Abstract
Human alternative open reading frames (HAltORF) is a publicly available and searchable online database referencing putative products of out-of-frame alternative translation initiation (ATI) in human mRNAs. Out-of-frame ATI is a process by which a single mRNA encodes independent proteins, when distinct initiation codons located in different reading frames are recognized by a ribosome to initiate translation. This mechanism is largely used in viruses to increase the coding potential of small viral genomes. There is increasing evidence that out-of-frame ATI is also used in eukaryotes, including human, and may contribute to the diversity of the human proteome. HAltORF is the first web-based searchable database that allows thorough investigation in the human transcriptome of out-of-frame alternative open reading frames with a start codon located in a strong Kozak context, and are thus the more likely to be expressed. It is also the first large scale study on the human transcriptome to successfully predict the expression of out-of-frame ATI protein products that were previously discovered experimentally. HAltORF will be a useful tool for the identification of human genes with multiple coding sequences, and will help to better define and understand the complexity of the human proteome.
Database URL: http://haltorf.roucoulab.com/.
Introduction
Each eukaryotic mRNA encoding a protein is usually associated with only one open reading frame (herein called reference ORF) or coding sequence (CDS) delineated by a start codon (most of the time AUG) and a stop codon, required to initiate and end translation, respectively. This simplistic view is however being challenged by the existence of at least two mechanisms resulting in increased protein diversity. In-frame alternative translation initiation (ATI) at downstream AUG codons allows the production of truncated protein isoforms with new functions or localization and is a well-characterized mechanism in eukaryotes (1,2). Out-of-frame ATI at the start codon of alternative ORFs (AltORFs) in the two other reading frames is a second mechanism producing proteins with an amino acid equence completely different from the reference protein. The nomenclature regarding reading frames used thereafter is the following (3). The +1 reading frame is determined by the coding sequence of the reference ORF for each transcript (independently of the gene or transcript). Hence, the annotated reference ORF is defined as frame +1, and there are two possible frames for AltORFs: frame +2 and frame +3.
The presence of overlapping ORFs and the use of out-of-frame ATI are well described in viruses (4–6) and provide small viral genomes with an increased coding capacity. In addition, a database referencing putative alternative ORFs in many prokaryotic genomes already exists (7). The role of out-of-frame ATI in eukaryotes has been overlooked. Yet, there is some evidence that proteins derived from AltORFs can affect physiological as well as pathological aspects of gene function. This is the case for the alternative protein ALEX encoded in the GNAS gene (8,9). In addition, we recently discovered the endogenous expression in human of an alternative protein product termed AltPrP which ORF(+3 reading frame) partially overlaps with the prion protein CDS (Figure 1) (10). Four other examples exist in human (11–14), which correspond to peptides that are targeted by anti-tumor responses in several types of cancers, and may thus serve as biomarkers or therapeutic targets (15). Interestingly, these AltORFs are all but one included within the reference ORF (11). This observation is critical since the expression of cDNAs composed solely of the CDS in experimental systems such as cultured cells may actually result in the expression of more than one protein (10). Consequently, co-expression of an alternative protein together with the reference protein in functional studies likely result in unnoticed confounding results. A database containing a list of all human mRNAs containing AltORFs overlapping with the reference ORF is important to identify potential genes with multiple CDS.
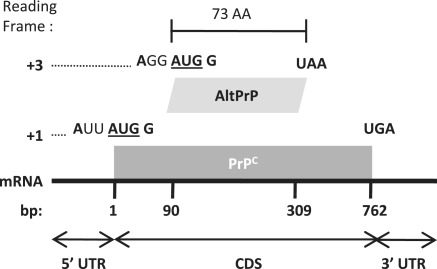
Figure 1.
AltPrP, a typical example of AltORFs in the HAltORF database. All mRNAs produced from the PRNP gene have the same reference ORF (nt 1–762, gray box) which encodes the prion protein (PrPC) in the +1 reading frame. An AltORF (white box) is present in the +3 reading frame (nt 90–309). Similar to all AltORFs present in the database, the alternative prion protein (AltPrP) encoding AltORF is entirely included in the CDS of the reference protein, and encodes a protein longer than 24 amino acids (minimum size threshold). Additionally, its AUG codon is in a different reading frame than the reference protein, and is located in an optimal Kozak context (shown in bold; consensus: A/GNNAUGG).
To our knowledge, three bioinformatics genome-wide studies aiming at the identification of AltORFs in mammals have been performed previously (16–18). However, none of them provided an online searchable option with links to GenBank and NCBI databases for further investigation. In one study, criteria such as conservation among species and a minimum length of 500 bp for the predicted AltORFs were used and only 40 putatively expressed AltORFs were referenced (16). In a more recent study, 138 potential dual coding transcripts were identified in human (18). In another study, a filter of a minimal length of 150 bp was applied and 1793 AltORFs were found to be conserved among rat, mouse and human (17). When the 1793 human AltORFs were filtered for the presence of an optimal Kozak context around the initiator AUG codon, known to be extremely important for efficient initiation of translation (19), this number dropped to 217 putative AltORFs. One objective of these three studies was to predict high confidence candidate AltORFs, and the highly stringent criteria used were extremely pertinent in this matter. However, they were unsuccessful in predicting the expression of two experimentally proven AltORFs, AltPrP and ALEX. For all these reasons, it is obvious that a less stringent and potentially more comprehensive large scale bioinformatics analysis of AltORFs in the human transcriptome and a publicly available and searchable online database of predicted AltORFs are lacking.
Human alternative open reading frames (HAltORF;http://haltorf.roucoulab.com/) is the first web-based searchable database that allows thorough investigation in the human transcriptome of AltORFs overlapping with annotated CDS, and putatively expressed by out-of-frame ATI. It is also the first large scale study on the human transcriptome to successfully predict the expression of AltPrP and ALEX, two experimentally discovered out-of-frame ATI protein products. HAltORF will be a useful tool for the identification of genes containing multiple CDS in human, and will help to better define and understand the complexity of the human proteome.
Database generation
The HAltORF database was built using a pipeline of Perl scripts that populate a MySQL database. All GenBank human mRNA and protein entries (release 37) were downloaded from the NCBI website (http://www.ncbi.nlm.nih.gov/), and each mRNA was associated with its reference protein. For each mRNA, in silico translation of the full sequence was performed using the Transeq software (20), and subsequent comparison of the results with the amino acid sequence of the reference protein allowed to map the translation start and stop sites coordinates of the reference ORF on its corresponding mRNA. The sequence 5′ of the translation start site of the reference ORF was then deleted. This action set the reading frame associated with the reference ORF in each mRNA to +1. The remaining sequence was then translated again using the Transeq software. All translation results equal to or above 24 amino acids, regardless of the reading frame, were stored in the database along with their start and stop sites coordinates. The arbitrary threshold of 24 amino acids was selected to reduce the database to an acceptable size, since we (data not shown) and other groups (16,17) noticed that the numbers of predicted AltORFs increases as the size threshold decreases. Additionally, the validation of the expression of smaller peptides by standard techniques, such as SDS–PAGE and western blots, would be technically too challenging. Next, based on a simplified consensus Kozak sequence (A/GNNATGG) known to be favorable for efficient translation initiation (19), we determined for each predicted ORF start site if it was located in a strong (perfect fit to the consensus) or weak (any other sequence) Kozak context. The last step was to select, in the CDS of each mRNA, the putative AltORFs that are the most likely to be expressed. To do so, we filtered the database using the following criteria: (i) ORFs had to be in the +2 or +3 reading frames to be selected, thus storing AltORFs, which are currently absent from existing protein databases; (ii) the predicted AltORFs had to possess a strong Kozak context around their AUG codon, to increase the chance of efficient translation initiation; (iii) the stop site of the AltORFs had to be located prior to the stop site of the reference ORFs, thus removing ORFs that are not entirely contained within the CDS of the reference protein. More details on the construction of the database are available on the HAltORF website. For a typical example of AltORFs found in this new database (Figure 1).
Database content
We identified 17 096 distinct predicted AltORFs in the CDS of 31 422 mRNAs (41.2% of total human mRNAs) transcribed from 8744 genes (42.5% of total human genes). A total of 14 195 (83%) are located in the +2 reading frame and 2901 (17%) are located in the +3 reading frame.
For each AltORF, the gene name and accession number of the mRNA in which it is encoded are provided. Other information can also be found, including the reference protein produced from the corresponding mRNA, the coordinates of the start and stop codon of both the reference ORF and the alternative ORF in the mRNA, and the predicted length and amino acid sequence of the alternative protein.
Web interface
The HAltORF database (http://haltorf.roucoulab.com/) can be searched by gene name or symbol, by mRNA or protein GenBank accession number, and by protein sequence (with a minimum of 5 amino acids). Detailed explanations on how to perform a search and how results are displayed are available on the website under the Documentation tab. The search results are summarized in a table containing information for each retrieved AltORF, including the gene symbol, mRNA and reference protein accession numbers, reading frames, the location of the reference and alternative ORFs on the mRNA sequence, and the alternative protein length (Figure 2). The nucleotide numbers indicating the location of the ORFs are the first nucleotide of the start codon, and the first nucleotide of the stop codon, respectively. If multiple transcript variants exist for a given gene, all variants containing an alternative ORF are listed. If a search by protein sequence is performed, the table includes a supplementary column displaying part of the alternative protein sequence matching the query sequence. For each retrieved alternative ORF, a detailed result page is accessible through a link and provides the user with basic information concerning the reference mRNA and protein. Links to the NCBI website are also provided to help the user retrieve supplementary information on the gene, mRNA and reference protein associated with the AltORF. The detailed result page also contains an alignment section where the reference and alternative protein sequences are aligned on the reference mRNA sequence (Figure 2). The complete HAltORF database can be freely downloaded in Microsoft Excel or FASTA format under the download tab. The complete MySQL data dump is also available in this section, thus providing developers with the possibility to predict other AltORFs using different parameters such as the length of AltORFs for example.
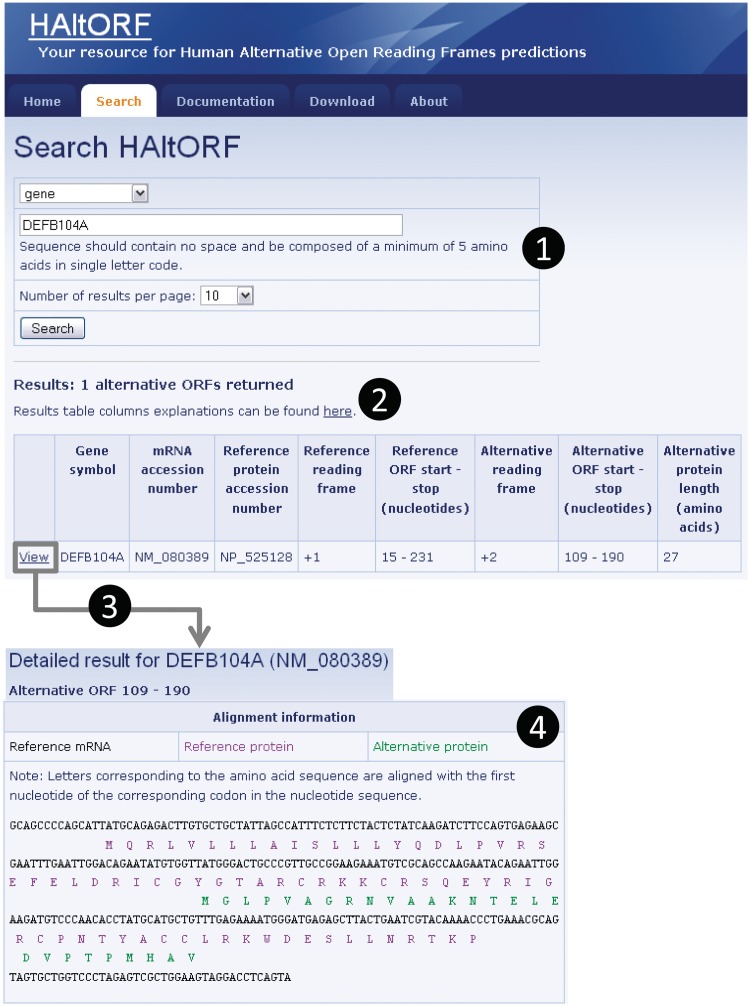
Figure 2.
Snapshot of a typical search and associated results pages. (1) Search by gene (DEFB104A, which encodes the β-defensin 104 protein). (2) The number of corresponding AltORFs is indicated, and details on each AltORF are summarized in a table. Although this is not the case for this particular example, note that for a single gene, all AltORFs present in each transcript variants would be listed. The reference ORF is by definition in the +1 frame, and the alternative ORFs is in the +2 frame in this example. The nucleotide numbers indicating the location of the ORFs are the first nucleotide of the start codon, and the first nucleotide of the stop codon, respectively. (3) A detailed result page is available for each AltORF through the ‘View’ link. (4) In the detailed result page, basic information on the gene and mRNA of origin as well as the associated reference protein are displayed along with links to GenBank for each of these items (not shown). An alignment of the reference (blue letters) and alternative (green letters) protein sequences on the reference mRNA sequence (black letters) is provided.
Relevance and research avenues
The number of predicted AltORFs present in HAltORF is much greater when compared to other studies (16–18). This can be explained by different reasons. In particular, we used a lower cut-off for the size of AltORFs, and chose not to consider criteria such as conservation among species and specific codon usage. However, in our approach, we have established several limits, including AUG initiation codons located in an optimal Kozak context. Expression from AUG codons in the absence of an optimal Kozak sequence or from non-traditional CUG sites (21,22) is also possible and may be included in further studies. Nevertheless, the reduced stringency of our approach resulted in the successful prediction of AltPrP and ALEX, two experimentally well-characterized out-of-frame ATI products. It is likely that at least one of the several functions previously attributed to the prion protein is actually catalyzed by AltPrP (10), and we expect that some paradoxical experimental results regarding the function of other genes might be explained by multiple coding as well. This example highlights the fact that conservation along evolution of an alternative ORF is not necessary to be biologically relevant since the initiation codon for AltPrP is present in higher order mammals but not in lower mammals, including rodents (10). In addition, the presence of ALEX in HAltORF, for which polymorphisms have been associated with inherited neurological problems and increased trauma-related bleeding tendency (9), indicates that HAltORF could be valuable for the identification of biologically important AltORFs in human genes with multiple CDS.
Last but not least, the complete database may help mass spectrometry services to identify the great proportion of unknown peptides in their data sets which cannot be currently matched to any protein in existing databases. Altogether, HAltORF will help in the meticulous exploration of this potential alternative proteome which has been largely overlooked to date.
Funding
The Canadian Institutes for Health Research to XR [grant number MOP-89881]. X.R. is a senior research scholar from the Fonds de la Recherche en Santé du Québec. Funding for open access charge: The Canadian Institutes for Health Research [grant number MOP-89881].
Conflict of interest. None declared.
References
- 1.Kochetov AV. Alternative translation start sites and their significance for eukaryotic proteome. Mol. Biol. (Mosk) 2006;40:788–795. [PubMed] [Google Scholar]
- 2.Kochetov AV. Alternative translation start sites and hidden coding potential of eukaryotic mRNAs. Bioessays. 2008;30:683–691. doi: 10.1002/bies.20771. [DOI] [PubMed] [Google Scholar]
- 3.Veloso F, Riadi G, Aliaga D, et al. Large-scale, multi-genome analysis of alternate open reading frames in bacteria and archaea. Omics. 2005;9:91–105. doi: 10.1089/omi.2005.9.91. [DOI] [PubMed] [Google Scholar]
- 4.Branch AD, Stump DD, Gutierrez JA, et al. The hepatitis C virus alternate reading frame (ARF) and its family of novel products: the alternate reading frame protein/F-protein, the double-frameshift protein, and others. Semin. Liver Dis. 2005;25:105–117. doi: 10.1055/s-2005-864786. [DOI] [PubMed] [Google Scholar]
- 5.Kozak M. Pushing the limits of the scanning mechanism for initiation of translation. Gene. 2002;299:1–34. doi: 10.1016/S0378-1119(02)01056-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Yamasaki K, Weihl CC, Roos RP. Alternative translation initiation of Theiler's murine encephalomyelitis virus. J. Virol. 1999;73:8519–8526. doi: 10.1128/jvi.73.10.8519-8526.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Pedroso I, Rivera G, Lazo F, et al. AlterORF: a database of alternate open reading frames. Nucleic Acids Res. 2008;36:D517–D518. doi: 10.1093/nar/gkm886. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Klemke M, Kehlenbach RH, Huttner WB. Two overlapping reading frames in a single exon encode interacting proteins: a novel way of gene usage. EMBO J. 2001;20:3849–3860. doi: 10.1093/emboj/20.14.3849. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Freson K, Jaeken J, Van Helvoirt M, et al. Functional polymorphisms in the paternally expressed XLalphas and its cofactor ALEX decrease their mutual interaction and enhance receptor-mediated cAMP formation. Hum. Mol. Genet. 2003;12:1121–1130. doi: 10.1093/hmg/ddg130. [DOI] [PubMed] [Google Scholar]
- 10.Vanderperre B, Staskevicius AB, Tremblay G, et al. An overlapping reading frame in the PRNP gene encodes a novel polypeptide distinct from the prion protein. FASEB J. 2011;25:2373–2386. doi: 10.1096/fj.10-173815. [DOI] [PubMed] [Google Scholar]
- 11.Oh S, Terabe M, Pendleton CD, et al. Human CTLs to wild-type and enhanced epitopes of a novel prostate and breast tumor-associated protein, TARP, lyse human breast cancer cells. Cancer Res. 2004;64:2610–2618. doi: 10.1158/0008-5472.can-03-2183. [DOI] [PubMed] [Google Scholar]
- 12.Ronsin C, Chung-Scott V, Poullion I, et al. A non-AUG-defined alternative open reading frame of the intestinal carboxyl esterase mRNA generates an epitope recognized by renal cell carcinoma-reactive tumor-infiltrating lymphocytes in situ. J. Immunol. 1999;163:483–490. [PubMed] [Google Scholar]
- 13.Rosenberg SA, Tong-On P, Li Y, et al. Identification of BING-4 cancer antigen translated from an alternative open reading frame of a gene in the extended MHC class II region using lymphocytes from a patient with a durable complete regression following immunotherapy. J. Immunol. 2002;168:2402–2407. doi: 10.4049/jimmunol.168.5.2402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Wang RF, Parkhurst MR, Kawakami Y, et al. Utilization of an alternative open reading frame of a normal gene in generating a novel human cancer antigen. J. Exp. Med. 1996;183:1131–1140. doi: 10.1084/jem.183.3.1131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Ho O, Green WR. Alternative translational products and cryptic T cell epitopes: expecting the unexpected. J. Immunol. 2006;177:8283–8289. doi: 10.4049/jimmunol.177.12.8283. [DOI] [PubMed] [Google Scholar]
- 16.Chung WY, Wadhawan S, Szklarczyk R, et al. A first look at ARFome: dual-coding genes in mammalian genomes. PLoS Comput. Biol. 2007;3:e91. doi: 10.1371/journal.pcbi.0030091. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Ribrioux S, Brungger A, Baumgarten B, et al. Bioinformatics prediction of overlapping frameshifted translation products in mammalian transcripts. BMC Genomics. 2008;9:122. doi: 10.1186/1471-2164-9-122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Xu H, Wang P, Fu Y, et al. Length of the ORF, position of the first AUG and the Kozak motif are important factors in potential dual-coding transcripts. Cell Res. 2010;20:445–457. doi: 10.1038/cr.2010.25. [DOI] [PubMed] [Google Scholar]
- 19.Kozak M. Point mutations define a sequence flanking the AUG initiator codon that modulates translation by eukaryotic ribosomes. Cell. 1986;44:283–292. doi: 10.1016/0092-8674(86)90762-2. [DOI] [PubMed] [Google Scholar]
- 20.Rice P, Longden I, Bleasby A. EMBOSS: the European molecular biology open software suite. Trends Genet. 2000;16:276–277. doi: 10.1016/s0168-9525(00)02024-2. [DOI] [PubMed] [Google Scholar]
- 21.Ivanov IP, Firth AE, Michel AM, et al. Identification of evolutionarily conserved non-AUG-initiated N-terminal extensions in human coding sequences. Nucleic Acids Res. 2011;39:4220–4234. doi: 10.1093/nar/gkr007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Ingolia NT, Lareau LF, Weissman JS. Ribosome profiling of mouse embryonic stem cells reveals the complexity and dynamics of mammalian proteomes. Cell. 2011;147:789–802. doi: 10.1016/j.cell.2011.10.002. [DOI] [PMC free article] [PubMed] [Google Scholar]