Abstract
Summary: RNA-seq, the application of next-generation sequencing to RNA, provides transcriptome-wide characterization of cellular activity. Assessment of sequencing performance and library quality is critical to the interpretation of RNA-seq data, yet few tools exist to address this issue. We introduce RNA-SeQC, a program which provides key measures of data quality. These metrics include yield, alignment and duplication rates; GC bias, rRNA content, regions of alignment (exon, intron and intragenic), continuity of coverage, 3′/5′ bias and count of detectable transcripts, among others. The software provides multi-sample evaluation of library construction protocols, input materials and other experimental parameters. The modularity of the software enables pipeline integration and the routine monitoring of key measures of data quality such as the number of alignable reads, duplication rates and rRNA contamination. RNA-SeQC allows investigators to make informed decisions about sample inclusion in downstream analysis. In summary, RNA-SeQC provides quality control measures critical to experiment design, process optimization and downstream computational analysis.
Availability and implementation: See www.genepattern.org to run online, or www.broadinstitute.org/rna-seqc/ for a command line tool.
Contact: ddeluca@broadinstitute.org
Supplementary information: Supplementary data are available at Bioinformatics online.
1 INTRODUCTION
RNA-seq is a highly parallelized sequencing technology that allows for comprehensive transcriptome characterization and quantification (Wang et al., 2009). As with all forms of parallelized sequencing, significant computational processing is required to unlock transcript abundance levels and other measures for biological interpretation (Garber et al., 2011). However, prior to the calculation of biologically relevant data such as transcript abundance, presence of novel isoforms and genotype identity, it is necessary to evaluate the performance of the RNA-seq experiment itself. Summary statistics and quality control scores provide insight into inherently complex data prior to downstream analysis.
Here we present RNA-SeQC, a metrics tool with application to two domains: experiment design and process optimization; and quality control prior to computational analysis. Metrics such as duplication rate, rRNA abundance, alignment rates, coverage continuity and correlation to reference expression profiles are highly informative during selection of experiment conditions and library construction methods (Levin et al., 2010). RNA-SeQC's multi-sample input feature allows for direct comparison across samples (Fig. 1). Additionally, a single-sample mode can be used to monitor samples on an ongoing basis to rapidly assess the quality of a particular sequencing run, and to monitor and optimize these measures in production over time and prior to downstream analysis. RNA-SeQC provides a suite of experiment quality measures, many of which are currently not provided by other available tools (Supplementary Material).
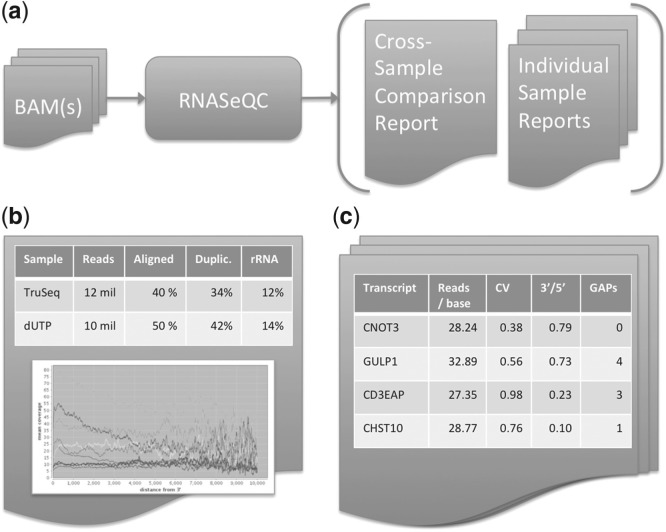
Fig. 1.
Overview of the RNA-SeQC process. (a) RNA-SeQC will work with one or more input samples to produce both a comparative summary across samples as well as a more detailed report for each sample. (b) The comparative summary report includes an extensive range of metrics (in addition to those shown) as well as coverage plots. (c) For each sample, additional reports quantify the coverage profile (variation, gaps, etc.) for individual transcripts
2 METRICS
RNA-SeQC provides three types of quality control metrics: Read Counts, Coverage and Correlation. A list and description of these metrics is shown below. RNA-SeQC is compatible with any alignment method that produces a specification-conforming BAM file (Li et al., 2009), with flags properly set. For additional information, usage and software requirements, see the GenePattern help document provided as Supplementary Material 1. Metrics reports are provided in HTML for human consumption, as well as tab-delimited text files for pipeline integration.
2.1 Read counts
The following metrics are generated by counting reads with particular characteristics. Rates are also provided, and are calculated as either per total reads or per aligned reads. Since the BAM format does support multiple alignments per read, this implementation ignores any read flagged as not being a primary alignment.
Total, unique and duplicate reads
Mapped reads and mapped unique reads
rRNA reads: counted in one of two modes: (i) interval mode where an interval file defines the location in the given alignment to which rRNA reads map; and (ii) BWA mode, where an independent Burrows–Wheeler Aligner (Li and Durbin, 2009) alignment to reference rRNA sequences is performed.
Transcript-annotated reads: intragenic (regions between genes), intergenic (within genes), exonic and intronic. These regions are defined in a user-specified GTF file (Supplementary Material). GENCODE annotations (Harrow et al., 2006) are used by default.
Expression profile efficiency: the ratio of exon-mapped reads to the total reads sequenced.
Expressed transcripts: count of transcripts with reads ≥1.
Strand specificity: to assess the performance of strand-specific library construction methods, the percentage of sense-derived reads is given for each end of the read pair. Whereas a non-strand-specific protocol would give values of 50%/50%, strand-specific protocols typically yield 99%/1% or 1%/99% for this metric.
2.2 Coverage
The following metrics are based on coverage: the number of reads that cover a given genomic position (in units of reads per base). RNA-SeQC quantifies the uniformity of coverage with several different metrics. To reflect the effect of expression level on these metrics, we select genes from three categories: low, middle and high expression genes (see Supplementary Material) and also report the average of these metrics for each gene set.
Mean coverage: the mean number of reads per base.
Mean coefficient of variation: the mean coefficient of variation across all transcripts.
5′/3′ Coverage: the mean per-base coverage for end regions of RNA transcripts. The length of the end region has a default value of 100 base pairs.
Gaps in coverage: a stretch of sequence of at least 5 base pairs having zero coverage. Both the number of gaps as well as the summed gap length across all transcripts in the set is reported.
Cumulative gap length: sum of gap lengths of all transcripts.
Downsampling: to normalize data to a specific total read count we enable an on-the-fly random reduction of reads to reach a user-defined number. This is useful for comparing certain statistics across datasets, e.g. gap metrics, which are not otherwise adjusted for depth.
GC bias: to assess effects of GC content on sequencing performance, all coverage metrics are additionally reported for three levels of transcript GC content: high, low and moderate (see Supplementary Material for default threshold settings).
Coverage plots: plots of coverage level versus base index, either for a single transcript or a set of transcripts.
2.3 Expression correlation
One of the most valuable ways to interpret the performance of an RNA-seq run is to compare the measured expression levels to a reference (Levin et al., 2010). RNA-SeQC provides RPKM-based estimation of expression levels (Mortazavi et al. 2008). When run with multiple samples, RNA-SeQC creates a matrix of correlations among all combinations, reporting the Spearman (rank based) and Pearson (quantity based) correlation coefficients. Optionally, an array based or RNA-seq reference expression profile can be provided for the correlation analysis. Correlation metrics are also provided for the different GC content stratifications to measure GC bias.
3 IMPLEMENTATION
Implemented in Java, RNA-SeQC is platform independent and requires no installation. For investigators who prefer a web interface to a command-line tool, this software can be run using the GenePattern web interface found at http://www.GenePattern.org (Reich et al., 2006).
Within the RNA-SeQC software package, Read Count metrics were implemented by inheriting from the ReadWalker class of the GATK software package (McKenna et al., 2010). Transcript annotations are bound to the walker in the RefGen format. This format is created on-the-fly from a user-provided GTF file. The program is designed to support the minimal GTF specification, but the GTF format used by GENCODE (Harrow et al., 2006) is recommended. For continuity of coverage calculations, the GATK's Depth of Coverage walker was used to calculate the number of bases at a given position in the genomic alignment. Finally, ribosomal RNA quantification is performed by realigning all reads to rRNA reference sequences using the Burrows–Wheeler Aligner (Li and Durbin, 2009).
Funding: Funded in part with Federal funds from the National Human Genome Research Institute, National Institutes of Health, Department of Health and Human under Contract No. HHSN268201000029C.
Conflict of Interest: none declared.
Supplementary Material
REFERENCES
- Garber M., et al. Computational methods for transcriptome annotation and quantification using RNA-seq. Nat. Meth. 2011;8:469–477. doi: 10.1038/nmeth.1613. [DOI] [PubMed] [Google Scholar]
- Harrow J., et al. GENCODE: producing a reference annotation for ENCODE. Genome Biol. 2006;7(Suppl. 1):S4.1–S4.9. doi: 10.1186/gb-2006-7-s1-s4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Levin J.Z., et al. Comprehensive comparative analysis of strand-specific RNA sequencing methods. Nat. Meth. 2010;7:709–715. doi: 10.1038/nmeth.1491. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li H., Durbin R. Fast and accurate short read alignment with Burrows-Wheeler transform. Bioinformatics. 2009;25:1754–1760. doi: 10.1093/bioinformatics/btp324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li H., et al. The sequence alignment/map (SAM) format and SAMtools. Bioinformatics. 2009;25:2078–2079. doi: 10.1093/bioinformatics/btp352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McKenna A., et al. The Genome Analysis Toolkit: a MapReduce framework for analyzing next-generation DNA sequencing data. Genome Res. 2010;20:1297–1303. doi: 10.1101/gr.107524.110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mortazavi A., et al. Mapping and quantifying mammalian transcriptomes by RNA-Seq. Nat. Meth. 2008;5:621–628. doi: 10.1038/nmeth.1226. [DOI] [PubMed] [Google Scholar]
- Reich M., et al. GenePattern 2.0. Nat. Genet. 2006;38:500–501. doi: 10.1038/ng0506-500. [DOI] [PubMed] [Google Scholar]
- Wang Z., et al. RNA-Seq: a revolutionary tool for transcriptomics. Nat. Rev. Genet. 2009;10:57–63. doi: 10.1038/nrg2484. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.