Abstract
Cotton fibers are single-celled extensions of the seed epidermis. They can be isolated in pure form as they undergo staged differentiation including primary cell wall synthesis during elongation and nearly pure cellulose synthesis during secondary wall thickening. This combination of features supports clear interpretation of data about cell walls and cellulose synthesis in the context of high throughput modern experimental technologies. Prior contributions of cotton fiber to building fundamental knowledge about cell walls will be summarized and the dynamic changes in cell wall polymers throughout cotton fiber differentiation will be described. Recent successes in using stable cotton transformation to alter cotton fiber cell wall properties as well as cotton fiber quality will be discussed. Futurec prospects to perform experiments more rapidly through altering cotton fiberwall properties via virus-induced gene silencing will be evaluated.
Keywords: cellulose, cotton fiber, elongation, Gossypium, pectin, transition stage, virus-induced gene silencing, xyloglucan
IMPORTANCE OF COTTON FIBER CELL WALLS
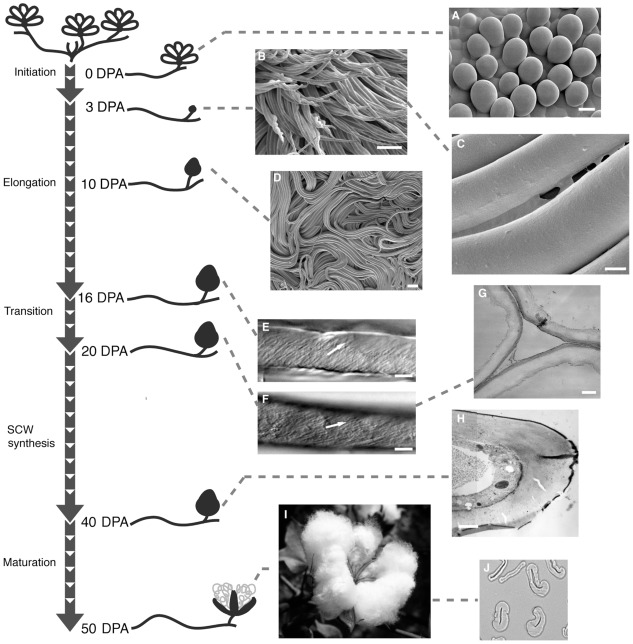
Cotton fiber, a highly elongated and thickened single cell of the seed epidermis, is the world’s most important natural textile fiber. This article will focus on allotetraploid Gossypium hirsutum, the source of most commercial cotton fiber, and fiber cell wall polymers that help to define cotton fiber quality. Cotton fibers initiate by swelling above the ovule surface near the day of flower opening (or anthesis). Days post anthesis (DPA) describes the temporal progression of fiber development, although genotype-by-environment interactions affect the rate of progression (see below). Fiber initials expand isodiametrically, which likely involves cell wall loosening, due to increased turgor pressure in selected epidermal cells. Over the next ~50 days the cotton fiber undergoes: (a) extreme elongation (to >2.5 cm) via primary wall synthesis; (b) transitional wall thickening and primary wall remodeling; and (c) secondary wall thickening via deposition of nearly pure cellulose. Finally, ill-defined “maturation” and cell death processes occur before the boll opens to reveal the fluffy fiber within the cotton fruit (or boll; Figure 1). Typically four carpels (or locules) of one fruit contain about 32 seeds and ~500,000 long cotton fibers (lint fiber; Bowman et al., 2001). A population of thick-walled but short fibers (fuzz fiber or linters) also exists; these are an important source of chemical cellulose for many industrial applications.
FIGURE 1.
A representation of the progression of cotton fiber development. The “stem” indicates the fastest developmental timeline when plants are grown under warm conditions. “Branches” show DPA with associated images of fiber. Cryo- field-emission SEM of (A) fiber initials on the ovule surface (bar = 10 μm); (B) twisting and elongating 3 DPA fibers (bar = 100 μm); (C) CFML stretched between two 3 DPA fibers (bar = 4 μm); (D) ordered bundles of fibers inside the boll (bar = 100 μm). Differential interference contrast micrographs indicating microfibril angle (as highlighted by arrows) in fiber at (E) 16 DPA and (F) 20 DPA (bars = 10 μm). TEM fiber cross-section show (G) an early stage of secondary wall thickening (bar = 300 μm); and (H) a more advanced stage of secondary wall deposition (bar = 1 μm). (I) Mature cotton boll and (J) cross-section of mature fiber viewed in the light microscope. SCW, secondary cell wall. Re-published with permission from The Cotton Foundation (http://www.cotton.org/foundation/index.cfm; Stiff and Haigler, in press).
In addition to defining morphogenesis, cotton fiber cell walls define the industrially important fiber quality parameters (Wakelyn et al., 2007). The fiber length generated through primary wall synthesis is essential for spinning yarn. Fiber strength is contributed by a thin “winding” cell wall layer (see below) and secondary wall cellulose, which also allows the fiber to dye deeply and absorb water. The amount of secondary wall cellulose versus fiber perimeter determines the desirable intermediary extent of secondary wall thickening, which is required for the fiber to collapse into the kidney bean shape that facilitates the spinning of cotton yarn. Cellulose biophysical properties such as degree of crystallinity, fibril angle, and degree of polymerization affect the performance of the fiber. The mature cotton fiber contains >90% (w/w) crystalline cellulose, including its cellulosic secondary wall surrounded by a ~200 nm cuticulated primary wall.
COTTON FIBER AS MODEL CELL
Cotton fiber is a powerful cell wall research model because it is an easily isolated single cell with distinct stages of cell wall synthesis. Other advantages include the ability to culture cotton ovules/fibers in vitro (Kim and Triplett, 2001). The expression of many genes in cotton fiber (Hovav et al., 2008) supports diverse cell wall-related developmental events. After ~6 DPA, frozen cotton fibers can be isolated from the developing seed by gentle grinding. At earlier DPA, fiber enriched RNA can be isolated by beating frozen ovules/fiber with glass beads (Taliercio and Boykin, 2007). Since initial large-scale efforts began a decade ago (Arpat et al., 2004), many studies to characterize the fiber transcriptome have occurred. Even earlier, cotton fiber transcriptomics led to the identification of the first (putative) plant cellulose synthase gene (Pear et al., 1996), a finding that was then genetically confirmed in arabidopsis (Arioli et al., 1998). Use of biochemical, molecular, and microscopic techniques to analyze cotton fiber showed that sucrose synthase is able to provide substrate, as UDP-glucose, to cellulose synthesis (Amor et al., 1995). Cryogenic electron microscopy and biochemical experiments showed that sucrose synthase was present inside and outside the cotton fiber plasma membrane (Salnikov et al., 2003; Brill et al., 2011), which could be related to callose (β-1, 3-glucan) and/or cellulose (β-1, 4-glucan) synthesis. Drug treatments of cotton fiber showed that cellulose fibril orientation, but not cellulose synthesis itself, depended on microtubules (Seagull, 1993), which was confirmed by live cell imaging in arabidopsis (Paredez et al., 2006). Biophysical analyses on cotton fiber revealed cellulose IV in primary walls (Chanzy et al., 1978), helped to define cellulose Iα and cellulose Iβ cellulose allomorphs (Atalla and Vanderhart, 1984), and demonstrated the higher molecular weight of secondary wall versus primary wall cellulose (Timpa and Triplett, 1993).
For secondary wall cellulose synthesis, cotton fibers adopted the genetic program of sclerenchyma cells of the plant body (e.g., xylem tracheary elements and interfascicular fibers) while shutting down the synthesis of the matrix polysaccharides and lignin (Haigler et al., 2009; Betancur et al., 2010). This finding was surprising given that cotton fibers are morphologically classified as trichomes. Nonetheless, the apparent orthologs of genes required for secondary wall synthesis in arabidopsis sclerenchyma cells are up-regulated at the onset of cotton fiber secondary wall deposition (Haigler et al., 2005, 2009; Al-Ghazi et al., 2009; Singh et al., 2009a; Hinchliffe et al., 2010). In contrast, genes encoding the secondary wall-related CESA isoforms are not up-regulated in arabidopsis leaf trichomes as shown by qRT–PCR, microarrays, analysis of CESA promoter::GUS reporter genes in transgenic arabidopsis, and observation of thick trichome walls in arabidopsis lines with non-functional secondary wall cellulose synthases (Betancur et al., 2010). This result is consistent with the contrast between the nearly pure cellulose in cotton fiber secondary walls versus a blend of traditionally defined primary and secondary wall components (including cellulose, pectin, mannan, and lignin) in mature trichome cell walls (Marks et al., 2008). Comparisons of developmentally regulated gene expression sets confirmed the traditional primary wall-related characteristics of arabidopsis leaf trichomes as contrasted with traditional secondary wall-related characteristics of cotton fiber at the onset of wall thickening (Betancur et al., 2010). The xylem-derived genetic program adopted by cotton fiber was present at the base of the land plant lineage with little change since that time (Haigler et al., 2009; Hinchliffe et al., 2010; Zhong et al., 2010), as also reflected in comparisons of gene expression within and between Gossypium species (Al-Ghazi et al., 2009; Rapp et al., 2010). Sclerenchyma cells, not leaf trichomes, provide the best analogies in the model plant arabidopsis for cell wall thickening in cotton fiber. This illustrates the flexible use of a “plant cell wall toolbox” to generate diverse cell walls particularly suited for their purpose (Betancur et al., 2010).
COTTON FIBER CELL WALL COMPOSITION
Cotton fiber cell wall composition changes dynamically throughout development. In young fiber between 0 and 2 DPA: (a) a pectin-rich, xyloglucan-depleted outer sheath develops; (b) an epitope is lost that is characteristic of (1–6)-β-D-galactan carrying arabinose (possibly contained in situ within arabinogalactan protein); and (c) an epitope appears that is characteristic of (1–4)-β-D-galactan, one of the possible side chains of rhamnogalacturonan I (Vaughn and Turley, 1999; Bowling et al., 2011). Also by 2 DPA, the synthesis of a cotton fiber middle lamella (CFML) begins. This thin adhesive outer primary cell wall layer is enriched in fucosylated- and non-fucosylated xyloglucan and homogalacturonan with no or relatively low esterification. The CFML serves to join adjacent fibers into tissue-like bundles that become highly organized and tightly packed within the confined boll space (Singh et al., 2009a). The inner cotton fiber primary wall contains ~22% crystalline cellulose fibrils surrounded by xyloglucan and pectin (Meinert and Delmer, 1977; Singh et al., 2009a). Glycome profiling of fiber wall extracts with numerous monoclonal antibodies recognizing plant polysaccharide epitopes (Pattathil et al., 2010) showed that xylan and acetylated mannan also exist in cotton fiber (Haigler et al., in preparation). Genes encoding expansin, a cell wall loosening protein, are expressed during cotton fiber elongation and associated with QTLs related to fiber length (Harmer et al., 2002; An et al., 2007). At the transition between primary and secondary wall deposition, transcriptionally-regulated cell wall degradative enzymes break down the CFML so that fibers are released as individuals – a phenomenon correlated with decreases in primary wall-related sugars, up-regulation of cell wall hydrolases, and reduction of pectin and xyloglucan molecular mass as secondary wall synthesis begins (Meinert and Delmer, 1977; Tokumoto et al., 2002, 2003; Guo et al., 2007; Singh et al., 2009a).
A distinct transition stage between primary and secondary wall deposition occurs in cotton fiber. During the transition, a unique “winding” cell wall layer, analogous to the S1 layer in wood fiber, is laid down over several days (Seagull, 1993). Fiber strength increases substantially at this time although wall thickening is minimal (Hsieh et al., 1995; Hinchliffe et al., 2011), presumably due to the differently oriented cellulose microfibrils in the winding layer compared to the primary wall. The winding layer may also fuse the primary and secondary cell walls together. Other developmental changes also occur at the transition stage: microtubules rearrange to support the changed direction of cellulose fibril deposition in the cell wall; respiration rate declines temporarily; concentrations of some metabolic sugars change; cellulose synthesis rate increases along with an increase to ~35% (w/w) in cellulose content; the CFML degrades; and the level of callose peaks within the cotton fiber cell wall. At the onset of secondary wall thickening, the rate of cellulose synthesis increases again and both microtubules and cellulose fibrils adopt a steep helix relative to the fiber axis (Meinert and Delmer, 1977; Maltby et al., 1979; Seagull, 1993; Martin and Haigler, 2004; Guo et al., 2007; Singh et al., 2009a; Abidi et al., 2010). The cotton fiber secondary wall ultimately becomes 3–6 μm thick.
CELL WALL-BASED CONTROLS OF COTTON FIBER MORPHOGENESIS AND QUALITY
Through comparing different cotton genotypes and/or experiments with cotton ovule/fiber culture, insights are emerging about how cell walls control cotton fiber morphogenesis and quality. As predicted given the positive correlation between pectin supply and cell expansion in plants (Boyer, 2009), signaling pathways (related to ethylene and C24:0 lignoceric acid) stimulate gene expression related to pectin biosynthesis as part of promoting fiber elongation (Qin et al., 2007; Pang et al., 2010). A family of recombinant inbred lines between Gossypium species with lower and higher fiber quality showed a correlation between expression of a pectin methylesterase gene and fiber quality (Al-Ghazi et al., 2009). Increasing the amount of de-esterified homogalacturonan reduced fiber elongation in transgenic cotton: up to 16% shorter fiber developed on plants with lower activity of pectate lyase, which can degrade de-esterified homogalacturonan (Wang et al., 2010a). In keeping with the above correlations, probing wild-type fiber cross-sections and cell wall extracts with antibodies provided evidence for increased de-esterification of homogalacturonan near the end of fiber elongation (Singh et al., 2009a). These data are consistent with the possibility of developmentally programmed regulation of fiber growth through cell wall rigidification as de-esterified homogalacturonan become cross-linked by calcium, a phenomenon that is well known in plants (Cosgrove, 2005; Boyer, 2009).
Related to xyloglucan, the family of xyloglucan endo-transglycosylase/hydrolase (XTH) genes with expression peaks during elongation has been explored in cotton fiber (Michailidis et al., 2009; Lee et al., 2010). The XTH proteins have one or both abilities to degrade xyloglucan irreversibly (xyloglucan endo-transglycosylase or XET activity) or to cleave and transfer chain ends between molecules (xyloglucan endo-hydrolase or XEH activity; Eklöf and Brumer, 2010). Both activities could possibly increase the plasticity of the primary wall and promote fiber elongation (Cosgrove, 2005). Consistent with this prediction, transgenic cotton plants constitutively over-expressing GhXTH1 showed a positive correlation between inheritance of the transgene and increased fiber length. The transgenic plants had about twofold higher XET activity and 15–20% longer fiber compared to wild-type cotton or null segregants under greenhouse or field conditions. Therefore, the transfer of xyloglucan chain ends between molecules was predicted to be a limiting factor for cotton fiber elongation (Lee et al., 2010). Similarly, XTH activity was lower in a short fiber mutant compared to wild-type cotton (Shao et al., 2011).
The timing of the transition to secondary wall deposition is: (a) affected by temperature and exogenous hormones in vitro (Roberts et al., 1992; Singh et al., 2009b); (b) differs between cultivars grown in the greenhouse (Abidi et al., 2010); and (c) correlates with differences in fiber bundle strength in two near-isogenic lines grown in the field (Hinchliffe et al., 2010). Field studies of the two near-isogenic lines showed that the time of onset of the transition stage was determined by the accumulation of a minimal number of heat units. Earlier entry into the transition stage resulted in higher fiber bundle strength, for reasons that remain to be discovered (Hinchliffe et al., 2011), and it is correlated with shorter fiber in transgenic cotton over-expressing GhPFN2, a profilin gene. Profilin is an actin bundling protein, and GhPFN2 is normally highly expressed between 15 and 24 DPA (Wang et al., 2010b). Correlating with these results, transgenic cotton plants with down-regulated expression of GhADF1, encoding an actin-depolymerizing factor, had a heritable increase in fiber length (+5.6%) and thicker secondary walls (Wang et al., 2009).
Under salt stress, higher cellulose content occurred in transgenic cotton over-expressing mustard annexin compared to wild-type, and genes encoding sucrose phosphate synthase, sucrose synthase, and cellulose synthase were up-regulated in leaves and fiber (Divya et al., 2010). These results correlate with increased fiber cellulose content when transgenic cotton over-expressing spinach sucrose phosphate synthase was grown in the growth chamber under cool night conditions (Haigler et al., 2007).
VIRUS-INDUCED GENE SILENCING AS AN EFFICIENT TOOL FOR TESTING COTTON GENE FUNCTION
Knock-downs of gene expression through virus-induced gene silencing (VIGS; Robertson, 2004) can occur without stable transformation, which is inefficient in cotton (Wilkins et al., 2004). In VIGS experiments, a modified virus transiently down-regulates (silences) a plant messenger RNA (mRNA; Ruiz and Baulcombe, 1998). During viral infection, the plant produces small-interfering RNAs (siRNAs) complementary to the viral genome (Hamilton and Baulcombe, 1999). These siRNAs bind to homologous target mRNAs and ultimately prevent translation of the target gene (Brodersen et al., 2008). When a fragment of a plant gene is inserted into a VIGS vector, both the viral transcripts and the targeted plant transcript(s) are silenced (Kumagai et al., 1995). Down-regulation of the targeted gene will ideally lead to a phenotype that reveals the gene’s function. For a possibly redundant gene family, a conserved fragment can be used to potentially knock-down the expression of several genes and increase the chances of observing a phenotype. VIGS has been successfully used in Nicotiana benthamiana to produce phenotypes by silencing cell wall biosynthetic genes (Burton et al., 2000; Hoffmann et al., 2004; Ahn et al., 2006; Zhu et al., 2010).
Two VIGS vectors have been developed for cotton: one from a DNA virus (Cotton leaf crumple virus, CLCrV; Tuttle et al., 2008) and another from a broad-host-range RNA virus (Tobacco rattle virus, TRV; Ye et al., 2010; Gao et al., 2011). These VIGS vectors are introduced into the plant by either agrobacterium (TRV and CLCrV) or particle bombardment (CLCrV only). Following inoculation, silencing initiates between 7 and 14 days post infection (dpi; TRV) or by 21 dpi (CLCrV). The extent of silencing from both CLCrV and TRV is inversely associated with growth temperature (Fu et al., 2006; Tuttle et al., 2008). Neither CLCrV-VIGS nor TRV-VIGS produced completely uniform silencing in all cultivars (Idris et al., 2010; Gao et al., 2011). Silencing a visible marker, such as PDS or GFP, along with the target gene may be necessary in some cases to facilitate phenotyping (Quadrana et al., 2011), and experiments are ongoing to employ this strategy in cotton fiber (Tuttle, 2011). There are two strategies for achieving VIGS in cotton fiber: (1) plants are inoculated early followed by translocation of the vector or siRNAs to the developing cotton fibers; or (2) isolated ovules are inoculated and cultured in vitro. Given the persistence of CLCrV-VIGS in cotton (Idris et al., 2010), the CLCrV vector is suitable for plant inoculation. For cultured ovules, only TRV-VIGS has been tested so far (Ye et al., 2010).
Consistent with the role of expansins as cell wall loosening agents (Cosgrove, 2005), down-regulation of GhExp1 in 16 DPA cotton fiber resulted in a short fiber phenotype (Tuttle, 2011; Tuttle et al., in preparation). This experiment proves the efficacy of VIGS to test the function of cell wall-related genes expressed in cotton fiber. We are continuing to investigate whether VIGS is effective throughout cotton fiber development, given that fibers undergo symplastic isolation during the mid-phase of elongation (Ruan et al., 2001) so that the spread of silencing into the fiber may be blocked (Voinnet et al., 1998). If the VIGS vectors cannot replicate in fiber, silencing a target gene may depend on the translocation of siRNAs into fiber from adjacent cells in the seed coat.
FUTURE PROSPECTS
Virus-induced gene silencing technology will synergize with cotton genome sequencing (Chen et al., 2007; Paterson et al., 2010; see http://www.phytozome.net/cotton.php) to make cotton fiber functional genomics more efficient. Cotton fiber transcriptomics provides many clues about important genes to test. Proteomics, metabolomics, and antibody probing of cell wall composition are in the early stages of revealing additional insights. Other areas that were not fully summarized here, such as investigation of cell wall-related signaling mechanisms and regulation of cotton fiber cellulose synthesis, also benefit from the clarity provided by staged development in single-celled cotton fiber. The rich history of Gossypium evolution and domestication, including living ancestral genotypes with different fiber developmental programs and properties (Wendel et al., 2009), allows insights about changes in cell wall properties under natural or human selection pressures. In summary, key elements are converging to make cotton fiber one of the most useful systems for cell wall and cellulose research. Further analysis of cotton fiber cell walls is relevant to improvement of this important textile fiber and to creating the next generation of crop plants for optimized production of biofuels and biomaterials.
Conflict of Interest Statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Acknowledgments
For financial support of related research, the authors thank Cotton Inc., Cary, NC, USA and the National Science Foundation (Grant Number IOS1025947). For collaboration to produce images, we thank Dr. Mark Grimson (Figures 1A–D), Dr. Utku Avci (Figure 1G), Dr. Vadim Salnikov (Figure 1H), and Drs. Eric Hequet and Bobby Wyatt (Figure 1J). We thank Dr. Richard Glick for technical support of our related research.
REFERENCES
- Abidi N., Hequet E., Cabrales L. (2010). Changes in sugar composition and cellulose content during the secondary cell wall biogenesis in cotton fibers. Cellulose 17 153–160 [Google Scholar]
- Ahn J., Verma R., Kim M., Lee J., Kim Y., Bang J., Reiter W., Pai H. (2006). Depletion of UDP-d-apiose/UDP-d-xylose synthases results in Rhamnogalacturonan-II deficiency, cell wall thickening, and cell death in higher plants. J. Biol. Chem. 281 13708–13716 [DOI] [PubMed] [Google Scholar]
- Al-Ghazi Y., Bourot S., Arioli T., Dennis E. S., Llewellyn D. J. (2009). Transcript profiling during fiber development identifies pathways in secondary metabolism and cell wall structure that may contribute to cotton fiber quality. Plant Cell Physiol. 50 1364–1381 [DOI] [PubMed] [Google Scholar]
- Amor Y., Haigler C. H., Johnson S., Wainscott M., Delmer D. P. (1995). A membrane-associated form of sucrose synthase and its potential role in synthesis of cellulose and callose in plants. Proc. Natl. Acad. Sci. U.S.A. 92 9353–9357 [DOI] [PMC free article] [PubMed] [Google Scholar]
- An C., Saha S., Jenkins J. N., Scheffler B. E., Wilkins T. A., Stelly D. M. (2007). Transcriptome profiling, sequence characterization, and SNP-based chromosomal assignment of the EXPANSIN genes in cotton. Mol. Genet. Genomics 279 539–553 [DOI] [PubMed] [Google Scholar]
- Arioli T., Peng L., Betzner A. S., Burn J., Wittke W., Herth W., Camilleri C., Höfte H., Plazinski J., Birch R., Cork A., Glover J., Redmond J., Williamson R. E. (1998). Molecular analysis of cellulose biosynthesis in Arabidopsis. Science 279 717–720 [DOI] [PubMed] [Google Scholar]
- Arpat A. B., Waugh M., Sullivan J. P., Gonzales M., Frisch D., Main D., Wood T., Leslie A., Wing R. A., Wilkins T. A. (2004). Functional genomics of cell elongation in developing cotton fibers. Plant Mol. Biol. 54 911–929 [DOI] [PubMed] [Google Scholar]
- Atalla R. H., Vanderhart D. L. (1984). Native cellulose: a composite of two distinct crystalline forms. Science 223 283–285 [DOI] [PubMed] [Google Scholar]
- Betancur L., Singh B., Rapp R. A., Wendel J. F., Marks M. D., Roberts A. W., Haigler C. H. (2010). Phylogenetically distinct cellulose synthase genes support secondary wall thickening in Arabidopsis shoot trichomes and cotton fiber. J. Integr. Plant Biol. 52 205–220 [DOI] [PubMed] [Google Scholar]
- Bowling A. J., Vaughn K. C., Turley R. B. (2011). Polysaccharides and glycoprotein distribution in the epidermis of cotton ovules during early fiber initiation and growth. Protoplasma 248 579–590 [DOI] [PubMed] [Google Scholar]
- Bowman D. T., van Esbroeck G. A., Van’t Hof J., Jividen G. M. (2001). Ovule fiber cell numbers in modern upland cottons. J. Cotton Sci. 5 81–83 [Google Scholar]
- Boyer J. S. (2009). Cell wall biosynthesis and the molecular mechanism of plant enlargement. Funct. Plant Biol. 36 383–394 [DOI] [PubMed] [Google Scholar]
- Brill E., van Thournout M., White R. G., Llewellyn D., Campbell P. M., Engelen S., Ruan Y.-L., Arioli T., Furbank R. T. (2011). A novel isoform of sucrose synthase is targeted to the cell wall during secondary cell wall synthesis in cotton fibre. Plant Physiol. 157 40–54 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brodersen P., Sakvarelidze-Achard L., Bruun-Rasmussen M., Dunoyer P., Yamamoto Y. Y., Sieburth L., Voinnet O. (2008). Widespread translational inhibition by plant miRNAs and siRNAs. Science 320 1185–1190 [DOI] [PubMed] [Google Scholar]
- Burton R. A., Gibeaut D. M., Bacic A., Findlay K., Roberts K., Hamilton A., Baulcombe D. C., Fincher G. B. (2000). Virus-induced silencing of a plant cellulose synthase gene. Plant Cell 12 691–706 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chanzy H., Imada K., Vuong R. (1978). Electron diffraction from the primary walls of cotton fibers. Protoplasma 94 299–306 [Google Scholar]
- Chen Z. J., Scheffler B. E., Dennis E., Triplett B., Zhang T., Chen X., Stelly D. M., Rabinowicz P. D., Town C., Arioli T., Brubaker C., Cantrell R., Lacape J.-M., Ulloa M., Chee P., Gingle A. R., Haigler C. H., Percy R., Saha S., Wilkins T., Wright R. J., Deynze A. V., Zhu Y., Yu S., Guo W., Abdurakhmonov I., Katageri I., Rahman M., Zafar Y., Yu J. Z., Kohel R. J., Wendel J., Paterson A. H. (2007). Towards sequencing cotton (Gossypium) genomes. Plant Physiol. 145 1303–1310 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cosgrove D. J. (2005). Growth of the plant cell wall. Nat. Rev. Mol. Cell Biol. 6 850–861 [DOI] [PubMed] [Google Scholar]
- Divya K., Jami S. K., Kirti P. B. (2010). Constitutive expression of mustard annexin, AnnBj1, enhances abiotic stress tolerance and fiber quality in cotton under stress. Plant Mol. Biol. 73 293–308 [DOI] [PubMed] [Google Scholar]
- Eklöf J. M., Brumer H. (2010). The XTH Gene Family: an update on enzyme structure, function, and phylogeny in xyloglucan remodeling. Plant Physiol. 153 456–466 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fu D. Q., Zhu B. Z., Zhu H. L., Zhang H. X., Xie Y. H., Jiang W. B., Zhao X. D., Luo K. B. (2006). Enhancement of virus-induced gene silencing in tomato by low temperature and low humidity. Mol. Cells 21 153–160 [PubMed] [Google Scholar]
- Gao X., Wheeler T., Li Z., Kenerley C. M., He P., Shan L. (2011). Silencing GhNDR1 and GhMKK2 compromises cotton resistance to Verticillium wilt. Plant J. 66 293–305 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guo G.-Y., Wang L.-J., Chen S.-P., Hu W.-L., Chen X.-Y. (2007). Gene expression and metabolite profiles of cotton fiber during cell elongation and secondary cell wall synthesis. Cell Res. 17 422–434 [DOI] [PubMed] [Google Scholar]
- Haigler C. H., Zhang D., Wilkerson C. G. (2005). Biotechnological improvement of cotton fibre. Physiol. Plant. 124:285–294 [Google Scholar]
- Haigler C. H., Singh B., Zhang D., Hwang S., Wu C., Cai W. X., Hozain M., Kang W., Kiedaisch B., Strauss R. E., Hequet E. F., Wyatt B. G., Jividen G. M., Holaday A. S. (2007). Transgenic cotton over-producing spinach sucrose phosphate synthase showed enhanced leaf sucrose synthesis and improved fiber quality under controlled environmental conditions. Plant Mol. Biol. 63 815–832 [DOI] [PubMed] [Google Scholar]
- Haigler C. H., Singh B., Wang G., Zhang D. (2009). “Genomics of cotton fiber secondary wall deposition and cellulose biogenesis,” in Genetics and Genomics of Cotton ed. Paterson A. H. (New York: Springer; ), 385–417 [Google Scholar]
- Harmer S. E., Orford S. J., Timmis J. N. (2002). Characterisation of six alpha-expansin genes in Gossypium hirsutum (upland cotton). Mol. Genet. Genomics 268 1–9 [DOI] [PubMed] [Google Scholar]
- Hamilton A. J., Baulcombe D. C. (1999). A species of small antisense RNA in posttranscriptional gene silencing in plants. Science 286 950–952 [DOI] [PubMed] [Google Scholar]
- Hinchliffe D. J., Meredith W. R., Delhorn C. D., Thibodeaux D. P., Fang D. D. (2011). Elevated growing degree days influence transition stage timing during cotton fiber development resulting in increased fiber-bundle strength. Crop Sci. 51 1683–1692 [Google Scholar]
- Hinchliffe D. J., Meredith. W. R. Yeater, K. M., Kim H. J., Woodward A. W., Chen Z. J., Triplett B. A. (2010). Near-isogenic cotton germplasm lines that differ in fiber-bundle strength have temporal differences in fiber gene expression patterns as revealed by comparative high-throughput profiling. Theor. Appl. Genet. 120 1347–1366 [DOI] [PubMed] [Google Scholar]
- Hoffmann L., Besseau S., Geoffroy P., Ritzenthaler C., Meyer D., Lapierre C., Pollet B., Legrand M. (2004). Silencing of hydroxycinnamoyl-coenzyme A shikimate/quinate hydroxycinnamoyltransferase affects phenylpropanoid biosynthesis. Plant Cell 16 1446–1465 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hovav R., Udall J. A., Hovav E., Rapp R., Flagel L., Wendel J. F. (2008). A majority of cotton genes are expressed in single-celled fiber. Planta 227 319–329 [DOI] [PubMed] [Google Scholar]
- Hsieh Y.-L., Honik D. D., Hartzell M. M. (1995). A developmental study of single fiber strength: greenhouse grown SJ-2 Acala cotton. Text. Res. J. 65 101–112 [Google Scholar]
- Idris A. M., Tuttle J. R., Robertson D., Haigler C. H., Brown J. K. (2010). Differential cotton leaf crumple virus-VIGS-mediated gene silencing and viral genome localization in different Gossypium hirsutum genetic backgrounds. Physiol. Mol. Plant Pathol. 75 13–22 [Google Scholar]
- Kim H. J., Triplett B. A. (2001). Cotton fiber growth in planta and in vitro. Models for plant cell elongation and cell wall biogenesis. Plant Physiol. 127 1361–1366 [PMC free article] [PubMed] [Google Scholar]
- Kumagai M. H., Donson J., della-Cioppa G., Harvey D., Hanley K., Grill L. K. (1995). Cytoplasmic inhibition of carotenoid biosynthesis with virus-derived RNA. Proc. Natl. Acad. Sci. U.S.A. 92 1679–1683 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee J., Burns T. H., Light G., Sun Y., Fokar M., Kasukabe Y., Fujisawa K., Maekawa Y., Allen R. D. (2010). Xyloglucan endotransglycosylase/hydrolase genes in cotton and their role in fiber elongation. Planta 232 1191–1205 [DOI] [PubMed] [Google Scholar]
- Maltby D., Carpita N. C., Montezinos D., Kulow Y., Delmer D. P. (1979). Beta-1, 3-glucan in developing cotton fibers – structure, localization, and relationship of synthesis to that of secondary wall cellulose. Plant Physiol. 63 1158–114 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marks M. D., Betancur L, Gilding, E., Chen F., Bauer S., Wenger J., Dixon R. A., Haigler C. H. (2008). A new method for isolating large quantities of Arabidopsis trichomes for transcriptome, cell wall and other types of analyses. Plant J. 56 483–492 [DOI] [PubMed] [Google Scholar]
- Martin L. K., Haigler C. H. (2004). Cool temperature hinders flux from glucose to sucrose during cellulose synthesis in secondary wall stage cotton fibers. Cellulose 11 339–349 [Google Scholar]
- Meinert M. C., Delmer D. P. (1977). Changes in biochemical composition of the cell wall of the cotton fiber during development. Plant Physiol. 59 1088–1097 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Michailidis G., Argiriou A., Darzentas N., A. Tsaftaris A. (2009). Analysis of xyloglucan endotransglycosylase/hydrolase (XTH) genes from allotetraploid (Gossypium hirsutum) cotton and its diploid progenitors expressed during fiber elongation. J. Plant Physiol. 166 403–416 [DOI] [PubMed] [Google Scholar]
- Pang C. Y., Wang H., Pang Y., Xu C., Jiao Y., Qin Y. M., Western T. L., Yu S. X., Zhu Y. X. (2010). Comparative proteomics indicates that biosynthesis of pectic precursors is important for cotton fiber and Arabidopsis root hair elongation. Mol. Cell. Proteomics 9 2019–2033 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Paredez A. R., Somerville C. R., Ehrhardt D. W. (2006). Visualization of cellulose synthase demonstrates functional association with microtubules. Science 312 1491–1495 [DOI] [PubMed] [Google Scholar]
- Paterson A., Rong J., Gingle A., Chee P., Dennis E., Llewellyn D., Dure L., Haigler C., Myers G., Peterson D., Rahman M., Zafar Y., Reddy U., Saranga Y., Stewart J., Udall J., Waghmare V., Wendel J., Wilkins T., Wright R., Zaki E., Hafez E., Zhu J. (2010). Sequencing and utilization of the Gossypium genomes. Trop. Plant Biol. 3 71–74 [Google Scholar]
- Pattathil S. K., Avci U., Baldwin D., Alton G. S., McGill J. A., Popper Z., Bootten T., Albert A., Davis R. H., Chennareddy C., Dong R., O’Shea B., Rossi R., Leoff C., Freshour G., Narra R., O’Neil M., York W. S., Hahn M. G. (2010). A comprehensive toolkit of plant cell wall glycan-directed monoclonal antibodies. Plant Physiol. 153 514–525 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pear J. R., Kawagoe Y., Schreckengost W. E., Delmer D. P., Stalker D. M. (1996). Higher plants contain homologs of the bacterial celA genes encoding the catalytic subunit of cellulose synthase. Proc. Natl. Acad. Sci. U.S.A. 93 12637–12642 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qin Y. M., Hu C. Y., Pang Y., Kastaniotis A. J., Hiltunen J. K., Zhu Y. X. (2007). Saturated very-long-chain fatty acids promote cotton fiber and Arabidopsis cell elongation by activating ethylene biosynthesis. Plant Cell 19 3692–3704 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Quadrana L., Rodriguez M. C., Lopez M., Bermudez L., Nunes-Nesi A., Fernie A. R., Descalzo A., Asis R., Rossi M., Asurmendi S., Carrari F. (2011). Coupling virus-induced gene silencing to exogenous green fluorescence protein expression provides a highly efficient system for functional genomics in Arabidopsis and across all stages of tomato fruit development. Plant Physiol. 156 1278–1291 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rapp R. A., Haigler C. H., Flagel L., Hovav R. H., Udall J. A., Wendel J. F. (2010). Gene expression in developing fibers of Upland cotton (Gossypium hirsutumL.) was massively altered by domestication. BMC Biol. 8 139 10.1186/1741-7007-8-139 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roberts E. M., Nunna R. R., Huang J. Y., Trolinder N. L., Haigler C. H. (1992). Effects of cycling temperatures on fiber metabolism in cultured cotton ovules. Plant Physiol. 100 979–986 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Robertson D. (2004). VIGS vectors for gene silencing: many targets, many tools. Annu. Rev. Plant. Biol. 55 495–519 [DOI] [PubMed] [Google Scholar]
- Ruan Y. L., Llewellyn D. J., Furbank R. T. (2001). The control of single-celled cotton fiber elongation by developmentally reversible gating of plasmodesmata and coordinated expression of sucrose and K+ transporters and expansin. Plant Cell 13 47–60 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ruiz M., Baulcombe D. (1998). Initiation and maintenance of virus-induced gene silencing. Plant Cell 10 937–946 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Salnikov V., Grimson M. J., Seagull R. W., Haigler C. H. (2003). Localization of sucrose synthase and callose in freeze substituted, secondary wall stage, cotton fibers. Protoplasma 221 175–184 [DOI] [PubMed] [Google Scholar]
- Seagull R. W. (1993). Cytoskeletal involvement in cotton fiber growth and development. Micron 24 643–660 [Google Scholar]
- Shao M. Y., Wang X. D., Ni M., Bibi N., Yuan S. N., Malik W., Zhang H. P., Liu Y. X., Hua S. J. (2011). Regulation of cotton fiber elongation by xyloglucan endotransglycosylase/hydrolase genes. Genet. Mol. Res. 10 3771–3782 [DOI] [PubMed] [Google Scholar]
- Singh B., Avci U., Eichler Inwood C. H., Grimson M. J., Landgraf J., Mohnen D., Sorenson I., Wilkerson C. G., Willats W. G. T., Haigler C. H. (2009a). A specialized outer layer of the primary cell wall joins elongating cotton fibers into tissue-like bundles. Plant Physiol. 150 684–699 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Singh B., Cheek H. D., Haigler C. H. (2009b). A synthetic auxin (NAA) suppresses secondary wall cellulose synthesis and enhances elongation in cultured cotton fiber. Plant Cell Rep. 28 1023–1032 [DOI] [PubMed] [Google Scholar]
- Stiff M. R., Haigler C. H. (in press). “Recent advances in cotton fiber development,” in Cotton Flowering and Fruiting, Cotton Physiology Book Series, eds, Oosterhuis D., Cothren T. (Cordova TN: The Cotton Foundation; ). [Google Scholar]
- Taliercio E. W., Boykin D. (2007). Analysis of gene expression in cotton fiber initials. BMC Plant Biol. 7 22 10.1186/1471-2229-7-22 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Timpa J. D., Triplett B. A. (1993). Analysis of cell-wall polymers during cotton fiber development. Planta 189 101–108 [Google Scholar]
- Tokumoto H., Wakabayashi K., Kamisaka S., Hoson T. (2002). Changes in the sugar composition and molecular mass distribution of matrix polysaccharides during cotton fiber development. Plant Cell Physiol. 43 411–418 [DOI] [PubMed] [Google Scholar]
- Tokumoto H., Wakabayashi K., Kamisaka S., Hoson T. (2003). Xyloglucan breakdown during cotton fiber development. J. Plant Physiol. 160 1411–1414 [DOI] [PubMed] [Google Scholar]
- Tuttle J. R. (2011). Development of a Virus-induced Gene Silencing System for Cotton and its Application for Functional Genomics in Fiber. Doctoral Dissertation, 201 p. Available at: http://www.lib.ncsu.edu/resolver/1840.16/7024 [Google Scholar]
- Tuttle J. R., Idris A. M., Brown J. K., Haigler C. H., Robertson D. (2008). Geminivirus-mediated gene silencing from cotton leaf crumple virus is enhanced by low temperature in cotton. Plant Physiol. 148 41–50 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vaughn K. C., Turley R. B. (1999). The primary walls of cotton fibers contain an ensheathing pectin layer. Protoplasma 209 226–237 [Google Scholar]
- Voinnet O., Vain P., Angell S., Baulcombe D. C. (1998). Systemic spread of sequence-specific transgene RNA degradation in plants is initiated by localized introduction of ectopic promoterless DNA. Cell 95 177–187 [DOI] [PubMed] [Google Scholar]
- Wakelyn P. J., Bertoniere N. R., French A. D., Thibodeaux D. P., Triplett B. A., Rousselle M. A., Goynes W. R., Jr., Edwards J. V., Hunter L., McAlister D. D., Gamble G. R. (2007). Cotton Fiber Chemistry and Technology. Boca Raton, FL: CRC Press, 162 p [Google Scholar]
- Wang H., Guo Y., Lv F., Zhu H., Wu S., Jiang Y., Li F., Zhou B., Guo W., Zhang T. (2010a). The essential role of GhPEL gene, encoding a pectate lyase, in cell wall loosening by depolymerization of the de-esterified pectin during fiber elongation in cotton. Plant Mol. Biol. 72 397–406 [DOI] [PubMed] [Google Scholar]
- Wang J., Wang H., Zhao P., Han L., Jiao G., Zheng Y., Huang S., Xia G. (2010b). Overexpression of a profilin (GhPFN2) promotes the progression of developmental phases in cotton fibers. Plant Cell Physiol. 51 1276–1290 [DOI] [PubMed] [Google Scholar]
- Wang H., Wang J., Gao P., Jiao G., Zhao P., Li Y., Wang G., Xia G. (2009). Down-regulation of GhADF1 gene expression affects cotton fibre properties. Plant Biotechnol. J. 7 13–23 [DOI] [PubMed] [Google Scholar]
- Wendel J. F., Brubaker C., Alvarez I., Cronn R., Stewart J. M. D. (2009). “Evolution and natural history of the cotton genus,” in Genetics and Genomics of Cotton, ed. Paterson A. H. (New York: Springer; ). [Google Scholar]
- Wilkins T. A., Mishra R., Tro-linder N. L. (2004). Agrobacterium-mediated transformation and regeneration of cotton. J. Food Agric. Envt. 2 179–187 [Google Scholar]
- Ye J., Chua N. H., Qu J., Gene Y. F., Bu Y. P. (2010). Virus Induced Gene Silencing (VIGS) for Functional Analysis of Genes in Cotton. Patent: WO/2010/144058 Publication Date: 16.12.2010 1–92 [Google Scholar]
- Zhong R., Lee C., Ye Z.-H. (2010). Evolutionary conservation of the transcriptional network regulating secondary wall biosynthesis. Trends Plant Sci. 15 625–632 [DOI] [PubMed] [Google Scholar]
- Zhu X., Pattathil S., Mazumder K., Brehm A., Hahn M. G., Dinesh-Kumar S. P., Joshi C. P. (2010). Virus-induced gene silencing offers a functional genomics platform for studying plant cell wall formation. Mol. Plant 3 818–833 [DOI] [PubMed] [Google Scholar]