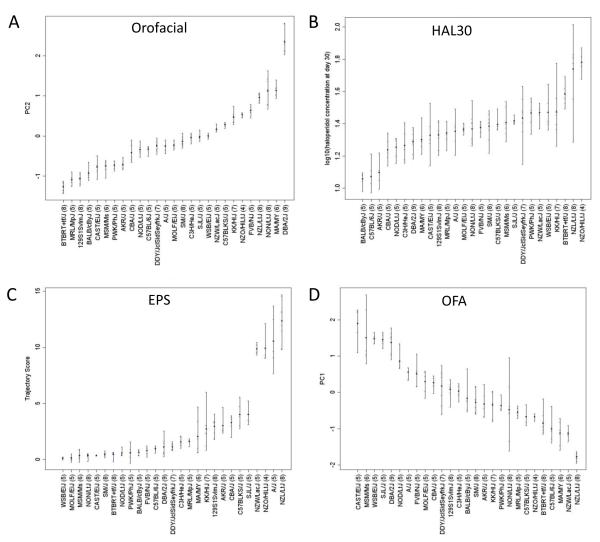
We first selected four phenotypes from Crowley et al (Crowley et al. 2010) with optimal properties for genetic analysis. To this end, we chose one trait with high heritability (all >0.8) for four domains of primary interest: VCMs (Orofacial), pharmacokinetics (HAL30), rigidity (EPS) and open field activity (OFA). Figure 1 shows the 27-strain distribution for each of these phenotypes. First, the five wild-derived strains (CAST, PWK, WSB, MSM, MOLF) were evenly distributed among Orofacial and HAL30, but tended to be less affected by haloperidol in the inclined screen and open field activity tests. Since this could cause spurious GWAS results owing to population stratification, we decided to run GWA for all traits with and without wild-derived strains. Second, strains derived from New Zealand (NZL, NZO, NZW) were susceptible to haloperidol-induced rigidity on the inclined screen test (Figure 1C) and had higher levels of haloperidol (Figure 1B). For EPS we still found an overabundance of GWAS peaks with p <1×10−5, suggesting residual stratification effects. To test this, we ran EMMA with the three New Zealand strains removed and found the top peak to be just 3.1×10−5 (Figure S4), confirming our suspicion. However, since we believe the high EPS susceptibility of the New Zealand strains is a true genetic effect, we decided to leave them in the analysis, calculate a permutation-based genome-wide significance level for each phenotype (see below) and focus follow-up analyses on the top 1% of SNP associated loci. Finally, we found a great deal of variability among the eight Collaborative Cross (Churchill et al. 2004) parental strains (A/J, C57BL6/J, 129S1, NOD, NZO, CAST, PWK, WSB) for each of our phenotypes, indicating that this population would be appropriate to confirm and extend our results.
Figure 1.
Phenotypic data from Crowley et al (Crowley et al. 2010) selected for genetic analysis. The strains are sorted, left to right, in order of increasing haloperidol response (or plasma drug level). The x-axis lists strain name (with the number of animals tested in parentheses) and y-axis gives the phenotypic value for A) Orofacial, B) HAL30, C) EPS and D) OFA.
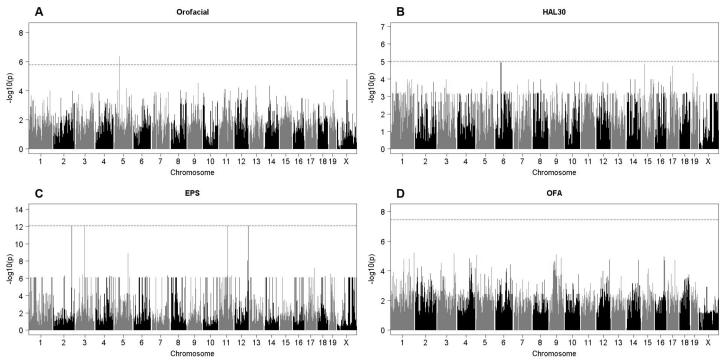
Association mapping (Figure 2)
Figure 2.
Genome-wide association results from EMMA for A) Orofacial, B) HAL30, C) EPS and D) OFA. Chromosome position is on the x-axis and the −log P value is on the y-axis. The dashed horizontal line indicates the permutation-based genome-wide significance level, which is unique for each phenotype.
(a) Orofacial was the only trait with an association exceeding genome-wide significance (Figure 2A, chr5:42.3-44.4 Mb, P=1.6×10−6). This association was robust to analytical method (Table 1). This region contains 12 known genes and 6 predicted genes, several of which are expressed in brain (i.e. Cpeb2, Bst1). A search of the literature did not reveal any immediate links between genes in this locus and haloperidol pharmacology, movement disorders or monoamine neurotransmission. The second most significant peak was on chrX: 86.6-86.8 Mb (P=1.7×10−5), near Pit2 (plasmacytoma expressed transcript 2) which is expressed in substantia nigra (Lagrue et al. 2010), a region thought to be affected in TD (Chen et al. 2011). The third most significant peak occurred on chr9:91.8-92.0 Mb (P=3.0×10−5), flanking two genes also expressed in brain motor control regions: Zic4 (zinc finger protein of the cerebellum 4) and Plscr1 (phospholipid scramblase 1). Finally, the fourth most significant peak (chr13:53.912-53.914 Mb, P=4.6×10−5) was adjacent to the dopamine receptor gene Drd1a, of great interest since haloperidol binds this receptor (Hsin-tung E and Simpson 2000) and down-regulates it in pre-frontal cortex (Lidow and Goldman-Rakic 1994). As with all of the Orofacial associations, this chr13 locus was seen with all four analytical methods (Table 1). The individual strain genotypes for the top SNPs listed in Table 1 are listed in Table S4.
Table 1.
The five most significant loci for each phenotype after genome-wide association mapping with 426,493 SNPs.
| Phenotype | Chr | Locus Start bp |
Locus End bp |
# SNPs |
Max SNP | EMMA1 p |
Wilcoxon p |
TreeQA p |
EMMA2 p |
Genes in Locus3 |
|---|---|---|---|---|---|---|---|---|---|---|
| Orofacial | 5 | 42736571 | 44487186 | 25 | rs32777671 | 4.47E-7 | 8.91E-7 | 1.00E-5 | 1.48E-5 |
Prom1, Bst1, Fgfbp1, 5730509K17Rik, [Pet2, Cpeb2, 4932429P05Rik, C1qtnf7], (A230054D04Rik, AK040452) |
| X | 86780367 | - | 1 | rs33843488 | 1.70E-5 | 4.57E-5 | 1.00E-5 | 2.75E-4 | (Pet2, 4932429P05Rik) | |
| 9 | 91831454 | 92024938 | 3 | rs30230732 | 3.02E-5 | 2.69E-5 | 3.98E-5 | 1.07E-4 | (Zic4, Plscr1) | |
| 13 | 53912176 | 53913808 | 2 | rs29249826 | 4.57E-5 | 6.61E-6 | 1.00E-5 | 2.29E-4 | (AK039269, Drd1a) | |
| 14 | 46701783 | - | 1 | rs46050648 | 4.57E-5 | 1.48E-4 | 6.03E-5 | 1.07E-3 | (mKIAA1705, Ubb) | |
| HAL30 | 6 | 50916357 | 51023936 | 3 | rs29752875 | 1.12E-5 | 6.76E-4 | 0.08 | 7.59E-5 | (AK145307, AK002748) |
| 15 | 23536278 | 25639040 | 22 | rs31631263 | 1.41E-5 | 2.29E-4 | 0.02 | 9.12E-5 |
Basp1, Myo10 , 9230109A22Rik, [BC10049], (Cdh18, 1810015C04Rik) |
|
| 17 | 44564946 | 45318710 | 27 | rs33465886 | 1.86E-5 | 1.70E-5 | 3.02E-5 | 1.15E-4 |
Supt3h, Osf2/Cbfa1, [Clic5, Runx2, Cdc5l], (Clic5, Cdc5l) |
|
| 19 | 21652218 | - | 1 | rs45713927 | 4.79E-5 | 2.69E-5 | 5.01E-5 | 2.51E-4 | (Gda, 1110059E24Rik) | |
| 17 | 28814308 | 28849916 | 2 | rs49358929 | 7.24E-5 | 4.37E-3 | 2.14E-4 | 2.82E-4 | Slc26a8, Mapk14, (Srpk1, Brpf3) | |
| EPS | 11 | 73156943 | 74300325 | 13 | rs29458500 | 8.13E-13 | 1.15E-4 | 6.91E-3 | 3.55E-11 |
Garnl4, Spata22, [Olfr cluster, AK145370], (Aspa, Pafah1b1) |
| 12 | 120978319 | 121006298 | 4 | rs33723656 | 8.13E-13 | 1.15E-4 | 7.59E-3 | 3.55E-11 | (Macc1, 4732474O15Rik) | |
| 2 | 157211478 | 157216167 | 4 | rs27307301 | 8.13E-13 | 1.15E-4 | 0.10 | 3.55E-11 | Manbal, (Ghrh, Src) | |
| 3 | 73919980 | 75832889 | 2 | rs31054510 | 8.13E-13 | 1.15E-4 | 7.94E-5 | 3.55E-11 | Pdcd10, Golim4, BC050789, (Bche, Fstl5) | |
| 5 | 115665863 | - | 1 | rs29569591 | 1.29E-9 | 5.01E-5 | 0.35 | 1.45E-7 | (Cabp1, Pop5) | |
| OFA | 1 | 196329578 | - | 1 | rs49505396 | 6.31E-6 | 4.47E-6 | 1.00E-5 | 3.72E-4 | (Camk1g, Plxna2) |
| 3 | 142811186 | 142821385 | 2 | rs30557323 | 7.08E-6 | 3.02E-5 | 1.00E-5 | 1.95E-4 | (AK035466, AK043679) | |
| 5 | 3989136 | - | 1 | rs31177638 | 8.91E-6 | 1.38E-5 | 1.00E-5 | 1.95E-4 | Akap9 (Mterf, Cyp51) | |
| 16 | 79380579 | - | 1 | rs4209251 | 1.12E-5 | 2.69E-5 | 3.02E-5 | 2.14E-4 | (Prss7, Ncam2) | |
| 1 | 154344352 | 155157963 | 4 | rs30691213 | 1.58E-5 | 2.04E-4 | 7.08E-4 | 1.35E-4 |
Glt25d2, Nmnat2, Lamc1 LOC66637, AK087784) |
EMMA was run using all 27 strains. SNPs are ranked by EMMA1 −log10(p), chromosome, and then position.
EMMA was run using 22 classical strains.
Un-bracketed genes are in the locus and contain a genotyped EMMA1 SNP with p< 10−4 within their transcript. Bracketed genes are in the locus but do not have a genotyped SNP with p< 10−4. Genes in parentheses are the upstream and downstream nearest genes to the locus: (upstream, downstream). Genes in bold are mentioned in the results.
(b) HAL30 (Figure 2B) had loci on chr6:50.92-51.02 Mb (P=1.1×10−5) and chr15:23.54-25.64 (P=1.4×10−5) that narrowly missed reaching genome-wide significance. Both of these peaks occurred in regions harboring genes of primarily unknown function. Three other suggestive peaks were seen, two on chr17 and one on chr19. The chr19 peak occurs within a liver-expressed anion exchanger, Slc26a8.
(c) EPS (Figure 2C) also had associations that narrowly missed permutation-based genome-wide significance, including four peaks of similar magnitude on chr 2, 3, 11 and 12. The significance threshold for EPS (8.7×10−13) was higher because this phenotype was essentially dichotomous: 4 of 27 strains showed high levels of rigidity and three of these are derived from New Zealand.
(d) OFA (Figure 2D) showed several peaks of similar height that failed to reach the significance threshold. Two well-known neurodevelopmental genes Ncam2 (neuronal cell adhesion molecule 2) and Plxna2 (plexin A2) were within these regions.
Pathway analysis
Since we analyzed complex genetic traits, and our sample size was relatively small, clear-cut identification of genome-wide significant loci was perhaps unlikely. Some proportion of the top loci could contain true signals that did not reach genome-wide significance due to low power. Therefore, we ran pathway analysis on the top 1% of genes in each phenotype. Table 2 and Figures S5-S8 describe these networks. Many of these networks have biological plausibility and some genes in these networks have known roles in movement disorders, neurotransmission and drug absorption, distribution, metabolism and excretion (ADME). For example, the Orofacial networks contained the primary receptor for haloperidol (dopamine receptor D2, Drd2), two glutamate receptors (Grin1, Grin2a), the Huntington’s disease gene (Htt), and Ncam1, as mentioned above.
Table 2.
The two most significant networks for each phenotype following pathway analysis.
| Phenotype | Network ID |
Gene Network* | Score | # of top 1% Genes |
Top Functions |
|---|---|---|---|---|---|
| Orofacial | 1 |
Afp, Ass1, Atr, Cd38, Cdkn1a, Cdkn2a, Chek1, Ciita, Cpox, Cyp19a1, Dgka, E2f1, Ezh2, Gata1, Hras, Htt, Il4, Itpr1, Mcm3, Mcm5, Pdgfrb, Plcb1, Pou2f3, Rb1, Runx2, Rxrg, Scmh1, Scn3b, Srm, Tp53, Trpc4, Ttk, Uqcrc1, Usp2 |
1E-14 | 11 | Cancer, Genetic Disorder, Reproductive System Disease |
| 2 |
Acat1, Adam10, Akap9, Arc, Bcar1, Cam2kb, Cdh2, Cltc, Dclk1, Dlg2, Dlg3, Dlg4, Dlgap1, Dlgap3, Drd2, Fyn, Grik2, Grin1, Grin2a, Hspa1a, Mpl, Ncam1, Nsf, Ntrk2, Penk, Pgk1, Plat, Prkaca, Rims1, Scn2a, Slc25a4, Sp1, Syngap1 |
1E-11 | 9 | Nervous System Development and Function, Cell-To-Cell Signaling and Interaction, Behavior |
|
| HAL30 | 1 |
Abcc3, Abr, C2, Coro2b, Cr1, Creb1, Dlg4, Fam162a, Foxn3, Glo1, Gstt2, Htt, Il4, Il5, Il1rap, Ints7, Kcnab1, Kcnk2, Mapk14, Mtdh, Ndufa3, Pfkp, Pld1, Prim1, Runx2, Sdf2l1, Ska1, Slain1, Slc4a4, St7, Tnf, Tnfsf11, Tp53, Uck2 |
1E-30 | 18 | Hematological System Development and Function, Cell Morphology, Growth and Proliferation |
| 2 | Fam3d, YBX2 | 1E-2 | 1 | Cellular Development | |
| EPS | 1 |
Abcc3, Abr, Adam10, Atp5a1, Atrx, Camp, Ccl4, Cd200r1, Clec7a, Cr1, Cyp17a1, Dlg4, Dlgap1, Erk, Esr2, Gapdh, Gnaq, Il6, Il1rap, Klra8, Lep, Lipc, Man2b2, Map3k1, Mpl, Mtus1, Pdia3, Prdx1, Slc25a4, Slc25a6, Slco1a2, Sp1 |
1E-16 | 11 | Hepatic System disease, Liver Cholestasis, Lipid metabolism |
| 2 |
Abcc3, Afp, Cdh1, Csmd1, Ctnnb1, Cyp7a1, Grik1, Hnf4a, Il1b, Ins1, Mtmr7, Nr1i2, Nr1i3, Pck1, Pitx2, Rhpn2, Setdb1, Sfrp1, Sult1a1, T, Wnt3a |
1E-10 | 7 | Neurological Disease, Genetic Disorder, Metabolic Disease |
|
| OFA | 1 |
Acat1, Akap9, Arc, Baiap2, Camk2b, Cldn18, Cltc, Coro2b, Dlg2, Dlg3, Dlg4, Dlgap1, Dlgap3, Foxp1, Gapdh, Gnao1, Grik2, Grin1, Grin2a, Homer1, Htt, Kcnab1, Mir122a, Ncam1, Nsf, Pkm2, Plat, Plcb1, Rims1, Rxrg, Syngap1, Trpc4 |
1E-14 | 11 | Behavior, Nervous System Development and Function, Cell-Cell Signaling and Interaction |
| 2 |
Atp1a1, Atr, Bbc3, Birc5, Ccl6, Cd81, Cdkn2a, Chek1, Eif4b, Ifng, Kcnj1, Klra8, Mbp, Mpl, Ncoa2, Pdgfrb, Pias1, Pik3cd, Rorc, Scmh1, Scn3b, Scnn1a, Scnn1g, Slc12a1, Slc28a1, Slc28a2, Slc29a1, Slc9a3, Stat1, Tgfa, Tnf, Tp53, Ttk |
1E-12 | 10 | Cell Death, Cell-mediated Immune Response, Cellular Development |
Bold = A gene within the top 1% of genes ranked by minimum p-value among all SNPs mapped to the gene. Underlined genes are mentioned in the results.
For HAL30, only one network was significantly associated with the phenotype and it acts in liver development and function. Data from humans indicate that about 40% of haloperidol is subject to reabsorption via enterohepatic recycling (Eddington and Young 1990; Froemming et al. 1989). Therefore, it is intriguing that at least one gene in this network, Abcc3 (multidrug resistance protein 3), is known to regulate biliary secretion. Slc4a4, also in this pathway, modulates renal tubular pH which is a critical factor in drug elimination via urine. EPS was also associated with a liver network, including the ADME-related genes Cyp17a1, Slco1a2 and Abcc3. The second network contained genes that are mutated in human neurological diseases: Grik1 (epilepsy), Wnt3a (neural tube defects), Mtmr7 (myotubular myopathy). The top network for OFA listed a number of genes that, when knocked-out in mice, result in abnormal behavior, including activity differences. These included the glutamate receptors Grik2, Grin1 and Grin2a, and the calcium/calmodulin-dependent protein kinase Camk2b (Mohn et al. 1999; Sakimura et al. 1995; Shaltiel et al. 2008; van Woerden et al. 2009).
Bioinformatic analysis
Table 3 highlights published mouse QTLs that are within 5 Mb of an EMMA SNP with p ≤ 0.0001; a 5 Mb window was used since most of these eQTLs were mapped using low resolution crosses (F2 or BXD). It is intriguing that all three pharmacodynamic phenotypes (Orofacial, EPS, OFA) share confidence intervals with at least 5 QTLs for compounds which act directly (haloperidol, cocaine, methamphetamine) or indirectly (ethanol) on dopaminergic neurotransmission.
Table 3.
A selection of previously identified QTL within 5 Mb of the top 1% of EMMA SNPs for each phenotype.
| Phenotype | QTL ID | QTL description1 | Chr | QTL Start bp | QTL End bp |
|---|---|---|---|---|---|
| Orofacial | Cocia6 | cocaine induced activation 6 | 5 | 37,920,888 | 37,921,029 |
| Alcp9 | alcohol preference locus 9 | 5 | 48,258,172 | 48,258,377 | |
| Hpic1 | haloperidol-induced catalepsy 1 | 9 | 86,440,120 | 86,440,252 | |
| Cocia11 | cocaine induced activation 11 | 13 | 54,579,545 | 54,579,665 | |
| Cosz2 | cocaine seizure 2 | 14 | 36,137,026 | 69,166,344 | |
| HAL30 | Chab5 | cholesterol absorption 5 | 19 | 18,750,890 | 18,751,053 |
| EPS | Dautb4 | dopamine uptake transporter binding 4 | 11 | 79,078,681 | 79,078,829 |
| Brmth3 | behavioral response to methamphetamine 3 | 5 | 104,668,024 | 104,668,218 | |
| Drb2 | dopamine receptor binding 2 | 5 | 104,668,024 | 104,668,218 | |
| Actd3 | activity-distance traveled 3 | 5 | 113,601,539 | 113,601,687 | |
| Chab7 | cholesterol absorption 7 | 5 | 112,514,368 | 112,514,484 | |
| Diht | dopamine-induced hypothermia | 5 | 115,413,178 | 115,413,490 | |
| OFA | Lore10 | loss of righting induced by ethanol 10 | 3 | 142,848,413 | 147,002,931 |
| Elorr2 | ethanol induced loss of righting response 2 | 3 | 142,848,413 | 142,848,587 | |
| Cocia12 | cocaine induced activation 12 | 16 | 76,817,838 | 76,817,961 | |
| Etia | ethanol induced activation | 16 | 80,517,264 | 80,517,373 | |
| Lore8 | loss of righting induced by ethanol 8 | 1 | 154,849,280 | 154,849,394 |
Chemicals that act directly or indirectly on the dopaminergic system are in bold.
We used the Sanger SNPs database to determine the functional consequence of each EMMA SNP with p ≤ 0.0001. Furthermore, since the Sanger database has full genome sequence for 13 of our 27 strains, we were also able to identify genomic regions where the strain distribution pattern matched our top EMMA SNPs (restricted to ± 40 kb from the EMMA SNP). In this way, we identified hundreds of additional variants linked to the genotyped SNP markers, including several putatively functional variants (see Table 4). With Orofacial, for example, we found that a VCM-protective haplotype on chr5 is linked to a SNP in the 3′ UTR of Nkx6-1, a gene that plays an essential role in midbrain dopaminergic neuron development (Prakash and Wurst 2006). In addition, we found that the strain with the highest level of haloperidol-induced VCMs (DBA/2J) has two variants within the 5′ UTR of striatum-expressed Plscr1 and a non-synonymous coding SNP within the predicted gene EG624120.
Table 4.
A sample of potentially functional Sanger SNPs occurring within 40kb, and sharing the same strain distribution pattern, as EMMA SNPs with p < 0.0001 for each phenotype.
| Phenotype | Gene | Chr | Position | Type | Variant1 | Sanger strains with variant |
|---|---|---|---|---|---|---|
| Orofacial | EG624120 | 5 | 44492593 | Non-synonymous | G>A | DBA/2J |
| Nkx6-1 | 5 | 102088274 | 3′ UTR | A>G | 129S1, AKR, BALBc/J, CAST, NOD, PWK |
|
| Plscr1 | 9 | 92144923 | 5′ UTR | T>C | DBA/2J | |
| Plscr1 | 9 | 92145121 | 5′ UTR | G>T | DBA/2J | |
| Cdkn2aipnl | 11 | 51790688 | 3′ UTR | A>G | DBA/2J, NZO | |
| HAL30 | EG432939 | 15 | 25344191 | Non-synonymous | A>G | 129S1, A/J, C3H, CAST, CBA, DBA/2J, NZO, PWK, WSB |
| Mapk14 | 17 | 28828610 | 5′ UTR | C>T | NZO | |
| Runx2 | 17 | 44873257 | Non-synonymous | T>C | 129S1, A/J, CAST, NOD, NZO, PWK, WSB |
|
| Supt3h | 17 | 44914140 | 5′ UTR | C>T | 129S1, A/J, CAST, NOD, NZO, PWK, WSB |
|
| EPS | Garnl4 | 11 | 74225827 | Non-synonymous | T>C | A/J, CAST, NZO, PWK, WSB |
| Spata22 | 11 | 73153799 | Non-synonymous | C>G | A/J, NZO | |
| OFA | Akap9 | 5 | 3968775 | Non-synonymous | T>G | A/J, C3H, CAST, CBA, DBA/2J, NOD, PWK, WSB |
| Lamc1 | 1 | 155096884 | Splice site | A>G | CAST, DBA/2J, NOD, WSB |
The reference strain, C57BL6/J, is listed first.
CNV analysis
CNVs are another form of genetic variation that could explain the heritability of these traits. We performed GWA with a set of 4,047 structural variants predicted to occur from Mouse Diversity Array data from these strains. Table S5 lists the five most significant CNVs for each phenotype after GWA and the corresponding genes with at least one exon lost or gained due to deletion or duplication. For Orofacial, we found a strong relationship between duplication of Mtch2 (mitochondrial carrier homolog 2) and increased haloperidol-induced VCMs. The five strains with this duplication all fall within the top 9 strains in a 27-strain ranking of the Orofacial variable. Duplication of Mtch2 is a plausible variant for contributing to VCM susceptibility, given its role in apoptosis (Zaltsman et al. 2010) and the possible role of neuroleptic-induced neuronal toxicity (via apoptosis) in TD (Galili et al. 2000; Mitchell et al. 2002; Skoblenick et al. 2006). With OFA, a deletion of Epha6 was found in the two strains with the greatest haloperidol-induced decrease in activity (NZL and NZW), and EPHA6 was associated with therapeutic response to the neuroleptic risperidone (Ikeda et al. 2010). Furthermore, its expression was significantly increased in mouse frontal cortex following chronic risperidone treatment (Ikeda et al. 2010).
Next, since SNPs are capable of tagging CNVs, we looked for overlap between CNVs and the top 1% of SNP-associated loci for each phenotype (Table S6). A total of 6 variants, all deletions, were found in the vicinity of the top SNPs, though only one of them resulted in loss of exons. For Orofacial, a deletion on chr9 in the SM/J strain (which showed average VCM susceptibility) was found to flank two genes mentioned above with expression in brain motor control regions: Zic4 and Plscr1.
Gene expression analysis
Gene expression data can be used to prioritize regions for follow-up (Aylor et al. 2011). Therefore, we sought to determine if brain gene expression correlated with any of the phenotypes examined or with SNPs within EMMA-significant loci. We collected whole brain tissue from mice that had been chronically treated with haloperidol for 120 days (the same mice used in (Crowley et al. 2010)). Whole brain tissue was used because the pathophysiology of EPS is thought to include several dispersed neuroanatomical regions (Koshikawa et al. 2011) and we found it more desirable to capture all regions rather than microdissect a single region. Since the Crowley et al study did not have a placebo-treated arm (but rather focused on differences before and after drug treatment) we did not have tissue from untreated animals to use as a comparison group. We deemed it a worthwhile experiment, however, since if haloperidol susceptibility was tightly linked to the expression level of a particular transcript (or a set of transcripts in a biological pathway) after haloperidol treatment, it should be detectable. We prioritized transcripts near: 1) top EMMA SNPs that correlated with gene expression in cis, 2) the top 1% of genes in terms of correlation between expression and phenotype, and 3) expression levels for the most interesting candidate genes derived from the SNP GWAS, CNV GWAS and pathway analysis.
First, we collected all probe sets within 1Mb of EMMA SNPs with p<0.0001, stratified gene expression values by SNP genotype and tested for a significant expression difference between the two genotypes. For each phenotype, a large number of genes showed evidence of cis regulation (see Table S7). For example, with Orofacial we tested 287 probe sets and 61 were consistent with cis-regulation at a false discovery rate of 5%. Among these genes were the dopamine receptor Drd1a, liver enzyme Cyp46a1, the cerebellar gene Zic4 mentioned above and also a related gene, Zic1. The other three phenotypes showed fewer immediately obvious candidates.
Second, we examined the relationship between gene expression of the top 1% of associated genes (Table S8) and phenotype. For Orofacial, this list included two synthetic enzymes and one receptor binding partner for the primary inhibitory neurotransmitter in the brain, GABA (Gad1, Gad2, Gabarapl2). HAL30 associated with several metabolic genes (i.e. Cyp2a22), EPS correlated with several neurotransmitter-related genes including the dopamine-transporting vesicular monoamine transporter 1 (Slc18a1) and OFA associated with a number of genes that show brain expression, though of less-obvious relevance to haloperidol.
Finally, we have identified many intriguing quantitative trait gene candidates through SNP GWAS, CNV GWAS and pathway analysis. Table S9 lists association between gene expression and phenotype for 27 such genes (the bold gene names in Tables 1, 2, S5). We searched for genes whose expression level is among the top 20% of genes in terms of association with the phenotype. First, two direct targets of haloperidol (Drd1 and Drd2) tend to correlate with Orofacial. Six other genes identified by genetic analysis were similarly linked to Orofacial, including the glutamate receptor gene Grin1. The HAL30 results are more difficult to interpret for the reasons mentioned above, though the renal transporter Slc4a4 fell within the top 20%. There was an interesting set of genes that we initially found with Orofacial, but whose expression was correlated more with EPS (Cpeb2, Pet2, Ncam1, Plscr1). This suggests the possibility of pleiotropic effects for these genes, regulating susceptibility for two distinct haloperidol ADRs. Finally, OFA had three candidate genes in the top 20%, including the risperidone-responsive gene Epha6.
Discussion
The purpose of this report was to investigate the genetics of several haloperidol-associated phenotypes with the eventual goal of improving understanding of the genomics of human TD. If VCMs are a reasonable analogue of TD, then it might be possible to accelerate discovery by using a design whereby mouse genetic mapping resources are used to screen the genomic search space to derive high-probability targets whose orthologs can be studied in human samples. In this way, the multiple testing burden is paid in a relatively inexpensive and experimentally tractable system and human samples are used only for testing candidate targets. To achieve this end, we selected optimal phenotypes from Crowley et al (Crowley et al. 2010), and took a comprehensive genomic analysis approach.
First, we mapped QTL for haloperidol-response using 27 inbred strains and 426,493 SNPs. This approach led to QTL of much higher resolution than with traditional mouse mapping methods, which have employed populations with lower diversity (i.e. F2 cross or BXD recombinant inbred lines) and lower density genotyping (~500 markers). For example, our largest locus spans just 2.1 Mb (HAL30 association on chr15), while the confidence intervals from traditional mouse QTL studies can easily cover 20 Mb and include hundreds of genes. Our top peaks pointed to a number of interesting genes, only one of which has ever been included in a TD candidate gene study (Drd1). For our three pharmacodynamic traits, we identified genes expressed in the striatum (Plscr1, Bche, Drd1), neurodevelopmental regulators (Ncam2, Plxna2), a key cerebellum gene (Zic4) and ~20 genes of currently unknown function.
Second, we performed pathway analysis using our top GWA loci. The top networks for each phenotype were consistent with the phenotypes, suggesting that our top loci contained true signal, but did not reach genome-wide significance due to low power. Also consistent with this idea is the presence of haloperidol’s primary receptor (Drd2) within the second Orofacial network. As mentioned in the results, pathway analysis revealed a number of additional genes that are plausible candidates for TD/EPS, including three glutamate receptors (Grik2, Grin1, Grin2a) and genes known to cause neurological disease (Grik1, Wnt3a, Mtmr7). As for haloperidol plasma levels, our overall pattern of results seem to suggest a role for enterohepatic recycling, perhaps via Abcc3 (multidrug resistance protein 3).
Third, we collected a wealth of bioinformatic data on the top peaks of interest. When we looked at the published mouse QTLs surrounding our top SNPs, it was reassuring how many of these were related to dopaminergic drug response. All three of our pharmacodynamic phenotypes shared confidence intervals with at least five QTLs for compounds which act on dopamine (haloperidol, cocaine, methamphetamine, ethanol), while HAL30 shared no QTLs with these compounds. This congruence is encouraging, but the ultimate goal of QTL studies is to identify the causal quantitative trait nucleotide (QTN) underlying the QTL (Mackay et al. 2009). Identification of the QTN has been the rate-limiting step in traditional mouse complex trait studies, owing in part to a lack of polymorphism data genome-wide. However, increasing amounts of genomic data in mice make this task straight-forward to search for variants with a strain distribution pattern consistent with causality. We used the Sanger database to identify hundreds of additional variants linked to the genotyped SNP markers, including several putatively functional variants. Of particular interest was a VCM-protective haplotype on chr5 linked to a SNP in the 3′ UTR of Nkx6-1, a gene required for midbrain dopaminergic neuron development (Prakash and Wurst 2006). It is conceivable that altered dopaminergic function via polymorphism in Nkx6-1 could predispose mice to haloperidol-induced adverse drug reactions.
Fourth, we tested whether CNVs affected these traits. We performed GWA with a set of 4,047 structural variants known to occur in these strains and also looked for overlap between CNVs and the top 1% of SNP-associated loci. In the CNV GWA, we found a strong relationship between duplication of the Mtch2 (mitochondrial carrier homolog 2) gene and increased haloperidol-induced VCMs. This is interesting given the role of Mtch2 in apoptosis and the literature implicating neuroleptic-induced neuronal toxicity (via apoptosis) in the development of TD (Galili et al. 2000; Mitchell et al. 2002; Skoblenick et al. 2006). In addition, the relationship between a deletion of Epha6 and haloperidol-induced decreased activity is intriguing given the results of a recent clinical study linking this gene to risperidone treatment response (Ikeda et al. 2010). Finally, it was notable that the liver-expressed gene Supt3h was deleted in two strains with high plasma drug levels.
Finally, we examined the relationship between our findings and brain gene expression following withdrawal from chronic haloperidol treatment. This extra step provided a plausible method for prioritizing candidate genes within our top loci for follow-up. For example, since Drd1a is located within one our top Orofacial loci and is cis-regulated, it is reasonable to hypothesize that gene regulatory variation in this gene could underlie the QTL.
In conclusion, we have identified ~50 genes that we expect to have high prior probabilities for association with haloperidol-induced TD. Furthermore, since the human genome contains ~25,000 genes, we have provided a logical rationale for focusing on just a small fraction (0.2%) of that genomic search space. As such, the multiple testing penalty is reduced 500-fold.
Acknowledgements
The mice used in this study were acquired as part of the Mouse Phenome Project, an ongoing international collaborative effort headquartered at The Jackson Laboratory (Bar Harbor, ME, USA). This work was supported by the Pharmacogenetics Research Network (U01 GM63340, PI Dr. McLeod), a NIMH/NHGRI Center of Excellence for Genome Sciences grant (P50 MH90338, PIs Drs. Fernando Pardo-Manuel de Villena and Sullivan), and the Mouse Behavioral Phenotyping Laboratory (NICHD P30 HD03110, PI Dr. Joseph Piven). Dr Sullivan was supported by MH080403, MH077139, and MH074027.
Footnotes
Financial Disclosures
The authors report no biomedical financial interests or potential conflicts of interest.
On-Line Resources
Phenotypic data from this project are available online via the Mouse Phenome Database (MPD; http://www.jax.org/phenome).
References
- Aberg K, Adkins DE, Bukszar J, Webb BT, Caroff SN, Miller del D, Sebat J, Stroup S, Fanous AH, Vladimirov VI, McClay JL, Lieberman JA, Sullivan PF, van den Oord EJ. Genomewide association study of movement-related adverse antipsychotic effects. Biol.Psychiatry. 2010;67:279–282. doi: 10.1016/j.biopsych.2009.08.036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aylor DL, Valdar W, Foulds-Mathes W, Buus RJ, Verdugo RA, Baric RS, Ferris MT, Frelinger JA, Heise M, Frieman MB, Gralinski LE, Bell TA, Didion JD, Hua K, Nehrenberg DL, Powell CL, Steigerwalt J, Xie Y, Kelada SN, Collins FS, Yang IV, Schwartz DA, Branstetter LA, Chesler EJ, Miller DR, Spence J, Liu EY, McMillan L, Sarkar A, Wang J, Wang W, Zhang Q, Broman KW, Korstanje R, Durrant C, Mott R, Iraqi FA, Pomp D, Threadgill D, Pardo-Manuel de Villena F, Churchill GA. Genetic analysis of complex traits in the emerging collaborative cross. Genome research. 2011 doi: 10.1101/gr.111310.110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bakker PR, van Harten PN, van Os J. Antipsychotic-induced tardive dyskinesia and the Ser9Gly polymorphism in the DRD3 gene: a meta analysis. Schizophr.Res. 2006;83:185–192. doi: 10.1016/j.schres.2006.01.010. [DOI] [PubMed] [Google Scholar]
- Barnes DE, Robinson B, Csernansky JG, Bellows EP. Sensitization versus tolerance to haloperidol-induced catalepsy: multiple determinants. Pharmacol Biochem Behav. 1990;36:883–887. doi: 10.1016/0091-3057(90)90094-x. [DOI] [PubMed] [Google Scholar]
- Barry WT, Nobel AB, Wright FA. Significance analysis of functional categories in gene expression studies: a structured permutation approach. Bioinformatics. 2005;21:1943–1949. doi: 10.1093/bioinformatics/bti260. [DOI] [PubMed] [Google Scholar]
- Blake JA, Bult CJ, Kadin JA, Richardson JE, Eppig JT. The Mouse Genome Database (MGD): premier model organism resource for mammalian genomics and genetics. Nucleic acids research. 2011;39:D842–848. doi: 10.1093/nar/gkq1008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bogue MA, Grubb SC. The Mouse Phenome Project. Genetica. 2004;122:71–74. doi: 10.1007/s10709-004-1438-4. [DOI] [PubMed] [Google Scholar]
- Chen S, Seeman P, Liu F. Antipsychotic drug binding in the substantia nigra: an examination of high metoclopramide binding in the brains of normal, Alzheimer’s disease, Huntington’s disease, and Multiple Sclerosis patients, and its relation to tardive dyskinesia. Synapse. 2011;65:119–124. doi: 10.1002/syn.20825. [DOI] [PubMed] [Google Scholar]
- Chipkin RE, Iorio LC, Coffin VL, McQuade RD, Berger JG, Barnett A. Pharmacological profile of SCH39166: a dopamine D1 selective benzonaphthazepine with potential antipsychotic activity. The Journal of pharmacology and experimental therapeutics. 1988;247:1093–1102. [PubMed] [Google Scholar]
- Churchill GA, Airey DC, Allayee H, Angel JM, Attie AD, Beatty J, Beavis WD, Belknap JK, Bennett B, Berrettini W, Bleich A, Bogue M, Broman KW, Buck KJ, Buckler E, Burmeister M, Chesler EJ, Cheverud JM, Clapcote S, Cook MN, Cox RD, Crabbe JC, Crusio WE, Darvasi A, Deschepper CF, Doerge RW, Farber CR, Forejt J, Gaile D, Garlow SJ, Geiger H, Gershenfeld H, Gordon T, Gu J, Gu W, de Haan G, Hayes NL, Heller C, Himmelbauer H, Hitzemann R, Hunter K, Hsu HC, Iraqi FA, Ivandic B, Jacob HJ, Jansen RC, Jepsen KJ, Johnson DK, Johnson TE, Kempermann G, Kendziorski C, Kotb M, Kooy RF, Llamas B, Lammert F, Lassalle JM, Lowenstein PR, Lu L, Lusis A, Manly KF, Marcucio R, Matthews D, Medrano JF, Miller DR, Mittleman G, Mock BA, Mogil JS, Montagutelli X, Morahan G, Morris DG, Mott R, Nadeau JH, Nagase H, Nowakowski RS, O’Hara BF, Osadchuk AV, Page GP, Paigen B, Paigen K, Palmer AA, Pan HJ, Peltonen-Palotie L, Peirce J, Pomp D, Pravenec M, Prows DR, Qi Z, Reeves RH, Roder J, Rosen GD, Schadt EE, Schalkwyk LC, Seltzer Z, Shimomura K, Shou S, Sillanpaa MJ, Siracusa LD, Snoeck HW, Spearow JL, Svenson K, Tarantino LM, Threadgill D, Toth LA, Valdar W, de Villena FP, Warden C, Whatley S, Williams RW, Wiltshire T, Yi N, Zhang D, Zhang M, Zou F. The Collaborative Cross, a community resource for the genetic analysis of complex traits. Nature genetics. 2004;36:1133–1137. doi: 10.1038/ng1104-1133. [DOI] [PubMed] [Google Scholar]
- Crane GE. Tardive dyskinesia in patients treated with major neuroleptics: a review of the literature. Am J Psychiatry. 1968;124(Suppl):40–48. doi: 10.1176/ajp.124.8S.40. [DOI] [PubMed] [Google Scholar]
- Crawley JN. Exploratory behavior models of anxiety in mice. Neurosci Biobehav Rev. 1985;9:37–44. doi: 10.1016/0149-7634(85)90030-2. [DOI] [PubMed] [Google Scholar]
- Crowley JJ, Adkins DE, Pratt AL, Quackenbush CR, van den Oord EJ, Moy SS, Wilhelmsen KC, Cooper TB, Bogue MA, McLeod HL, Sullivan PF. Antipsychotic-induced vacuous chewing movements and extrapyramidal side effects are highly heritable in mice. The pharmacogenomics journal. 2010 doi: 10.1038/tpj.2010.82. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dayalu P, Chou KL. Antipsychotic-induced extrapyramidal symptoms and their management. Expert Opin Pharmacother. 2008;9:1451–1462. doi: 10.1517/14656566.9.9.1451. [DOI] [PubMed] [Google Scholar]
- Eddington ND, Young D. Biliary excretion of reduced haloperidol glucuronide. Psychopharmacology. 1990;100:46–48. doi: 10.1007/BF02245788. [DOI] [PubMed] [Google Scholar]
- Fleischmann N, Christ G, Sclafani T, Melman A. The effect of ovariectomy and long-term estrogen replacement on bladder structure and function in the rat. J Urol. 2002;168:1265–1268. doi: 10.1016/S0022-5347(05)64637-X. [DOI] [PubMed] [Google Scholar]
- Froemming JS, Lam YW, Jann MW, Davis CM. Pharmacokinetics of haloperidol. Clinical pharmacokinetics. 1989;17:396–423. doi: 10.2165/00003088-198917060-00004. [DOI] [PubMed] [Google Scholar]
- Fujita PA, Rhead B, Zweig AS, Hinrichs AS, Karolchik D, Cline MS, Goldman M, Barber GP, Clawson H, Coelho A, Diekhans M, Dreszer TR, Giardine BM, Harte RA, Hillman-Jackson J, Hsu F, Kirkup V, Kuhn RM, Learned K, Li CH, Meyer LR, Pohl A, Raney BJ, Rosenbloom KR, Smith KE, Haussler D, Kent WJ. The UCSC Genome Browser database: update 2011. Nucleic acids research. 2011;39:D876–882. doi: 10.1093/nar/gkq963. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Galili R, Mosberg, Gil-Ad I, Weizman A, Melamed E, Offen D. Haloperidol-induced neurotoxicity--possible implications for tardive dyskinesia. Journal of neural transmission. 2000;107:479–490. doi: 10.1007/s007020070089. [DOI] [PubMed] [Google Scholar]
- Goldstein H. Multilevel statistical models. In: Searle S, Casella G, McCulloch C, editors. Variance Components. Wiley; New York: 1995. [Google Scholar]
- Harrill AH, Watkins PB, Su S, Ross PK, Harbourt DE, Stylianou IM, Boorman GA, Russo MW, Sackler RS, Harris SC, Smith PC, Tennant R, Bogue M, Paigen K, Harris C, Contractor T, Wiltshire T, Rusyn I, Threadgill DW. Mouse population-guided resequencing reveals that variants in CD44 contribute to acetaminophen-induced liver injury in humans. Genome research. 2009;19:1507–1515. doi: 10.1101/gr.090241.108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Herken H, Erdal ME, Boke O, Savas HA. Tardive dyskinesia is not associated with the polymorphisms of 5-HT2A receptor gene, serotonin transporter gene and catechol-o-methyltransferase gene. Eur.Psychiatry. 2003;18:77–81. doi: 10.1016/s0924-9338(03)00005-1. [DOI] [PubMed] [Google Scholar]
- Hsin-tung E, Simpson G. Kaplan and Sadocks’s Comprehensive Textbook of Psychiatry. Lippincott, Williams and Wilkins; Philadelphia: 2000. Medication-induced movement disorders; pp. 2265–2271. [Google Scholar]
- Hsin-Tung E, Simpson G. Medication-induced movement disorders. In: Kaplan HI, Sadock BJ, editors. Comprehensive Textbook of Psychiatry. Lippincott, Williams and Wilkins; Philadephia, PA: 2000. [Google Scholar]
- Hsu F, Kent WJ, Clawson H, Kuhn RM, Diekhans M, Haussler D. The UCSC Known Genes. Bioinformatics. 2006;22:1036–1046. doi: 10.1093/bioinformatics/btl048. [DOI] [PubMed] [Google Scholar]
- Ikeda M, Tomita Y, Mouri A, Koga M, Okochi T, Yoshimura R, Yamanouchi Y, Kinoshita Y, Hashimoto R, Williams HJ, Takeda M, Nakamura J, Nabeshima T, Owen MJ, O’Donovan MC, Honda H, Arinami T, Ozaki N, Iwata N. Identification of novel candidate genes for treatment response to risperidone and susceptibility for schizophrenia: integrated analysis among pharmacogenomics, mouse expression, and genetic case-control association approaches. Biological psychiatry. 2010;67:263–269. doi: 10.1016/j.biopsych.2009.08.030. [DOI] [PubMed] [Google Scholar]
- Joreskog K. A general approach to confirmatory maximum likelihood factor analysis. Psychometrika. 1969;34:183–202. [Google Scholar]
- Kaiser R, Tremblay PB, Klufmoller F, Roots I, Brockmoller J. Relationship between adverse effects of antipsychotic treatment and dopamine D(2) receptor polymorphisms in patients with schizophrenia. Mol.Psychiatry. 2002;7:695–705. doi: 10.1038/sj.mp.4001054. [DOI] [PubMed] [Google Scholar]
- Kang HM, Zaitlen NA, Wade CM, Kirby A, Heckerman D, Daly MJ, Eskin E. Efficient control of population structure in model organism association mapping. Genetics. 2008;178:1709–1723. doi: 10.1534/genetics.107.080101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koshikawa N, Fujita S, Adachi K. Behavioral pharmacology of orofacial movement disorders. Int Rev Neurobiol. 2011;97:1–38. doi: 10.1016/B978-0-12-385198-7.00001-1. [DOI] [PubMed] [Google Scholar]
- Lagrue E, Abe H, Lavanya M, Touhami J, Bodard S, Chalon S, Battini JL, Sitbon M, Castelnau P. Regional characterization of energy metabolism in the brain of normal and MPTP-intoxicated mice using new markers of glucose and phosphate transport. Journal of biomedical science. 2010;17:91. doi: 10.1186/1423-0127-17-91. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lai IC, Wang YC, Lin CC, Bai YM, Liao DL, Yu SC, Lin CY, Chen JY, Liou YJ. Negative association between catechol-O-methyltransferase (COMT) gene Val158Met polymorphism and persistent tardive dyskinesia in schizophrenia. J.Neural Transm. 2005;112:1107–1113. doi: 10.1007/s00702-004-0252-6. [DOI] [PubMed] [Google Scholar]
- Lee HJ, Kang SG, Choi JE, Paik JW, Kim YK, Kim SH, Lee MS, Joe SH, Jung IK, Kim L. No association between dopamine D4 receptor gene -521 C/T polymorphism and tardive dyskinesia in schizophrenia. Neuropsychobiology. 2007;55:47–51. doi: 10.1159/000103576. [DOI] [PubMed] [Google Scholar]
- Lerer B, Segman RH, Tan EC, Basile VS, Cavallaro R, Aschauer HN, Strous R, Chong SA, Heresco-Levy U, Verga M, Scharfetter J, Meltzer HY, Kennedy JL, Macciardi F. Combined analysis of 635 patients confirms an age-related association of the serotonin 2A receptor gene with tardive dyskinesia and specificity for the non-orofacial subtype. Int.J.Neuropsychopharmacol. 2005;8:411–425. doi: 10.1017/S1461145705005389. [DOI] [PubMed] [Google Scholar]
- Lidow MS, Goldman-Rakic PS. A common action of clozapine, haloperidol, and remoxipride on D1- and D2-dopaminergic receptors in the primate cerebral cortex. Proceedings of the National Academy of Sciences of the United States of America. 1994;91:4353–4356. doi: 10.1073/pnas.91.10.4353. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mackay TF, Stone EA, Ayroles JF. The genetics of quantitative traits: challenges and prospects. Nature reviews. Genetics. 2009;10:565–577. doi: 10.1038/nrg2612. [DOI] [PubMed] [Google Scholar]
- Matsumoto C, Shinkai T, Hori H, Ohmori O, Nakamura J. Polymorphisms of dopamine degradation enzyme (COMT and MAO) genes and tardive dyskinesia in patients with schizophrenia. Psychiatry Res. 2004;127:1–7. doi: 10.1016/j.psychres.2004.03.011. [DOI] [PubMed] [Google Scholar]
- McKusick VA. Mendelian Inheritance in Man and its online version, OMIM. American journal of human genetics. 2007;80:588–604. doi: 10.1086/514346. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mitchell IJ, Cooper AC, Griffiths MR, Cooper AJ. Acute administration of haloperidol induces apoptosis of neurones in the striatum and substantia nigra in the rat. Neuroscience. 2002;109:89–99. doi: 10.1016/s0306-4522(01)00455-9. [DOI] [PubMed] [Google Scholar]
- Mohn AR, Gainetdinov RR, Caron MG, Koller BH. Mice with reduced NMDA receptor expression display behaviors related to schizophrenia. Cell. 1999;98:427–436. doi: 10.1016/s0092-8674(00)81972-8. [DOI] [PubMed] [Google Scholar]
- Muller DJ, Schulze TG, Knapp M, Held T, Krauss H, Weber T, Ahle G, Maroldt A, Alfter D, Maier W, Nothen MM, Rietschel M. Familial occurrence of tardive dyskinesia. Acta Psychiatr Scand. 2001;104:375–379. doi: 10.1034/j.1600-0447.2001.00401.x. [DOI] [PubMed] [Google Scholar]
- Muthén B, Muthén L. Traditional latent variable modeling using Mplus: Mplus Short course notes. Los Angeles, CA: 2003. [Google Scholar]
- O’Callaghan E, Larkin C, Kinsella A, Waddington JL. Obstetric complications, the putative familial-sporadic distinction, and tardive dyskinesia in schizophrenia. Br J Psychiatry. 1990;157:578–584. doi: 10.1192/bjp.157.4.578. [DOI] [PubMed] [Google Scholar]
- Pan F, McMillan L, Pardo-Manuel De Villena F, Threadgill D, Wang W. TreeQA: quantitative genome wide association mapping using local perfect phylogeny trees. Pac Symp Biocomput. 2009:415–426. [PMC free article] [PubMed] [Google Scholar]
- Patsopoulos NA, Ntzani EE, Zintzaras E, Ioannidis JP. CYP2D6 polymorphisms and the risk of tardive dyskinesia in schizophrenia: a meta-analysis. Pharmacogenet Genomics. 2005;15:151–158. doi: 10.1097/01213011-200503000-00003. [DOI] [PubMed] [Google Scholar]
- Prakash N, Wurst W. Genetic networks controlling the development of midbrain dopaminergic neurons. The Journal of physiology. 2006;575:403–410. doi: 10.1113/jphysiol.2006.113464. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reynolds GP, Templeman LA, Zhang ZJ. The role of 5-HT2C receptor polymorphisms in the pharmacogenetics of antipsychotic drug treatment. Prog. Neuropsychopharmacol. Biol. Psychiatry. 2005;29:1021–1028. doi: 10.1016/j.pnpbp.2005.03.019. [DOI] [PubMed] [Google Scholar]
- Rusyn I, Gatti DM, Wiltshire T, Kleeberger SR, Threadgill DW. Toxicogenetics: population-based testing of drug and chemical safety in mouse models. Pharmacogenomics. 2010;11:1127–1136. doi: 10.2217/pgs.10.100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sakimura K, Kutsuwada T, Ito I, Manabe T, Takayama C, Kushiya E, Yagi T, Aizawa S, Inoue Y, Sugiyama H, et al. Reduced hippocampal LTP and spatial learning in mice lacking NMDA receptor epsilon 1 subunit. Nature. 1995;373:151–155. doi: 10.1038/373151a0. [DOI] [PubMed] [Google Scholar]
- Shaltiel G, Maeng S, Malkesman O, Pearson B, Schloesser RJ, Tragon T, Rogawski M, Gasior M, Luckenbaugh D, Chen G, Manji HK. Evidence for the involvement of the kainate receptor subunit GluR6 (GRIK2) in mediating behavioral displays related to behavioral symptoms of mania. Molecular psychiatry. 2008;13:858–872. doi: 10.1038/mp.2008.20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Simpson GM. Long-acting, antipsychotic agents and extrapyramidal side effects. Dis Nerv Syst. 1970;31(Suppl):12–14. [PubMed] [Google Scholar]
- Skoblenick KJ, Castellano JM, Rogoza RM, Dyck BA, Thomas N, Gabriele JP, Chong VZ, Mishra RK. Translocation of AIF in the human and rat striatum following protracted haloperidol, but not clozapine treatment. Apoptosis: an international journal on programmed cell death. 2006;11:663–672. doi: 10.1007/s10495-006-5698-6. [DOI] [PubMed] [Google Scholar]
- Soares-Weiser K, Fernandez HH. Tardive dyskinesia. Seminars in neurology. 2007a;27:159–169. doi: 10.1055/s-2007-971169. [DOI] [PubMed] [Google Scholar]
- Soares-Weiser K, Fernandez HH. Tardive dyskinesia. Semin Neurol. 2007b;27:159–169. doi: 10.1055/s-2007-971169. [DOI] [PubMed] [Google Scholar]
- Tandon R, Belmaker RH, Gattaz WF, Lopez-Ibor JJ, Jr., Okasha A, Singh B, Stein DJ, Olie JP, Fleischhacker WW, Moeller HJ. World Psychiatric Association Pharmacopsychiatry Section statement on comparative effectiveness of antipsychotics in the treatment of schizophrenia. Schizophr Res. 2008;100:20–38. doi: 10.1016/j.schres.2007.11.033. [DOI] [PubMed] [Google Scholar]
- Tomiyama K, McNamara FN, Clifford JJ, Kinsella A, Koshikawa N, Waddington JL. Topographical assessment and pharmacological characterization of orofacial movements in mice: dopamine D(1)-like vs. D(2)-like receptor regulation. Eur J Pharmacol. 2001;418:47–54. doi: 10.1016/s0014-2999(01)00908-6. [DOI] [PubMed] [Google Scholar]
- Turrone P, Remington G, Kapur S, Nobrega JN. The relationship between dopamine D2 receptor occupancy and the vacuous chewing movement syndrome in rats. Psychopharmacology (Berl) 2003;165:166–171. doi: 10.1007/s00213-002-1259-z. [DOI] [PubMed] [Google Scholar]
- Turrone P, Remington G, Nobrega JN. The vacuous chewing movement (VCM) model of tardive dyskinesia revisited: is there a relationship to dopamine D(2) receptor occupancy? Neuroscience and biobehavioral reviews. 2002;26:361–380. doi: 10.1016/s0149-7634(02)00008-8. [DOI] [PubMed] [Google Scholar]
- Van Prooijen J, Van Der Kloot WA. Confirmatory analysis of exploratively obtained factor structures. Educational and Psychological Measurement. 2001;51:777–792. [Google Scholar]
- van Woerden GM, Hoebeek FE, Gao Z, Nagaraja RY, Hoogenraad CC, Kushner SA, Hansel C, De Zeeuw CI, Elgersma Y. betaCaMKII controls the direction of plasticity at parallel fiber-Purkinje cell synapses. Nature neuroscience. 2009;12:823–825. doi: 10.1038/nn.2329. [DOI] [PubMed] [Google Scholar]
- Waddington JL, Cross AJ, Gamble SJ, Bourne RC. Spontaneous orofacial dyskinesia and dopaminergic function in rats after 6 months of neuroleptic treatment. Science. 1983;220:530–532. doi: 10.1126/science.6132447. [DOI] [PubMed] [Google Scholar]
- Wang K, Li M, Hadley D, Liu R, Glessner J, Grant SF, Hakonarson H, Bucan M. PennCNV: an integrated hidden Markov model designed for high-resolution copy number variation detection in whole-genome SNP genotyping data. Genome research. 2007;17:1665–1674. doi: 10.1101/gr.6861907. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang H, Ding Y, Hutchins LN, Szatkiewicz J, Bell TA, Paigen BJ, Graber JH, de Villena FP, Churchill GA. A customized and versatile high-density genotyping array for the mouse. Nat Methods. 2009a;6:663–666. doi: 10.1038/nmeth.1359. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang H, Ding Y, Hutchins LN, Szatkiewicz J, Bell TA, Paigen BJ, Graber JH, de Villena FP, Churchill GA. A customized and versatile high-density genotyping array for the mouse. Nature methods. 2009b;6:663–666. doi: 10.1038/nmeth.1359. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yassa R, Ananth J. Familial tardive dyskinesia. Am J Psychiatry. 1981;138:1618–1619. doi: 10.1176/ajp.138.12.1618. [DOI] [PubMed] [Google Scholar]
- Zaltsman Y, Shachnai L, Yivgi-Ohana N, Schwarz M, Maryanovich M, Houtkooper RH, Vaz FM, De Leonardis F, Fiermonte G, Palmieri F, Gillissen B, Daniel PT, Jimenez E, Walsh S, Koehler CM, Roy SS, Walter L, Hajnoczky G, Gross A. MTCH2/MIMP is a major facilitator of tBID recruitment to mitochondria. Nature cell biology. 2010;12:553–562. doi: 10.1038/ncb2057. [DOI] [PMC free article] [PubMed] [Google Scholar]