Abstract
BACKGROUND
Analysis of chromosomal aberrations or single-gene disorders from rare fetal cells circulating in the blood of pregnant women requires verification of the cells’ genomic identity. We have developed a method enabling multiple analyses at the single-cell level that combines verification of the genomic identity of microchimeric cells with molecular genetic and cytogenetic diagnosis.
METHODS
We used a model system of peripheral blood mononuclear cells spiked with a colon adenocarcinoma cell line and immunofluorescence staining for cytokeratin in combination with DNA staining with the nuclear dye TO-PRO-3 in a preliminary study to define candidate microchimeric (tumor) cells in Cytospin preparations. After laser microdissection, we performed low-volume on-chip isothermal whole genome amplification (iWGA) of single and pooled cells.
RESULTS
DNA fingerprint analysis of iWGA aliquots permitted successful identification of all analyzed candidate microchimeric cell preparations (6 samples of pooled cells, 7 samples of single cells). Sequencing of 3 single-nucleotide polymorphisms was successful at the single-cell level for 20 of 32 allelic loci. Metaphase comparative genomic hybridization (mCGH) with iWGA products of single cells showed the gains and losses known to be present in the genomic DNA of the target cells.
CONCLUSIONS
This method may be instrumental in cell-based noninvasive prenatal diagnosis. Furthermore, the possibility to perform mCGH with amplified DNA from single cells offers a perspective for the analysis of nonmicrochimeric rare cells exhibiting genomic alterations, such as circulating tumor cells.
Microchimerism, the presence of a small number cells that are genetically distinct from those of the host individual, has been linked to autoimmune diseases (1), but it has also been a basis for the quest for cell-based noninvasive prenatal diagnosis. Although methods used for enriching rare cells remove the bulk of the background cells, processed samples remain a mixture of target cells and a large majority of nontarget cells (2–7).
The analytical definition of rare cells solely on the basis of a biochemical parameter entails the risk of contaminating the target cell population (8). This problem even applies to an excellent marker, such as the embryonic hemoglobin produced by nucleated red blood cells, a subpopulation of fetal microchimeric cells present in the blood of pregnant women (9). Fluorescence in situ hybridization, a powerful tool for diagnosis, also not a reliable tool in the search of rare cells because it may yield false-positive signals (2). Furthermore, Y chromosome–specific fluorescence in situ hybridization obviously does not detect female fetal cells. Nevertheless, individual identification of the genomic origin of particular target cells is imperative for cell-based noninvasive prenatal diagnosis. Such unambiguous identification is feasible via DNA fingerprint analysis single cells, as has been shown with candidate target cells preenriched and defined on the basis of biochemical markers (10). Identification of the genomic origin of single cells by DNA fingerprint analysis is independent of sex and cell type; however, the exhaustion of the available DNA for target cell identification impedes further analysis of the cells. There is clearly a need for a method that allows both genomic identification and molecular genetic and cytogenetic analysis of the same cell.
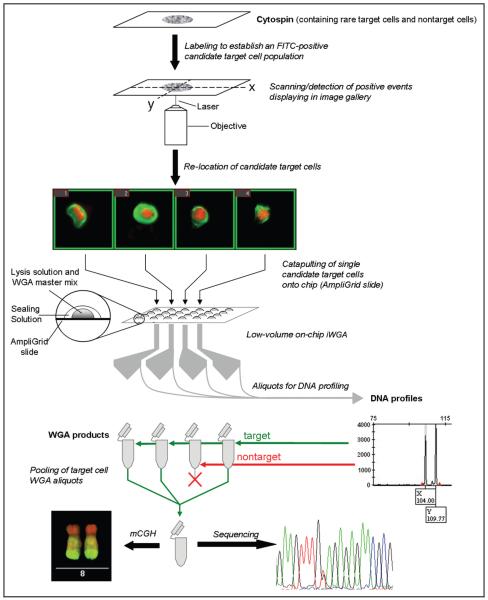
We present a whole-genome amplification (WGA)4 method that allows multiple molecular genetic and cytogenetic analysis of single cells while covering a wide range of resolution. For this purpose, we have adapted our previously reported method of low-volume on-chip DNA fingerprint analysis (10) to isothermal WGA (iWGA). As a proof of principle, we used preparations of peripheral blood mononuclear cells spiked with cells from a carcinoma cell line to mimic both a microchimeric sample (e.g., fetal cells present in the blood of pregnant women) and a sample containing a few cells with chromosomal imbalances on a background of chromosomally balanced cells (e.g., circulating tumor cells). After defining candidate target cells on the basis of a biochemical marker, semiautomated detection, and isolation, we performed low-volume on-chip iWGA. We assessed the suitability of the iWGA products for DNA fingerprint analysis (a post hoc genomic identification of the candidate cells that yields a postidentification pool of verified amplicons), as well as for sequencing and metaphase comparative genomic hybridization (mCGH) (Fig. 1).
Fig. 1. Concept of the establishment and aim of a postidentification pool of whole-genome amplified DNA.
The genomic identity of preselected candidate target cells is verified by DNA fingerprint analysis by using aliquots of single-cell WGA products. The remainder of the WGA products remains available for genetic analyses, such as sequencing or CGH. FITC, fluorescein isothiocyanate.
Materials and Methods
CELLULAR METHODOLOGY
Detailed information regarding cell preparations, staining, and virtual enrichment of candidate target cells is provided in the Text 1 file in the Data Supplement that accompanies the online version of this article at http://www.clinchem.org/content/vol57/issue7. In brief, the candidate microchimeric cell population in Cytospin preparations was defined by immunofluorescence staining for cytokeratin (11). Combination with the nuclear dye TO-PRO-3 was chosen for the rare cell–detection software (Metafer P module RCDetect; Metasystems) to identify the HT-29 cells on the basis of a preestablished set of parameters, as described previously (10).
LASER MICRODISSECTION
As described in the Text 1 file in the online Data Supplement, the position information of the candidate cells was fed into the software for laser microdissection and pressure catapulting (PALM; Zeiss MicroImaging). The catapulted cells were collected at the reaction sites of chemically modified chips (AmpliGrid AG240F; LTF Labortechnik).
iWGA OF CANDIDATE CELLS
Reaction sites containing microdissected samples were charged with 0.75 μL of a cell-lysis mix (AmpliGrid Cell Extraction Kit; LTF Labortechnik) and overlaid with 5 μL of PCR oil (Sealing Solution; LTF Labortechnik) as recommended by the manufacturer. After cell-lysis treatment at 75 °C and enzyme inactivation at 95 °C for 5 min and 2 min, respectively, iWGA of the lysed samples was performed with the illustra GenomiPhi V2 DNA Amplification Kit (GE Healthcare). For this purpose, we added 0.5 μL of sample buffer (illustra GenomiPhi V2 DNA Amplification Kit) containing 1× AdvaBlue PCR dye (LTF Labortechnik) to lysed samples and denatured the sample at 95 °C for 3 min. We added another 0.5 μL containing a mixture of reaction buffer and enzyme mix (10:1; illustra GenomiPhi V2 DNA Amplification Kit) and performed iWGA at 30 °C for 3 h. The enzyme was then inactivated at 65 °C for 10 min. All thermal-incubation steps were carried out on a slide cycler (AmpliSpeed ASC400D; Advalytix). iWGA products (approximately 1.5 μL) were collected from reaction sites, diluted with 8 μLofwater, and stored at 4 °C until further use. We used 1-μL aliquots of diluted iWGA products to perform DNA fingerprint analysis and single-nucleotide polymorphism (SNP) sequencing.
DNA FINGERPRINTING
We used the PowerPlex 16 System (Promega) for this analysis. iWGA aliquots were placed on AmpliGrid reaction sites and allowed to air-dry. The respective sites were charged with 1 μL of a multiplex PCR master mix, consisting of 0.032 μL Ampli-Taq Gold DNA Polymerase (Applied Biosystems), 0.1 μL Gold Star 10× Buffer, 0.1 μL Powerplex 16 10× Primer Pair Mix, and 0.768 μL nuclease free water (all from Promega), and covered with 5 μL of Sealing Solution. Amplification was performed on the AmpliSpeed ASC400D via the PCR, as recommended by the manufacturer. The PCR products were then collected from the reactive sites and forwarded for PCR cleanup (Wizard SV Gel & PCR Clean-Up System; Promega) according to the manufacturer’s recommendations. DNA profiles were compiled from electropherograms (3730 DNA Analyzer; Applied Biosystems) with GeneMapper 4.0 software and analyzed as previously described (10). We defined PCR performance by the quotient of the number of detected products and the theoretical maximum of possible PCR products, calculated from heterozygous loci only. Allele dropout (ADO) was defined as the ratio of the number of heterozygous loci yielding single peaks to the total number of heterozygous loci. Amplification failure was calculated from the number of loci lacking PCR peaks per the total number of loci (equations 1–3):
| 1 |
| 2 |
| 3 |
where npeak is number of peaks detected at heterozygous loci only, nmax. peaks is the maximum number of peaks at heterozygous loci only, nlocus, single peak is the number of heterozygous loci yielding only a single peak, nheterozygous locus is the total number of heterozygous loci, nlocus, no peaks is the number of loci lacking peaks, and nmax. loci is the total number of analyzed loci.
SNP SEQUENCING
Sequencing data were retrieved from reactions that used iWGA products fed directly into the PCR for SNP detection. We analyzed the following SNPs known to be heterozygous in HT-29 cells: BRAF5 (v-raf murine sarcoma viral oncogene homolog B1) c.1799 T>A; APC (adenomatous polyposis coli) c.2557 T>G; and PIK3CA (phosphoinositide-3-kinase, catalytic, α polypeptide) c.1345 C>A. In brief, SNP loci were amplified with 0.5 μL of iWGA products and 0.5 μL of the respective primers [APC (forward/reverse), 5′-CAC GACGTTGTAAAACGAAGGAAGCATTATGGGACAT GG-3′/5′-TTCCATGACTTTGGCAATCTG-3′; BRAF (forward/reverse), 5′-TCATCCTAACACATTTCAAG CC-3′/5′-CACGACGTTGTAAAACGACTTTGTGAAT ACTGGGAACTATGAAA-3′; PIK3CA (forward/reverse), 5′-CACGACGTTGTAAAACGACAAGTGCCT TTTCCAATCAATC-3′/5′-TTCATCATAAATTCCTGAA GCTC-3′], 6 μL HotStar Taq Master Mix Kit (Qiagen), and 5 μL PCR-grade water. After an initial denaturation step at 95 °C for 15 min, the PCR was cycled 35 times at 95 °C, 57 °C, and 72 °C for 45 s each before a final extension step at 72 °C for 5 min and cooling to 8 °C. For the sequencing reaction, we added 0.5 μL of the SNP PCR products to 1 μL sequencing buffer (ABI BigDye Terminator v3.1 Cycle Sequencing Kit; Applied Biosystems), 0.5 μL primer M13, and 7.0 μL PCR-grade water. The sequencing reaction was performed with 25 cycles of denaturation at 95 °C for 30 s, annealing at 50 °C for 15 s, and extension at 60 °C for 4 min; samples were then cooled to 8 °C. The sequencing products were then purified by vacuum filtration (Millipore cleanup plates) and forwarded to analysis on an ABI Prism 3130xl Genetic Analyzer using SeqScape analysis software (Applied Biosystems).
CYTOGENETIC ANALYSIS
For mCGH (12), we reamplified and purified the initial iWGA products. We transferred 1 μL of the iWGA product for an in-tube Φ29 reamplification with the illustra GenomiPhi V2 DNA Amplification Kit (GE Healthcare) and subsequently purified the reamplified samples with the QIAamp DNA Micro Kit (Qiagen), according to the manufacturers’ protocols. The purified samples were recovered from the spin columns with 30 μL of water. Before labeling, we assessed the DNA yield with a NanoDrop Spectrophotometer (Thermo Fisher Scientific) and forwarded the required amount of DNA for restriction enzyme digestion with AluI and RsaI (Promega) at 37 °C for 2 h. We then used the BioPrime DNA Labeling System (Invitrogen) to label 300 ng of the amplified sample DNA (single cells) and “low cell” reference DNA (obtained from 10 cells of normal cultured lymphocytes). Random primers (octamers) were annealed to the denatured DNA template and extended with Klenow fragment in the presence of biotin-16-dUTP (Roche Diagnostics) for sample DNA and digoxigenin-11-dUTP (Roche Diagnostics) for reference DNA. Samples were concentrated with Microcon YM-30 spin columns (Millipore). In a single tube, 45 μL biotin-labeled sample and 45 μL digoxigenin-labeled reference DNA were added to 80 μL human Cot-1 DNA (Roche Diagnostics), 5 μL salmon testes DNA (Sigma-Aldrich), 17.5 μL 3 mol/L sodium acetate (pH 5.2; Sigma-Aldrich), and 481 μL of cold absolute ethanol. After gentle mixing, the samples were precipitated overnight at –20 °C.
For CGH analysis, metaphase spreads were prepared according to standard procedures from stimulated peripheral blood lymphocytes obtained from a healthy male proband (46,XY). Chromosomal metaphase preparations were incubated with RNase A [Roche Diagnostics; 100 μg/mL in 2× standard saline citrate (SSC) (0.30 mol/L NaCl and 0.030 mol/L sodium citrate)] in a moist chamber at 42 °C for 30 min, rinsed 3 times with 2× SSC, and digested with 30 mg/L pepsin (Sigma-Aldrich; diluted in 0.01 mol/L HCl) at 37 °C for 1 min, 45 s. The slides were rinsed 3 times with 1× PBS (8 g/L NaCl, 0.2 g/L KCl, 0.2 g/L KH2PO4, 1.37 g/L Na2HPO4 · 2H2O); dehydrated in an ascending ethanol series; denatured in 700 mL/L formamide containing 2× SSC (pH 7.15) at 73 °C for 1 min, 50 s; dehydrated in an ascending series of ice-cold ethanol; and dried at 42 °C on a heating plate. The precipitated mixture of sample and reference DNA was washed with 700 mL/L ethanol, dried at 42 °C, and incubated with formamide and 300 g/L dextran sulfate at 42 °C while shaking at 600 rpm on an incubator (Thermomixer compact; Eppendorf) for 30 min. For denaturation, the DNA mixture was incubated at 78 °C for 7 min, preannealed at 42 °C for 30 min, and then placed on slides containing the denatured chromosomes. Hybridization was performed at 37 °C for 48 h in a water bath.
After hybridization, the slides were washed 3 times with 4× SSC (0.60 mol/L NaCl and 0.060 mol/L sodium citrate) containing 2 mL/L Tween 20 at 42 °C, washed 3 times with 1× SSC (0.15 mol/L NaCl and 0.015 mol/L sodium citrate) at 60 °C, and blocked with 30 g/L BSA (in 4× SSC containing 2 mL/L Tween 20). For detection of biotin and digoxigenin, the samples were incubated with Cy3.5-labeled avidin (5 ng/μL) and fluorescein isothiocyanate–labeled antidigoxigenin (2 ng/μL) antibodies diluted in 10 g/L BSA for 50 min. The slides were washed 3 times in 4× SSC containing 2 mL/L Tween 20, counterstained with 4′,6-diamidino-2-phenylindole dihydrochloride (Sigma-Aldrich) to allow for chromosome identification, and mounted in phenylenediamine mounting medium. For image capture and processing of CGH data, we used a Leica DMRXA microscope (Leica) equipped with Leica Q-FISH and Leica Q-CGH. The diagnostic thresholds used to score losses and gains were 0.8 and 1.2.
For comparison purposes, we isolated genomic DNA from bulk cultured HT-29 cells and performed mCGH according to standard protocols.
Results
VIRTUAL PREENRICHMENT OF CANDIDATE TARGET CELLS
Cells positive for both cytokeratin and TO-PRO-3 were found on all slides (see Fig. 1 in the online Data Supplement). Selected cells were chosen from the image gallery of the rare cell–detection software and forwarded to laser microdissection and pressure catapulting onto AmpliGrid slides.
DNA FINGERPRINT ANALYSIS OF iWGA SAMPLES
DNA fingerprinting of iWGA aliquots was performed with nonstained HT-29 cells at the level of 10 pooled cells and 5 pooled cells (see Table 1 in the online Data Supplement). The respective samples yielded PCR products at all loci. The PCR performance was 97% (range, 96%–100%) and 94% (range, 92%–96%) for the template levels of 10 cells and 5 cells, respectively. ADO occurred in 5% (range, 0%–8%) at the 10-cell level and in 13% (range, 8%–15%) at the 5-cell level (Table 1; see Table 2 in the online Data Supplement). We did not detect allele dropin or erroneous amplification (see Table 1 in the online Data Supplement).
Table 1.
Quality assessment of DNA fingerprint analysis on the basis of aliquots derived from iWGA.
| Ten HT-29 cellsa |
Five HT-29 cellsb |
Isolated single cellsc |
|||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Sample ID | I | II | III | A | B | C | 1 | 2 | 3 | 4 | 5 | 6 | 7 |
| Amplification failure, %d | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 13 | 0 | 6 | 0 |
| ADO, %e | 8 | 8 | 0 | 8 | 15 | 15 | 23 | 15 | 0 | 54 | 0 | 23 | 15 |
| PCR performance, % | 96 | 96 | 100 | 96 | 92 | 92 | 88 | 92 | 100 | 58 | 100 | 81 | 92 |
Ten nonstained HT-29 cells were pooled for each sample (I–III).
Five nonstained HT-29 cells were pooled for each sample (A–C).
Single cells (1–7) isolated from the artificial mixture that were positive for cytokeratin and TO-PRO-3.
Locus lacking PCR products (peaks).
Heterozygous locus yielding 1 of 2 peaks.
We compiled the PCR profiles (Fig. 2; see Fig. 2 in the online Data Supplement for the full profile) for 7 single cells double-positive for anticytokeratin and TO-PRO-3 staining (see Table 1 and Fig. 1 in the online Data Supplement). The DNA fingerprint analysis of the respective iWGA aliquots yielded a mean PCR performance of 87.4% (range, 58%–100%). Amplification failure and ADO were observed in 2.7% (range, 0%–13%) and 19% (range, 0%–54%), respectively (Table 1; see Table 2 in the online Data Supplement). Erroneous PCR and allele dropin were not detected (see Table 1 in the online Data Supplement).
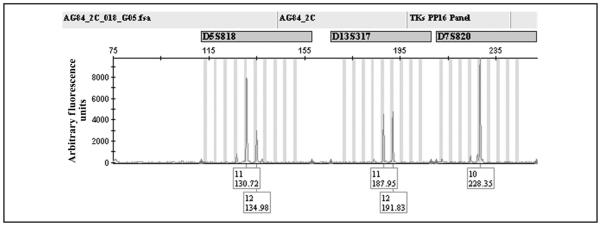
Fig. 2. Detail of DNA profile obtained from iWGA products of a single cell labeled with anticytokeratin fluorescein isothiocyanate and TO-PRO-3, including 3 examples of STR loci.
The DNA profile was obtained from cell no. 5 (designated cell in Fig. 1 in the online Data Supplement). After the initial iWGA round, the amplified products were diluted to a volume of 10 μL. Aliquots of 1 μL were transferred for DNA fingerprint analysis with on-chip multiplex PCR. See Fig. 2 in the online Data Supplement for the full DNA profile for cell no. 5.
The generated DNA profiles showed unambiguous data from samples at all template levels (see Table 1 in the online Data Supplement). Loci were informative in 44 of 48 instances at the 10-cell level, in 41 of 48 cases at the 5-cell level, and in 99 of 109 instances at the single-cell level. Summarizing the allelic pattern across each sample yielded an allocation rate of 100%.
SEQUENCING OF iWGA SAMPLES
Sequencing of 3 SNPs known to be heterozygous in HT-29 cells produced successful PCR results for all tested SNP loci at the 10-and 5-cell levels (9 of 9 reactions each) and for 16 of 18 reactions at the single-cell level (Table 2; see Table 3 in the online Data Supplement). The heterozygous signals were assessed quantitatively, and the signal ratios ranged from subheterozygous (wild type–mutant ratio, 1:4) to superheterozygous (wild type–mutant ratio, 5:1) owing to preferential amplification in the course of iWGA. The ADO rates were similar across the reactions at all levels [ranging between 56% (5 of 9 loci) and 67% (6 of 9 and 12 of 16 loci, respectively], yielding PCR performances of 72% (10-cell level), 67% (5-cell level), and 63% (single-cell level) (see Table 3 in the online Data Supplement).
Table 2.
SNPs detected by sequencing iWGA products of pooled cells and single cells.
| Ten HT-29 cellsa |
Five HT-29 cellsb |
Isolated single cellsc |
Pools of isolated single cellsd |
|||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Sample ID | I | II | III | A | B | C | 1 | 2 | 3 | 4 | 5 | 6 | a | b |
| APC (c.2557 G>T) | G/Te | G/–f | G/– | G/– | G/– | G/– | G/T | G/– | –/T | –/T | G/T | –/–g | –/T | G/T |
| BRAF (c.1799 T>A) | –/A | T/Ah | T/A | T/– | T/Ah | T/Ah | T/– | T/– | –/A | –/– | –/A | –/A | T/– | T/A |
| PIK3CA (c.1345 C>A) | –/A | –/A | C/Ai | –/A | –/A | C/A | –/A | C/Ah | C/Ai | –/A | C/– | –/A | C/Ai | iaj |
| Successful PCR, n/n maxk | 3/3 | 3/3 | 3/3 | 3/3 | 3/3 | 3/3 | 3/3 | 3/3 | 3/3 | 2/3 | 3/3 | 2/3 | 3/3 | 2/2 |
| ADO, n/nmax | 2/3 | 2/3 | 1/3 | 3/3 | 2/3 | 1/3 | 2/3 | 2/3 | 2/3 | 2/2 | 2/3 | 2/2 | 2/3 | 0/2 |
| PCR performance, n/nmax | 4/6 | 4/6 | 5/6 | 3/6 | 4/6 | 5/6 | 4/6 | 4/6 | 4/6 | 2/4 | 4/6 | 2/4 | 4/6 | 4/4 |
Ten nonstained HT-29 cells were pooled for each sample (I–III).
Five nonstained HT-29 cells were pooled for each sample (A–C).
Single cells (1–6) positive for cytokeratin and TO-PRO-3 were isolated by microdissection.
After iWGA, the PCR products of 5 single cells (nos. 1, 2, 3, 5, and 7) (a) and 3 single cells (nos. 2, 3, and 4) (b) were pooled for analysis.
G/T, T/A, and C/A indicate a heterozygous SNP locus yielding both alleles.
G/–, –/T, –/A, T/–, and C/– indicate a heterozygous SNP showing ADO.
–/–, PCR failure.
Wild type–heterozygote ratio of 5:1.
Subheterozygous (wild typ–heterozygote ratio of 1:4).
ia, Inefficient analysis because of a too-high background.
n/nmax, Number of peaks at heterozygous loci/maximum number of peaks at heterozygous loci.
In 2 additional approaches, we pooled the iWGA products of 5 single cells and the products of 3 single cells (Table 2; see Table 3 in the online Data Supplement) after DNA fingerprint analysis revealed the cells to be of HT-29 origin. Sequencing data from pooled iWGA aliquots of 5 single cells yielded DNA sequences for all 3 SNP loci, with 1 SNP locus (PIK3CA) displaying heterozygosity and the other 2 SNP loci displaying ADO. The analysis of the pooled iWGA aliquots for 3 single cells showed heterozygous signals for 2 of the 3 SNP loci (APC and BRAF). The PIK3CA locus could not be evaluated because of high background levels.
CHROMOSOMAL CGH FROM iWGA SAMPLES
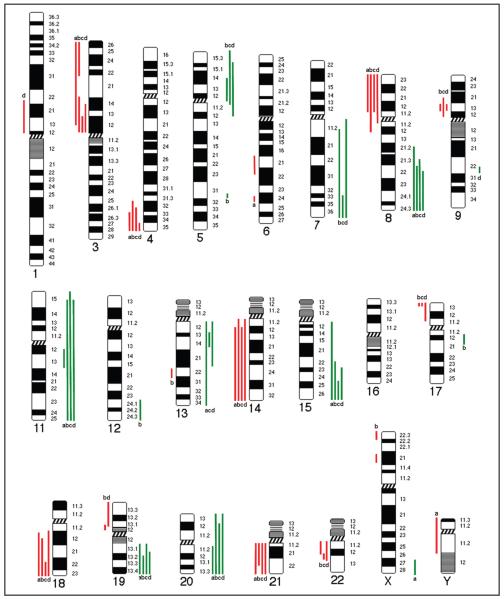
To check the concordance between the CGH profiles, we compared chromosomal CGH data of iWGA aliquots from 3 single cells double-positive for cytokeratin and TO-PRO-3 with non-preamplified DNA isolated from cultured HT-29 cells (bulk HT-29 genomic DNA) (Fig. 3; see Fig. 3 in the online Data Supplement). Balanced CGH profiles of all samples were obtained for chromosomes 2 and 10. With respect to gains and losses, the summarized data for these 4 samples coincide within varying ranges for 11 of 20 DNA sequences (on chromosomes 3p, 4qter, 8p, 8q, 11q, 14, 15, 18q, 19q, 20q, 21). At chromosome 13, two of the single-cell samples gave the same result as the bulk HT-29 genomic DNA. For 7 of 20 DNA sequences, the results for single cells deviated from the (balanced) data obtained for bulk HT-29 genomic DNA, with gains shown for 5p, 7q, 11q, and 20p and losses shown for 9p, 17p, and 22. Alteration of DNA in the CGH profile at the level of the bulk genomic DNA but not at the single-cell level was seen only for the long arm of chromosome 6. Alterations seen only in 1 single-cell preparation were detected at chromosomes 1p (cell no. 5, loss), 5q (cell no. 2, gain), 9q (cell no 5, gain), 12q (cell no. 2, gain), 13q (cell no. 2, loss), and 17q (cell no. 2, gain). Single cell no. 3 deviated from the other 2 single-cell samples at 19p by showing a balanced CGH profile, as seen in the bulk HT-29 CGH profile.
Fig. 3. Comparison of mCGH profiles.
The karyogram shows the chromosomal gains (green lines) and losses (red lines) in non-preamplified DNA from bulk HT-29 cells (a) and in 3 cytokeratin and TO-PRO-3 double-positive single cells (b–d) after iWGA amplification (see Fig. 3 in the online Data Supplement for CGH profiles). The typical gains and losses characterizing HT-29 cells as seen for chromosomes 4qter, 8q, and 8p are represented in both bulk and single-cell profiles. Gains and losses not common to all samples partly reflect the heterogeneity at the single-cell level [see Kawai et al. (16 )].
Discussion
The objective of this work was to create a process for linking unambiguous identification of single cells to subsequent analyses covering a wide range of resolutions. This process expands on a recently described method that permits automated target cell detection and virtual enrichment, microdissection, and identification of the genomic origin of haploidentical single cells by means of DNA fingerprinting (10). Our implementation of an iWGA step between the laser microdissection of cells and DNA fingerprinting created the potential to perform multiple analyses from a particular cell. The aliquots of the iWGA products permitted both sex- and cell type–independent identification of the cell’s genomic origin and the analysis of specific genetic features.
One purpose of DNA fingerprint analysis of iWGA products was to assess the quality of the DNA amplified from single cells. Furthermore, we wanted to know whether the quality of the respective DNA profiles would still be acceptable for verifying the genomic origin of a particular isolated candidate target cell. Such results are a prerequisite for generating pools of amplification products for further molecular genetic and cytogenetic analysis. We found DNA fingerprinting to be compatible with the incorporation of a preceding iWGA step. PCR performance was high and unaffected by potential disturbances due to fixation or labeling. The data rule out contamination because the allelic patterns are unambiguous. The fact that DNA fingerprinting was successful not only demonstrated its feasibility in combination with iWGA but also documented the respective loci to be correctly amplified. Thus, DNA fingerprint analysis provides a benefit in 2 ways: It allows unambiguous allocation of cells from microchimeric samples to one of 2 possible genotypes, and it may be used as a quality control. The amplification of degraded DNA would produce partial DNA profiles. The advantage of detecting and eliminating degraded samples at this stage of the process is welcome.
Only amplification methods that do not implicate DNA fragmentation may yield full DNA profiles. WGA methods that use restriction enzymes may cause amplification failure in the markers of the DNA fingerprinting assay. Random fragmentation is even less applicable because this approach lacks any possibility to predict the sites of fragmentation. The iWGA method we used leads to comparatively less-effective amplification. Therefore, we carried out a double round of iWGA, which might contribute, however, to some decrease in the signal-to-noise ratio in mCGH.
The detection of heterozygous SNPs was comparatively less effective. Although a complete (heterozygous) SNP profile of HT-29 cells could be obtained by summarizing the data from 2 to 4 single cells, sequencing of pooled iWGA samples did not produce a full SNP “profile.” The reason for this effect is not clear. The fact that both molecular genetic methods rely on intact DNA makes DNA degradation unlikely to be the main cause of this effect. As discussed by Rohlin et al. (13), the lowest abundance of mutant alleles that can be detected by Sanger sequencing varies between 15% and 50%. In addition, we cannot rule out that differences in the chromosomal locations of the respective loci (short tandem repeats and SNPs) or in the performance of the iWGA might have influenced the PCR performance.
The CGH data correspond well with published profiles for the HT-29 cell line, despite some additional gains and losses seen in single-cell CGH analyses. Using reference DNA from iWGA results for pooled microdissected cells, we obtained interpretable profiles for all 3 iWGA samples from single cells. The findings of 11 of 20 alterations in the non-preamplified genomic DNA of bulk HT-29 cells and in all 3 single-cell mCGH profiles are in accord with published data (14); however, we did not detect the reported gain at chromosome 3q. The reported findings for chromosomes 5p, 7, 11, 17p, and 22 could be retrieved in our data obtained from single cells (gains at chromosomes 7 and 11) but not from bulk cells (gains only at 7q and 11q). At chromosome 13, one single cell lacked the published gain of DNA. None of our CGH data retrieved the gain of 18p, whereas all CGH profiles showed the loss of 18q. A gain on chromosome 17q was detected for one of the isolated single cells, a result that may reflect the heterogeneity of this cell line, because this gain has also been reported elsewhere (15). That report includes a loss of 6q, which we found in the CGH profile for bulk cells but not for single cells. In addition and in contrast to the cited publications, we detected gains of 9p and 20p at the single-cell level but not in the CGH profile for the bulk cells.
Some aberrations may be attributed to inherent problems of iWGA with rare cells. Differences among the single-cell CGH profiles (chromosomes 1p, 5q, 9q, 12q, 13p, 17q, 19p, X) were sometimes due to data just beyond signal thresholds (cell line/single cell: 6q; single cell/single cell: 1p, 5p, 9q, Xq). In other cases, the chromosomal profile was sigmoid, thereby exceeding (12p, 13p) or missing (19q) the ratio limits. At other loci (5p, 7q), cell heterogeneity may be the reason for the averaging out of the gains and losses seen at the single-cell level but not at the level of the non-preamplified bulk cells. The results in other cases may be due to a heterogeneity seen only at the single-cell level, such as that reported by Kawai et al. (16). Stochastic effects leading to unbalanced amplification are known to occur during amplification at low DNA template concentrations. Reducing the iWGA time would reduce these effects, thereby smoothing the profile; on the other hand, it would probably also lead to insufficient DNA quantities.
In combination with virtual preenrichment, we believe this approach to be promising for detecting chromosomal aberrations in cell-based noninvasive prenatal diagnosis. Furthermore, it may also be helpful in the analysis of circulating tumor cells, for which mCGH may be expected to be a useful tool for discriminating circulating tumor cells from nontumor cells, thereby permitting identification at the cytogenetic level. Further efforts will be taken to expand the applicability of this method toward its use in array CGH analysis. Thus far, our attempts to align the respective procedures have not been successful.
We have demonstrated the preselection of candidate target cells and single-cell iWGA to be compatible with multiple downstream molecular genetic and cytogenetic analyses. The identification of the genomic affiliation of cells from microchimeric samples can be performed unambiguously. Sequencing of iWGA aliquots at sites of heterozygous SNPs proved to be applicable, yet such sequencing is not free from ADO. The detection of chromosomal imbalances for the purpose of cell-based noninvasive prenatal diagnosis promises to be feasible, because new methods for efficient enrichment of fetal cells have been reported (5). With these considerations taken together, we have followed a holistic approach that enables manifold molecular genetic and cytogenetic analyses of single and pooled identified cells, which we expect to be feasible for the analysis of microchimeric cells. This approach may also help in the analysis of other types of rare cells, such as circulating tumor cells, and in forensic medicine.
Supplementary Material
Acknowledgments
Research Funding: P. Sedlmayr, European Commission (Network of Excellence, grant no. FP6–503243), and the Austrian Federal Ministry for Transport, Innovation and Technology together with the Austrian Science Fund (grant no. TRP 17-B18).
Expert Testimony: None declared.
Role of Sponsor: The funding organizations played no role in the design of study, choice of enrolled patients, review and interpretation of data, or preparation or approval of manuscript.
Footnotes
Nonstandard abbreviations: WGA, whole-genome amplification; iWGA, isothermal WGA; mCGH, metaphase comparative genomic hybridization; SNP, single-nucleotide polymorphism; ADO, allele dropout; SSC, standard saline citrate.
Human genes: BRAF, v-raf murine sarcoma viral oncogene homolog B1; APC, adenomatous polyposis coli; PIK3CA, phosphoinositide-3-kinase, catalytic, α polypeptide.
Authors’ Disclosures or Potential Conflicts of Interest: Upon manuscript submission, all authors completed the Disclosures of Potential Conflict of Interest form. Potential conflicts of interest:
Employment or Leadership: P. Sedlmayr, Medical University of Graz.
Consultant or Advisory Role: None declared.
Stock Ownership: None declared.
Honoraria: None declared.
References
- 1.Nelson JL. Your cells are my cells. Sci Am. 2008;298:64–71. [PubMed] [Google Scholar]
- 2.Christensen B, Philip J, Kolvraa S, Lykke-Hansen L, Hromadnikova I, Gohel D, et al. Fetal cells in maternal blood: a comparison of methods for cell isolation and identification. Fetal Diagn Ther. 2005;20:106–12. doi: 10.1159/000082432. [DOI] [PubMed] [Google Scholar]
- 3.Guetta E, Gutstein-Abo L, Barkai G. Trophoblasts isolated from the maternal circulation: in vitro expansion and potential application in non-invasive prenatal diagnosis. J Histochem Cytochem. 2005;53:337–9. doi: 10.1369/jhc.4B6426.2005. [DOI] [PubMed] [Google Scholar]
- 4.Hennerbichler S, Kroisel PM, Zierler H, Pertl B, Wintersteiger R, Dohr G, Sedlmayr P. Fetal nucleated red blood cells in peripheral blood of pregnant women: detection and determination of location on a slide using laser-scanning cytometry. Prenat Diagn. 2003;23:710–5. doi: 10.1002/pd.668. [DOI] [PubMed] [Google Scholar]
- 5.Huang R, Barber TA, Schmidt MA, Tompkins RG, Toner M, Bianchi DW, et al. A microfluidics approach for the isolation of nucleated red blood cells (NRBCs) from the peripheral blood of pregnant women. Prenat Diagn. 2008;28:892–9. doi: 10.1002/pd.2079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Mavrou A, Kolialexi A, Antsaklis A, Korantzis A, Metaxotou C. Identification of fetal nucleated red blood cells in the maternal circulation during pregnancy using anti-hemoglobin-epsilon antibody. Fetal Diagn Ther. 2003;18:309–13. doi: 10.1159/000071971. [DOI] [PubMed] [Google Scholar]
- 7.Vona G, Beroud C, Benachi A, Quenette A, Bonnefont JP, Romana S, et al. Enrichment, immunomorphological, and genetic characterization of fetal cells circulating in maternal blood. Am J Pathol. 2002;160:51–8. doi: 10.1016/S0002-9440(10)64348-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Weichel W, Irlenbusch S, Kato K, Radbruch A. Sorting of rare cells. In: Radbruch A, editor. Flow cytometry and cell sorting. Springer-Verlag; Berlin: 1992. pp. 159–67. [Google Scholar]
- 9.Ponnusamy S, Mohammed N, Ho SS, Zhang HM, Chan YH, Ng YW, et al. In vivo model to determine fetal-cell enrichment efficiency of novel noninvasive prenatal diagnosis methods. Prenat Diagn. 2008;28:494–502. doi: 10.1002/pd.2009. [DOI] [PubMed] [Google Scholar]
- 10.Kroneis T, Gutstein-Abo L, Kofler K, Hartmann M, Hartmann P, Alunni-Fabbroni M, et al. Automatic retrieval of single microchimeric cells and verification of identity by on-chip multiplex PCR. J Cell Mol Med. 2010;14:954–69. doi: 10.1111/j.1582-4934.2009.00784.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Gangnus R, Langer S, Breit E, Pantel K, Speicher MR. Genomic profiling of viable and proliferative micrometastatic cells from early-stage breast cancer patients. Clin Cancer Res. 2004;10:3457–64. doi: 10.1158/1078-0432.CCR-03-0818. [DOI] [PubMed] [Google Scholar]
- 12.James LA. Comparative genomic hybridization as a tool in tumour cytogenetics. J Pathol. 1999;187:385–95. doi: 10.1002/(SICI)1096-9896(199903)187:4<385::AID-PATH290>3.0.CO;2-5. [DOI] [PubMed] [Google Scholar]
- 13.Rohlin A, Wernersson J, Engwall Y, Wiklund L, Bjork J, Nordling M. Parallel sequencing used in detection of mosaic mutations: comparison with four diagnostic DNA screening techniques. Hum Mutat. 2009;30:112–20. doi: 10.1002/humu.20980. [DOI] [PubMed] [Google Scholar]
- 14.Knutsen T, Padilla-Nash HM, Wangsa D, Barenboim-Stapleton L, Camps J, McNeil N, et al. Definitive molecular cytogenetic characterization of 15 colorectal cancer cell lines. Genes Chromosomes Cancer. 2010;49:204–23. doi: 10.1002/gcc.20730. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Ghadimi BM, Sackett DL, Difilippantonio MJ, Schrock E, Neumann T, Jauho A, et al. Centrosome amplification and instability occurs exclusively in aneuploid, but not in diploid colorectal cancer cell lines, and correlates with numerical chromosomal aberrations. Genes Chromosomes Cancer. 2000;27:183–90. [PMC free article] [PubMed] [Google Scholar]
- 16.Kawai K, Viars C, Arden K, Tarin D, Urquidi V, Goodison S. Comprehensive karyotyping of the HT-29 colon adenocarcinoma cell line. Genes Chromosomes Cancer. 2002;34:1–8. doi: 10.1002/gcc.10003. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.